Abstract
In the developing muscle, a pool of myogenic progenitor cells is formed and maintained. These resident progenitors provide a source of cells for muscle growth in development and generate satellite cells in the perinatal period. By the use of conditional mutagenesis in mice, we demonstrate here that the major mediator of Notch signaling, the transcription factor RBP-J, is essential to maintain this pool of progenitor cells in an undifferentiated state. In the absence of RBP-J, these cells undergo uncontrolled myogenic differentiation, leading to a depletion of the progenitor pool. This results in a lack of muscle growth in development and severe muscle hypotrophy. In addition, satellite cells are not formed late in fetal development in conditional RBP-J mutant mice. We conclude that RBP-J is required in the developing muscle to set aside proliferating progenitors and satellite cells.
Keywords: muscle differentiation, myogenic progenitors, Notch signaling
Myogenesis is a tightly regulated process that is essential in muscle development and regeneration. During mammalian development, phases of embryonic and fetal myogenic differentiation lead to the formation and growth of skeletal muscles. In the postnatal and adult organism, skeletal muscle grows and regenerates by the myogenic differentiation of stem cells, the satellite cells (1). Muscle progenitor cells during development or satellite cells in the adult initiate myogenic differentiation as a result of the activation of myogenic determination factors like Myf5 and MyoD and form myoblasts (for reviews, see refs. 2–4). Mononucleated myoblasts begin to express muscle-specific proteins and fuse to form multinucleated myotubes, the constituents of mature skeletal muscle.
Skeletal muscle and satellite cells of the body and the extremities derive from the somites, segmental derivatives of the paraxial mesoderm (5–10). As the somite matures, myogenic progenitor cells become confined to the dermomyotome that expresses the transcription factor Pax3 (paired box protein 3). After myogenesis is initiated, a resident progenitor population that expresses Pax3 and Pax7 is maintained in the developing muscle (7–9). Late in fetal development, the progenitor population generates satellite cells, which are marked by the expression of Pax7 (7–9). Some, but not all, satellite cells also express Pax3 (11). Thus, in a developing or adult muscle, a pool of undifferentiated cells is preserved that has the potential to undergo myogenic differentiation. The molecular mechanism used to set aside this population of progenitor cells is not understood.
The Notch signaling pathway is highly conserved in evolution and plays important roles during development and in the adult. Notch signals regulate diverse processes, including maintenance of progenitors, cell fate decisions, proliferation, and differentiation (for reviews, see refs. 12–14). Notch signaling is initiated by the interaction of the Notch receptor (Notch 1–4 in mammals) with its ligand (Delta-like 1, 3, and 4 and Jagged 1 and 2 in mammals). Ligand binding results in proteolytic cleavage of the receptor and releases the Notch intracellular domain, which interacts directly with the primary mediator of Notch signaling, the RBP-J (Rbpsuh) transcription factor. In the absence of Notch signals, RBP-J is associated with corepressors and represses transcription. The Notch intracellular domain displaces corepressors from RBP-J, allows the recruitment of coactivators, and induces the activation of target genes like Hes-1 (15, 16). A wealth of data demonstrates the importance of various components of the Notch signaling pathway in somitogenesis (reviewed in refs. 17 and 18). Notch signaling is also essential for the establishment of rostral and caudal identities in the somite (19–21). In addition, Notch signals have been implicated in regulating postnatal muscle regeneration. Aged muscle has an impaired ability to regenerate because of the decreased induction of Delta-like-1 upon injury, and forced activation of Notch restores the regenerative capacity by regulating stem cell activation, proliferation, and self-renewal (22). Furthermore, ectopic activation of Notch in satellite cell culture or in the chicken embryo interferes with myogenic differentiation (23–27). Notch signaling inhibits myogenesis by RBP-J, activating the expression of the transcription factor Hes1; Hes1 encodes a transcriptional repressor that in turn suppresses MyoD (15, 28). In addition, RBP-J-independent mechanisms may also contribute to Notch function (29, 30).
We used conditional mutagenesis of RBP-J to assess Notch functions in muscle differentiation. The Lbx1cre transgene allowed us to elicit recombination of the floxed RBP-J gene (31) in migrating muscle progenitors that generate hypaxial muscles of the limbs, tongue, and diaphragm. In addition, a Pax3cre allele (32) was used to investigate RBP-J function in nonmigrating hypaxial and epaxial muscle progenitors. This approach circumvented the midgestation lethality associated with null mutations in RBP-J and allowed the analysis of RBP-J functions in muscle differentiation. Our data show that RBP-J is essential to maintain a resident pool of muscle progenitor cells and to prevent their differentiation. We also show that RBP-J is essential to set aside satellite cells late in fetal development.
