Abstract
Horizontal gene transfer (HGT) is thought to play an important role in the evolution of species and innovation of genomes. There have been many convincing evidences for HGT for specific genes or gene families, but there has been no estimate of the global extent of HGT. Here, we present a method of identifying HGT events within a given protein family and estimate the global extent of HGT in all curated protein domain families (≈8,000) listed in the Pfam database. The results suggest four conclusions: (i) for all protein domain families in Pfam, the fixation of genes horizontally transferred is not a rampant phenomenon between organisms with substantial phylogenetic separations (1.1–9.7% of Pfam families surveyed at three taxonomic ranges studied show indication of HGT); (ii) however, at the level of domains, >50% of Archaea have one or more protein domains acquired by HGT, and nearly 30–50% of Bacteria did the same when examined at three taxonomic ranges. But, the equivalent value for Eukarya is <10%; (iii) HGT will have very little impact in the construction of organism phylogeny, when the construction methods use whole genomes, large numbers of common genes, or SSU rRNAs; and (iv) there appears to be no strong preference of HGT for protein families of particular cellular or molecular functions.
Keywords: protein domain family, protein sequence family, lateral gene transfer
One of the new important concepts that emerged from a large number of genomic sequences in the last decade is that of horizontal gene transfer (HGT): gene transfer among organisms of different species. HGT has been found to have occurred in all three domains: Archaea, Bacteria, and Eukarya. The concept of HGT has been evoked to interpret various evolutionary processes ranging from speciation and the adaptation of organisms to uncertainties in phylogenetic inference of the tree of life (1–9). Although HGT has been regarded as a driving force in the innovation and evolution of genomes, especially in prokaryotes, its extent and impact on the evolutionary process and phylogeny of organisms or species remains controversial (8–10).
There have been several methods developed to detect HGT, including (i) difference between gene trees derived from a limited number of gene families and the reference trees such as the small-subunit ribosomal RNA (SSU rRNA) tree (11–13) or whole genome tree (14); (ii) unexpectedly high sequence similarity of a gene from two distant genomes compared with those among homologous genes in closely related genomes (15); and (iii) unusual nucleotide composition or codon usages of a gene compared with the rest of the genes within a genome (16, 17). Many factors affect the detection of HGT, such as lineage-specific gene loss (18, 19), unequal rates of base substitution (1), loss of signal due to amelioration processes (16), and others (1, 15).
It has been suggested that HGT may have been “rampant” in primitive genomes (6, 20), but, for modern organisms, it may not be a dominant factor in speciation, because HGT has less effect on overall genome phylogeny (10, 21).
There have been many convincing evidences for HGT for specific genes or gene families, but there has been no estimate of the global extent of HGT in terms of protein domains. Here, we present a statistical method to identify the member(s) in a protein family that may have joined the family by HGT events and examine the global extent of HGT events for all protein domain families of known curated sequences at various ranges of taxonomic levels.
A protein (sequence) domain is a functionally independent unit in protein sequence. The gene coding for it often behaves like a modular genetic element that transfers within or between genomes, sometimes forming a new gene coding for a multiple domain protein (22–24). Because the fixation of a new gene during evolution depends mostly on its advantage for survival, we focus on HGT of the genetic module coding for the sequence domains, rather than the entire genes. At present, there are ≈1.2 million curated protein domain sequences from three domains of life (Archaea, Bacteria, and Eukarya) in the Pfam (release 16.0) (25).
Results
The Phylogenetic Tree of All Organisms Represented by the Pfam Protein Domains.
The first step in our method requires constructing a phylogenetic tree of the organisms represented by all protein domains in Pfam. Many approaches have been developed recently to construct phylogeny of organisms by using a set of selected gene families or whole genomes, and it was found that it is practically the same as that constructed from SSU rRNA sequences, suggesting that HGT does not alter significantly the SSU rRNA-based tree (19, 26).
