Abstract
The superfamily of ferritin-like proteins has recently expanded to include a phylogenetically distinct class of proteins termed DPS-like (DPSL) proteins. Despite their distinct genetic signatures, members of this subclass share considerable similarity to previously recognized DPS proteins. Like DPS, these proteins are expressed in response to oxidative stress, form dodecameric cage-like particles, preferentially utilize H2O2 in the controlled oxidation of Fe2+, and possess a short N-terminal extension implicated in stabilizing cellular DNA. Given these extensive similarities, the functional properties responsible for the preservation of the DPSL signature in the genomes of diverse prokaryotes have been unclear. Here, we describe the crystal structure of a DPSL protein from the thermoacidophilic archaeon Sulfolobus solfataricus. Although the overall fold of the polypeptide chain and the oligomeric state of this protein are indistinguishable from those of authentic DPS proteins, several important differences are observed. First, rather than a ferroxidase site at the subunit interface, as is observed in all other DPS proteins, the ferroxidase site in SsDPSL is buried within the four-helix bundle, similar to bacterioferritin. Second, the structure reveals a channel leading from the exterior surface of SsDPSL to the bacterioferritin-like dimetal binding site, possibly allowing divalent cations and/or H2O2 to access the active site. Third, a pair of cysteine residues unique to DPSL proteins is found adjacent to the dimetal binding site juxtaposed between the exterior surface of the protein and the active site channel. The cysteine residues in this thioferritin motif may play a redox active role, possibly serving to recycle iron at the ferroxidase center.
Oxidative stress, in which reactive oxygen species (ROS1) react indiscriminately with DNA, proteins and lipids, is a universal phenomenon experienced by organisms in all domains of life. Cumulative damage from ROS is now thought to contribute to numerous disease states and a multitude of reports in the primary literature describe the role of oxidative stress in human diseases. Thus, understanding molecular responses to oxidative stress is of significant medical importance. Although much work has been done to characterize the management of oxidative stress in eukaryotes and bacteria, the oxidative stress response in archaea is poorly understood. However, elucidation of the protective mechanisms utilized in archaea is certain to provide insight into the diversity of molecular responses to oxidative stress across all domains of life.
Numerous mechanisms have evolved to minimize and repair the damaging effects of ROS. These include enzymes such as superoxide dismutase and superoxide reductase, which convert the superoxide ion into molecular oxygen (O2) and/or hydrogen peroxide (H2O2). Hydrogen peroxide is generated in other ways as well and is, in fact, a natural product of the cell. Although hydrogen peroxide is a relatively moderate oxidant, its interaction with Fe2+ through the Fenton reaction results in the production of the more destructive hydroxyl radical (eq I). The combination of Fe2+ and H2O2 thus represents a lethal combination for life, and mechanisms for the elimination of H2O2 are essential.
| (I) |
Catalase represents one means per destruction of unwanted H2O2, where disproportionation of H2O2 results in the production of water and molecular oxygen. An alternative strategy is the use of various peroxidase activities, including glutathione and thioredoxin dependent peroxidases, to reduce H2O2 to H2O while avoiding the production of molecular oxygen.
A third strategy for minimizing hydroxyl radical production by the Fenton reaction is to minimize the concentration of free ferrous ion. This strategy is employed in the mineralization reactions catalyzed by ferritin, bacterioferritin, and DPS proteins (DNA binding protein from starved cells), all members of the ferritin superfamily (1–3). In the case of ferritin and bacterioferritin, ferrous ion and oxygen react at the ferroxidase center, eventually leading to the deposition of a benign insoluble ferric oxide core within the hollow, spherically shaped protein particles.
DPS catalyzes a similar reaction. However, in contrast to ferritin, DPS clearly prefers H2O2 as the oxidant (4, 5). Thus, through reaction with H2O2, the activity of DPS simultaneously mitigates the toxicity of H2O2 through a 2-electron reduction to form water, while at the same time, it inactivates the Fe2+ by oxidation to Fe3+. Recent work indicates that bacterioferritin can also utilize H2O2 as an oxidant (5, 6); hence, bacterioferritin will also catalyze the simultaneous consumption of both Fenton reactants.
The secondary and tertiary structures in DPS molecules are largely homologous to those of ferritin and bacterioferritin (7). The structural core of these proteins is a highly conserved four-helix bundle, composed of two consecutive helix-turn-helix motifs. The two helix-turn-helix motifs are related to each other by pseudo 2-fold symmetry and are connected to each other by an extended loop that runs the length of the four helix bundle. In structurally characterized DPS proteins, a fifth helix is found within this loop, where it helps to define the 2-fold symmetric subunit interface (7–9).
Although DPS, ferritin, and bacterioferritin all serve to protect the organism from oxidative damage by sequestering intracellular ferrous iron, the active site at which iron is bound and oxidized in DPS is different from that of ferritin and bacterioferritin. In ferritin and bacterioferritin, iron is found in a di-iron carboxylate binding motif located within the four-helix bundle. In contrast, the structurally characterized DPS proteins show an iron binding site located at the interface between two subunits related by 2-fold symmetry (5, 10, 11). A second difference between bacterioferritin and DPS proteins is the nature of their oligomeric assemblies. While ferritin and bacterioferritin are typically found as 24-mers, with 432 point group symmetry, DPS is found as a dodecamer (12-mer), with 23 point group symmetry (7).
We have previously reported the isolation and characterization of DPS-like (DPSL) proteins from Sulfolobus solfataricus (SsDPSL) (12) and Pyrococcus furiosus (13) and described their role as antioxidant proteins in vitro. On the basis of phylogenetic analysis, these proteins, along with eight others, were shown to form a monophyletic cluster that is distinct from all other protein subclasses within the ferritin superfamily. Biochemical characterization of these DPSL proteins reveals dodecameric assemblies that preferentially utilize H2O2 in the controlled oxidation of Fe2+ (12), and an N-terminal extension similar to that present in DPS molecules, which by analogy, is thought to mediate an interaction with DNA. On the basis of the observed antioxidant properties, oligomeric state, and N-terminal extension of the prototypical proteins from Pyrococcus furiosus and Sulfolobus solfataricus, this subclass of proteins was termed DPS-like (12).
To date, the nature of the ferroxidase center in these DPSL proteins has been less clear. Analogous to authentic DPS proteins, candidate residues for an intersubunit di-iron binding site in the DPSL proteins have been suggested (12). However, residues corresponding to a bacterioferritin-like ferroxidase center are also found within the sequences of all DPSL proteins (13). Thus, it is not surprising that DPSL proteins are classified within Pfam (14) as a bacterioferritin-like subfamily (CD 1052.2) that lacks residues involved in heme binding.
Given the extensive biochemical similarities between DPS and DPSL proteins, the functional properties responsible for preservation of the DPSL signature across a diverse group of prokaryotes are unclear. So too the nature of the ferroxidase site itself. In an effort to illuminate the basis for this distinct phylogenetic signature, we have undertaken structural studies of DPSL proteins. Here, we describe the structure of SsDPSL, the DPS-like protein from the hyperthermophilic archaeon Sulfolobus solfataricus.
MATERIAL AND METHODS
SsDPSL Expression in E. coli
The dpsl gene from S. solfataricus (P2) (12, 15) was cloned into the pET-30a(+) vector (Novagen, Madison, WI) and transformed into BL21-(DE3) or B834(DE3) E. coli for protein expression (Novagen). Cells were grown in a 10 L fermenter (New Brunswick Scientific BF-2000) at 37 °C perfused with air at 8 L/min and mixing at 300 rpm. When the cells reached AU600 = 2.0, protein expression was induced with 0.5 mM IPTG. Antifoam A concentrate (Sigma) was added as required, typically <0.5 mL per fermenter run. After 4–6 h, cells were harvested by centrifugation (6000g for 10 min), washed with lysis buffer (50 mM bis-tris-Cl at pH 6.5, 100 mM NaCl) and frozen at −20 °C. Media for native protein expression contained 3.5 g/L of KH2PO4, 5.0 g/L of K2HPO4, 3.5 g/L of (NH4)HPO4, 0.5 g/L of MgSO4·7H2O, 30 g/L of dextrose, 5 g/L of yeast extract, 1.6 mg/L of FeCl3·6H2O, 0.2 mg/L of CoCl2, 0.1 mg/L of CuCl2, 0.2 mg/L of ZnCl2·4H2O, 0.2 mg/L of NaMoO4·2H2O, and 0.05 mg/L of H3BO3. Media for Selenomethionine protein was SelenoMet Mix (Athena Enzyme Systems) supplemented with 50 mg/L of Selenomethionine.