Results
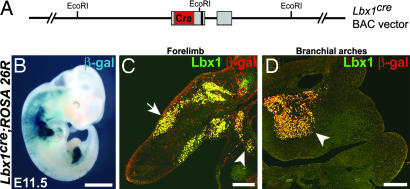
To investigate the role of RBP-J in muscle differentiation by conditional mutagenesis, a transgenic mouse line was constructed that expresses Cre-recombinase under the control of Lbx1 genomic sequences (Fig. 1A; see also Materials and Methods). The endogenous Lbx1 gene is expressed in long-range migrating muscle progenitor cells (33, 34). By the use of ROSA26R reporter mice, we showed that Lbx1cre introduced efficiently recombination in muscle progenitor cells that migrate to the limbs and branchial arches (Fig. 1 B–D); our subsequent analysis indicated that recombination in progenitor cells that move to the diaphragm (arrowhead in Fig. 1C) was incomplete (see Materials and Methods). Introduction of a mutation in the RBP-J gene by Lbx1cre in mice (RBP-Jflox/flox; Lbx1cre animals, subsequently referred to as RBP-J/Lbx1cre mice) did not interfere with migration of muscle progenitor cells. At embryonic day (E)10.5, we observed comparable numbers of Pax3+ or Lbx1+ cells in the limbs (Fig. 2 A and B). Cells that initiated the expression of the muscle determination gene MyoD were not observed in limbs of control and RBP-J/Lbx1cre mutant mice at this stage (Fig. 2 A and B).
Fig. 1.
Recombination introduced by the Lbx1cre transgene. (A) Schematic display of the Lbx1cre transgene. In the modified 144-kb BAC clone, cre-recombinase (red), was fused to the ATG initiation codon of Lbx1 and replaced Lbx1 coding sequences (gray boxes); the vector was used to generate the transgenic Lbx1cre mouse strain. (B–D) Lbx1cre-induced recombination was monitored in embryos (E11.5) carrying the ROSA26R reporter; recombination was assessed by X-Gal staining (B) or by the use of antibodies to detect Lbx1 (green) or β-galactosidase (red) (C and D). (C) Longitudinal section on the forelimb; muscle progenitors in the limb and in the stream moving to the diaphragm are indicated by arrow and arrowhead, respectively. (D) Section of the branchial arches; the arrowhead points to muscle progenitors that subsequently generate the tongue muscle. (Scale bars: B, 2 mm; C and D, 250 μm.)
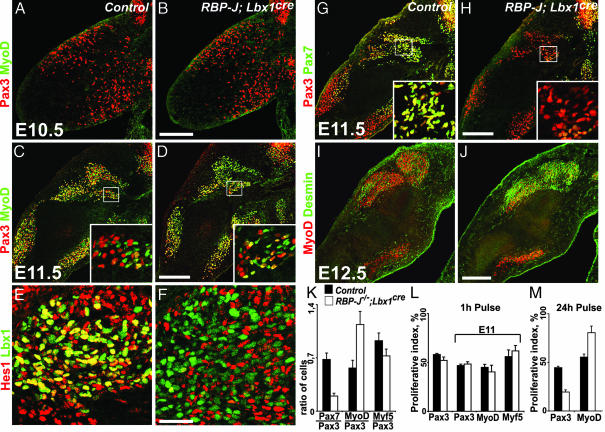
Fig. 2.
Development of myogenic cells in the limb of RBP-J/Lbx1cre mice. Immunohistological analysis of myogenic cells in developing limbs of control and RBP-J/Lbx1cre mice. Myogenic cells were analyzed at E10.5 (A and B), E11.5 (C–H), and E12.5 (I and J) by using the indicated antibodies. Insets (C and D, G and H) show the boxed areas at higher magnification. (K) Ratios of Pax7/Pax3, MyoD/Pax3, and Myf5/Pax3 cells observed at E11.5. (L) Proliferation was assessed by BrdU-labeling; shown are the proportion of Pax3+, MyoD+, and Myf5+ cells labeled 1 h after BrdU injection at E10.5 or E11.5. (M) BrdU was injected at E10.5, and the proportions of Pax3+ or MyoD+ cells that incorporated BrdU were assessed after a 24-h chase. (Scale bars: A–D and G–J, 200 μm; E and F, 50 μm.)
At subsequent developmental stages, changes in myogenic differentiation were apparent in mutant mice. The majority of muscle progenitor cells in the limb of control mice coexpress Pax3 and Lbx1 at E11.5. Pax3+ and Lbx1+ progenitor cells were present in RBP-J/Lbx1cre mice, but their overall number was reduced (Fig. 2 C and D). At E11.5, cells that initiated the expression of the muscle determination factor MyoD can be observed in control and conditional mutant mice. MyoD+ cells were more abundant in RBP-J/Lbx1cre mice, and the ratio of MyoD+/Pax3+ cells was increased (Fig. 2 C and D; for quantification, see Fig. 2K). The ratio of Myf5+/Pax3+ cells, however, was not significantly changed (Fig. 2K). Hes1 is a direct target gene of RBP-J, and its expression is activated in the presence of the Notch intracellular domain. Immunohistological and in situ hybridization analysis demonstrated that Hes1 is expressed in myogenic and mesenchymal cells of the limb at E11.5 (Fig. 2E and data not shown). We observed that many Lbx1+ cells coexpressed Hes1 in control mice, and that Hes1 expression was markedly down-regulated in Lbx1+ cells of RBP-J/Lbx1cre mice (Fig. 2F). In limbs of control mice, many Pax3+ and Lbx1+ progenitors, particularly those that locate to the proximal limb, coexpress Pax7 (Fig. 2G). Interestingly, Pax7+ cells were rare in the limbs of RBP-J/Lbx1cre mice, and those present contained low levels of the Pax7 protein (Fig. 2 H and K).
