Abstract
Reelin and glutamic acid decarboxylase 67 (GAD67) expression down-regulation in GABAergic interneurons of mice exposed to protracted treatment with l-methionine (MET) is attributed to RELN and GAD67 promoter cytosine-5-hypermethylation. This process recruits various transcription repressor proteins [methyl-CpG binding protein (MeCP2) and histone deacetylases (HDACs)] leading to formation of transcriptionally inactive chromatin. Here, we tested the hypothesis that RELN and GAD67 promoter cytosine-5-hypermethylation induced by a protracted MET treatment is reversible and that repeated administration of HDAC inhibitors influences this process by an activation of DNA-cytosine-5-demethylation. In the frontal cortices of mice receiving MET (5.2 mmol/kg twice a day for 7 days) and killed at 1, 2, 3, 6, and 9 days during MET washout, we measured RELN (base pairs −414 to −242) and GAD67 (base pairs −1133 to −942) promoter methylation and MeCP2 bound to methylated cytosines of RELN (base pairs −520 to −198) and GAD67 (base pairs −446 to −760) promoters. Levels of RELN and GAD67 promoter hypermethylation induced by 7 days of MET treatment declines by ≈50% after 6 days of MET withdrawal. When valproate (VPA) (2 mmol/kg) or MS-275 (0.015–0.12 mmol/kg), two structurally unrelated HDAC inhibitors, was given after MET treatment termination, VPA and MS-275 dramatically accelerated RELN and GAD67 promoter demethylation in 48–72 h. At these doses, VPA and MS-275 effectively increased the binding of acetylhistone-3 to RELN and GAD67 promoters, suggesting that histone-3 covalent modifications modulate DNA demethylation in terminally differentiated neurons, supporting the view that, directly or indirectly, HDAC inhibitors may facilitate DNA demethylation.
Keywords: DNA demethylation, histone deacetylase inhibitors, valproate, MS-275
The down-regulation of glutamic acid decarboxylase 67 (GAD67) and reelin expressed in telencephalic GABAergic neurons is a characteristic and prominent neuropathological alteration observed in patients with schizophrenia (SZ) and bipolar (BP) disorder with psychosis (1–8).
GAD67 is one of the two molecular forms of GABA-synthesizing enzymes and plays an important role in maintaining GABA steady-state levels at GABAergic synapses (9). Reelin is a 400-kDa glycoprotein synthesized by cortical GABAergic neurons. On secretion into the extracellular matrix, reelin adheres to dendritic shafts and spines of pyramidal neurons (10–12). Previous experiments have suggested that reelin may play a role in the regulation of event-related protein synthesis in dendritic spines (11). Reelin has also been implicated in synaptic plasticity and learning and memory functions (10–15).
The down-regulation of reelin expression is probably responsible for the decrease in dendritic spine density observed in the frontal cortex (FC) of heterozygous reeler mice and perhaps also in the prefrontal cortex (PFC) of SZ and BP disorder patients (12). Hence, a deficit in GABAergic transmission because of a decrease of GAD67 and reelin expression may cause a disruption of the intermittent high-frequency synchronization bursts of cortical pyramidal neuron firing that occurs during cortical function, and an alteration of this firing may play a role in thought disorders and in working memory impairments expressed by SZ and BP patients (16).
Several lines of evidence suggest that epigenetic mechanisms, including the increase of promoter cytosine-5-methylation at specific CpG islands, are probably associated with reelin and GAD67 expression down-regulation in psychiatric patients. For example, there is evidence that CpG islands embedded in the RELN promoter are hypermethylated in the PFC of SZ patients (17–19).
In the PFC, GABAergic neurons of SZ and BP patients show (i) an increased expression of DNA methyltransferase 1 (DNMT1), the enzyme that catalyzes DNA (cytosine-5) methylation at CpG island-enriched promoter regions (8, 20); and (ii) an increased level of S-adenosylmethionine (SAM) (21), which functions as a methyl donor in DNMT1-mediated cytosine-5-methylation reactions.
We have also reported that the increase of SAM brain content elicited by protracted l-methionine (MET) treatment in mice causes the RELN and GAD67 promoter hypermethylation that is probably operative in the transcriptional down-regulation of these genes in the FC and perhaps other brain areas of psychotic patients (19, 22–25).
Recent studies suggest that in addition to the increased levels of DNMTs and SAM, an active DNA-cytosine-5-demethylase (DNA demethylase) may play an important regulatory role in determining the level of gene-specific cytosine methylation in differentiated somatic cells and also in terminally differentiated neurons (26–28). Thus, to study whether hypermethylated RELN and GAD67 promoters in the brain can be demethylated by the action of a DNA demethylase, we tested in mice (i) whether RELN and GAD67 promoter hypermethylation induced by protracted MET administration is reversible on MET withdrawal, (ii) and whether the demethylation process can be induced by 2-propylpentanoate (valproate, VPA) or by N-(2-aminophenyl)-4-[N-(pyridin-3-yl-methoxycarbonyl)aminomethyl]benzamide (MS-275), which is also known to inhibit HDACs. VPA is a weak HDAC inhibitor, whereas MS-275, which is structurally unrelated to VPA, is a potent and selective HDAC inhibitor that enters the brain (29).
In this paper, we report that VPA and MS-275, in doses that increase brain nucleosomal acetylhistone-3 (29), also accelerate RELN and GAD67 promoter demethylation, presumably by activating a putative DNA demethylase.