Protein Purification
Cells were resuspended in lysis buffer (10 mL/g cells) supplemented with 0.002 mg/mL of DNase, 0.05 mg/mL of RNaseA, 1 mg/mL of lysozyme and incubated for 30 min at room temperature. The slurry was passed through a microfluidizer (Microfluidics Corp. 110L), and cellular debris was removed by centrifugation (20 000g for 20 min). The supernatant was heated at 65 °C for 10 min, cooled on ice, and then centrifuged as before to remove denatured proteins. Protein in the supernatant was precipitated with ammonium sulfate (0.603 g (NH4)2SO4 per mL of supernatant) and removed by centrifugation (20 000g for 20 min). The precipitated protein was resuspended in lysis buffer, concentrated to 10 mg/mL by ultrafiltration, and loaded onto a Superose 6 size-exclusion column (Amersham Biosciences) equilibrated with 20 mM bis-tris-Cl at pH 6.5 and 100 mM NaCl. Elution of the protein was monitored at 280 and 260 nm. Protein was concentrated by ultrafiltration to 25 mg/mL for crystallization. Protein concentration was determined by Bradford assay (16) using bovine serum albumin as a standard. For elemental analysis, 200 μL of purified SsDPSL at 1 mg/mL was digested in a 5% HNO3 solution. Samples were then injected into an Ar plasma for analysis by inductively coupled plasma-mass spectrometry (ICP-MS) with species and concentrations determined from the observed mass/charge ratios and appropriate standards.
Quantification of Free Cysteine
Ellman’s reagent (5,5′-dithio-bis-(2-nitrobenzoic acid) or DTNB) was used per supplier’s (Pierce) instructions to quantify the number of solvent accessible cysteine residues (17). Briefly, SsDPSL at 0.3 or 0.6 mg/mL (14 or 28 μM monomer equivalents) was incubated with 180 μM DTNB for 15 min at 25 °C in 0.1 M sodium phosphate at pH 8.0 and 1 mM EDTA with and without 6 M guanidine HCl prior to determining AU at 412 nm. The extinction coefficients used for DTNB were 14 150 M−1cm−1 and 13 700 M−1cm−1 for the native and 6 M guanidine experiments, respectively.
Crystallization and Data Collection
SsDPSL was crystallized at 22 °C by hanging drop vapor diffusion. Drops were assembled with 2 μL of protein mixed with 2 μL of well solution: 0.5 M LiCl, 9% PEG 6000, 10 mM CaCl2, and 50 mM Tris-Cl at pH 8.5. Crystals typically appeared in 3–10 days. Selenomethionine incorporated crystals were grown under identical conditions. Synthetic mother liquor containing 25% PEG 400 was used as a cryoprotectant and introduced by dialysis prior to plunge freezing in liquid nitrogen.
A three-wavelength MAD data set to 3.1 Å resolution was collected at the selenium K-edge (edge, peak and remote wavelengths) at SSRL beamline 9-1 (Table 1). Two additional SAD data sets were collected at SSRL beamline 9-2; a 2.4 Å resolution data set was collected at the peak wavelength of the zinc K-edge, and a 3.8 Å resolution data set was collected at the peak wavelength of the iron K-edge. These zinc- and iron-edge data sets were collected from the same crystal. Data were integrated and reduced in space group I213 using the HKL2000 software package (18).
Table 1.
Data Collection
| crystal 1
|
crystal 2
|
|||||
|---|---|---|---|---|---|---|
| space group | I213 | I213 | ||||
| unit cell | 212.76 | 213.04 | ||||
| parameters a = b = c (Å) | ||||||
| data | Sea | Sea | Sea | Znb | Zna | Fea |
| collection | inflection | peak | remote | peak | peak | peak |
| wavelength (Å) | 0.97924 | 0.97902 | 0.91837 | 1.28237 | 1.28237 | 1.75943 |
| resolution rangec | 30.00–3.23 (3.35–3.23) | 30.00–3.23 (3.35–3.23) | 30.00–3.23 (3.35–3.23) | 30.00–2.40 (2.49–2.40) | 30.00–2.40 (2.49–2.40) | 30.00–2.91 (3.01–2.91) |
| observations | 223064 | 222900 | 222668 | 447265 | 447028 | 181251 |
| unique reflections | ||||||
| observed: | 49515 | 49484 | 49470 | 56599 | 107242 | 45597 |
| possible: | 49864 | 49833 | 49870 | 62679 | 122005 | 68670 |
| redundancy | 4.5 | 4.5 | 4.5 | 7.9 | 4.2 | 4.0 |
| data coverage(%) | 99.3 (99.7) | 99.3 (99.7) | 99.2 (99.3) | 90.3 (61.0) | 87.9 (54.1) | 66.4 (11.0) |
| average(I)/sigma(I) | 10.5 (3.4) | 10.5 (3.4) | 10.3 (3.4) | 17.4 (2.7) | 16.1 (2.5) | 17.3 (2.0) |
| Rsym (%)d | 9.0 (26.1) | 9.1 (26.6) | 9.3 (26.7) | 5.7 (28.3) | 4.6 (26.3) | 4.7 (28.0) |
Bijvoets separate.
Bijvoets merged.
Number in parentheses refer to the highest resolution shell.
Rsym = 100*∑h∑i|Ii(h) − 〈I(h)〉|/∑hI(h), where Ii(h) is the ith measurement of the h reflection, and 〈I(h)〉 is the average value of the reflection intensity.
Structure Determination and Refinement
SOLVE (19) was used to identify positions of the selenium substructure and for calculation of the initial phases. Thirty selenomethionine sites were found. RESOLVE (20) was used to build an initial model with eight partial monomers per asymmetric unit. The dodecameric SsDPSL particles are centered on crystallographic 3-fold axes with each SsDPSL particle contributing four monomers to the asymmetric unit. The partial models of the eight monomers were then superimposed upon each other with LSQKAB (21), and the program O (22) was used to build a composite monomer. This model was then rotated and translated back to the original eight monomer positions in the asymmetric unit with PDBSET (21). This new model (containing eight composite monomers) was refined with REFMAC5 (23, 24) against our best data, the 2.4 Å resolution data set collected at the peak wavelength of the zinc K-edge. The subsequent electron density map was of excellent quality and was used to manually rebuild the model. Additional rounds of manual model building and refinement resulted in a final model with an Rcryst = 19.9% and an Rfree = 22.2% (Table 2). The refinement included the use of noncrystallographic symmetry (NCS) constraints and temperature, libration, and screw (TLS) tensors. NCS refinement used loose restraints on both main chain and side chain atoms and a single span for each monomer. TLS refinement defined each monomer as one rigid body. The final model has good stereochemistry, with no residues in the disallowed regions of the Ramachandran plot (25).
Table 2.
Refinement
| resolution range (Å) | 30.00–2.40 |
| Rcryst(%)/Rfree(%)a | 19.9/22.2 |
| coordinate error (Å)b | 0.188 |
| real space CCc | 0.942 |
| RMSD from ideality | |
| bonds (Å)/angles (deg) | 0.007/0.898 |
| Ramachandran plotd | 93.6/5.6/0.8 |
| most favored, additional, generously allowed (%) average residual B values (Å2) | 26.265 |
Rcryst = ∑||Fo| − |Fc||/∑|Fo|, where Fo and Fc are the structure factor amplitudes from the data and the model, respectively. Rfree is calculated similarly, using 5% of the structure factors held back as a test set.
Based on maximum likelihood.
Correlation coefficient (CC) is between the model and the 2mFo − DFc electron density map.
Calculated using PROCHECK (25).
All eight monomers in the model have interpretable electron density for residues 8–174. The electron density map was better at the N-terminus of most monomers, allowing one or two additional residues to be built. The model contains the following residues for each monomer: A (6–174), B (8–174), C (6–174), D (7–174), M (6–174), N (7–174), O (6–174), and P (7–174). Mass spectrometry shows that all 188 residues are present in the recombinant SsDPSL protein; thus, the missing N- and C-terminal residues not present in the model are presumably disordered in the crystal.
Structural comparisons were performed using the DALI (26) and VAST (http://www.ncbi.nlm.nih.gov/Structure/VAST/vastsearch.html) servers. Figures were generated with PyMOL (27) and SPOCK (28).
Coordinates
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 2CLB.
RESULTS
The structure of SsDPSL was determined by multiwavelength anomalous dispersion at the selenium edge with selenomethionine incorporated protein (Table 1). The final structure was refined against a 2.4 Å data set, yielding an Rcryst of 19.9% and an Rfree of 22.2% (Table 2). As predicted by the previous biochemical characterization (12), the SsDPSL monomer assembles into a dodecameric cage with 23 symmetry. Measured along the 3-fold axis, the external diameter of the dodecamer is 94 Å, whereas the diameter of the internal cavity is 55 Å.
The asymmetric unit contains eight monomers that belong to two different dodecamers. The two dodecamers sit adjacent to one another in the crystal lattice, each centered on a crystallographic 3-fold axis. Monomers A, B, C, and D are contributed by the first dodecamer and monomers M, N, O, and P by the second. The overall fold of the eight monomers is identical with little apparent difference between them when they are superimposed. The average RMSD between Cα positions in equivalent residues in the eight monomers is 0.04 Å, with a maximum Cα deviation of 0.22 Å for Asp97.
Monomer Fold
Not surprisingly, the tertiary structure of the SsDPSL molecule shows strong resemblance to DPS, revealing a decorated four helix bundle, with the N- and C-termini at opposite ends of the bundle (Figure 1). The four-helix bundle, composed of helices A, B, C, and D can be described as pair a of helix-turn-helix motifs related by a pseudo 2-fold axis that lies perpendicular to the long axis of the helical bundle. Consequently, helices B and C are connected by a long loop (Leu84–Asp112) that runs the length of the helical bundle. In comparison to the A helices of ferritin, bacterioferritin, and DPS, the A helix in SsDPSL is slightly bent, with the bend centered around Tyr42. As in the structures of authentic DPS molecules, SsDPSL also has a short BC helix (Ile92–Ile98) in the middle of this loop. The BC helix is oriented perpendicular to the long axis of the four-helix bundle and lies on the exterior of the dodecamer at the 2-fold interface (Figure 1).