When the limbs of control and mutant mice were compared at subsequent stages (E12.5), we observed a marked reduction in the number of Pax3+ or Lbx1+ progenitor cells, as well as a reduction in the number of cells that expressed MyoD in RBP-J/Lbx1cre mice (Fig. 2 I and J and data not shown). Pax7 was present in limbs of control mice but not detectable in RBP-J/Lbx1cre mice (not shown). Desmin is an intermediate filament protein whose expression is initiated early during myoblast differentiation. Desmin+ cells were not detectable in the limbs of control and RBP-J/Lbx1cre mice at E11.5. Compared with control mice, we observed more widespread desmin and myogenin expression in RBP-J/Lbx1cre mice at E12.5 (Fig. 2 I and J and data not shown). We conclude that more myogenic progenitor cells initiated differentiation at E11.5. Furthermore, more cells that progressed in myogenic differentiation and expressed desmin or myogenin were observed in the limb of RBP-J/Lbx1cre than in control mice at E12.5. This was accompanied by a reduction in the number of Pax3+/Lbx1+ progenitor cells.
We also assessed the proliferation capacity of muscle progenitor cells in control and RBP-J/Lbx1cre mice and observed that similar proportions of Pax3+ cells had incorporated BrdU one hour after BrdU injection at E10.5 or E11.5 (Fig. 2L). Proliferative activities of MyoD+ and Myf5+ cells were also similar at E11.5 (Fig. 2L). TUNEL staining did not reveal changes in cell death in developing limbs at E11.5 (not shown). A pulse–chase experiment in which BrdU was injected 24 h before analysis at E11.5 demonstrated that a larger proportion of BrdU+ cells expressed MyoD, and a smaller proportion expressed Pax3 (Fig. 2M). Thus, proliferating muscle progenitor cells in the limb that are labeled by BrdU injection at E10.5 were less likely to give rise to a Pax3+ progenitor in RBP-J/Lbx1cre mice than in control mice. In contrast, they were more likely to generate cells that initiate myogenic differentiation and express MyoD.
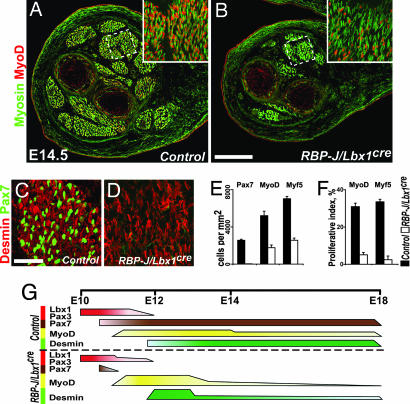
Differentiated muscle groups in the limbs can be discerned by using skeletal muscle-specific myosin antibodies at E14.5 and were present in control and RBP-J/Lbx1cre mice. However, the size of the muscle groups was markedly reduced in conditional mutant mice (Fig. 3 A and B). Progenitor cells that expressed Pax7 were present in muscle of control mice but were not observed in RBP-J/Lbx1cre mice (Fig. 3 C–E). Other markers (Pax3 and Lbx1) useful for the identification of progenitor cells at E10–E12.5 were not expressed in the limbs of control and conditional mutant mice at E14.5 (data not shown). MyoD and Myf5 act as determination factors only at the onset of myogenesis and are down-regulated after myoblasts reach a postmitotic state and fuse. Cells that expressed MyoD or Myf5 were associated with muscle fibers in control and conditional mutant mice, but their numbers were reduced in the mutants (Fig. 3 A, B, and E; the expression of various markers is summarized in Fig. 3G). In addition, BrdU labeling demonstrated that the proliferative capacity of MyoD+ and Myf5+ cells was reduced at this stage in the RBP-J/Lbx1cre animals, indicating that these differentiating cells had acquired a postmitotic state (Fig. 3F). We conclude that myofiber formation had occurred by E14.5 in the limbs of RBP-J/Lbx1cre mice, but muscles were small, and progenitor cells were no longer present.
Fig. 3.
Differentiated muscle groups in the distal limb of RBP-J/Lbx1cre mice. (A–D) Immunohistological analysis of muscle groups in the distal limb of control (A and C) and RBP-J/Lbx1cre (B and D) mice at E14.5 by using the indicated antibodies. (E) Quantification of Pax7+ and MyoD+ or Myf5+ cells in control and RBP-J/Lbx1cre mice. Shown are the numbers of cells/mm2. (F) Quantification of proliferating MyoD+ and Myf5+ cells in distal limb muscles of control and RBP-J/Lbx1cre mice. BrdU was injected 1 h before the analysis at E14.5. Displayed are the proportions of MyoD+ and Myf5+ cells that incorporated BrdU. (G) Summary of the expression of various markers used to identify myogenic progenitors and differentiating myogenic cells in limbs of control (Upper) and RBP-J/Lbx1cre (Lower) mice during development. Bar thickness indicates cell numbers at particular stages that express the indicated proteins. (Scale bars: A and B, 250 μm; C and D, 50 μm.)