Results
MET Increases RELN and GAD67 Promoter Cytosine-5-Methylation.
We assessed DNA cytosine-5 methylation by using a methylation-specific PCR (MSP) technique to measure the methylated and unmethylated cytosines in CpG island-enriched promoter regions of RELN (sequence from −414 to −242 bp), GAD67 (sequence from base pair −1133 to base pair −942), and GAD65 (sequence from base pair 3512 to base pair 3664) (refs. 30 and 31 and GenBank accession no. AB032757, respectively). Because the MPS analysis is not quantitative and provides only a comparative assessment, the methylated/unmethylated status of RELN and GAD67 promoters was independently quantified by determining the amount of RELN (sequence from base pair −520 to base pair −198) and GAD67 (sequence from base pair −760 to base pair −446) CpG island-enriched promoter fragments associated with MeCP2 by using ChIP with a specific MeCP2 antibody coupled to a competitive PCR (25).
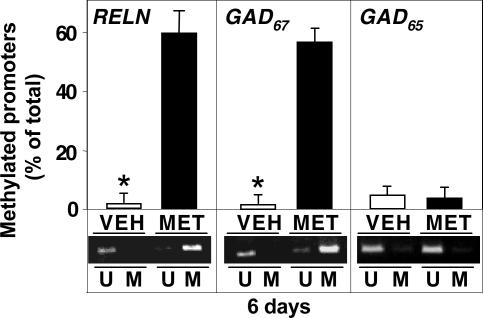
As shown in Fig. 1, the FC of mice receiving MET (5.2 mmol/kg) s.c. twice daily for 6 days shows a significant and robust increase in methylated cytosine and a concomitant decrease of unmethylated cytosine residues in CpG sites of RELN and GAD67 promoters. In contrast, this MET treatment failed to change the levels of methylated or unmethylated cytosines in the CpG sites of the GAD65 promoter region that were analyzed.
Fig. 1.
MET-induced increase of RELN and GAD67 but not GAD65 promoter methylation in mouse FC. MET (filled bars) (5.2 mmol/kg s.c. twice a day) or vehicle (VEH) (open bars) was administered to mice for 6 days. MSP was used to evaluate the percentage of methylated or unmethylated RELN, GAD67, or GAD65 promoter CpG sites (A302 ratio of methylated DNA/unmethylated DNA). At the bottom is a representative agarose gel electrophoresis showing increased RELN and GAD67 methylated promoters (M) and decreased unmethylated promoters (U) in MET-treated mice. The data represent the mean ± SE of three to five mice. ∗, P < 0.01 vs. MET-treated group (Student's t test). The last injection of MET was administered 2 h before decapitation.
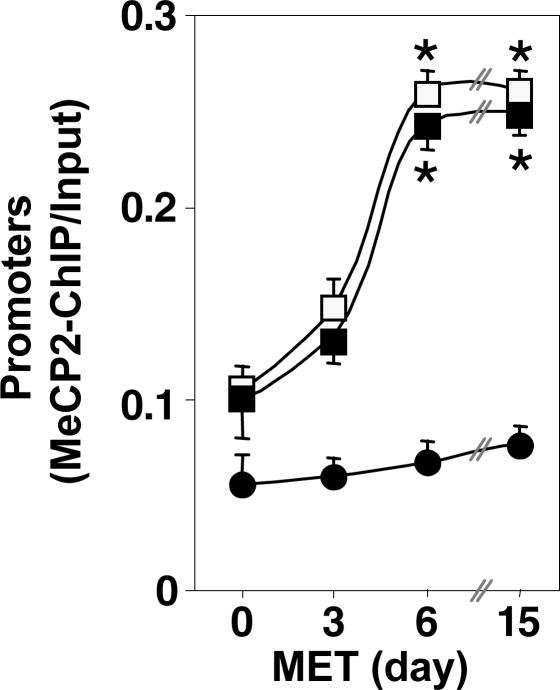
The results reported in Fig. 2 are consistent with the data of Fig. 1 and show that in the FC of mice treated with MET (5.2 mmol/kg s.c. twice daily), RELN and GAD67 promoters express a time-dependent increase of MeCP2 binding that reaches its maximum levels within 6 days of MET treatment.
Fig. 2.
MET treatment induces a time-related increase in the amount of FC RELN and GAD67 but not GAD65 promoter immunoprecipitated with MeCP2 antibody. RELN, GAD67, and GAD65 promoter fragments were immunoprecipitated with MeCP2 antibody and were quantified by using competitive PCR with internal standards (25). Depicted are the ratios between the amount of RELN (□), GAD67 (■), and GAD65 (●) promoters immunoprecipitated with MeCP2 antibody (MeCP2-ChIP) and the amount of the corresponding promoter fragments in the initial nonimmunoprecipitated extract (input). MET was given at dose of 5.2 mmol/kg s.c. twice a day; the last injection of MET was administered 2 h before decapitation. The data represent the mean ± SE of three to five mice. ∗, P < 0.01 when 0- and 3-day are compared with 6- and 15-day MET-treated groups (ANOVA followed by Dunnett's test).
MET-Induced RELN and GAD67 Promoter Methylation Is Reversed After MET Withdrawal.