Figure 1.
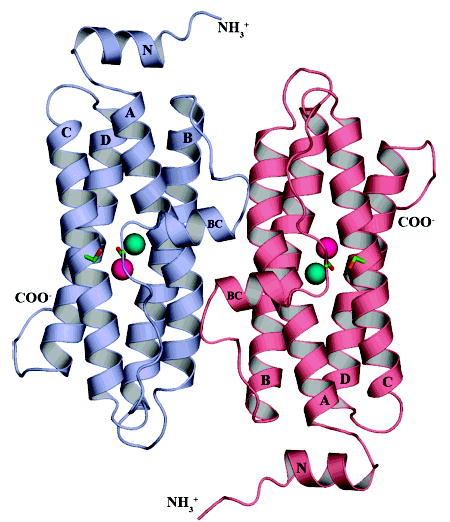
DPS-like protein from Sulfolobus solfataricus. The ribbon diagram depicts 2 of the 12 subunits in the SsDPSL dodecamer viewed from the exterior surface of the dodecamer. The overall fold of the SsDPSL subunit is most similar to that of authentic DPS molecules. It is composed of a 4-helix bundle (helices A, B, C, and D), common to members of the ferritin superfamily, decorated by two additional helices. The BC helix is found in all DPS structures to date, whereas the N-terminal helix (helix N) is observed only in the L. lactis DPS structure (29). Each chain contains a ferroxidase site buried within the core of the four-helix bundle. In contrast, metal binding sites in authentic DPS dodecamers are found at the 2-fold interface. The ferroxidase site contains a mixture of metals at both the A- and B-sites (cyan and pink, respectively). The side chains for Cys101 and Cys126, which are adjacent to the ferroxidase site, are also shown in green (carbon) and orange (sulfur).
Additional ordered residues are seen at the N- and C-terminal ends of the four helix bundle. The model begins at the N-terminus with a short stretch of solvent exposed random coil present on the exterior of the dodecamer (Gln6–Val10), and then leads into a short α-helix (helix N, Val11–Lys18). An equivalent N-terminal helix is observed in only one previous DPS structure, that from Lactococcus lactis (29). In the SsDPSL protein, helix N runs across the N-terminal end of the bundle such that the termini of helices A and B lie on one side and C and D on the other (Figure 1). A four-residue loop then joins helix N to helix A of the four-helix bundle. After threading its way through the four-helix bundle, the polypeptide then exits helix D, where we see density for seven additional residues (Tyr168–His174). These residues are present as a random coil that extends into the interior of the dodecamer. The last 14 residues of the protein were not resolved in the electron density map.
Bacterioferritin-Like Metal Binding Site
Significant concentrations of both iron and zinc were identified in recombinant SsDPSL protein by ICP-MS. The analysis indicates approximately nine iron atoms, two zinc atoms, and 0.5 manganese atoms per dodecamer. Accordingly, two strong peaks corresponding to metal atoms were apparent in the electron density maps for each SsDPSL monomer. Interestingly, the metals are not at the dimer interface, as has been observed in all other DPS structures. Instead, the metals in SsDPSL are buried within the core of the four-helix bundle, as is seen for ferritin and bacterioferritin. These metals are coordinated by two histidine residues and four acidic residues (His73, His159, Glu37, Asp70, Glu124, and Glu156) in a canonical di-iron carboxylate metal binding motif that is most similar to that seen in bacterioferritin (Figures 2 and 3).
Figure 2.
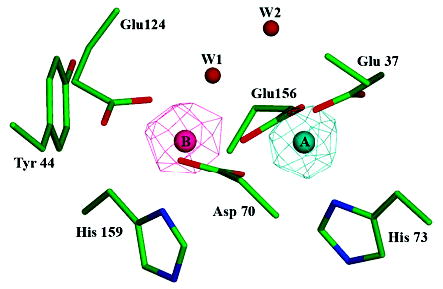
Bacterioferritin-like dimetal binding site. Residues Glu37, Asp70, His73, Glu124, Glu156, and His159 of SsDPSL form a dimetal binding site within the core of the four-helix bundle. Side chain atoms are colored in green (carbon), blue (nitrogen), and red (oxygen). Anomalous difference electron density maps (mesh) indicate asymmetric binding of the iron and zinc atoms, with Zn2+ binding preferentially to the A site (cyan) and Fe3+ to the B site (pink). The iron edge anomalous difference map is contoured at 8 σ (pink mesh), whereas the zinc edge anomalous difference map is contoured at 15 σ (cyan mesh). When contoured at lower levels, however, it is clear that each site contains a mixed population of both iron and zinc. Water W1 shows strong coordination to the B site metals but only weak coordination, at best, to the A site.
Figure 3.
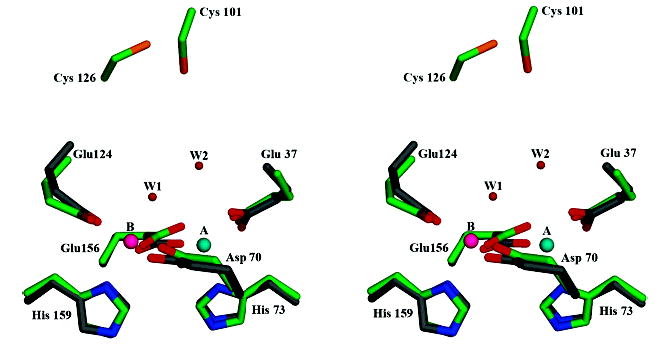
Superposition of the SsDPSL and E. coli bacterioferritin ferrioxidase sites. Residues composing the dimetal binding site of SsDPSL are superimposed on those of E. coli bacterioferritin (1BCF) (33). Atom types are colored as in Figure 2; however, the carbon atoms of the bacterioferritin side chains are in gray. The cysteine pair of SsDPSL is also depicted and can be seen above the dimetal binding site. For clarity, only residues in SsDPSL are labeled.
In an effort to differentiate between iron and zinc in the electron density maps, additional X-ray diffraction data were collected at the iron and zinc absorption edges. The resulting anomalous difference maps indicate a mixture of both iron and zinc at each of the metal binding sites. However, the relative populations of the Fe3+ and Zn2+ ions at the two sites apparently differ and are, thus, nonequivalent in this regard (Figure 2). Nonequivalence at these sites, generally referred to as the A and B sites (9), has been observed previously (30–32). The zinc edge anomalous difference map shows a strong 24 σ peak at site A and a weaker 8 σ peak at site B, indicating significantly higher Zn2+ occupancy at the A site compared to that of the B site. In contrast, the iron edge anomalous difference map shows a 10 σ peak at B and a 4 σ peak at A, indicating higher Fe3+ occupancy at the B site as opposed to the A site. Importantly, model refinement indicates that both A and B sites are at or near full occupancy with respect to iron and/or zinc. For example, the best Rfree value, a measure of agreement between the observed and predicted diffraction patterns, is obtained with Fe3+ at full occupancy in the B site and either Zn2+ or Fe3+ at full occupancy in the A site. In reality, however, each site probably contains a mixture of metals. These include iron, zinc and manganese present in the particle as purified from E. coli as well as calcium and associated impurities present in the crystallization mother liquor to facilitate crystal growth.
The A site metals are coordinated by Glu37, Asp70, His73, and Glu156, whereas the B site metals are coordinated by Asp70, Glu124, Glu156, His159, and a bound water, W1 (Table 3). The distance between the two metal centers is 3.7 Å. A second water, W2, is also found in the vicinity of the dimetal binding site. However, it is too far from the A site (3.8 Å) to coordinate the metal. Instead, W2 forms a hydrogen-bonded bridge between W1 and Glu37. In addition, the carboxylates at each end of the active site (Glu37 and Glu124) are stabilized by hydrogen bonds to aromatic residues (Trp131 to Glu37 and Tyr44 to Glu124). Overall, the active site in SsDPSL is remarkably similar to that seen in bacterioferritins (32–34). More detailed comparisons can be made upon completion of DPSL structures with defined metal content.
Table 3.
Active Site Interatomic Distancesa
| His73ND1 | Glu37OE1 | Glu37OE2 | Asp70OD1 | Glu156OE1 | Cys101SG | |
|---|---|---|---|---|---|---|
| Metal A | 2.16 (2.08–2.27) | 2.18 (2.10–2.23) | 2.38 (2.32–2.45) | 2.21 (2.15–2.24) | 2.07 (2.01–2.13) | 8.63 (8.35–8.77) |
| His159ND1 | Glu124OE1 | Glu124OE2 | Asp70OD2 | Glu156OE2 | Cys126SG | |
|---|---|---|---|---|---|---|
| Metal B | 2.51 (2.43–2.54) | 2.27 (2.23–2.32) | 2.54 (2.49–2.60) | 2.55 (2.47–2.61) | 2.32 (2.21–2.49) | 10.29 (10.19–10.40) |
| Metal A | Metal B | Cys101SG | Cys126SG | |||
|---|---|---|---|---|---|---|
| Water W1 | 3.46 (3.34–3.63) | 2.44 (2.32–2.55) | 6.49 (6.40–6.64) | 8.22 (8.13–8.34) | ||
| Water W2 | 3.76 (3.37–3.94) | 4.93 (4.68–5.13) | 5.05 (4.72–5.40) | 7.26 (6.93–7.63) |
The average distance (Å) is reported for all eight subunits in the asymmetric unit, with ranges in parentheses.
