Abstract
The protective antigen component of anthrax toxin forms a homoheptameric pore in the endosomal membrane, creating a narrow passageway for the enzymatic components of the toxin to enter the cytosol. We found that, during conversion of the heptameric precursor to the pore, the seven phenylalanine-427 residues converged within the lumen, generating a radially symmetric heptad of solvent-exposed aromatic rings. This “φ-clamp” structure was required for protein translocation and comprised the major conductance-blocking site for hydrophobic drugs and model cations. We conclude that the φ clamp serves a chaperone-like function, interacting with hydrophobic sequences presented by the protein substrate as it unfolds during translocation.
Anthrax toxin is composed of three nontoxic proteins, which combine on eukaryotic cell surfaces to form toxic, noncovalent complexes. [See (1) for a review.] Protective antigen (PA), the protein translocase component, binds to a cellular receptor and is activated by a furin-family protease. The resulting 63-kD receptor-bound fragment, PA63, self-assembles into the prepore, which is a ring-shaped homoheptamer (Fig. 1A). The prepore then forms complexes with the two ~90-kD enzymatic components, lethal factor (LF) and edema factor (EF). These complexes are endocytosed and delivered to an acidic compartment (2). There, the prepore undergoes an acidic pH-dependent conformational rearrangement (3) to form an ion-conducting, cation-selective, transmembrane pore (4), allowing bound LF and EF to translocate into the cytosol.
Fig. 1.
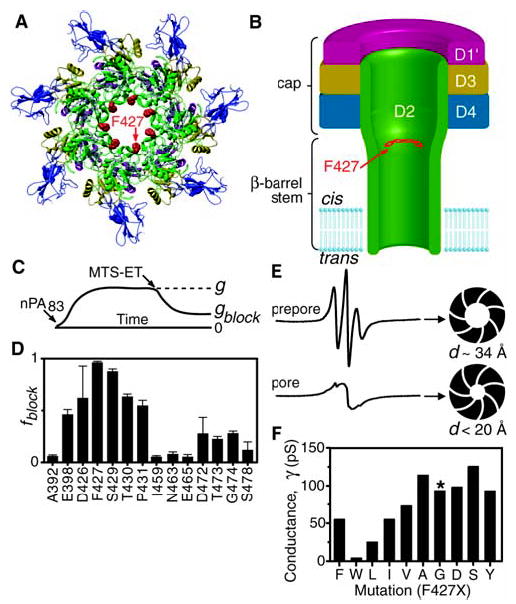
Structural models of a lumen-facing phenylalanine heptad. (A) A ribbons rendering of the PA63 prepore (27), viewed axially, where domain 4 is proximal. Domains are colored: D1′ (magenta), D2 (green), D3 (gold), and D4 (blue). F427 (red, space filling) is modeled into the structure. (B) Hypothetical cross section of the PA63 channel, or pore, colored as in (A). The membrane-spanning tube is the 14-stranded β barrel from domain 2 (5, 6). (C) Illustration of the effect of MTS-ET modification on Cys-substituted mutants of PA63 in macroscopic conductance studies. Conductance, g, is determined from the current, I, and Δψ as g = I/Δψ. (D) Fraction of conductance blocked ( fblock) by MTS-ET modification (28) in domain 2 cap residues [as in (C), where fblock = 1 − gblock/g] (table S2). Error bars show means + σSE (n = 3). (E) EPR spectra of PA63 heptamers uniformly labeled at F427C with a Cys-reactive nitroxide spin label in the prepore state at pH 8.5 (upper spectrum) and the pore state at pH 6 (lower spectrum). Approximate luminal diameters, d, are based on the observed spin-spin interactions. (F) Unitary conductance, γ, of single PA63 channels, with indicated substitutions at F427. Channels formed by F427G PA63 (*) initially opened to a conductance of 90 pS, but, unlike any of the other channels, flickered to 60 and 30 pS substates. γ values are accurate to at least ±10%, except for F427L and F427W, which are accurate to ±20%.