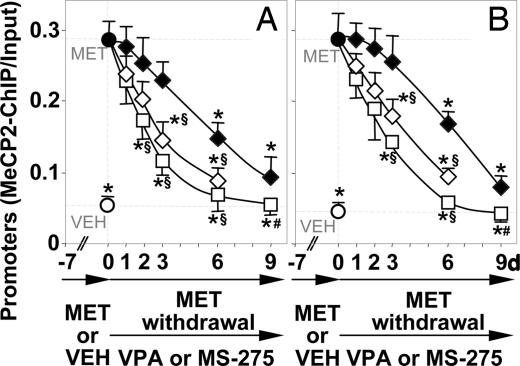
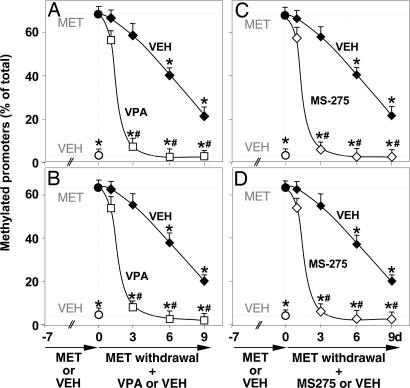
In mice receiving MET for 7 days, the hypermethylation of RELN and GAD67 promoters can be reversed after MET withdrawal. Fig. 3 shows that the increased recruitment of MeCP2 to the hypermethylated RELN and GAD67 promoters elicited by 7 days of MET treatment declines slowly, reaching a 50% reduction after 6 days and approaching steady-state control values only after 9 days of MET withdrawal. Fig. 4 shows that after MET withdrawal, the cytosine-5-methylation at CpG islands of RELN and GAD67 promoters, measured by the MSP technique, declines with a time course similar to that of the MeCP2 binding.
Fig. 3.
VPA and MS-275 facilitate the reversal of the MET-induced increase in the amount of RELN and GAD67 promoter immunoprecipitated with MeCP2 antibodies. Mice were first treated with MET (5.2 mmol/kg s.c. twice a day, for 7 days) (days −7 to 0) to obtain an increase of MeCP2 binding to the hypermethylated RELN (A) or GAD67 (B) promoters in FC. Then, after MET (●) treatment termination (day 0), vehicle (VEH ◆), VPA (□) (2 mmol/kg s.c. twice a day), and MS-275 (◇) (0.12 mmol/kg s.c. once a day) were given to mice for 1, 2, 3, 6, and 9 days. The data represent the mean ± SE of three to five mice. ∗, P < 0.01 vs. 7-day MET-treated mice (●); #, P < 0.01 vs. 3, 6, and 9 days VEH groups; §, P < 0.01 when 3- and 6-day VPA- or MS-275-treated mice are compared with 3- and 6-day VEH-treated groups (ANOVA followed by Dunnett's test).
Fig. 4.
VPA and MS-275 facilitate the reversal of the MET-induced hypermethylation of RELN and GAD67 promoters. Mice were first treated for 7 days (days −7 to 0) with MET (5.2 mmol/kg s.c. twice a day) to obtain hypermethylation of RELN (A and C) and GAD67 (B and D) promoters, or for seven days with VEH (○). Then, after MET treatment termination (day 0), VPA (□, A and B) (2 mmol/kg s.c. twice a day), MS-275 (◆, C and D) (0.060 mmol/kg s.c. once a day), or vehicle (VEH ◆) was administered for 1, 3, 6, and 9 days. MSP was used to evaluate the percentage of methylated promoters. The data represent the mean ± SE of three to five mice. ∗, P < 0.01 vs. 7 days MET-treated mice (●); #, P < 0.01 when VPA- or MS-275-treated mice are compared with the corresponding VEH groups (ANOVA followed by Dunnett's test).
As in previous studies (22, 24, 25), protracted daily MET treatments and the consequent RELN and GAD67 promoter hypermethylation are associated with an ≈50% reduction of reelin and GAD67 expression (Fig. 5). The expression down-regulation of these two proteins is reversed 7 days after MET withdrawal (Fig. 5).
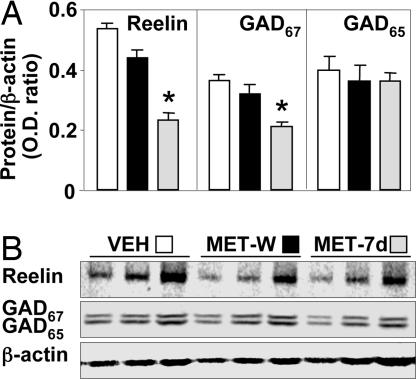
Fig. 5.
Reversal of MET-induced reelin and GAD67 expression down-regulation after MET withdrawal. (A) Average reelin, GAD67, and GAD65 protein levels (OD ratios of reelin, GAD67, and GAD65 to β-actin) in FC of mice treated with MET (5.2 mmol/kg s.c. twice a day) for 7 days (MET-7d), shaded bars; VEH, open bars; or after 7 days of MET withdrawal (MET-W), filled bars. ∗, P < 0.05 vs. VEH group (Student's t test). (B) Typical Western blots of reelin and GAD67/GAD65 with three serial dilutions of the same sample. The intensity of β-actin determined on the same blot was used for a comparative estimation of sampling variation. The data represent the mean ± SE of three to five mice.
VPA and MS-275 Injections After MET Withdrawal Accelerate RELN and GAD67 Promoter Demethylation.
Two chemically unrelated HDAC inhibitors, VPA and MS-275, were tested for their ability to accelerate RELN and GAD67 promoter demethylation.