As one might expect, a search for structurally similar proteins using the DALI or VAST servers indicates greatest similarity (based on Z-scores) to DPS structures. However, it also provides structure based alignments with other members of the ferritin-like superfamily, including bacterioferritin and ferritin, as well as other proteins containing a similar dimetal binding site buried within a four helix bundle, including manganese catalase and ribonucleotide reductase. For E. coli bacterioferritin, the DALI server identifies 136 structurally equivalent residues with 20% sequence identity, whose Cα atoms superpose with an RMSD of 2.0 Å. For manganese catalase from Lactobacillus plantarum (33, 35), 131 structurally equivalent Cα atoms showing 18% sequence identity superimpose with an RMSD of 1.9 Å. When the superposition is restricted to metal binding residues, the E. coli bacterioferritin and manganese catalase from Lactobacillus plantarum superimpose on the active site residues of SsDPSL with an RMSD of 0.5 and 0.7 Å, respectively (Figure 3). In light of this close similarity to bacterioferritin, it is not surprising that SsDPSL is a member of Pfam CD1052.2, a group of bacterioferritin-like proteins (14). To our knowledge, SsDPSL is the first protein from this family for which a structure has been determined.
Active Site Solvent Channel
An important feature of SsDPSL is the presence of a boot shaped solvent channel leading from the exterior of the dodecamer to the ferroxidase center within each subunit (Figure 4A and B). The channel leads to the metal B site, makes a turn of approximately 90°, and then continues along the longitudinal axis of the 4-helix bundle, creating a polar cavity directly adjacent to the dimetal site. At least two water molecules are present in the cavity, W1 and W2 discussed above, suggesting that the active site is solvent accessible and might allow ferrous ions, hydrogen peroxide, or water to access the active site from the exterior of the dodecameric particle. A channel providing access to the ferroxidase site is also seen in structures of bacterioferritin (32, 36).
Figure 4.
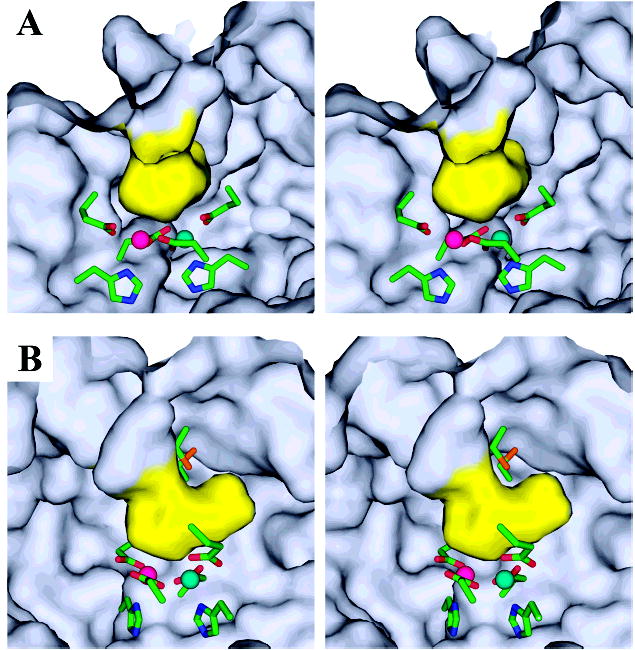
Active site channel. (A) The ferroxidase center lies adjacent to a boot shaped, solvent filled channel that opens to the outside surface of the dodecamer. The van der Waals surface of the active site cavity (yellow) is shown as viewed from inside the monomer from a position behind the heel of the boot shaped cavity. The line of sight is approximately down the longitudinal axis of the four-helix bundle, looking toward the N-terminal end. In this orientation, the exterior of the dodecamer is up, and the internal cavity of the dodecamer is down. The conserved cysteine pair is hidden from view behind the upper surface of the active site channel (the tongue of the boot). The dimetal binding site lies beneath the channel (the heel of the boot). Metal sites A and B are depicted in cyan and pink, respectively. Active site residues are colored by atom: carbon, green; nitrogen, blue; oxygen, red and sulfur, orange. (B) The relative proximity of the cysteine pair (boot laces) to the dimetal binding site is more apparent in this view, in which the ferroxidase center has been rotated 60° about the vertical axis.
Cysteine Pair
The SsDPSL monomer contains three cysteine residues, Cys101, Cys126, and Cys135. Importantly, Cys101 and Cys126 are found together, juxtaposed between the exterior surface of the particle and the boot shaped channel that leads to the ferroxidase center. Thus, the sulfur atom of Cys126 is solvent exposed on the exterior surface of the particle, whereas the side chain of Cys101 reaches in toward the solvent channel. The arrangement is such that the cysteine pair and the metals of the ferroxidase center are found on opposite sides of the channel (approximately 9.5 Å apart), separated only by the intervening solvent molecules (Figures 3 and 4B). Importantly, the sulfur atoms of Cys101 and Cys126 are immediately adjacent to each other. However, the distance between the sulfur atoms in the refined structure is 2.6 Å, significantly greater than the mean disulfide bond distance of 2.0 ± 0.1 Å. Thus, despite the presence of some bridging electron density between these sulfur atoms, it appears that they are present in the reduced form. We note, however, that this 2.6 Å sulfur–sulfur distance has been observed before in situations where a labile disulfide bond is cleaved in the presence of synchrotron radiation (37), raising the possibility that the cysteine residues were in the oxidized, disulfide bonded state prior to X-ray exposure at the synchrotron.
To address the oxidation state of the cysteine residues prior to exposure to X-rays, we probed freshly purified SsDPSL protein for the presence of free sulfhydryl groups with Ellman’s reagent (DTNB) (17). These experiments indicate approximately 0.15 DTNB reactive cysteine residues per subunit in the native conformation and 0.85 cysteine residues per subunit in protein denatured with 6 M guanidine-HCl. This suggests the presence of a single DTNB reactive residue that becomes accessible only upon denaturation of the protein, presumably Cys135. This lone cysteine lies partially solvent exposed on the interior of the particle at the C-terminal end of helix C; it is likely to be inaccessible to DTNB in the native state. These data are, thus, consistent with a disulfide bond between Cys101 and Cys126. Importantly, this implies an intact disulfide bond prior to synchrotron data collection, with subsequent bond cleavage as a result of X-ray exposure. Because the disulfide bond is cleaved by synchrotron radiation after the crystal has been frozen, we believe that the structure presented here is most similar to that of SsDPSL with an intact disulfide. Conformational changes normally associated with the reduction of the disulfide bond in solution are probably prevented by the vitreous ice surrounding the protein in the frozen crystal, that is, the oxidized structure, has been frozen in place. Thus, conformational changes associated with disulfide bond cleavage are probably not reflected in the structure described here.
The SsDPSL protein is a member of Pfam CD 1052.2 (14), a bacterioferritin-like family that has not been extensively characterized. We thus used a position specific iterated (PSI) BLAST search to identify additional DPSL homologues that might help to illuminate the function of this protein family (Figure 5). SsDPSL was used as the initial seed in the PSI-BLAST search to identify the remaining members of Pfam CD 1052.2. These 10 proteins were then used to seed the second round, followed by a third round in which default cutoff values were accepted. This search identified 27 proteins with strong homology to SsDPSL, all exhibiting E values less than 10−46 (Figure 5). Residues corresponding to the Cys101–Cys126 pair in SsDPSL were present in all but one of these 27 proteins. The exception is a sequence from an uncultured crenarchaeote, 4B7, wherein one of the two cysteines is mutated to a glycine. Athough a single nucleotide sequencing error could account for this glycine, 4B7 also lacks an otherwise strictly conserved tyrosine residue that is found in bacterioferritin, the SsDPSL protein (Tyr44), and its remaining homologues. For these reasons, the uncultured crenarchaeote is not included in Figure 5.
Figure 5.