The reconstruction of the phylogenetic tree of organisms, with all species covering ≈1.2 million protein domain sequences in the Pfam is not practical, and thus we simplified the tree structure by using representative taxa: To obtain representative taxa, we examined the taxonomic origins of all organisms from which all protein sequences in Pfam (release 16.0) are derived and extracted their taxonomic identifications at three ranges of taxonomic levels (second to fourth hierarchical level listed in the Pfam) as described in Materials and Methods (see Fig. 1). In most cases, the second, third, and fourth levels correspond to phylum, the range between phylum and order, and the range between order and genus, respectively. Although the number of protein members per family in the Pfam varied considerably from 2 to 49,343, the number of unique representative taxa per family ranged only from 1 to 191 at the three taxonomic ranges. There is good correlation among the numbers of unique representative taxa, nonredundant species, and family size (Fig. 2), suggesting that selected taxonomic ranges are representative, and sampling bias resulting from them would not affect the estimation of the global extent of HGT. Thus, we used these representative taxa to reconstruct the phylogenetic tree of organisms for each taxonomic range.
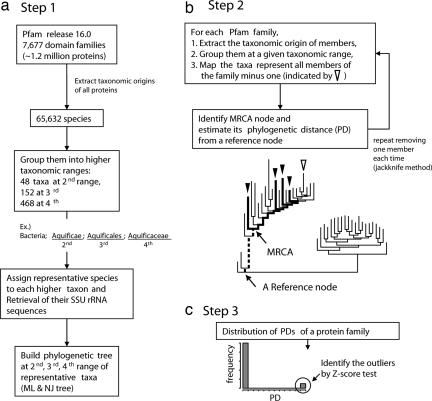
Fig. 1.
Flow diagram for the detection of HGT in a protein domain family divided into three steps. (a) Step 1: Building the phylogenetic tree of representative taxa covering taxonomic origins of organisms for all protein members in the Pfam (see Materials and Methods for details). (b) Step 2: Mapping all organisms (minus one) represented by the members of a given Pfam family onto the tree, identifying the MRCA node, and estimating its PD from a reference node. This process is repeated each time, removing different organism (jackknife operation). We define the CAG as the gene from which all genes coding for the member proteins of a subset of a protein domain family are derived and assume that the CAG resided in the organism at MRCA node. The PD of the MRCA is defined by the branch length (dashed line) between MRCA node and the reference node. (c) Step 3: Calculate the variance of the values of PDs obtained from the jackknife method and test the presence or absence of outliers from monomodal distribution (by Z-score test) of the PDs. The protein families with outliers are considered as the candidate families containing the members (outliers) that have undergone HGT.
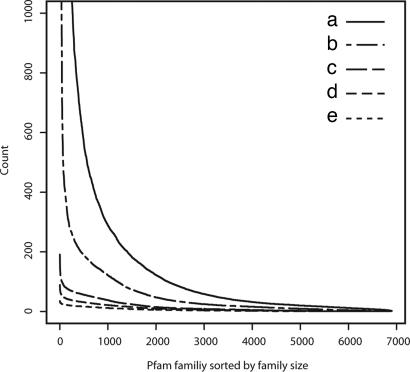
Fig. 2.
The 6,883 protein families used in this study are sorted by the family size, i.e., number of members, and listed in decreasing order (horizontal axis). On the vertical axis, the counts of various quantities in a given family are shown: number of family members (a), number of nonredundant species (b), number of representative taxa at fourth taxonomic range (c), at third taxonomic range (d), and at second taxonomic range (e) in the Pfam (release 16) (25). The Spearman's rank-correlation coefficients vary from 0.81 [between number of members (a) and representative taxa at second taxonomic range (e)] to 0.90 [between number of members (a) and nonredundant species (b)].
Bimodal Distribution of Phylogenetic Distances of Most Recent Common Ancestor (MRCA) Organisms [Containing Common Ancestral Genes (CAGs) of a Protein Family] as an Indication of HGT.
As a first approximation, according to the Pfam, the totality of the curated known protein domains is grouped into 7,677 protein domain families based on protein sequence similarity. Because homologous protein sequences imply common evolutionary origin, for each protein domain family, we assumed that there was a CAG from which the genes coding for all members of that family were derived and that the CAG resided in the MRCA organism in the phylogenetic tree of organisms.