In a first group of experiments, VPA and MS-275 were given in doses that (i) maximally acetylate nucleosomal histones in the brain cortex and hippocampus (29), and (ii) effectively increase the acetylhistone-3 bound to RELN and GAD67 promoters in the FC (29). As shown in Figs. 3 and 4, VPA (2 mmol/kg) and MS-275 (0.060 mmol/kg) given to mice for 1–6 days after termination of a 7-day treatment with MET dramatically (in 48–72 h) accelerated the reversal of FC RELN and GAD67 CpG island methylation measured (i) as the time course of MeCP2 disassociation from the methylated promoter region of both genes determined by using the quantitative ChIP technique (Fig. 3), and (ii) by using the MSP technique (Fig. 4).
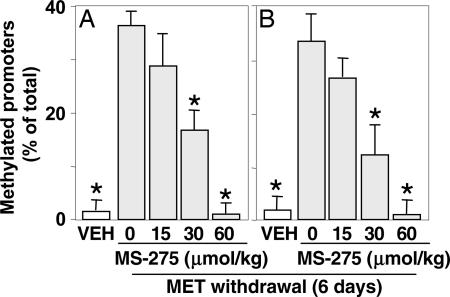
In a second group of experiments, we observed that MS-275 stimulates RELN and GAD67 demethylation in a dose-dependent manner (Fig. 6) and that this stimulation can be related to the extent of the increase of acetylhistone-3 at RELN and GAD67 promoters (29).
Fig. 6.
MS-275 facilitates the reversal of MET-induced hypermethylation of RELN (A) and GAD67 (B) promoters in a dose-dependent manner. Mice were first treated for 7 days with VEH (open bars) or MET (5.2 mmol/kg s.c. twice a day) to obtain hypermethylation of RELN or GAD67 promoters. After termination of MET treatment, various doses of MS-275 (from 0 to 0.060 mmol/kg s.c.) (shaded bars) were administered once a day for 6 days. MSP was used to evaluate the percentage of methylated RELN or GAD67 promoters. The data represent the mean ± SE of three to five mice. ∗, P < 0.05 when MS-275-treated mice are compared with mice that received MET but did not receive MS-275.
Importantly, we also observed that a single injection of VPA immediately after termination of MET treatment facilitates RELN promoter demethylation (a reduction of ≈50%) after 3 days of MET withdrawal. However, as expected, in this case the extent of RELN promoter demethylation is not as large as that obtained by repeatedly injecting VPA (see Figs. 3 and 4).
MET, at a dose (5.2 mmol/kg s.c. twice a day for 7 days) that induces RELN and GAD67 promoter hypermethylation in the FC (Fig. 1) also exerts a similar effect in the hippocampus. In the hippocampus, VPA also facilitates RELN promoter demethylation with a time course similar to that found in the FC (MeCP2 ChIP/Input) for RELN promoters: VEH = 0.09 ± 0.005, MET = 0.31 ± 0.035, VEH (MET withdrawal for 3 days) = 0.29 ± 0.018, and VPA (MET withdrawal for 3 days) = 0.08 ± 0.003. However the liver, which expresses a high basal RELN promoter methylation (ratio of MeCP2 ChIP/Input ≈ 0.8), seems to be unaffected.
Procainamide, in Doses That Inhibit MET-Induced RELN and GAD67 Promoter Cytosine-5-Methylation, Fails to Alter the Time Course of Their Demethylation.
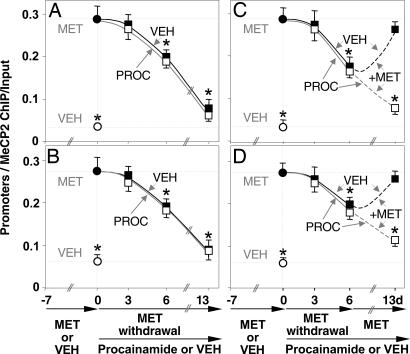
To establish whether the acceleration of RELN and GAD67 promoter demethylation induced by VPA and MS-275 is related to a reduction of DNMT1 activity, we inhibited this methylation process by administering procainamide (a competitive inhibitor of DNA methyltransferases) (32). In preliminary experiments, we established that procainamide (0.29 mmol/kg, four times a day) completely prevents the RELN and GAD67 promoter hypermethylation induced by 7 days of treatment with MET (not shown). Fig. 7 A and B shows that mice receiving procainamide (0.29 mmol/kg four times a day) during MET withdrawal exhibited an apparent time course of RELN and GAD67 promoter demethylation comparable to that of mice treated with a vehicle during MET withdrawal. To test whether the dose of procainamide used in these experiments was able to effectively block DNMT1 activity, after 7 days of MET withdrawal we reexposed mice (previously challenged with either procainamide or vehicle for 7 days during MET withdrawal) to MET (5.2 mmol/kg, twice a day). As shown in Fig. 7 C and D, MET reexposure elicits the reappearance of RELN and GAD67 promoter methylation in vehicle-treated but not in procainamide-treated mice. This finding suggests that procainamide, at the doses used in these experiments, is capable of blocking DNMT1 methylation activity.
Fig. 7.