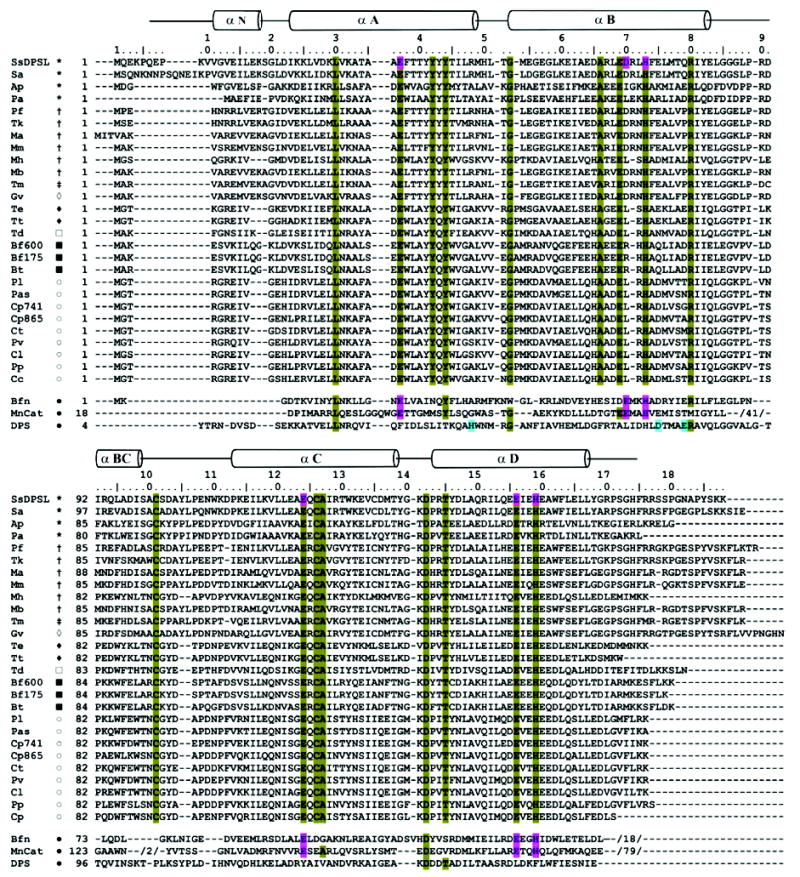
SsDPSL sequence alignments. Position specific iterated (PSI) BLAST (62, 63) was used to identify 26 SsDPSL homologues. The alignments reveal strong sequence conservation within the ferroxidase center and for the cysteine pair (Cys101 and Cys126). The Figure also presents structure based alignments between SsDPSL and structures representative of bacterioferritin (Bfn: E. coli, 1BCF (33)), DPS (DPS: E. coli, 1DPS (7)), and manganese catalase (MnCat: L. plantarum, 1JKU (35)) (bottom). Secondary structural elements for SsDPSL are indicated above the sequence. Conserved residues involved in metal coordination are found at structurally equivalent positions in the SsDPSL protein, E. coli bacterioferritin, and L. plantarum manganese catalase (pink) but not in E. coli DPS (1DPS). Conversely, the E. coli DPS metal binding site residues (blue) are not conserved in SsDPSL. Residues conserved among the DPSL homologues are highlighted in olive; this includes residues involved in metal coordination and the cysteine pair. The canonical dimetal carboxylate motif within helix B is D/ExxH, with two residues separating the acidic residue and the histidine. Two SsDPSL homologues do not satisfy this spacing; both Ap and Pa have 3 residues separating the acidic residue and the histidine at the first metal site. This and the presence of adjacent acidic residues result in local misalignment by ClustalW within helix B. However, the individual sequence alignments from PSI-BLAST do correctly align the D/ExxH motif in the B helix. Members of Pfam CD1052.2 used to seed the search are denoted as follows: SsDPSL, Sulfolobus solfataricus DPS-like protein; Pf, Pyrococcus furiosus DPS-like protein; Tm, Thermotoga maritima hypothetical protein TM0560; Mb, Methanosarcina barkeri ferritin-like protein; Ap, Aeropyrum pernix hypothetical protein APE1457; Pa, Pyrobaculum aerophilum hypothetical protein PAE2701; Tt, Thermoanaerobacter tengcongensis hypothetical protein TTE2230; Ct, Chlorobium tepidum hypothetical protein CT1328; Bt, Bacteroides thetaiotaomicron hypothetical protein BT3823. The 18 additional homologues identified by PSI-BLAST are from Sa, Sulfolobus acidocaldarius; Tk, Thermococcus kodakarensis; Ma, Methanosarcina acetivorans; Mm, Methanococcus maripaludis S2; Mh, Methanospirillum hungatei; Gv, Gloeobacter violaceus; Te, Thermoanaerobacter ethanolicus; Td, Thiomicrospira denitrificans; Bf600, Bacteroides fragilis gi:52217600; Bf175, Bacteroides fragilis gi:60494175; Pl, Pelodictyon luteolum; Pas, Prosthecochloris aestuarii; Cp741, Chlorobium phaeobacteroides gi:67939741; Cp865, Chlorobium phaeobacteroides gi:67934865; Pv, Prosthecochloris vibrioformis; Cl, Chlorobium limicola; Pp, Pelodictyon phaeoclathratiforme; and Cc, Chlorobium chlorochromatii. Symbols indicate taxa: (*) crenarchaeotes; (†) euryarchaeotes; (‡) thermotogales; (⋄) cyanobacteria; ((♦) eubacteria; (□) e-proteobacteria; (▪) CFB group bacteria; (○) green sulfur bacteria; (•) bacteria.
Although each of the SsDPSL homologues has an E value of 10−46 or less, the E value rises to 10−22 for the next most similar sequence, a putative bacterioferritin from Xanthomonas campestris, and with a single exception, both cysteine residues are absent in all sequences with E values greater than 10−22. Thus, the occurrence of these cysteine residues in the primary sequence is highly coupled, suggesting that they act in concert and are consistent with disulfide bond formation. Therefore, the combination of a ferroxidase-like active site and the conserved cysteine pair, a thio-ferritin motif, are defining elements of the DPSL family of proteins.
Although the occurrence of disulfide bonds in intracellular proteins is generally rare, there is strong evidence that hyperthermophiles, including Sulfolobus, do make use of intracellular disulfides to confer protein stability at extreme temperatures (38–40). Thus, it is possible that the Cys101–Cys126 pair plays a structural role. However, the occurrence of the cysteine-pair motif also extends to a number of mesophilic organisms. For example, the motif is found in the genomes of all green sulfur bacteria sequenced to date as well as in several Bacteroides, close relatives to the green sulfur bacteria. The prevalence of the cysteine pair in the green sulfur bacteria and other mesophilic organisms argues against a structural role; the need for thermostability is absent in these organisms, and the general occurrence of intracellular disulfides are correspondingly uncommon. What then is the function of the conserved cysteine pair? One attractive possibility is that the cysteine pair is redox active, capable of cycling between reduced and oxidized forms, during which it contributes directly to the antioxidant properties of this family of proteins.
N-Terminal 3-Fold Interface
In DPS and the SsDPSL dodecamer, the N-terminal end of the four-helix bundles come together at one end of the 3-fold axis in a set of interactions that are similar to the 3-fold interactions in ferritin (7). Thus, interactions formed by the N-terminal end of the four-helix bundle are ferritin-like. In contrast, interactions at the opposite end of the 3-fold axis involve the C-terminal end of the four-helix bundle (7). The 3-fold interactions at the C-terminal end of the four-helix bundle are unique to DPS and DPS-like dodecamers.
Hydrophilic pores are found at the N-terminal 3-fold interfaces in ferritin, bacterioferritin, and DPS. The interior surface surrounding the pore, and the pore itself, generally show strong negative charge. In contrast, the exterior surfaces frequently create a mix of negative and positive potential, at least in mammalian ferritin. This serves to create an electrostatic field that is thought to direct ferrous ions into the interior of the protein shell (41). For the SsDPSL particle, however, surface electrostatics indicate a local positive potential surrounding the exterior of the 3-fold axis (Figure 6A), and a relatively constricted, negatively charged pore. The external positive charge is contributed by lysine residues present in the N-terminal α-helix and the extended N-terminus (Lys9 and Lys18).
Figure 6.
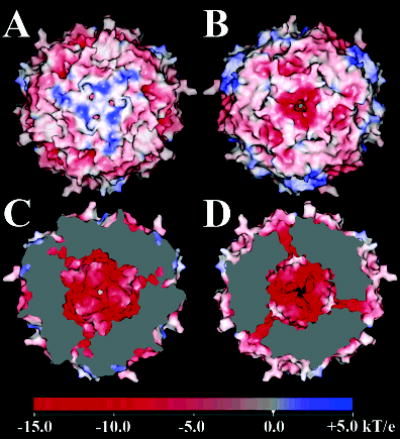
Surface electrostatic potential of the SsDPSL dodecamer. (A) Exterior surface surrounding the N-terminal 3-fold interface. The positive surface potential, indicated in blue, is imparted by basic residues present in the N-terminal helix and the N-terminus. Three small pores opening from the N-terminal channel are immediately adjacent to the center of symmetry. (B) Exterior surface potential surrounding the C-terminal 3-fold pore. The surrounding surface is acidic, with a strong negative potential (red). Relative to panel A, the dodecamer has been rotated 180° about the vertical axis. (C) Interior surface at the N-terminal 3-fold interface. Relative to panel A, the dodecamer has been rotated 180° about the vertical axis, and the clipping plane has been positioned to cut away a portion of the particle, revealing the negatively charged (red) internal surface surrounding the N-terminal pore (center). An oblique view of the surfaces lining the three symmetry related N-terminal pores is also apparent, seen as channels of negative potential (red) connecting the interior and exterior surfaces. Unlike the C-terminal channel (below), the N-terminal channels do not lie completely within the clipping plane; hence, a contiguous surface from the interior to the exterior of the dodecamer is not apparent. (D) Interior surface surrounding the C-terminal pore. Relative to panel B, the dodecamer has been rotated 180° about the vertical axis, and the clipping plane has been positioned to cut away a portion of the particle, revealing the negatively charged (red) internal surface surrounding the acidic C-terminal pore (center). An oblique view of the surfaces lining the three symmetry related C-terminal pores is also seen, with red channels of negative potential connecting the interior and exterior surfaces. Electrostatic potentials were generated with SPOCK (28), using a probe radius of 1.4 Å, a temperature of 353 K, an ionic strength of 0.15 M, and protein and solvent dielectric constants of 4 and 80, respectively.