In the second step in our method, for a given protein domain family, we mapped all organisms represented by the members of the family on the phylogenetic tree of organisms reconstructed from SSU rRNA, each time leaving one test organism out (the jackknife operation, Fig. 1), identified the MRCA node representing the organism containing the CAG, and estimated the phylogenetic distance (PD) defined as the branch length between MRCA node and a common reference point in the tree. Each jackknife operation generates one PD; thus, the number of PDs equals that of the representative taxa of the family.
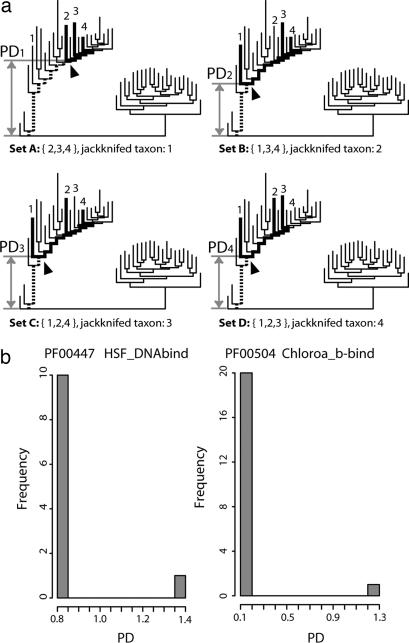
The third step in our method analyzes the distribution of the PDs for each family (Fig. 1). If the distribution of the PDs is not homogeneous, we assume that there have been one or more possible HGT events during evolution in this family at the given taxonomic range we tested. Fig. 3a shows a hypothetical example, where PD2, PD3, and PD4 have the same length, but PD1 is an outlier (see also Fig. 3b for an actual example). For those with heterogeneous distribution of PDs, we applied a statistical method (robust Z test) to identify the outlier organism(s), whose phylogeny deviates significantly from the rest of the population generating aberrant PDs. We interpret that the outlier organism(s) acquired the gene coding for a protein belonging to the protein family by HGT. This operation was repeated for all protein domain families in the Pfam to obtain the extent of the global HGT events that have been fixed in present-day organisms.
Fig. 3.
Detection of HGT candidates. (a) A simplified hypothetical example of a jackknife operation: If all members of a given protein family belong to four taxa (marked 1–4), the PDs (the dashed lines and scaled gray arrow bars) of MRCAs containing CAGs (indicated by arrowheads) can be calculated from four different subsets, in which one taxon of four was jackknifed out at a time. In this example, set A shows an aberrant PD from others: PD1 is much longer than the remaining three PDs, which have the same short branch length. Thus, we regard this family as a candidate family with HGT. (b) Actual examples for two Pfam families: The histograms showing the number of MRCAs (frequency) vs. PD values obtained from jackknife operation. Both examples show a bimodal distribution, which suggest that both families experienced HGT events. The histogram on the right presents a wider variation in PD distribution. Both clearly show the significant outliers (Z scores ≥3) due to HGT.
The determination of the node of a MRCA of a CAG and its PD in the tree may be sensitive to the topology of the tree (phylogeny reconstruction method) as well as the distribution pattern of taxa in the tree. Therefore, we carried out the jackknife operations; first, in two independent tree topologies constructed, one by maximum likelihood (ML) and the other by the neighbor-joining (NJ) method and, second, at the three taxonomic ranges as described in Materials and Methods.