Procainamide prevents the MET-induced increase in the amount of FC RELN and GAD67 promoter immunoprecipitated with MeCP2 antibody but fails to facilitate its reversal after MET withdrawal. In the first group of experiments (A and B), mice were first treated with MET (5.2 mmol/kg s.c. twice a day for 7 days) (●) to obtain a robust hypermethylation of RELN and GAD67 promoters, or VEH (twice a day for 7 days) (○). After MET withdrawal, one group of mice received VEH (■) and one group received procainamide (□) (0.29 mmol/kg s.c. four times a day) for 13 days. Measurements of RELN promoters (A) and GAD67 promoters (B) are reported. In the second group of experiments (C and D), after MET withdrawal mice were first treated with VEH (■), or procainamide (□) for 7 days, both groups received MET again, together with VEH (■) or procainamide (□) for an additional 7 days [measurements of RELN promoters (C) and GAD67 promoters (D) are reported]. The data represent mean ± SE of three to five mice. ∗, P < 0.01 vs. MET-treated group at day 0 (ANOVA followed by Dunnett's test). Methods were as in Fig. 2.
Discussion
DNA cytosine-5-methylation established during somatic cell division and differentiation is a very stable DNA covalent modification faithfully maintained throughout the lifespan of a cell. However, recent data (26–28, 33) suggest that in differentiated cells, including terminally differentiated neurons, DNA methylation marking may also undergo rapid changes that are probably mediated by the action of a putative DNA demethylase. Hence, differentiated neurons not only are able to express a maintenance cell type-specific DNA methylation pattern that is a form of genetic inventory at the cellular level but also are likely to express a rapid DNA demethylation process as an aspect of a transient epigenomic response to intracellular or extracellular signals or drugs.
The present study, using two independent but complementary methods to measure promoter cytosine methylation, provides evidence that in the FC of the adult mouse, the robust CpG island hypermethylation of RELN and GAD67 promoters induced by protracted MET treatment is slowly reversed after MET withdrawal, in ≈9 days.
Direct measurements of brain SAM exclude the hypothesis that after MET withdrawal, the long time course of RELN and GAD67 promoter demethylation is because of an accumulation of FC SAM elicited by 7 days of MET pretreatment. In fact, measurement of FC SAM 12 h after 7 days of MET treatment (22, 24) showed only a slight increase of this methyl group donor. Furthermore, preliminary data suggest that expression of FC DNMT1, which is the most abundant form of DNMT mRNA expressed in the mouse brain, is not changed by the protracted MET treatment schedule adopted in our experiments.
To definitively establish whether the RELN and GAD67 promoter demethylation process in MET-treated mice is slow because of a direct or indirect increase of DNMT activity, we used procainamide in doses and treatment schedules that effectively inhibited DNMT1 activity (Fig. 7). Procainamide failed to accelerate the demethylation of RELN and GAD67 promoters (Fig. 7).
Taken together, the available data suggest that direct or indirect changes in DNMT activity are not responsible for the long time course of RELN and GAD67 promoter demethylation.
In the mammalian neocortex, RELN and GAD67 are expressed in GABAergic neurons but not in pyramidal neurons or glial cells (12). Because the protracted administration of MET induces RELN and GAD67 expression down-regulation in GABAergic neurons and because after protracted administration, MET withdrawal results in a reversal of this down-regulation (Fig. 5), it is probable that at least in part, methylation and demethylation of RELN and GAD67 promoters may occur in differentiated nondividing GABAergic neurons. Hence, under physiological conditions in the CNS, RELN and GAD67 promoter demethylation does not appear to be a passive process dependent on cell replication or differentiation.
The experiments with VPA and MS-275 strongly suggest that DNA demethylation can be dramatically activated by increasing histone acetylation. In fact, VPA and MS-275 administered to mice after the termination of protracted MET treatment elicit a demethylation of RELN and GAD67 promoters in 48–72 h (Figs. 3 and 4). At the doses used here, VPA and MS-275 both increase the content of nuclear acetylhistone-3 bound to RELN and GAD67 promoters by 40–50% (29), suggesting that a putative histone code modification may participate in the modulation of DNA demethylation by these chemically unrelated drugs.
The action of VPA and MS-275 on RELN and GAD67 promoter demethylation could be elicited by (i) an inhibition of DNMT activity or (ii) the activation of a DNA demethylase.
Two sets of data suggest that the accelerated RELN and GAD67 promoter demethylation induced by treatment with VPA or MS-275 may be directly or indirectly related to an induction of DNA demethylase rather than to a reduced DNMT1 activity: (i) reduction of DNMT1 activity with procainamide fails to activate demethylation of the hypermethylated RELN and GAD67 promoters, and (ii) VPA fails to block SAM biosynthesis, and in fact may even favor its biosynthesis (22, 24). Hence, we believe that in mice the mechanism by which the recovery of RELN and GAD67 expression is accelerated by VPA or MS-275 after MET withdrawal could be related to the induction of a putative DNA demethylase activity. The data agree with a previous report of Detich et al. (28), demonstrating that VPA triggers a replication-independent active demethylation of DNA.
Because the biochemical nature of a putative DNA demethylase in mammalian cells is not yet clearly established (26–28, 33, 34), it is impossible to determine whether the accelerated RELN and GAD67 promoter demethylation elicited in the mouse FC by HDAC inhibitors is the result of DNA demethylase activation or of an induction of DNA demethylase biosynthesis. In the future, the characterization of an inducible active demethylation mechanism in the mammalian brain offers an attractive possibility of discovering new classes of drugs that are active on epigenetic mechanisms by modulating the function of putative DNA demethylases. Obviously, these drugs will be extremely valuable in future tests of pharmacological treatment of SZ or BP disorder patients.