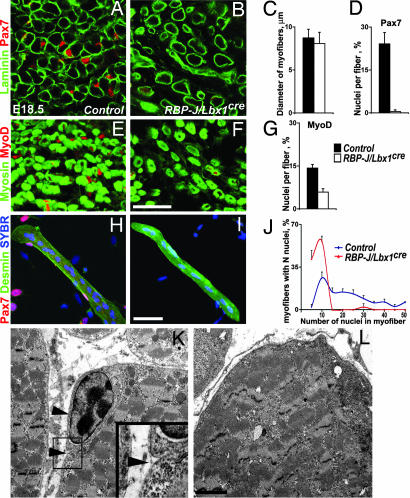
Mature myofibers surrounded by a basal membrane appear late in fetal development. Satellite cells can be discerned by their location below the basal lamina of myofibers and their expression of Pax7 (Fig. 4A). Mature myofibers of comparable diameter were present in limbs of control and RBP-J/Lbx1cre mice at E18.5, but we observed a reduced fiber density in conditional mutants (Fig. 4 A–C). Notably, the fibers of the RBP-J/Lbx1cre mice were devoid of Pax7+ cells (Fig. 4 B and D). MyoD+ nuclei in the muscle of the RBP-J/Lbx1cre mutants were still detectable but compared with control mice, the number of MyoD+ nuclei/fiber was reduced (Fig. 4 E–G). BrdU injection experiments indicated that at E18.5, all MyoD+ cells in limb muscles had reached a postmitotic state in RBP-J/Lbx1cre, but not in control mice (not shown). We isolated single fibers from fetal muscle and confirmed the absence of Pax7+ satellite cells in fiber preparations of conditional mutant mice (Fig. 4 H and I). This experiment also demonstrated that the numbers of nuclei in myofibers were reduced in RBP-J/Lbx1cre compared with control mice (Fig. 4J). In addition, we used electron microscopy to confirm the absence of satellite cells in muscle of RBP-J/Lbx1cre mice (Fig. 4 K and L). Thus, we observed not only a deficit in Pax7 expression but also a complete lack of satellite cells in the limbs of RBP-J/Lbx1cre mice. Migrating muscle progenitors also generate the intrinsic tongue muscle; immunohistological and electron microscopic analysis indicated that tongue muscle was similarly affected in RBP-J/Lbx1cre mice [supporting information (SI) Fig. 6].
Fig. 4.
Satellite cells are absent in the limb of RBP-J/Lbx1cre mice. (A and B) Immunohistological analysis of muscle in distal limbs of control (A) and RBP-J/Lbx1cre (B) mice at E18.5 by using antibodies against laminin (green) and Pax7 (red). (C) Quantification of the myofiber diameter in control and RBP-J/Lbx1cre mice; the outline of myofibers was visualized by using anti-laminin antibodies. (D) Quantification of the number of Pax7+ cells/myofiber in control and RBP-J/Lbx1cre mice. (E and F) Immunohistological analysis of limb muscle in control (E) and RBP-J/Lbx1cre (F) mice at E18.5 by using skeletal muscle-specific myosin (green) and MyoD (red) antibodies. (G) Quantification of the number of MyoD nuclei/myofiber in control and RBP-J/Lbx1cre mice. Immunohistological analyses of single muscle fibers from control (H) and RBP-J/Lbx1cre (I) mice at E18.5 by using desmin (green) and Pax7 (red) antibodies. A nuclear counterstain (SYBR) is shown in blue. (J) Quantification of the number of nuclei/myofiber in control and RBP-J/Lbx1cre mice. (K and L) Ultrastructure of limb muscle from control (K) and RBP-J/Lbx1cre (L) mice at E18.5. In control mice, satellite cells are separated from myofibers by plasma membranes and locate below the basal membrane (arrowheads). In RBP-J mutants, satellite cells were not detected. (Scale bars: A–I, 50 μm; K and L, 2 μm.)