The PA63 pore (Fig. 1B) is believed to consist of a mushroom-shaped structure, with a globular cap connected to a β-barrel stem that is ~100 Å long (5, 6). A model of the 14-strand β barrel reveals its lumen, which is ~15 Å wide and can only accommodate structure as wide as an α helix (7). The narrow pore creates a structural bottleneck, requiring that the catalytic factors, LF and EF, unfold in order to be translocated (8, 9). The destabilization energy required to unfold the tertiary structure of LF and EF originates partly from the acidic pH in endosomes, which causes their N-terminal domains (LFN and EFN) to become molten globules (MG) (7). A positive membrane potential [+Δψ (10)], when coupled with these acidic pH conditions, is sufficient to drive LFN through PA63 pores formed in planar lipid bilayers (9). To enter the narrow confines of the ~15-Å-wide lumen, LFN must shed its residual tertiary structure and convert from the MG form to an extended, “translocatable” conformation (7). How does a solvent-filled pore mediate the disassembly of an MG protein, packed, albeit loosely, with hydrophobically dense stretches of polypeptide? We surmised that an interaction surface inside the pore might facilitate further unfolding of the MG to the extended, translocatable form.
By cysteine-scanning mutagenesis coupled to [2-(trimethylammonium) ethyl] methanethiosulfonate (MTS-ET) modification (5) (Fig. 1C), we identified residues that line the lumen of PA63 in the globular cap portion of domain 2, the pore-forming domain (Fig. 1D). F427 (11) was the most hydrophobic residue identified in the otherwise hydrophilic pore lining. It is absolutely conserved in homologous toxins (fig. S1), and mutating it blocks protein translocation (12, 13). F427→C427 (F427C) channels were most strongly affected by MTS-ET modification, implying that F427 is prominent and solvent-exposed within the lumen.
To address how the seven F427 residues were arranged within the lumen of the PA63 pore, we used electron paramagnetic resonance spectroscopy (EPR) to measure the proximity of nitroxide spin labels attached to F427C. The EPR spectrum of the spin-labeled prepore showed weak spin-spin interactions (Fig. 1E). This observation was consistent with the crystal structure of the prepore, in which F427 lies in a disordered loop (2β10 to 2β11) near the lumen, such that neighboring F427 residues are 15 to 20 Å apart (Fig. 1A). Upon conversion to the pore state by acidification to pH 6, a saturating spin-spin interaction appeared, indicating that the spin probes had converged and were separated by less than 10 Å (Fig. 1E). Consistent with this, single-channel ion conductances were roughly inversely proportional to the size of the substitution at 427 (Fig. 1F), as predicted from a cylindrical pore conductance model (14). Channels with large aliphatic (Leu) or aromatic (Trp) residues at 427 showed smaller conductances than channels with Ala or Phe, their smaller respective counterparts. Thus, as the pore forms, the seven phenylalanines create a narrow aromatic iris, or “ring of rings,” within the lumen (Fig. 1B and supporting online material text).
In planar lipid bilayers, LFN binds to wild-type (WT) PA63 pores, blocking ion conductance by >95% (at +20 mV and ~10 nM LFN; Fig. 2C); the first 21 residues of LFN’s cation-rich, flexible N terminus are essential for this macroscopic blocking effect (15). At the single-channel level, the N terminus of LFN bound stably within the PA63 channel and prevented passage of hydrated K+ ions, as manifested by a continuously closed state (Fig. 2A). When F427A PA63 was similarly assayed, LFN was only able to block macroscopic conductance by ~50% (Fig. 2C). In single-channel recordings, the partial block observed for F427A PA63 channels resulted from a dynamic “flickering” between an open state, multiple partly closed substates, and a fully closed state (Fig. 2A). Histogram analysis of a single-channel recording of F427A PA63 (Fig. 2B) was consistent with macroscopic experiments, in that the average conductance was ~50% of the open channel conductance (Fig. 2C). We infer that LFN is bound to the domain 1′ surface of the F427A channel, but its flexible N terminus, which enters the channel first (15), is not stably bound within the lumen and is unable to block conductance effectively. LFN blocked PA63 pores more effectively with large aromatic or aliphatic residues at position 427 than with small or hydrophilic residues (fig. S2). Thus the heptad of F427 residues is integral to binding LFN’s N terminus, leading us to term the site the “φ clamp.”