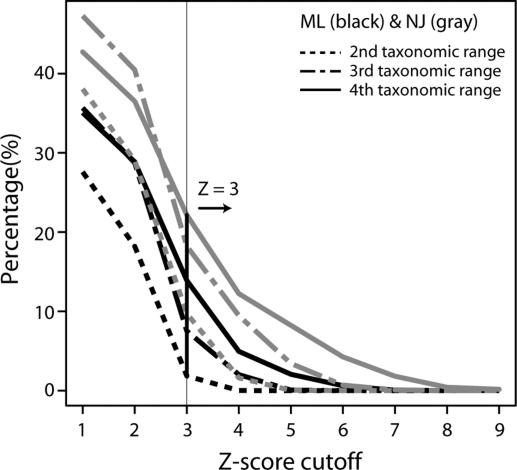
Among 6,883 Pfam families (7,677 minus 794 families from viruses, environmental samples, or other ambiguously annotated organisms), ≈28–47% of families showed heterogeneous (nonmonomodal) distribution of the PDs of MRCAs containing CAGs of the protein families, depending on the tree-building methods and the taxonomic ranges used (Fig. 4). We identified significant outliers from monomodal distribution of the PDs in two ways: (i) we used Z score to test whether the deviation of outlier PDs is statistically significant. We set the Z score cutoff as the inflection point of Z score plot, Z = 3 as seen in Fig. 4. However, at second and third taxonomic ranges, there are fewer taxa (fewer samples) than fourth range and, therefore, giving smaller Z-scores for the same deviation, the Z-score cutoff at Z ≥ 3 gave significant discrimination in general (P < 0.001) in a statistical analysis; (ii) we accepted an HGT event only when PD distribution significantly deviates from monomodal distribution using both ML and NJ trees. Thus, we regarded a given protein family as a true positive candidate of HGT only when the Z score is ≥3 in both tree topologies. Under these criteria, we find 1.1–9.7% of the Pfam families are detected for HGT at three taxonomic rages. It is worth pointing out that this method identifies one or a few HGT candidates in a protein family only when all other members in the family concur.
Fig. 4.
The percentage of protein families with HGT according to various Z-score cutoffs and tree topology. The percentages are presented at various taxonomic ranges by dashed (second level), dot-dashed (third level), or solid (fourth level) lines by using ML (black) or NJ (gray) trees. We set the common inflection point of the plot at Z = 3, which was used as a criterion for identifying outliers from monomodal distribution of PDs.
Global Extent of HGT in Three Domains of Life at Three Taxonomic Ranges.
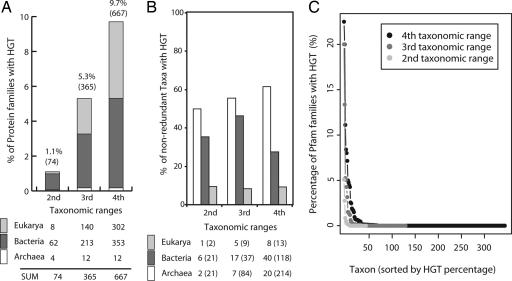
The extent of HGT varied from 1.1% (74 families) at the second, 5.3% (365 families) at the third, to 9.7% (667 families) at the fourth taxonomic range [Fig. 5a; see also supporting information (SI) Table 1]. Because we identified the outliers (HGT candidates) by the jackknife method, taxonomic origins of the outliers can infer that the destination of HGT happened in a given protein family. When we classified the destination of HGT candidates into three domains of life to which the outliers belong, the ratio of HGT in three domains of life was distributed unequally at three taxonomic ranges (Fig. 5a, gray colored regions in the bar plot). However, many of taxonomic origins of outliers classified into the same domain of life were overlapped at the three taxonomic ranges we used. For instance, the majority of HGT events in Eukarya were detected in specific taxa such as Fungi. Thus, we removed the redundant taxonomic origins at various taxonomic ranges and reclassified them into three domains of life according to the destination of HGT. After removal of duplicated taxonomic sources, we found the majority of HGT, ≈30–60% at three taxonomic ranges, happened mostly in Archaea and Bacteria, indicating that HGT is a more frequent event in Archaea and Bacteria across the taxonomic ranges.
Fig. 5.