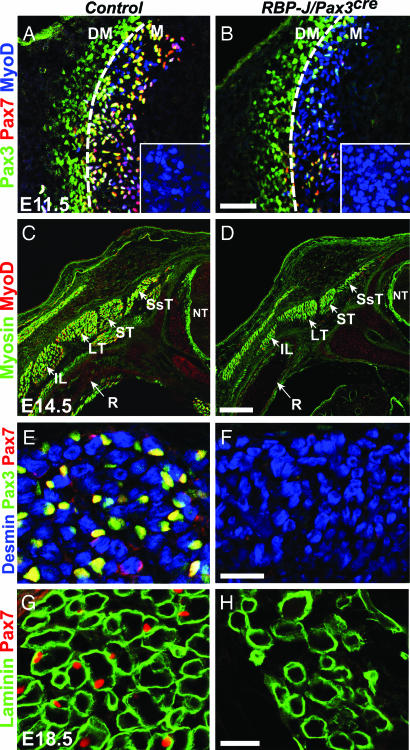
To extend the functional analysis of RBP-J to include also nonmigrating muscle progenitor cells, we used a Pax3cre allele to mutate RBP-J in the dermomyotome (32). The myotome can be discerned by the presence of MyoD+ cells and is populated in control mice at E11.5 by Pax3+/Pax7+ progenitor cells that derive from the dermomyotome (ref. 7; see also Fig. 5A; the dashed line indicates the boundary between the dermomyotome and the myotome). Pax3+/Pax7+ progenitor cells in the myotome were reduced in number in RBP-J/Pax3cre mice at E11.5 (Fig. 5B). The density of MyoD+ cells in the myotome, however, was increased in RBP-J/Pax3cre compared with control mice (Fig. 5 A and B Insets). TUNEL staining indicated that apoptosis rates were similar in myotomes of control and RBP-J/Pax3cre mice at E11.5 (180 ± 12 and 168 ± 15 TUNEL + cells/mm2 in control and mutant myotomes, respectively), indicating that cell death could not account for the reduction in the number of Pax3+/Pax7+ cells. The myotome generates deep muscles of the back. At E14.5, residual back muscles were observable in conditional mutant mice, but these were small and devoid of Pax7+ and Pax3+ cells (Fig. 5 C–F). Pax7+ satellite cells could not be discerned at E18.5 in residual muscle fibers of the back in RBP-J/Pax3cre mice (Fig. 5 G and H). Furthermore, intercostal and diaphragm muscles were small and devoid of Pax7+ cells in the RBP-J/Pax3cre mice, and the appearance of limb muscles was similar to that observed in RBP-J/Lbx1cre mice (SI Fig. 7). We conclude, therefore, that RBP-J is essential for the maintenance of progenitor cells and for formation of satellite cells in epaxial and hypaxial muscle compartments.
Fig. 5.
Myotome and myotome-derived muscle in RBP-J/Pax3cre mice. (A and B) Immunohistological analysis of the dermomyotome and myotome in control and RBP-J/Pax3cre mice at E11.5 by using Pax3 (green), Pax7 (red), and MyoD (blue) antibodies. The stippled lines indicate the border between myotome (M) and dermomyotome (DM). Insets (A and B) display magnifications of the myotome, and demonstrate a higher density of MyoD+ cells in the myotome of mutant mice. (C–F) Analysis of back muscle in control and RBP-J/Pax3cre mice at E14.5 by using the indicated antibodies. (G and H) Analysis of back muscle in control and RBP-J/Pax3cre mice at E18.5 by using laminin (green) and Pax7 (red) antibodies. Neural tube (NT), rib (R), and deep muscles of the back, semispinalis thoracis (SsT), spinalis thoracis (ST), longissimus thoracis (LT), ilicostalis lumborum (IL), are indicated. (Scale bars: A and B, 100 μm; C and D, 250 μm; and E–H, 25 μm.)
Discussion
During muscle development, a balance between progenitor cell proliferation and differentiation ensures the maintenance of progenitors and muscle growth. Various growth factors can enhance proliferation and delay myogenic differentiation (35–37). Ectopic activation of Notch signaling is known to interfere with muscle differentiation in the chicken embryo and suppresses myogenic differentiation in culture (23–28). Forced activation of Notch enhances regenerative capacity of adult muscle, which was attributed to an enhanced muscle stem cell activation, proliferation and self-renewal (38). By using the cre/loxP system to introduce a conditional mutation, we show here that RBP-J, the major transcriptional mediator of Notch signals, is essential to maintain muscle progenitor cells in an undifferentiated state. In these conditional RBP-J mutant mice, muscle progenitors undergo myogenic differentiation in an uncontrolled and premature manner. In addition, we show that RBP-J is required to set aside satellite cells late in development of the muscle.
RBP-J and Myogenic Differentiation.
Myogenic differentiation in normal development is a process that occurs over many days. We observed pronounced changes in myogenic differentiation, as assessed by MyoD and desmin expression. MyoD is present in proliferating and postmitotic myoblasts, whereas desmin is expressed in differentiating myoblasts and myotubes. In the limbs of control mice, the first wave of MyoD+ myoblasts appears at E11.5, but MyoD+ cells can be observed during the entire fetal period. MyoD+ cells appeared on schedule in the limbs of RBP-J/Lbx1cre mice, but their number was increased at early (E11.5) and reduced at late (E14.5 and E18.5) stages. Desmin-expressing myoblasts appeared on schedule in the limbs of RBP-J/Lbx1cre mice. Their number was increased at early stages, but desmin+ muscle groups were subsequently smaller (see Fig. 3G for a summary). We conclude, therefore, that differentiation occurs on schedule in the limbs of RBP-J/Lbx1cre mice, but the number of differentiating cells is increased at early stages. RBP-J controls directly the expression of Hes1, and Hes1 is known to suppress MyoD (15, 28). A loss of MyoD repression is in accordance with the increased myogenic differentiation in RBP-J/Lbx1cre conditional mutant mice. In contrast, the number and the proliferative index of Myf5+ cell were unchanged in RBP-J/Lbx1cre mice at early stages, indicating that Myf5 expression is not controlled by Notch signaling.
RBP-J, Myogenic Progenitors, and Satellite Cells.