Fig. 2.
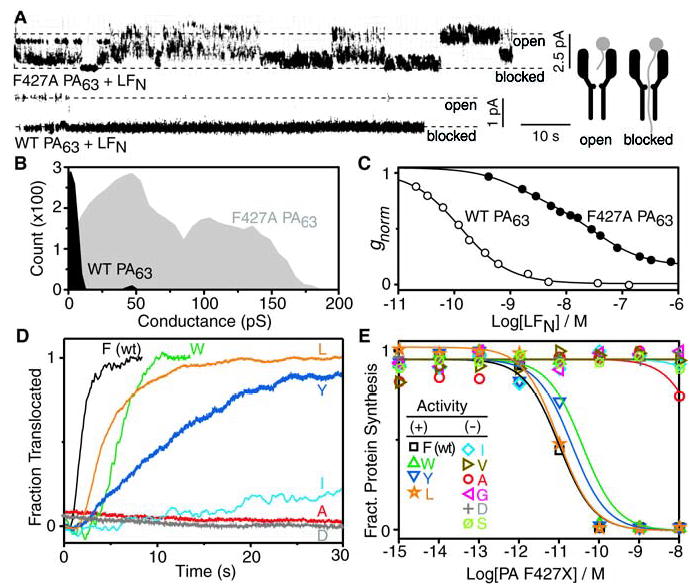
LFN conductance block and translocation studies with PA63 mutated at F427. (A) Current record for a single F427A PA63 channel (upper panel) exhibiting partial block with 7 nM LFN in the cis compartment. A similar single-channel record for a WT channel (lower panel) shows complete block with 7 nM LFN in the cis compartment [Δψ = +20 mV (10), pH 5.5]. A diagram (right) showing the “blocked” channel corresponds to LFN’s N terminus binding within the pore. (B) Histogram of the single-channel conductances for the LFN block in (A) for WT (black) and F427A PA63 (gray). (C) Concentration dependence of the LFN conductance block for WT (○) and F427A PA63 channels (•) under macroscopic conditions (Δψ = +20 mV, pH 5.5). Normalized conductance (gnorm) curves are fitted to a single- and two-site binding model forWT and F427A channels, respectively, where the WT equilibrium dissociation constant (KD) is ~150 pM. (D) Planar lipid bilayer macroscopic conductance records of LFN translocation through WT and the indicated F427 mutant channels [where pHcis = 5.5, pHtrans = 6.5 (29)]. After PA63-induced channel formation reached steady state, 20 nM LFN was added to the cis compartment (under a Δψ of +1 to +10 mV). When the conductance block reached steady state, the cis compartment was perfused, and translocation (as manifested by the rise in conductance) was initiated by increasing Δψ to +30 mV. Plotted records were normalized as the fraction translocated, excepting F427A and F427D channels, in which no appreciable translocation occurred. (E) Cellular translocation assay of F427 PA mutants (12), where translocation is reported by detecting the inhibition of protein synthesis by the domain from diphtheria toxin, which is fused to the C terminus of LFN. Mutants are classified as active (+) or inactive (−).