In preliminary experiments, we observed that at therapeutically relevant concentrations, antipsychotic drugs such as sulpiride and clozapine increase the brain content of acetylated histones and also accelerate DNA demethylation.† Because there is strong evidence for a down-regulation of RELN expression mediated by the hypermethylation of CpG islands embedded in the SZ promoter region, the possibility that some antipsychotics can activate DNA demethylases and may also reactivate the expression of RELN and GAD67 opens new horizons regarding the pharmacological treatment of SZ and related disorders.
Materials and Methods
Animals and Drug Administration.
Adult male Swiss Albino mice (Harlan, Indianapolis, IN), of 20–22 g body weight, were separated into vehicle (VEH), MET (Sigma, St. Louis, MO), VPA (Sigma), MS-275 (29), procainamide (Sigma), MET + VEH, MET + VPA, MET + MS-275, and MET + procainamide-treated groups as described in Results and the figure legends. MET, VPA, and procainamide were dissolved in H2O (0.1 ml/20 g of body weight). MS-275 was dissolved with a drop of glacial acetic acid brought to pH 6 with the addition of NaOH. The doses of VPA and MS-275 were selected based on previous studies showing that at these doses, VPA and MS-275 elicit maximum histone acetylation and prevent MET-induced RELN and GAD67 promoter methylation (29).
Western Blot Analysis.
Reelin, GAD67, or GAD65 was extracted from the FC samples homogenized in Laemmli buffer (100 μl/10 mg of tissue). Proteins were separated by SDS/PAGE (7.5% acrylamide gel for reelin and 10–20% gradient for GAD65 and GAD67) and blotted overnight onto Hybond ECL nitrocellulose membranes (Amersham Pharmacia, Piscataway, NJ). The membrane blots were reacted for 6 h at 25°C with (i) G-10 anti-reelin monoclonal antibody, diluted 1:5,000 (16); and (ii) anti-GAD67/GAD65 rabbit polyclonal antibody (Chemicom, Temecula, CA), diluted 1:2,000. The intensity of β-actin immunofluorescence was determined on the same blot with FITC-labeled anti-β-actin mouse monoclonal antibody, diluted 1:3,000 (clone AC-15; Sigma–Aldrich, St. Louis, MO), and used for a comparative estimation of the protein amount applied to the gels.
ChIP.
Approximately 10 mg of FC tissue was used for this procedure. Tissue slices (0.3 × 0.3 mm) were incubated at 37°C for 15 min with 400 μl of PBS containing 1% formaldehyde, supplemented with protease inhibitors (1 mM PMSF, 1 μg/ml aprotinin, and 1 μg/ml pepstatin) (Upstate Cell Signaling Solutions, Lake Placid, NY) to crosslink MeCP2 with the target genomic DNAs. After being washed six times with cold PBS containing protease inhibitors, slices were homogenized in 200–400 μl of SDS lysis buffer (supplied by ChIP kit; Upstate Cell Signaling Solutions) with minor modifications as described in ref. 25.
Measurements of RELN, GAD67, and GAD65 Promoter Fragments by Quantitative Competitive PCR.
Primer pair design.
For routine purposes, we performed PCR amplification reactions of CpG-rich RELN promoter regions from base pair −520 to base pair −198 (forward primer, 5′-CGCGCGCGGGGCACCGTC-3′; reverse primer, 5′-AGAGACCGACGGGCTGCC-3′). We PCR amplified the GAD67 promoter regions from base pair −760 to base pair −446 (forward primer, 5′-AGCGGCACTCGTGCGTGTTATTAA-3′ and reverse primer, 5′-TGTTGGGTGAGGGCAAGGGAAAAT-3′). The GAD65 promoter region was amplified by using forward primer 5′-TCTCTTCAGCCGTCAGTCAAAACC-3′ and reverse primer 5′-CACGTGTGCATCGATTGGCTCATT-3′.
Quantification of target promoters by competitive PCR.
To quantify RELN, GAD67, and GAD65 target promoter sequences by PCR analyses, we used appropriate internal standards. The internal standards were designed by deleting 100- to 150-bp fragments from the middle of the target gene sequence and were generated by an overlap extension PCR with internal deletion primers (25).
The amplification products of the internal standard and of the target gene were separated on agarose gels. The A302 ratio of target internal standard/target DNA equal to one gives the concentration of target gene promoters present in the sample (25). PCRs were carried out by using the GC melt PCR system (BD Biosciences Clontech, Palo Alto, CA). The thermocycler protocol included an initial denaturation cycle (1 min at 95°C), 28–30 cycles of denaturation (30 sec at 94°C), annealing (1 min at 60°C), and extension (2 min at 68°C), followed by a final extension cycle (3 min at 68°C), terminating at 15°C. DNA signals on agarose gels (1.5–2%) were visualized and quantified by using Kodak (Rochester, NY) software (EDAS290).
MSP.