The augmented myogenic differentiation observed in the limbs of RBP-J/Lbx1cre mice was accompanied by a rapid depletion of the progenitor pool. During normal development, progenitors that maintain proliferative capacity are set aside. These provide a cellular source that allows muscle growth over a prolonged period in development. In the limbs, such progenitors express Pax3, Lbx1, and Pax7 at early stages (E10–E12.5) and only Pax7 at late stages (E13 to birth). We observed that the early Pax3+ or Lbx1+ progenitors appear on schedule and in normal numbers in limbs of RBP-J/Lbx1cre mice. Subsequently, their number is, however, reduced, because a larger proportion of progenitor cells initiated myogenic differentiation early. At late developmental stages, the numbers and proliferative index of Myf5+ and MyoD+ cells were reduced. The pronounced reduction in cells that initiate myogenic differentiation at late developmental stages and the pronounced reduction of muscle mass in RBP-J/Lbx1cre animals appears thus to result from the premature depletion of the progenitor pool. Interestingly, Pax7 expression is massively down-regulated already at E11.5 in the limbs of mutant mice, and a reduction in progenitor numbers due to differentiation cannot account for this pronounced change.
Progenitor cells are set aside to become satellite cells in the late fetal period, and electron microscopic as well as immunohistological analyses demonstrated that satellite cells were not present in limbs of the RBP-J/Lbx1cre mice. We previously characterized Lbx1 mutant mice that display a migratory deficit in myogenic progenitor cells, which results in the appearance of only few progenitors in the limbs, and in the formation of small muscle groups (39). Pax7+ satellite cells, however, were associated with the remaining limb muscles (SI Fig. 8), indicating that a reduction in progenitor numbers and/or muscle size does not impede satellite cell formation.
Similarities in RBP-J Function in Hypaxial and Epaxial Muscle.
Lbx1cre induced mutations of RBP-J demonstrated an essential role of RBP-J in the maintenance of myogenic progenitors that derive from migratory cells. To assess whether RBP-J has a similar function in other types of muscle progenitors, we used a Pax3cre allele. Pax3cre introduces mutations in myogenic progenitors in the dermomyotome (32). At E11.5, progenitor cells that delaminate from the dermomyotome populate the myotome and can be discerned by the expression of Pax7 and Pax3 (7–9). In the developing myotome of RBP-J/Pax3cre mice, only few Pax3+/Pax7+ progenitors were observable at E11.5. This was accompanied by an increased density of MyoD+ cells in the myotome, indicating that progenitor cells had differentiated prematurely. The myotome subsequently generates muscles of the deep back, which contain resident progenitors and satellite cells that express Pax3 and Pax7. Pax3+/Pax7+ cells were absent at E14.5 in deep back muscles, and Pax7+ satellite cells were not observed at E18.5 in RBP-J/Pax3cre mice. We conclude that epaxial and hypaxial muscle compartments require RBP-J to maintain progenitor cells and to generate satellite cells. Mastermind acts as transcriptional coactivator of RBP-J; the RBP-J mutation and the expression of dominant-negative mastermind result in similar myogenic phenotypes (J.A.E., unpublished observations).
Notch signals maintain progenitor cells not only in the developing muscle but also in other organs like the nervous system, pancreas, and intestine (for reviews, see refs. 40–42). RBP-J is the major transcriptional mediator of Notch signals, but not all RBP-J functions depend on Notch. Recently, similar changes in muscle development to those reported here were described in mice that carry a hypomorph Delta-like-1 allele, indicating that we observed a Notch-dependent function of RBP-J (43). In neural progenitors, Notch signals induce, by RBP-J, the expression of Hes1 and Hes5 and suppress proneural genes, thus maintaining a pool of progenitor cells. Proneural genes, however, induce the expression of the Notch ligand Delta-like-1 in differentiating cells, resulting in up-regulated Notch signaling and suppressed differentiation of neighboring cells (44). Parallels to the function of Notch/RBP-J in muscle progenitors are apparent, where RBP-J, by its control of Hes1, represses MyoD. Notch ligands are expressed by myoblasts and/or myotubes (25), indicating that signals provided by differentiating myogenic cells control the maintenance of progenitors.
Materials and Methods
Generation of an Lbx1cre Transgenic Mouse Strain.
A 144-kb BAC clone RP23–188J8 (RZPD, Berlin, Germany) containing Lbx1 was modified by using homologous recombination in bacteria (45). Cre sequences were fused to the initiating ATG codon of Lbx1, replacing exon 1 sequences. In addition, a neomycin (neo) cassette flanked by FRT sites was inserted for selection, and neo was subsequently removed by transient Flpe expression in bacteria. The linearized Lbx1cre-BAC was injected into pronuclei of fertilized eggs, and transgenic founders were screened for Cre expression in ROSA26R mice (46). By using the Lbx1cre(TG3) transgene, we observed recombination in migrating muscle progenitors. Analysis of RBP-J/Lbx1cre mice demonstrated pronounced changes in the size of limb and tongue muscles, but the diaphragm muscle was mildly affected. We detected many RBP-J-positive cells in the diaphragm, indicating that recombination was incomplete. However, the diaphragm muscle was very small and devoid of Pax7+ cells in RBP-J/Pax3cre mice. RBP-J/Lbx1cre mice were born at expected Mendelian ratios, but did not suckle and died within the first postnatal day. RBP-J/Pax3cre mice were born at expected ratios but did not move or breath and died shortly after birth.
Immunohistochemistry and Electron Microscopy.