To probe the role of the φ clamp in polypeptide translocation, we measured the rate of LFN translocation through PA63 channels formed in planar lipid bilayers (9). LFN was added to the cis compartment, blocking conductance. After perfusing to remove unbound LFN, we stepped Δψ to a higher positive voltage and monitored the rate of translocation by the increase in channel conductance as LFN traversed the pore (Fig. 2D). With the most active substitutions at F427, LFN translocated at rates that trended as Phe > Leu ≈ Trp > Tyr (Fig. 2D). Other small aliphatic or hydrophilic substitutions, including Asp (an isosteric, hydrophilic control for Leu), were inefficient at promoting translocation. The most active residue, therefore, was Phe; the more hydrophilic aromatic, Tyr, was five times less active. These in vitro translocation studies recapitulated results of cellular assays of toxin action (Fig. 2E). Preference at the φ-clamp site for the γ-branched Leu over its β-branched isomer, Ile, correlates with the fact that aromatic residues also place more hydrophobic surface nearer to the center of the lumen. Thus the φ-clamp structure represents an active site, requiring either an aromatic surface or a more centrally oriented aliphatic surface to catalyze translocation efficiently.
Considering the hydrophobicity of the φ clamp, we hypothesized that this site may also be the binding site for hydrophobic cations, such as tetrabutylammonium (TBA) (Fig. 3A) (16). This hypothesis is consistent with studies suggesting that the TBA site is within the cap of the PA63 channel, cis to the extended β barrel (6). TBA’s affinity for F427A channels was greatly reduced from that of WT (4000- fold, or ~5 kcal mol−1; Fig. 3D). Large aliphatic residues at 427 were unable to recover the TBA block (Fig. 3E). F427L channels, for example, had a ~4 kcal mol−1 reduction in affinity for TBA. The least defective substitution was F427Y, which reduced the TBA block fourfold, or ~0.8 kcal mol−1. The recovery of the TBA block in F427Y channels was notable, because Tyr is substantially more hydrophilic than Phe. We infer that the TBA blocking mechanism also includes cation-π interactions, which occur when aromatic residues interact with cations through their delocalized, negative π-electron clouds (17).
Fig. 3.
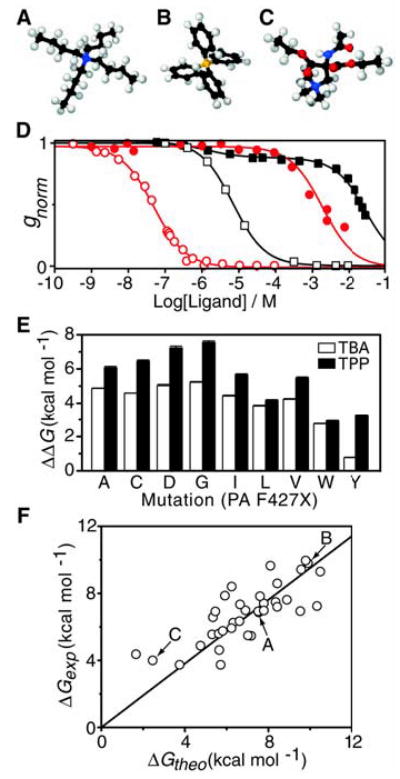
QAP conductance block at the φ-clamp site. (A) TBA, (B) TPP, and (C) a hydrophilic analog of TBA (2-acetylamino-2,2-bis-ethoxy-carbonyl- ethyl)-trimethyl-ammonium, colored by atom: C (black), N (blue), O (red), H (white), and P (orange). (D) QAP ions were added symmetrically to the cis and trans compartments (Δψ = +20 mV, pH 5.5), and gnorm was recorded once QAP binding reached equilibrium. For TPP block, single-site binding models were fit for WT PA63 (red ○), KD = 46 ± 2 nM (±SD), and F427A PA63 channels (red •), KD = 1.7 ± 0.2 mM. For TBA block, a single-site binding model was fit for WT PA63 (black □), KD = 7.3 ± 0.3 μM, but a two-site binding model was required for F427A PA63 (black ▪), such that the KD values of the major amplitude (88%) and minor amplitude (12%) were 30 ± 2 mM and 1.5 ± 0.7 μM, respectively. (E) Binding free energy change, , of TBA (white bars) and TPP (black bars) block for F427 mutants. (F) Model compound blocking studies of WT PA63 channels, performed as in (D). Experimental binding energies (ΔGexp = RT ln KD) were correlated to the expected binding energies (ΔGtheo) composed of solvation (ΔGs) (18) and aromatic enhancement (ΔGaro) energies as well as an offset, c, where ΔGtheo = ΔGs + ΔGaro + c. ΔGaro = αn, where α was fit to 0.7 ± 0.3 kcal mol−1 per aromatic ring, using the number of aromatic rings per compound, n. c was 0.8 ± 0.4 kcal mol−1 (table S1). The linear fit, ΔGexp = ΔGtheo × m (35 compounds), had a slope, m, of 0.95 ± 0.03 and total error, σSD, of 1.2 kcal mol−1. Arrows indicate compounds in (A), (B), and (C).