The exterior opening to the channel is partially occluded by three symmetry related Tyr139 side chains (Figure 7A). The tyrosine side chains split the exterior end of a larger pore into three smaller symmetry related arms that open to the outside (Figure 6A). Electron density in the distal portion of these tyrosine side chains is weak, indicating a possible rotation of the side chain about the Cα–Cβ bond. Such movement might serve to modulate the dimensions of the opening to this channel, reminiscent of dynamics suggested to be important in ferritin (42). An ordered water molecule is found in each of the three smaller arms. These arms converge deeper within the pore, where three additional tyrosine side chains, contributed by Tyr146, are found to form the walls of a central channel. An additional ordered water is observed to interact with each subunit inside the central channel. The channel then opens to the interior of the dodecamer through a 3.0 Å opening (Figure 6C). Overall, the partially occluded opening to the exterior with its surrounding positive charge suggests similarity to the somewhat unique 3-fold channel in Desulfovibrio desulfuricans bacterioferritin (32, 36).
Figure 7.
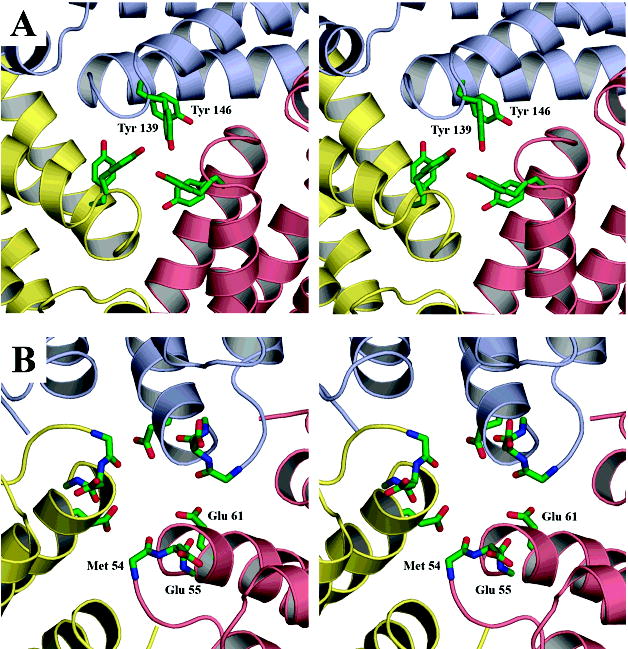
N- and C-terminal pores. Subunits related by the 3-fold symmetry of the particle are colored in yellow, pink, and blue. (A) N-terminal pore. Tyr139 forms a partial cap over the exterior end of the N-terminal channel, whereas the side chains of Tyr146 (behind Tyr139) help to form the walls of the channel. (B) C-terminal pore. Three successive layers of negative charge line the C-terminal pore. The first layer, on the exterior of the particle, is composed of three Glu55 side chains. The second layer comprises the carbonyl groups of Glu55 and Met54. The side chains of Met54 have been omitted for clarity. The third layer is contributed by the side chains of Glu61 on the interior of the dodecamer. In both views, the line of sight is along the 3-fold axis, looking from the exterior toward the interior. Atom colors are carbon, green; nitrogen, blue; and oxygen, red.
C-Terminal 3-Fold Interface
A channel through the shell of the SsDPSL dodecamer is also present at the C-terminal 3-fold interface (Figure 6B and D and 7B). Electrostatic calculations mapped to the surface demonstrate strong negative potential surrounding the channel. The channel itself also shows strong negative potential, it is lined with three successive rings of negative charge. First, a trio of symmetry related carboxylates, contributed by Glu55, forms a layer on the outside edge of the pore. Working inward, a second layer of negative charge is contributed by the six carbonyls of Met54 and Glu55. This ring of carbonyl oxygen atoms is well positioned to interact with desolvated ferrous ions that might pass through the channel. The carboxylate moiety of Glu61 contributes the final layer of negative charge as the pore empties into the interior of the dodecamer.
Only weak electron density is seen for the Glu55 side chains, suggesting that they sample multiple conformations. In contrast, positions of the other atoms lining the channel are better resolved, with the three side chains of Glu61 working to coordinate a water molecule in the middle of the channel as it empties into the cavity of the dodecamer. Halfway through the channel, the Cαatoms of Gly58 form a constriction point, resulting in a pore with a minimal diameter of 4.2 Å. The presence of an acidic pore at the C-terminal interface in the SsDPSL particle is in direct contrast to the situation for DPS, where the more acidic pore is found at the N-terminal or ferritin-like 3-fold interface.
Conserved Tyrosine Residues
In addition to the thioferritin motif, composed of the six residues coordinating the metal ions and the neighboring cysteine pair, the multiple sequence alignment identifies 10 additional conserved residues in the DPSL protein family (Figure 5). In the SsDPSL protein, two of these residues, Tyr42 and Tyr44, fall within a larger constellation of four consecutive tyrosine residues (Tyr41–Tyr44). Although the tyrosines at positions 41 and 43 are not strictly conserved, all of the sequences displaying the thioferritin motif contain at least three of these four tyrosine residues. Interestingly, the four tyrosine residues radiate from the A helix, tracing a path from the ferroxidase active site toward the 2-fold symmetric subunit interface.
A structural role is suggested for Tyr 42 because it is hydrogen bonded across the subunit interface to the carbonyl oxygen of Pro89. In contrast, Tyr44 is more likely to play a functional role. Tyr44 is found at the periphery of the ferroxidase active site. A similarly positioned tyrosine side chain is found in the active sites of bacterioferritin, Mn catalase, and ribonucleotide reductase. In ribonucleotide reductase, this residue is observed to form a functionally essential tyrosyl free radical (43). Mutation of the corresponding tyrosine residue in ferritin does slow the rate of iron oxidation; however, it is not essential for the activity of the ferroxidase center (44–46). Thus, the exact role of this residue in bacterioferritin and SsDPSL is not clear.
Though not strictly conserved, Tyr41 is also of interest. The Tyr41 side chain is positioned immediately adjacent to the 2-fold axis, resulting in a pair of Tyr41 side chains that interact across the 2-fold interface of SsDPSL. The equivalent position in bacterioferritin is occupied by a heme group. Because Tyr44 is at the edge of the ferroxidase site while Tyr41 is at the 2-fold symmetric interface, these tyrosines serve to connect a pair of ferroxidase centers to each other via the 2-fold center of symmetry (Tyr44–Tyr41·Tyr41′–Tyr44′). Thus, these residues might represent electron transfer pathways leading to or from the ferroxidase center or might mimic the roles of the active site tyrosine residue and the intersubunit heme group in bacterioferritin.
DISCUSSION
DPS structures show varying numbers of ordered residues at their N-termini. However, an N-terminal α-helix has only been observed once before, in the recently determined structure of DPS from Lactococcus lactis (29). Lysines or other residues in this N-terminal region are implicated in mediating DNA binding in L. lactis and E. coli DPS (3, 29). In the SsDPSL structure, there are three lysines in the N-terminal region, two of which are present in the model. These lysines may mediate DNA binding to the SsDPSL protein, although further work is required to test this hypothesis. The presence of the N-terminal α-helix in L. lactis DPS, and now in the SsDPSL protein, suggests a mechanism for DNA recognition by these proteins. Specifically, the N-terminal residues on the exterior of the dodecameric particles may adopt α-helical structures that interact with the major groove in DNA. In contrast, a role for the C-terminus of the SsDPSL protein in DNA binding is seemingly ruled out because the C-terminal tail projects into the interior of the particle. This is a distinct difference between the SsDPSL structure and the E. coli DPS structure, where all C-terminal residues of the protein are found on the exterior of the dodecamer. Although the C-terminal residues of the two L. lactis DPS models are on the exterior of the dodecamer, there are an additional 6–8 disordered residues at the C-terminus that are not present in these models (29).
The mineralization reaction catalyzed by bacterioferritin and DPS results in the simultaneous consumption of H2O2 and Fe2+, thus inhibiting hydroxyl radical production by the Fenton reaction. Accordingly, transcription of the dpsl gene in S. solfataicus is increased in response to elevated levels of H2O2 (12). However, contrary to expectations, elevated iron levels reduce transcription of dpsl, whereas transcription is increased when S. solfataricus is placed in iron deficient media (12). On the surface, this behavior seems inconsistent with mineralization as the primary role for this enzyme, suggesting an additional function for the SsDPSL protein, perhaps a peroxidase- or catalase-like activity that would serve to reduce H2O2. In this regard, the structural similarity of the dinuclear active site in SsDPSL with bacterioferritin, or even Mn catalase, may be relevant.