MSP was performed as described in ref. 35. Briefly, genomic DNA was obtained from brain tissue by using a routine DNA extraction protocol. Approximately 1 μg of DNA in a volume of 50 μl was denatured in 0.3 M NaOH for 20 min at 42°C followed by adding 1,200 μl of freshly prepared bisulfite solution [3 M sodium bisulfite (Sigma) and 10 mM hydroquinone (Sigma), pH 5.0]. The reaction was carried out in the dark for 12 h at 50°C. After bisulfite treatment, the DNA was desalted by using the QIAexII (Qiagen, Valencia, CA) purification kit, followed by desulfonation with 3 M NaOH. DNA was then precipitated with ethanol after the addition of 1 μg of glycogen at −20°C overnight. The primer pairs used for methylated or unmethylated RELN, GAD67, and GAD65 promoters were designed with MethPrimer provided at www.urogene.org. For detecting methylated CpG islands in RELN promoters, the pairs used were 5′-GGGAGGGTTTTAGGATAAGC-3′ (forward) and 5′-CCGAACTCATAACTACCCGTT-3′ (reverse), and for the unmethylated ones, 5′-GGAGGGTTTTAGGATAAGTGT-3′ (forward) and 5′-ACCCCAAACTCATAACTACCCAT-3′ (reverse) were used. These primers are complementary to the CpG islands of the RELN promoter region included between base pairs −414 and −242: for detecting methylated GAD67, the pairs used were 5′-GTTTTGTTTTTAAAGGATGAGAAAC-3′ (forward) and 5′-AAAAATTTCCACCAAAAAACG-3′ (reverse) whereas 5′-TTTGTTTTTAAAGGATGAGAAATCG-3′ (forward) and 5′-AAAATTTCCACCAAAAAACACC-3′ (reverse) were used for unmethylated ones. These primers are complementary to CpG islands of GAD67 promoters included between base pairs −1133 and −942: for GAD65, 5′-GTTTTTGGGGTTATTTAGGTTAGC-3′ (forward) and 5′-TCGTAAAATACGAATCAAATACGAA-3′ (reverse) were used for methylated DNA, and 5′-TTTTTGGGGTTATTTAGGTTAGTGT-3′ (forward) and 5′-TCATAAAATACAAATCAAATACAAA-3′ (reverse) were used for unmethylated ones, which are complementary to the GAD65 promoter region between base pairs 3512 and 3664. PCR amplification was carried out by using the GCmelt PCR kit (BD Biosciences Clontech). The cycling conditions were one cycle at 95°C for 1 min, 33 cycles at 94°C for 40 sec, 60°C for 50 sec, 68°C for 1 min, followed by a final 10-min extension at 68°C. Ten microliters of each PCR product was loaded onto 3% agarose gels, stained with ethidium bromide, and directly visualized under UV illumination. Data were presented as the ratio of optical density of the methylated band over the unmethylated ones. In the preliminary studies, we established that the ratio of unmethylated to methylated PCR-amplified products is maintained in a linear range between 28 and 33 cycles. Hence, for routine experiments, 30 cycles were used. For each sample analyzed, the assays, including extraction, bisulfate treatment, and PCR, were repeated at least three times. Genomic DNA methylated with SssI methylase in vitro served as a positive control for the methylated cytosines in the RELN promoters. Genomic DNA served as an additional control to indicate that the primers for methylated RELN and GAD67 did not amplify the unmethylated DNA.
Statistical Analysis.
All results were expressed as mean ± SE. Student's t test and one-way ANOVA followed by the Dunnett's multiple comparison test were used to assess the significance of the differences between groups. The criterion for significance was P < 0.05 or 0.01. The criterion for significance was adjusted for multiple tests on the same data set.
Acknowledgments
We thank Dr. F. M. Benes (McLean Hospital/Harvard Medical School, McLean, MA) and Dr. B. L. Roth (University of North Carolina, Chapel Hill Medical School, Chapel Hill, NC) for constructive criticisms and suggestions in the preparation of the manuscript. This work was supported in part by National Institute of Mental Health Grants MH 071667 (to E.C.), MH 070855 (to A.G.), and MH 62682 (to D.R.G.).
Abbreviations
- GAD67
glutamic acid decarboxylase 67
- MET
l-methionine
- MeCP2
methyl-CpG binding protein
- HDAC
histone deacetylase
- SZ
schizophrenia
- BP
bipolar
- FC
frontal cortex
- PFC
prefrontal cortex
- DNMT1
DNA methyltransferase 1
- SAM
S-adenosylmethionine
- VPA
valproate
- MSP
methylation-specific PCR.
Footnotes
The authors declare no conflict of interest.
Simonini, M., Dong, E., Grayson, D. R., Costa, E., Guidotti, A., 36th Annual Neuroscience Meeting, Oct 14–18, 2006, Atlanta, GA, abstr. 712.5.