Immunohistology was performed on 12-μm cryosections of tissues fixed in 4% paraformaldehyde for 2 h. The following antibodies were used: mouse anti-skeletal fast myosin (Sigma, St. Louis, MO), rabbit or mouse anti-desmin (Sigma), rabbit anti-MyoD (Santa Cruz Biotechnology, Santa Cruz, CA), guinea pig anti-Lbx1 (47), rat anti-Hes1 (MBL, Woburn, MA), rabbit anti-laminin (CAPPEL, Solon, OH), mouse anti-Pax7 (Developmental Studies Hybridoma Bank, Iowa City, IA), rat anti-Pax3 (M. Goulding, Salk Institute, La Jolla, CA), rabbit anti-Pax3 (48), goat anti-β-galactosidase (CAPPEL), and secondary antibodies conjugated with biotin, Cy2, Cy3, or Cy5 (Dianova, Hamburg, Germany). SYBR green I (Molecular Probes, Eugene, OR) was used as a nuclear stain. For Hes1 antibody staining the Cy3-TSA Fluorescence System (PerkinElmer Life Sciences, Wellesley, MA) was used. For BrdU pulse–chase experiments, BrdU (75 μg/g body weight; Sigma) was injected i.p. into pregnant females 1 or 24 h before dissection of embryos; BrdU+ nuclei were identified by using anti-BrdU antibodies (Sigma). Apoptosis was examined by TUNEL staining by using an Apop-Tag fluorescein in situ apoptosis detection kit (Chemicon, Hampshire, U.K.). For electron microscopy, E18.5 mice were perfused with 4% paraformaldehyde. Forelimbs were postfixed with 2.5% glutaraldehyde (24 h), treated with 1% osmium tetroxide (3 h), dehydrated, and embedded in Poly/Bed 812 (Polysciences, Warrington, PA). Ultrathin sections were stained with uranyl acetate and lead citrate.
Myofibers were isolated from muscle tissue of E18 embryos; tissue was dissociated by using NB4 collagenase (0.3 mg/ml, Serva, Heidelberg, Germany; 40 min, 37°C). Single myofibers were plated on coverslips coated with BD Matrigel (BD Biosciences, Franklin Lakes, NJ). After 20-h culture, myofibers were fixed for 10 min with 4% paraformaldehyde and analyzed by immunohistochemistry.
Supplementary Material
Acknowledgments
We thank Walter Birchmeier, Alistair Garratt, and Thomas Müller (Max Delbrück Center for Molecular Medicine) for critically reading the manuscript. We thank Tasuku Honjo (Kyoto University, Kyoto, Japan) for RBP-Jflox/flox mice and Martyn Goulding (The Salk Institute) for Pax3 antibodies. We also thank Achim Gossler (Medizinische Hochschule, Hannover, Germany) for sharing unpublished data before submission of this manuscript. Particular thanks go to Margaret Buckingham (Pasteur Institute, Paris, France) for valuable advice. This work was supported by grants from the Deutsche Forschungsgemeinschaft, Bundesministerium für Bildung und Forschung, and the European Union (Myores) (to C.B.), and by NIH P01 HL075215 (to J.A.E.).
Abbreviation
- En
embryonic day n.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610647104/DC1.
References
- 1.Buckingham M. Curr Opin Genet Dev. 2006;16:525–532. doi: 10.1016/j.gde.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Arnold HH, Braun T. Curr Top Dev Biol. 2000;48:129–164. doi: 10.1016/s0070-2153(08)60756-5. [DOI] [PubMed] [Google Scholar]
- 3.Buckingham M. Curr Opin Genet Dev. 2001;11:440–448. doi: 10.1016/s0959-437x(00)00215-x. [DOI] [PubMed] [Google Scholar]
- 4.Parker MH, Seale P, Rudnicki MA. Nat Rev Genet. 2003;4:497–507. doi: 10.1038/nrg1109. [DOI] [PubMed] [Google Scholar]
- 5.Ordahl CP, Le Douarin NM. Development (Cambridge, UK) 1992;114:339–353. doi: 10.1242/dev.114.2.339. [DOI] [PubMed] [Google Scholar]
- 6.Christ B, Ordahl CP. Anat Embryol (Berl) 1995;191:381–396. doi: 10.1007/BF00304424. [DOI] [PubMed] [Google Scholar]
- 7.Relaix F, Rocancourt D, Mansouri A, Buckingham M. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 8.Gros J, Manceau M, Thome V, Marcelle C. Nature. 2005;435:954–958. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- 9.Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomes D, Tajbakhsh S. Genes Dev. 2005;19:1426–1431. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schienda J, Engleka KA, Jun S, Hansen MS, Epstein JA, Tabin CJ, Kunkel LM, Kardon G. Proc Natl Acad Sci USA. 2006;103:945–950. doi: 10.1073/pnas.0510164103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, Buckingham M. J Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis J. Semin Cell Dev Biol. 1998;9:583–589. doi: 10.1006/scdb.1998.0266. [DOI] [PubMed] [Google Scholar]