Unlike ferritin, the ferroxidase center in bacterioferritin appears to be stable in the oxidized form; it does not spontaneously return to the apo form (30, 47). Importantly, recent work (6, 47) demonstrates that in addition to oxygen, bacterioferritin is also able to utilize hydrogen peroxide as an oxidant for Fe2+ at the ferroxidase center, resulting in the production of a μ-oxo-bridged diferric center and water. This reaction is illustrated by eq II, where FC indicates iron bound at the ferrroxidase center, as opposed to the core or elsewhere.
| (II) |
Further, Baaghil et al. (2) suggest that electrons produced by the ongoing mineralization reaction in the bacterioferritin core are funneled back to the ferroxidase center, where they reduce the μ-oxo-bridged diferric species to a diferrous center (eq III).
| (III) |
The sum of eqs (II) and (III) is thus eq IV.
| (IV) |
In this proposed mechanism (2), the growing iron oxide core is the site of overall Fe2+ oxidation, but oxygen reduction occurs only at the ferroxidase center.
The presence of Fe3+ in the ferroxidase center of SsDPSL suggests that its ferroxidase center may also be stable in the oxidized form, and like bacterioferritin, it may also react with hydrogen peroxide to produce a μ-oxo-bridged diferric center and water. However, in the presence of hydrogen peroxide and a general absence of Fe2+, conditions that most strongly up-regulate expression of the SsDPSL protein, the mineralization reaction cannot occur. This suggests the presence of an alternative source of electrons to reduce the μ-oxo-bridged diferric species back to the diferrous center. One possible source is the conserved cysteine pair. Oxidation of the two cysteines to form the disulfide results in the production of two electrons and two protons, as shown in eq V, where R indicates Cys101 and Cys126 near the ferroxidase center.
| (V) |
The sum of eqs III and V result in eq VI, with regeneration of the diferrous active site.
| (VI) |
A thiol–disulfide exchange reaction could then regenerate the reduced cysteine at the SsDPSL active site (eq VII).
| (VII) |
The sum of these reactions is then eq VIII, in which hydrogen peroxide is reduced to water with concomitant oxidation of a cellular thiol pool (R′SH).
| (VIII) |
Thus, SsDPSL could conceivably function as a peroxidase, in which redox active cysteine residues serve to funnel electrons to the ferroxidase center, recycling the iron. With respect to the cysteine residues, this potential mechanism is somewhat analogous to that in peroxiredoxins such as alkyl hydroperoxidase C (AhpC) (48, 49), the thioredoxin-linked peroxidase activity of Tpx (50, 51), or the thioredoxin dependent activity of bacterioferritin comigratory proteins (52). A number of thioredoxin, glutaredoxin, and ferredoxin type molecules have been annotated in the Sulfolobus solfataricus genome; this includes thioredoxins a, b, and c, and a protein disulfide oxidoreductase (15). Although speculative, this scheme is consistent with increased expression of SsDPSL in response to hydrogen peroxide under iron-limiting conditions. It also suggests a functional basis for conservation of the cysteine pair.
Alternatively, SsDPSL might function as a catalase, perhaps with manganese or some other metal at the ferroxidase center, or analogous to the thioredoxin and ferredoxin dependent thiol-peroxidases, the cysteine residues might confer a peroxidase activity upon SsDPSL that is independent of the ferroxidase center. Finally, the cysteine residues might play an important regulatory role, perhaps allowing the enzyme to sense the redox state of the cell in order to modulate the activity of the enzyme or its ability to bind and protect DNA.
The occurrence of the thioferritin motif in Bacteroides fragilis (Figure 5) is of particular interest. B. fragilis is a strict anaerobe that comprises approximately 1–2% of the normal intestinal flora in humans. However, it is isolated from 81% of anaerobic clinical infections (53). Bacteroides are among the most aerotolerant of anaerobes, able to tolerate atmospheric concentrations of oxygen for up to 3 days (54). During initiation of an intra-abdominal infection, oxygen tolerance is believed to allow the bacteria to survive in the oxygenated tissue of the abdominal cavity until E. coli and other synergistic organisms are able to reduce the redox potential at the site of infection. Oxygen tolerance is also key to surviving the production of reactive oxygen by the host immune system (55), and in B. fragilis, oxygen stress is known to induce expression of numerous proteins, including catalase (56–59), various peroxidases (60), an authentic DPS (59), and ferritin (61). It will be interesting to determine whether the DPS-like protein in B. fragilis is also upregulated in response to hydrogen peroxide. If so, it may contribute to the extreme aerotolerance, and thus the infectivity, of this organism.
In summary, structural studies of SsDPSL reveal a dodecameric DPS-like assembly that houses a bacterioferritin-like dimetal binding site with a proximal cysteine pair. Residues constituting the dimetal binding site and the cysteine pair, together, comprise a thioferritin motif that marks membership in this family of proteins. Although SsDPSL certainly plays a role in the antioxidant response of S. solfataricus, further work will be required to elucidate the molecular details associated with this unique combination of structural features. Thus, this work clearly illustrates the need for additional biochemical and genetic investigations into the role of the DPSL family of proteins. It also demonstrates the great value inherent in studies of oxidative stress in Archaea.
Acknowledgments
We thank Seth Staples and Lars Liepold for valuable assistance with the ICP-MS and LC/MS analyses.
Footnotes
This work was supported by the National Institutes of Health (DK57776) and the National Aeronautics and Space Administration (NAG5-8807). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. The Macromolecular Diffraction Laboratory at Montana State University was supported, in part, by a grant from the Murdock Foundation.
The atomic coordinates and structure factors have been deposited with the Protein Data Bank under accession code 2CLB.
Abbreviations: DPS, DNA binding protein from nutrient starved cells; DPSL, DPS-like proteins; ROS, reactive oxygen species; IPTG, isopropyl β-D-thiogalactopyranoside; ICP-MS, inductively coupled plasma mass spectrometry; DTNB, 5,5′-dithio-bis-(2-nitrobenzoic acid); MAD, multiwavelength anomalous dispersion; SAD, single wavelength anomalous dispersion; NCS, noncrystallographic symmetry; TLS, translation, libration, and screw rotation tensors; RMSD, root-mean-square deviation.
References
- 1.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.Baaghil S, Lewin A, Moore GR, Le Brun NE. Core formation in Escherichia coli bacterioferritin requires a functional ferroxidase center. Biochemistry. 2003;42:14047–14056. doi: 10.1021/bi035253u. [DOI] [PubMed] [Google Scholar]
- 3.Ceci P, Cellai S, Falvo E, Rivetti C, Rossi GL, Chiancone E. DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus. Nucleic Acids Res. 2004;32:5935–5944. doi: 10.1093/nar/gkh915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao GH, Ceci P, Ilari A, Giangiacomo L, Laue TM, Chiancone E, Chasteen ND. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells: A ferritin-like DNA-binding protein of Escherichia coli. J Biol Chem. 2002;277:27689–27696. doi: 10.1074/jbc.M202094200. [DOI] [PubMed] [Google Scholar]
- 5.Ilari A, Latella MC, Ceci P, Ribacchi F, Su M, Giangiacomo L, Stefanini S, Chasteen ND, Chiancone E. The unusual intersubunit ferroxidase center of Listeria innocua Dps is required for hydrogen peroxide detoxification but not for iron uptake. A study with site-specific mutants. Biochemistry. 2005;44:5579–5587. doi: 10.1021/bi050005e. [DOI] [PubMed] [Google Scholar]
- 6.Bou-Abdallah F, Lewin AC, Le Brun NE, Moore GR, Chasteen ND. Iron detoxification properties of Escherichia coli bacterioferritin. Attenuation of oxyradical chemistry. J Biol Chem. 2002;277:37064–37069. doi: 10.1074/jbc.M205712200. [DOI] [PubMed] [Google Scholar]
- 7.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 8.Chiancone E, Ceci P, Ilari A, Ribacchi F, Stefanini S. Iron and proteins for iron storage and detoxification. Biometals. 2004;17:197–202. doi: 10.1023/b:biom.0000027692.24395.76. [DOI] [PubMed] [Google Scholar]
- 9.Lewin A, Moore GR, Le Brun NE. Formation of protein-coated iron minerals. J Chem Soc, Dalton Trans. 2005:3597–3610. doi: 10.1039/b506071k. [DOI] [PubMed] [Google Scholar]
- 10.Zeth K, Offermann S, Essen LO, Oesterhelt D. Iron-oxo clusters biomineralizing on protein surfaces: Structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states. Proc Natl Acad Sci USA. 2004;101:13780–13785. doi: 10.1073/pnas.0401821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ilari A, Stefanini S, Chiancone E, Tsernoglou D. The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nat Struct Biol. 2000;7:38–43. doi: 10.1038/71236. [DOI] [PubMed] [Google Scholar]
- 12.Wiedenheft B, Mosolf J, Willits D, Yeager M, Dryden K, Young M, Douglas T. An archaeal antioxidant: Characterization of a Dps-like protein from Sulfolobus solfataricus. Proc Natl Acad Sci USA. 2005;102:10551–10556. doi: 10.1073/pnas.0501497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramsay B, Wiedenheft B, Allen M, Gauss GH, Martin Lawrence C, Young M, Douglas T. Dps-like protein from the hyperthermophilic archaeon Pyrococcus furiosus. J Inorg Biochem. 2006;100:1061–8. doi: 10.1016/j.jinorgbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CCY, Clausen IG, Curtis BA, De Moors A, Erauso G, Fletcher C, Gordon PMK, Heikamp-de Jong I, Jeffries AC, Kozera CJ, Medina N, Peng X, Thi-Ngoc HP, Redder P, Schenk ME, Theriault C, Tolstrup N, Charlebois RL, Doolittle WF, Duguet M, Gaasterland T, Garrett RA, Ragan MA, Sensen CW, Van der Oost J. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Ellman GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter C, Sweet R, editors. Macromolecular Crystallography, Part A. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 19.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr Sect D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr, Sect D. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr Sect D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr Sect A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 24.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr Sect D. 2001;57:122–33. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 26.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 27.DeLano WL. DeLano Scientific. 2002. [Google Scholar]
- 28.Christopher, J. A. (1998).