Occurrence of HGT at various taxonomic ranges and the distribution in three domains of life. (a) The percentage of protein families that acquired at least one member by HGT event at each taxonomic range. The numbers in parentheses indicate the numbers of protein families (of 6,883 families), of which at least one member joined the family by HGT. The distribution of them in three domains of life was obtained by counting of the target taxa of HGT. (b) The percentage of organisms that acquired at least one protein domain gene by HGT in three domains of life. After removing redundant taxonomic origins from the distribution shown in a, the relative percentages of nonredundant organisms that were the targets of HGT in each domain of life were shown at different taxonomic ranges. The number of nonredundant taxa identified as outliers is shown under the plot, and the numbers in parentheses indicate the number of representative nonredundant taxa examined at different taxonomic ranges. Sampling for Archaea is too small to be reliable, especially at the second taxonomic range. (c) The distribution of the percentages of Pfam families with HGT in taxa at three taxonomic ranges. The percentage of each taxon at a given taxonomic range is indicated by light gray (second taxonomic range), dark gray (third taxonomic range), and black (fourth taxonomic range) circles and lines. There are two extreme outliers with ≥15% HGT that belong to Bacteria (Bacteria Actinobacteria Rubrobacteridae) and Eukarya (Eukarya Fungi Microsporidia Unikaryonidae) and might be a bias due to small sample size.
Examined at domain level, Fig. 5b reveals that a very large fraction of Archaea and Bacteria (represented by all protein domains in Pfam) was the targets of HGT. It reveals that, at three taxonomic ranges, >50% of Archaea has one or more protein domain acquired by HGT and ≈30–50% of Bacteria did the same. However, the equivalent fraction for Eukarya is <10%. The extent of HGT in an individual taxon has important relevance in constructing the organism phylogeny. At a given taxonomic range, we estimated the percentage of HGT in a given taxon from the number of Pfam families with HGT candidates in the taxon divided by the number of all Pfam families belonging to the taxon. The range of the percentage Pfam families with HGT varies from 0% to 22.5% at various taxonomic ranges, with the overwhelming majority of taxa showing very small or negligible extent of HGT. The average percentages of HGT in representative taxa are 0.31% (fourth taxonomic range), 0.46% (third taxonomic range) and 0.22% (second taxonomic range), respectively (Fig. 5c). This observation strongly suggests that HGT has very little impact in the construction of organism phylogeny, when the construction methods use whole genomes, large numbers of common genes, or SSU rRNAs.
Propensity of HGT Among Different Gene Categories.
Jain et al. (4) suggested that HGT may have occurred preferentially among the operational genes (those that maintain cell growth such as metabolism-related genes) than the among informational genes (such as those genes involved in transcription and translation), whereas Nakamura et al. (17) observed that only parts of genes in functional categories such as mobile element, cell surface, DNA binding, and pathogenicity-related, were preferred.
To examine functional propensity in HGT of protein domains, we identified the functional categories of all 7,677 protein domain families in the Pfam by assigning Gene Ontology (GO) terms (27), which are controlled vocabularies for gene annotation. For simplicity, the terms of GO slim, a simplified version of the original GO, are used in three major categories (molecular function, biological process, and cellular component). According to Fisher's exact test (28), there was no strong preference of HGT events more than particular GO slim categories except in a few subcategories. We found a few marginal preferences (P < 0.001) for subcategories of molecular functions such as helicase activity (GO:0004386) at the second taxonomic range, nucleic acid binding (GO:0003676) at the third range, oxidoreductase activity (GO:0016491) at the fourth range, and a marginal preference for extracellular region (GO:0005576) at the third and fourth ranges in the GO cellular component category. These data reconfirmed previous observations that HGT was biased toward cell surface and DNA binding functions (17) and, for example, that the helicase domain integrated into reverse gyrase has undergone HGT (29), but the biases are marginal. Thus, as a first approximation, our estimation suggests that HGT is nearly neutral to all genes, suggesting that a random HGT process is followed by selection due to environment or other factors.
Discussion
Previous studies on the detection of HGT were confined to testing of a phylogenetic incongruency of a limited number of orthologous gene clusters within narrow taxonomic ranges with respect to entire organismic phylogeny (14, 30) or measuring the bias in the nucleotide composition of genes in a limited number of completed genomic sequences (16, 17). In our study, we used all curated protein sequences grouped into ≈7,000 protein domain families to assess a global extent of HGT.