We next examined a library of 35 quaternary ammonium and phosphonium ion (QAP) compounds to establish the nature of the φ-clamp binding interaction. Aliphatic QAP compounds (table S1) blocked conductance according to a solvent-accessible surface area solvation energy model (18); i.e., compounds with more hydrophobic surface blocked more effectively (Fig. 3F). Thus, a hydrophilic analog of TBA that is comparable in size, but functionalized with hydrophilic amide and ester groups (Fig. 3C), blocked 140-fold more weakly than TBA (table S1). We found that WT channels preferred tetraphenylphosphonium (TPP) (Fig. 3B) to TBA by 160-fold (Fig. 3D). Broadly across the QAP library, the φ clamp preferred aromatic moieties by ~0.7 kcal mol−1 per aromatic ring (Fig. 3F and table S1). Specifically, the conductance block observed for the polyaromatic, 4-aminoquinolone drug, quinacrine (19), was reduced ~1000-fold in F427A channels (fig. S3), indicating the φ-clamp site may be exploited in the development of channel-blocking drugs. Thus the correlation observed for the diverse QAP compound library (Fig. 3F and table S1) indicates that the φ clamp does not, in and of itself, recognize specific geometric or steric features of the substrates. Instead, the WT φ clamp recognizes substrates primarily by nonspecific hydrophobic interactions (20), although its negative π-clouds also contribute electrostatically through aromatic-aromatic, π-π and cation-π interactions.
In determining a model for translocation, we disfavored a priori relationships used to describe metal ion-conducting channels, because metal ion throughput (Fig. 1F) did not correlate with protein transport (Fig. 2, D and E), and the steric constraints imposed by the φ-clamp site did not impede protein translocation. The WT φ-clamp site should create a hydrophobic and steric energy barrier for bulky, hydrophilic, or charged residues in the protein substrate. Paradoxically, although channels containing narrower, aromatically lined φ-clamp sites were less ion conducting, they translocated LFN orders of magnitude more rapidly than channels made wider, more hydrophilic, and more ion conducting at the φ-clamp site.
We therefore considered a chaperone model in which the φ-clamp’s phenyl rings directly interacted with the translocating polypeptide. This view is supported, because the φ clamp forms a narrow iris (Fig. 1E) that can effectively grasp the translocating polypeptide chain (Fig. 2A, diagram). Furthermore, model compound studies indicate that the φ clamp recognizes substrates through the hydrophobic effect, enhanced by aromatic-aromatic, π-π, and cation-π interactions (Fig. 3 and table S1). Similarly, in the potassium ion channel from Streptomyces lividans (KcsA), a hydrophobic cavity defines the TBA blocking site and corresponds to a docking site for a hydrophobically dense peptide sequence (21). In globular proteins, like LF, hydrophobically dense peptide segments occur periodically along the translocating chain (Fig. 4A). As these segments unwind from the MG protein, they would be expected to bind favorably to the φ clamp, causing translocation to be blocked kinetically by the unfavorable hydrophilic barrier in the Ser/Thr-rich β barrel (Fig. 4B). Thus the φ-clamp energy well might be expected to hinder translocation, causing polypeptide segments bound at the site to “pause.” However, as the φ-clamp site actually catalyzes polypeptide translocation, it must reduce some other larger barrier, such as the unfolding of the substrate protein. By analogy to the KcsA channel’s selectivity filter, which provides sequential rings of hydrophilic carbonyl oxygen atoms that mimic the inner hydration shell of a K+ ion (22), we propose that the φ-clamp site creates an environment that mimics the hydrophobic core of the unfolding MG protein. This would reduce the energetic penalty of exposing hydrophobic sequences to solvent or the hydrophilic lumen of the channel. The PA63 pore would thereby function as a Brownian ratchet, enabling the unwound leading segment of a translocating protein to move through the channel, and the trailing part of the protein to more readily unfold (Fig. 4C).