- 29.Stillman TJ, Upadhyay M, Norte VA, Sedelnikova SE, Carradus M, Tzokov S, Bullough PA, Shearman CA, Gasson MJ, Williams CH, Artymiuk PJ, Green J. The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding. Mol Microbiol. 2005;57:1101–1112. doi: 10.1111/j.1365-2958.2005.04757.x. [DOI] [PubMed] [Google Scholar]
- 30.Le Brun NE, Andrews SC, Guest JR, Harrison PM, Moore GR, Thomson AJ. Identification of the ferroxidase centre of Escherichia coli bacterioferritin. Biochem J. 1995;312:385–392. [PMC free article] [PubMed] [Google Scholar]
- 31.Treffry A, Zhao ZW, Quail MA, Guest JR, Harrison PM. Dinuclear center of ferritin: Studies of iron binding and oxidation show differences in the two iron sites. Biochemistry. 1997;36:432–441. doi: 10.1021/bi961830l. [DOI] [PubMed] [Google Scholar]
- 32.Macedo S, Romao CV, Mitchell E, Matias PM, Liu MY, Xavier AV, LeGall J, Teixeira M, Lindley P, Carrondo MA. The nature of the di-iron site in the bacterioferritin from Desulfovibrio desulfuricans. Nat Struct Biol. 2003;10:285–290. doi: 10.1038/nsb909. [DOI] [PubMed] [Google Scholar]
- 33.Frolow F, Kalb AJ, Yariv J. Structure of a unique 2-fold symmetric haem-binding site. Nat Struct Biol. 1994;1:453–460. doi: 10.1038/nsb0794-453. [DOI] [PubMed] [Google Scholar]
- 34.Swartz L, Kuchinskas M, Li H, Poulos TL, Lanzilotta WN. Redox-dependent structural changes in the Azotobacter vinelandii bacterioferritin: new insights into the ferroxidase and iron transport mechanism. Biochemistry. 2006;45:4421–4428. doi: 10.1021/bi060146w. [DOI] [PubMed] [Google Scholar]
- 35.Barynin VV, Whittaker MM, Antonyuk SV, Lamzin VS, Harrison PM, Artymiuk PJ, Whittaker JW. Crystal structure of manganese catalase from Lactobacillus plantarum. Structure. 2001;9:725–738. doi: 10.1016/s0969-2126(01)00628-1. [DOI] [PubMed] [Google Scholar]
- 36.Carrondo MA. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 2003;22:1959–1968. doi: 10.1093/emboj/cdg215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts BR, Wood ZA, Jonsson TJ, Poole LB, Karplus PA. Oxidized and synchrotron cleaved structures of the disulfide redox center in the N-terminal domain of Salmonella typhimurium AhpF. Protein Sci. 2005;14:2414–2420. doi: 10.1110/ps.051459705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beeby M, O’Connor BD, Ryttersgaard C, Boutz DR, Perry LJ, Yeates TO. The genomics of disulfide bonding and protein stabilization in thermophiles. PLoS Biol. 2005;3:e309. doi: 10.1371/journal.pbio.0030309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mallick P, Boutz DR, Eisenberg D, Yeates TO. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc Natl Acad Sci USA. 2002;99:9679–9684. doi: 10.1073/pnas.142310499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Connor BD, Yeates TO. GDAP: a web tool for genome-wide protein disulfide bond prediction. Nucleic Acids Res. 2004;32:W360–W364. doi: 10.1093/nar/gkh376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Douglas T, Ripoll DR. Calculated electrostatic gradients in recombinant human H-chain ferritin. Protein Sci. 1998;7:1083–1091. doi: 10.1002/pro.5560070502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin W, Takagi H, Pancorbo B, Theil EC. “Opening” the ferritin pore for iron release by mutation of conserved amino acids at interhelix and loop sites. Biochemistry. 2001;40:7525–7532. doi: 10.1021/bi002509c. [DOI] [PubMed] [Google Scholar]
- 43.Reichard P, Ehrenberg A. Ribonucleotide reductase—a radical enzyme. Science. 1983;221:514–519. doi: 10.1126/science.6306767. [DOI] [PubMed] [Google Scholar]
- 44.Treffry A, Bauminger ER, Hechel D, Hodson NW, Nowik I, Yewdall SJ, Harrison PM. Defining the roles of the threefold channels in iron uptake, iron oxidation and iron-core formation in ferritin: a study aided by site-directed mutagenesis. Biochem J. 1993;296:721–728. doi: 10.1042/bj2960721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Treffry A, Zhao Z, Quail MA, Guest JR, Harrison PM. Iron(II) oxidation by H chain ferritin: evidence from site-directed mutagenesis that a transient blue species is formed at the dinuclear iron center. Biochemistry. 1995;34:15204–15213. doi: 10.1021/bi00046a028. [DOI] [PubMed] [Google Scholar]
- 46.Fetter J, Cohen J, Danger D, Sanders-Loehr J, Theil EC. The influence of conserved tyrosine 30 and tissue-dependent differences in sequence of ferritin function: use of blue and purple F(III) species as reporters of ferroxidation. J Biol Inorg Chem. 1997;2:652–661. [Google Scholar]
- 47.Yang X, Le Brun NE, Thomson AJ, Moore GR, Chasteen ND. The iron oxidation and hydrolysis chemistry of Escherichia coli bacterioferritin. Biochemistry. 2000;39:4915–4923. doi: 10.1021/bi992631f. [DOI] [PubMed] [Google Scholar]
- 48.Poole LB. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys. 2005;433:240–254. doi: 10.1016/j.abb.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 49.Wood ZA, Schroder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 50.Wan XY, Zhou Y, Yan ZY, Wang HL, Hou YD, Jin DY. Scavengase p20: a novel family of bacterial antioxidant enzymes. FEBS Lett. 1997;407:32–36. doi: 10.1016/s0014-5793(97)00302-5. [DOI] [PubMed] [Google Scholar]
- 51.Zhou Y, Wan XY, Wang HL, Yan ZY, Hou YD, Jin DY. Bacterial scavengase p20 is structurally and functionally related to peroxiredoxins. Biochem Biophys Res Commun. 1997;233:848–852. doi: 10.1006/bbrc.1997.6564. [DOI] [PubMed] [Google Scholar]
- 52.Jeong W, Cha MK, Kim IH. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/Alkyl hydroperoxide peroxidase C (AhpC) family. J Biol Chem. 2000;275:2924–2930. doi: 10.1074/jbc.275.4.2924. [DOI] [PubMed] [Google Scholar]
- 53.Werner H. Differentiation and medical importance of saccharolytic intestinal anaerobes. Arzneim -Forsch. 1974;24:340–343. [PubMed] [Google Scholar]
- 54.Tally FP, Stewart PR, Sutter VL, Rosenblatt JE. Oxygen tolerance of fresh clinical isolates. J Clin Microbiol. 1975;1:161–164. doi: 10.1128/jcm.1.2.161-164.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Storz G, Zheng M. Oxidative Stress. In: Storz G, Hengge-Aronis R, editors. Bacterial Stress Responses. ASM Press; Washington, D.C.: 2000. pp. 47–60. [Google Scholar]
- 56.Rocha ER, Smith CJ. Regulation of Bacteriodes fragilis katB mRNA by oxidative stress and carbon limitation. J Bacteriol. 1997;179:7033–7039. doi: 10.1128/jb.179.22.7033-7039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rocha ER, Smith CJ. Biochemical and genetic analyses of a catalase from the anaerobic bacterium Bacteroides fragilis. J Bacteriol. 1995;177:3111–3119. doi: 10.1128/jb.177.11.3111-3119.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rocha ER, Selby T, Coleman JP, Smith CJ. Oxidative stress response in an anaerobe, Bacteroides fragilis: a role for catalase in protection against hydrogen peroxide. J Bacteriol. 1996;178:6895–6903. doi: 10.1128/jb.178.23.6895-6903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rocha ER, Owens G, Jr, Smith CJ. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J Bacteriol. 2000;182:5059–5069. doi: 10.1128/jb.182.18.5059-5069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herren CD, Rocha ER, Smith CJ. Genetic analysis of an important oxidative stress locus in the anaerobe Bacteroides fragilis. Gene. 2003;316:167–175. doi: 10.1016/s0378-1119(03)00759-5. [DOI] [PubMed] [Google Scholar]
- 61.Rocha ER, Smith CJ. Transcriptional regulation of the Bacteroides fragilis ferritin gene (ftnA) by redox stress. Microbiology. 2004;150:2125–2134. doi: 10.1099/mic.0.26948-0. [DOI] [PubMed] [Google Scholar]
- 62.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]