The method does not distinguish orthologs from paralogs in a protein family, because it is based on the identification of MRCAs containing CAGs of a protein family in the tree and because both orthologs and paralogs share their ancestry at gene level. Furthermore, because we are examining the distribution pattern of all PDs of MRCAs containing CAGs of all members in a given protein domain family, the artifacts resulting from different tree-building procedures does not affect seriously the detection of a global trend of HGT. In addition, this method does not depend on a single observation of high homology of a gene between two distant organisms but, rather, identifies one or a few organisms among all organisms (represented by the members of a protein family) as the targets of HGT only when all other organisms in the family concur. Because the method is very conservative, the resolution for detection of HGT at the gene level may be lower than other methods, but it may provide a more reliable view of the global trend of HGT in overall protein domain.
An earlier study of the global extent of HGT by Ge et al. (14) examined 297 clusters of orthologous genes (COGs) from 40 genomes and found that 33 COGs (11.1%) show indication of HGT. In our study, of 6,883 Pfam families from 345 representative taxa, 753 families (10.9%) show the HGT indications across different taxonomic ranges. Despite the similarity in the percentage, we found that the correspondence between the COGs and Pfam families that have undergone HGT by the two methods is low. This may be due to fundamental differences in the methods used, differences in sampling size, and possibly other reasons: For example, Ge et al. compares individual gene tree with the whole genome tree to find incongruency due to HGT, whereas we used the organism tree only and identified organisms that contain protein genes (of a protein family) arrived by HGT as agreed by all remaining member organisms representing the protein family. Our method detects HGT between distant organisms, whereas their methods may detect HGT between close organisms; and our method employs 345 taxa covering the entire Pfam families, whereas their method uses only 40 microbial genomes and covers a limited number of COGs.
Daubin et al. (19) observed that a subset of HGTs resulting from homologous recombination is limited to a very closely related taxonomic unit and suggested that HGT among closely related species eventually makes cohesive taxonomic groups. Similarly, metagenomic analysis in natural environments revealed that there is commonly a higher genomic diversity but a limited rRNA variation in the microbial community (31, 32). This observation led to the suggestion of a new taxonomic unit such as ribotypes or ecotypes of species that are microdiverse clusters having a little rRNA variation but diverse genomic diversity (31–33). This enlarged taxonomic unit implies that HGT among closely related organisms is a frequent event but that not all HGT eventually affects the speciation of organisms. Thus, HGTs might blur the boundary of closely related species but have less impact on the phylogeny of distant taxa in the long run. These observations are consistent with our observation that HGT (as manifested in the protein domain families) is not rampant at the taxonomic ranges we examined and has no strong correlation to specific functional categories, suggesting that the ongoing HGT at present is largely random and relatively neutral, and only a small portion of all HGT events may be fixed and contribute to the evolution of genomes and speciation of distant organisms.
Materials and Methods
Reconstruction of the Phylogenetic Tree of Representative Taxa.
We used the Pfam (release 16.0) as a reference database that clusters all curated protein sequences (≈1.2 million protein sequences in release 16.0) into 7,677 protein sequence domain families. After removal of the families represented exclusively by the proteins from viruses, environmental samples, or other ambiguous sources, the number of the families used in our analysis was reduced to 6,883. To make the phylogenetic tree of the organisms represented by these protein families, we extracted the taxonomic origins of all members of the 6,883 Pfam families and assembled an organism set of 65,532 nonredundant species after removal of duplicated origins. When we grouped them further into representative taxa using the second to fourth levels of taxonomic hierarchy listed in the Pfam, they could be regrouped into 468 taxa at fourth, 152 at third, and 48 at second taxonomic ranges. The scientific classifications of representative taxa for the second, third, and fourth hierarchy in Pfam correspond to phylum, the range between phylum and order, and the range between order and genus, respectively. (SI Tables 2 and 3). Of these, the gene sequences of SSU rRNA (16S rRNA of prokaryotes or 18S rRNA of eukaryotes) of 345 taxa were available from the European ribosomal RNA database (34). Among the SSU rRNA sequences belonging to each of 345 taxa, we chose the longest SSU rRNA sequence (but not <1,200 bases) to represent each taxon. Then, we used these prealigned representative SSU rRNA sequences in the database to construct the phylogenetic tree of organisms (see Fig. 1a for a flow diagram).