- 13.Artavanis-Tsakonas S, Rand MD, Lake RJ. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 14.Lai EC. Development (Cambridge, UK) 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 15.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 16.Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Development (Cambridge, UK) 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 17.Giudicelli F, Lewis J. Curr Opin Genet Dev. 2004;14:407–414. doi: 10.1016/j.gde.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 18.Pourquié O. Annu Rev Cell Dev Biol. 2001;17:311–350. doi: 10.1146/annurev.cellbio.17.1.311. [DOI] [PubMed] [Google Scholar]
- 19.Hrabe de Angelis M, McIntyre J, II, Gossler A. Nature. 1997;386:717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi Y, Koizumi K, Takagi A, Kitajima S, Inoue T, Koseki H, Saga Y. Nat Genet. 2000;25:390–391. doi: 10.1038/78062. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi Y, Inoue T, Gossler A, Saga Y. Development (Cambridge, UK) 2003;130:4259–4268. doi: 10.1242/dev.00629. [DOI] [PubMed] [Google Scholar]
- 22.Conboy IM, Conboy MJ, Smythe GM, Rando TA. Science. 2003;302:1575–1577. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- 23.Kopan R, Nye JS, Weintraub H. Development (Cambridge, UK) 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 24.Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 25.Delfini MC, Hirsinger E, Pourquié O, Duprez D. Development (Cambridge, UK) 2000;127:5213–5224. doi: 10.1242/dev.127.23.5213. [DOI] [PubMed] [Google Scholar]
- 26.Hirsinger E, Malapert P, Dubrulle J, Delfini MC, Duprez D, Henrique D, Ish-Horowicz D, Pourquié O. Development (Cambridge, UK) 2001;128:107–116. doi: 10.1242/dev.128.1.107. [DOI] [PubMed] [Google Scholar]
- 27.Conboy IM, Rando TA. Dev Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 28.Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. J Biol Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 29.Shawber C, Nofziger D, Hsieh JJ, Lindsell C, Bogler O, Hayward D, Weinmaster G. Development (Cambridge, UK) 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 30.Wilson-Rawls J, Molkentin JD, Black BL, Olson EN. Mol Cell Biol. 1999;19:2853–2862. doi: 10.1128/mcb.19.4.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanigaki K, Han H, Yamamoto N, Tashiro K, Ikegawa M, Kuroda K, Suzuki A, Nakano T, Honjo TK. Nat Immunol. 2002;3:443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- 32.Engleka KA, Gitler AD, Zhang M, Zhou DD, High FA, Epstein JA. Dev Biol. 2005;280:396–406. doi: 10.1016/j.ydbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Jagla K, Dolle P, Mattei MG, Jagla T, Schuhbaur B, Dretzen G, Bellard F, Bellard M. Mech Dev. 1995;53:345–36. doi: 10.1016/0925-4773(95)00450-5. [DOI] [PubMed] [Google Scholar]
- 34.Dietrich S, Abou-Rebyeh F, Brohmann H, Bladt F, Sonnenberg-Riethmacher E, Yamaai T, Lumsden A, Brand-Saberi B, Birchmeier C. Development (Cambridge, UK) 1999;126:1621–1629. doi: 10.1242/dev.126.8.1621. [DOI] [PubMed] [Google Scholar]
- 35.Amthor H, Christ B, Weil M, Patel K. Curr Biol. 1998;8:642–652. doi: 10.1016/s0960-9822(98)70251-9. [DOI] [PubMed] [Google Scholar]
- 36.Scaal M, Bonafede A, Dathe V, Sachs M, Cann G, Christ B, Brand-Saberi B. Development (Cambridge, UK) 1999;126:4885–4893. doi: 10.1242/dev.126.21.4885. [DOI] [PubMed] [Google Scholar]
- 37.Anakwe K, Robson L, Hadley J, Buxton P, Church V, Allen S, Hartmann C, Harfe B, Nohno T, Brown AM, et al. Development (Cambridge, UK) 2003;130:3503–3514. doi: 10.1242/dev.00538. [DOI] [PubMed] [Google Scholar]
- 38.Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- 39.Brohmann H, Jagla K, Birchmeier C. Development (Cambridge, UK) 2000;127:437–445. doi: 10.1242/dev.127.2.437. [DOI] [PubMed] [Google Scholar]
- 40.Edlund H. Nat Rev Genet. 2002;3:524–532. doi: 10.1038/nrg841. [DOI] [PubMed] [Google Scholar]
- 41.Petersen PH, Tang H, Zou K, Zhong W. Dev Neurosci. 2006;28:156–168. doi: 10.1159/000090761. [DOI] [PubMed] [Google Scholar]
- 42.Radtke F, Clevers H. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- 43.Schuster-Gossler K, Cordes R, Gossler A. Proc Natl Acad Sci USA. 2007;104:537–542. doi: 10.1073/pnas.0608281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis J. Curr Opin Neurobiol. 1996;6:3–10. doi: 10.1016/s0959-4388(96)80002-x. [DOI] [PubMed] [Google Scholar]
- 45.Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 46.Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Dev Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- 47.Vasyutina E, Stebler J, Brand-Saberi B, Schulz S, Raz E, Birchmeier C. Genes Dev. 2005;19:2187–2198. doi: 10.1101/gad.346205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Liu KC, Jin F, Lu MM, Epstein JA. Development (Cambridge, UK) 1999;126:2495–2503. doi: 10.1242/dev.126.11.2495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.