References
- 1.Benes FM, Vincent SL, Alsterberg G, Bird ED, SanGiovanni JP. J Neurosci. 1992;12:924–929. doi: 10.1523/JNEUROSCI.12-03-00924.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbarian S, Kim JJ, Potkin SG, Hagman JO, Tafazzoli A, Bunney WE, jr, Jones EG. Arch Gen Psychiatry. 1995;52:258–266. doi: 10.1001/archpsyc.1995.03950160008002. [DOI] [PubMed] [Google Scholar]
- 3.Guidotti A, Auta J, Davis JM, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson DR, Impagnatiello F, Pandey G, Pesold C, Sharma R, et al. Arch Gen Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- 4.Benes MF, Berretta S. Neuropsychopharmacology. 2001;25:1–27. doi: 10.1016/S0893-133X(01)00225-1. [DOI] [PubMed] [Google Scholar]
- 5.Lewis DA, Hashimoto T, Volk DW. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- 6.Eastwood SL, Harrison PJ. Am J Psychiatry. 2006;163:540–542. doi: 10.1176/appi.ajp.163.3.540. [DOI] [PubMed] [Google Scholar]
- 7.Akbarian S, Huang HS. Brain Res Brain Res Rev. 2006;52:293–304. doi: 10.1016/j.brainresrev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Veldic M, Caruncho HM, Liu WS, Davis J, Satta R, Grayson DR, Guidotti A, Costa E. Proc Natl Acad Sci USA. 2004;101:348–353. doi: 10.1073/pnas.2637013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soghomonian JJ, Martin DL. Trends Pharmacol Sci. 1998;19:500–505. doi: 10.1016/s0165-6147(98)01270-x. [DOI] [PubMed] [Google Scholar]
- 10.Liu WS, Pesold C, Rodriguez MA, Carboni G, Auta J, Lacor P, Larson J, Condie BG, Guidotti A, Costa E. Proc Natl Acad Sci USA. 2001;98:3477–3482. doi: 10.1073/pnas.051614698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong E, Caruncho H, Liu WS, Smalheiser NR, Grayson DR, Costa E, Guidotti A. Proc Natl Acad Sci USA. 2003;100:5479–5484. doi: 10.1073/pnas.1031602100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa E, Davis J, Grayson DR, Guidotti A, Pappas GD, Pesold C. Neurobiol Dis. 2001;8:723–742. doi: 10.1006/nbdi.2001.0436. [DOI] [PubMed] [Google Scholar]
- 13.Weeber EJ, Beffert U, Jones C, Christian JM, Forster E, Sweatt JD, Herz J. J Biol Chem. 2002;277:39944–39952. doi: 10.1074/jbc.M205147200. [DOI] [PubMed] [Google Scholar]
- 14.Carboni G, Tueting P, Tremolizzo L, Sugaya I, Davis J, Costa E, Guidotti A. Neuropharmacology. 2004;46:1070–1081. doi: 10.1016/j.neuropharm.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Larson J, Hoffman JS, Guidotti A, Costa E. Brain Res. 2003;971:40–46. doi: 10.1016/s0006-8993(03)02353-9. [DOI] [PubMed] [Google Scholar]
- 16.Spencer KM, Nestor PG, Perlmutter R, Niznikiewicz MA, Klump MC, Frumin M, Shenton ME, McCarley RW. Proc Natl Acad Sci USA. 2004;101:17288–17293. doi: 10.1073/pnas.0406074101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grayson DR, Chen Y, Costa E, Dong E, Guidotti A, Kundakovic M, Sharma RP. Pharmacol Ther. 2006;111:272–286. doi: 10.1016/j.pharmthera.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, Shafa R, Glatt SJ, Nguyen G, Ponte JF, et al. Am J Med Genet B. 2005;16:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- 19.Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, Costa E. Proc Natl Acad Sci USA. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veldic M, Guidotti A, Maloku E, Davis JM, Costa E. Proc Natl Acad Sci USA. 2005;102:2152–2157. doi: 10.1073/pnas.0409665102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guidotti A, Ruzicka WB, Grayson DR, Veldic M, Pinna G, Davis JM, Costa E. NeuroReport. 2007;18:57–60. doi: 10.1097/WNR.0b013e32800fefd7. [DOI] [PubMed] [Google Scholar]
- 22.Tremolizzo L, Carboni G, Ruzicka WB, Mitchell CP, Sugaya I, Tueting P, Sharma R, Grayson DR, Costa E, Guidotti A. Proc Natl Acad Sci USA. 2002;99:17095–17100. doi: 10.1073/pnas.262658999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noh JS, Sharma RP, Veldic M, Salvacion AA, Jia X, Chen Y, Costa E, Guidotti A, Grayson DR. Proc Natl Acad Sci USA. 2005;102:1749–1754. doi: 10.1073/pnas.0409648102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tremolizzo L, Doueiri MS, Dong E, Grayson DR, Davis J, Pinna G, Tueting P, Rodriguez-Menendez V, Costa E, Guidotti A. Biol Psychiatry. 2005;57:500–509. doi: 10.1016/j.biopsych.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 25.Dong E, Agis-Balboa RC, Simonini MV, Grayson DR, Costa E, Guidotti A. Proc Natl Acad Sci USA. 2005;102:12578–12583. doi: 10.1073/pnas.0505394102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szyf M. Biochemistry (Moscow) 2005;70:533–549. doi: 10.1007/s10541-005-0147-7. [DOI] [PubMed] [Google Scholar]
- 27.Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 28.Detich N, Bovenzi V, Szyf M. J Biol Chem. 2003;278:27586–27592. doi: 10.1074/jbc.M303740200. [DOI] [PubMed] [Google Scholar]
- 29.Simonini MV, Camargo LM, Dong E, Maloku E, Veldic M, Costa E, Guidotti A. Proc Natl Acad Sci USA. 2006;103:15847–15892. doi: 10.1073/pnas.0510341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Royaux I, Rouvroit C, D'Arcangelo G, Demirov D, Goffinet A. Genomics. 1997;46:240–250. doi: 10.1006/geno.1997.4983. [DOI] [PubMed] [Google Scholar]
- 31.Szabo G, Katarova Z, Kortvely E, Greenspan R, Urban Z. DNA Cell Biol. 1996;15:1081–1091. doi: 10.1089/dna.1996.15.1081. [DOI] [PubMed] [Google Scholar]
- 32.Lee BH, Yegnasubramanian S, Lin X, Nelson WG. J Biol Chem. 2005;280:40749–40756. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weaver IC, Meaney MJ, Szyf M. Proc Natl Acad Sci USA. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kress C, Thomassin H, Grange T. Proc Natl Acad Sci USA. 2006;103:11112–11117. doi: 10.1073/pnas.0601793103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]