To avoid potential errors coming from tree topology and sampling bias, we constructed the trees using two independent tree-building procedures: NJ and ML at a lower taxonomic range (fourth level). For the NJ tree, we made 100 bootstrapped replicates of alignments and computed distance matrices based on an F84 model of sequence evolution using the PHYLIP package (35). The consensus tree of the bootstrapped replicates was obtained by majority rule. Because this consensus tree did not give information about branch lengths, we recalculated the branch lengths using the ML method without changing the tree topology of NJ. The ML tree was built under the assumption of constant rate with the F84 model.
For the trees of higher taxonomic ranges (second and third), we calculated a distance matrix of all-against-all pair-wise distances of taxa (leaves) from the branch lengths of the tree obtained at the fourth range and collapsed the distance matrix by averaging rows and columns belonging to the same taxonomic groups (SI Fig. 6). Then, the tree was reconstructed by using the NJ method at each taxonomic range. For all tree-building procedures described here, we used Bacteria (Aquifex pyrophilus) as an outgroup (the reference node). All procedures were carried out by using the PHYLIP package and ad hoc programs in our laboratory.
Statistical Test of Potential HGT from the Distribution of PDs of MRCAs.
Once we obtained the distribution of the PDs for each Pfam family by using a jackknife method, we tested its congruency (or monomodality) by using a statistical test for outliers. The outliers of distribution were identified by a slight modification of robust Z test, Z = (PDmax − PDmed)/SD, where PDmax is the maximum PD, PDmed is the median of the distribution of PDs, and SD is the standard deviation of distribution.
If there are outliers from the jackknife test, the distribution of the PD would be nonhomogeneous (Fig. 2b). We used the median value rather than the mean value of sample distribution because the median is more robust for detection of outliers. The Z score also depends on the size of the sample population: a higher Z score at the same variance when the size of the sample becomes larger.
GO Assignment of the Protein Domain Families with HGT.
To categorize HGT candidates by GO terms (27), we extracted the ontological terms of three organizing principles (biological process, molecular function, and cellular component) from the Pfam database and simplified them to GO slim terms, which are simplified terms of a higher hierarchical order in GO. To test whether there is a dependency between HGT candidates and their GO slim categories, we used two-sided Fisher's exact test by making 2 × 2 contingency table in which the table elements are built as in Ge et al. (14). The null hypothesis for Fisher's exact test was that HGT candidates and GO slim categories are associated with each other.
Supplementary Material
Acknowledgments
We thank Drs. Jingtong Hou, Gregory Sims, and Se-Ran Jun for our weekly discussions on the subjects of this work as well as other related subjects and Drs. George Church, Doug Brutlag, James Lake, Eugene Koonin, and Norman Pace, whose comments helped us to sharpen our thoughts and clarify ambiguous points in our early version of the manuscript. This work was supported by National Institutes of Health Grant GM62412.
Abbreviations
- CAG
common ancestral gene
- GO
gene ontology
- HGT
horizontal gene transfer
- ML
maximum likelihood
- MRCA
most recent common ancestor
- NJ
neighbor joining
- PD
phylogenetic distance
- SSU rRNA
small-subunit ribosomal RNA.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611557104/DC1.