Fig. 4.
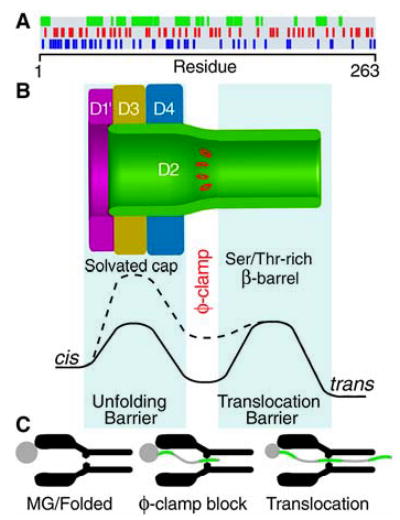
A model of φ-clamp catalyzed protein translocation. (A) Chemical complexity of a PA63 substrate, LFN, in which residues are colored by functionality: hydrophobic (green), greater than −1.75 kcal mol−1 in solvation energy (18) after applying a 10-residue running-window average; cationic (blue); and anionic (red). (B) The PA63 pore with a luminal φ-clamp site (red) is structurally imposed on an energy-well diagram for a hydrophobic stretch of polypeptide sequence from LFN [see (A)]. The φ-clamp site is the anticipated well, separating the unfolding barrier on the cis side in the solvent-filled cap from a translocation barrier on the trans side in the solvophilic, Ser/Thr-rich β barrel. Energy diagrams are for WT and F427A PA63 (solid and broken lines, respectively), where the mutation simultaneously reduces the hydrophobic-mediated stabilization imparted by the φ-clamp site and raises the barrier to unfolding hydrophobic sequences from the protein substrate. (C) Intermediate states for a stepwise, Brownian ratchet unfolding and translocation mechanism, such that hydrophobically dense polypeptide segments (green) interact with the φ-clamp constriction.
The results presented here show that PA63 is not merely a passive conduit through which proteins electrophorese; rather, it actively engages the translocating substrate via the φ clamp. This paradigm may be relevant to the functions of other polymer-translocating channels containing exposed hydrophobic sites, such as the “hydrophobic gasket” and “aromatic slide,” identified in the structures of the protein secretase (23) and maltoporin (24) channels, respectively.
Supplementary Material
References
- 1.Collier RJ, Young JA. Annu Rev Cell Dev Biol. 2003;1945 doi: 10.1146/annurev.cellbio.19.111301.140655. [DOI] [PubMed] [Google Scholar]
- 2.Friedlander AM. J Biol Chem. 1986;2617123 [PubMed] [Google Scholar]
- 3.Miller CJ, Elliott JL, Collier RJ. Biochemistry. 1999;3810432 doi: 10.1021/bi990792d. [DOI] [PubMed] [Google Scholar]
- 4.Blaustein RO, Koehler TM, Collier RJ, Finkelstein A. Proc Natl Acad Sci USA. 1989;862209 doi: 10.1073/pnas.86.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson EL, Huynh PD, Finkelstein A, Collier RJ. Biochemistry. 1998;373941 doi: 10.1021/bi972657b. [DOI] [PubMed] [Google Scholar]
- 6.Nassi S, Collier RJ, Finkelstein A. Biochemistry. 2002;411445 doi: 10.1021/bi0119518. [DOI] [PubMed] [Google Scholar]
- 7.Krantz BA, Trivedi AD, Cunningham K, Christensen KA, Collier RJ. J Mol Biol. 2004;344739 doi: 10.1016/j.jmb.2004.09.067. [DOI] [PubMed] [Google Scholar]
- 8.Wesche J, Elliott JL, Falnes PO, Olsnes S, Collier RJ. Biochemistry. 1998;3715737 doi: 10.1021/bi981436i. [DOI] [PubMed] [Google Scholar]
- 9.Zhang S, Udho E, Wu Z, Collier RJ, Finkelstein A. Biophys J. 2004;873842 doi: 10.1529/biophysj.104.050864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.We define the membrane potential as Δψ = ψcis − ψtrans, where ψtrans = 0 mV. The cis side is the side to which PA63 and LFN are added.
- 11.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
- 12.Sellman BR, Nassi S, Collier RJ. J Biol Chem. 2001;2768371 doi: 10.1074/jbc.M008309200. [DOI] [PubMed] [Google Scholar]
- 13.Mutation of several other sites in domain 2 were also shown to block translocation, but these prevented pore formation (12, 25).
- 14.Hille B. J Gen Physiol. 1968;51199 doi: 10.1085/jgp.51.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S, Finkelstein A, Collier RJ. Proc Natl Acad Sci USA. 2004;10116756 doi: 10.1073/pnas.0405754101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaustein RO, Finkelstein A. J Gen Physiol. 1990;96905 doi: 10.1085/jgp.96.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zacharias N, Dougherty DA. Trends Pharmacol Sci. 2002;23281 doi: 10.1016/s0165-6147(02)02027-8. [DOI] [PubMed] [Google Scholar]
- 18.Wesson L, Eisenberg D. Protein Sci. 1992;1227 doi: 10.1002/pro.5560010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orlik F, Schiffler B, Benz R. Biophys J. 2005;881715 doi: 10.1529/biophysj.104.050336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kauzmann W. Adv Protein Chem. 1959;141 doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 21.Zhou M, Morais-Cabral JH, Mann S, MacKinnon R. Nature. 2001;411657 doi: 10.1038/35079500. [DOI] [PubMed] [Google Scholar]
- 22.Doyle DA, et al. Science. 1998;28069 doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 23.Van den Berg B, et al. Nature. 2004;42736 [Google Scholar]
- 24.Schirmer T, Keller TA, Wang YF, Rosenbusch JP. Science. 1995;267512 doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 25.Mourez M, et al. Proc Natl Acad Sci USA. 2003;10013803 [Google Scholar]
- 26.Krantz BA, Melnyk RA, Collier RJ, Finkelstein A. unpublished data. [Google Scholar]
- 27.Lacy DB, Wigelsworth DJ, Melnyk RA, Harrison SC, Collier RJ. Proc Natl Acad Sci USA. 2004;10113147 doi: 10.1073/pnas.0405405101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Materials and methods are available as supporting material on Science Online.
- 29.Translocation of LFN (and full-length LF) was greatly accelerated when the pH on the trans side was greater than that on the cis side (26).
- 30.R.J.C. is cofounder, member of the scientific advisory board, and equity holder in PharmAthene, Inc., a startup company that investigates countermeasures against anthrax and other bioterrorism agents. We thank K. J. Oh for EPR data acquisition; R. Ross at the New England Research Center of Excellence Biomolecule Production Core as well as R. Pimental, L. Greene, and H. Lin for their assistance in purifying and generating PA mutants; and W. Hubbell, T. Sosnick, and A. Johnson for useful discussions. This work was supported by a National Research Service Award fellowship, AI062204 (B.A.K.), and NIH grants, AI022021 (R.J.C.) and GM29210 (A.F.).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