References
- 1.Syvanen M. Annu Rev Genet. 1994;28:237–261. doi: 10.1146/annurev.ge.28.120194.001321. [DOI] [PubMed] [Google Scholar]
- 2.Pennisi E. Science. 1998;280:672–674. doi: 10.1126/science.280.5364.672. [DOI] [PubMed] [Google Scholar]
- 3.Doolittle WF. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 4.Jain R, Rivera MC, Lake JA. Proc Natl Acad Sci USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochman H, Lawrence JG, Groisman EA. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 6.Woese CR. Proc Natl Acad Sci USA. 2002;99:8742–8747. doi: 10.1073/pnas.132266999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris JK, Kelley ST, Spiegelman GB, Pace NR. Genome Res. 2003;13:407–412. doi: 10.1101/gr.652803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown JR. Nat Rev Genet. 2003;4:121–132. doi: 10.1038/nrg1000. [DOI] [PubMed] [Google Scholar]
- 9.Gogarten JP, Townsend JP. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 10.Kurland CG, Canback B, Berg OG. Proc Natl Acad Sci USA. 2003;100:9658–9662. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith MW, Feng DF, Doolittle RF. Trends Biochem Sci. 1992;17:489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- 12.McGowan C, Fulthorpe R, Wright A, Tiedje JM. Appl Environ Microbiol. 1998;64:4089–4092. doi: 10.1128/aem.64.10.4089-4092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich MW. J Bacteriol. 2002;184:278–289. doi: 10.1128/JB.184.1.278-289.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge F, Wang L-S, Kim J. PLoS Biol. 2005;3:e316. doi: 10.1371/journal.pbio.0030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin EV, Makarova KS, Aravind L. Annu Rev Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence JG, Ochman H. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura Y, Itoh T, Matsuda H, Gojobori T. Nat Genet. 2004;36:760–766. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- 18.Mirkin B, Fenner T, Galperin M, Koonin E. BMC Evol Biol. 2003;3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daubin V, Moran NA, Ochman H. Science. 2003;301:829–832. doi: 10.1126/science.1086568. [DOI] [PubMed] [Google Scholar]
- 20.Woese C. Proc Natl Acad Sci USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurland CG. EMBO Rep. 2000;1:92–95. doi: 10.1093/embo-reports/kvd042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doolittle RF, Bork P. Sci Am. 1993;269(4):50–56. doi: 10.1038/scientificamerican1093-50. [DOI] [PubMed] [Google Scholar]
- 23.Amit Fliess BM, Ron Unger. Proteins. 2002;48:377–387. doi: 10.1002/prot.10156. [DOI] [PubMed] [Google Scholar]
- 24.Tordai H, Nagy A, Farkas K, Banyai L, Patthy L. FEBS J. 2005;272:5064–5078. doi: 10.1111/j.1742-4658.2005.04917.x. [DOI] [PubMed] [Google Scholar]
- 25.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer ELL, et al. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snel B, Huynen MA, Dutilh BE. Annu Rev Microbiol. 2005;59:191–209. doi: 10.1146/annurev.micro.59.030804.121233. [DOI] [PubMed] [Google Scholar]
- 27.Camon E, Magrane M, Barrell D, Binns D, Fleischmann W, Kersey P, Mulder N, Oinn T, Maslen J, Cox A, Apweiler R. Genome Res. 2003;13:662–672. doi: 10.1101/gr.461403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher RA. J R Stat Soc. 1922;85:87–94. [Google Scholar]
- 29.Forterre P, Bouthier De La Tour C, Philippe H, Duguet M. Trends Genet. 2000;16:152–154. doi: 10.1016/s0168-9525(00)01980-6. [DOI] [PubMed] [Google Scholar]
- 30.Novichkov PS, Omelchenko MV, Gelfand MS, Mironov AA, Wolf YI, Koonin EV. J Bacteriol. 2004;186:6575–6585. doi: 10.1128/JB.186.19.6575-6585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J, Sarma-Rupavtarm R, Distel DL, Polz MF. Science. 2005;307:1311–1313. doi: 10.1126/science.1106028. [DOI] [PubMed] [Google Scholar]
- 32.Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, Polz MF. Nature. 2004;430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]
- 33.Giovannoni S. Nature. 2004;430:515–516. doi: 10.1038/430515a. [DOI] [PubMed] [Google Scholar]
- 34.Wuyts J, Perriere G, Van de Peer Y. Nucleic Acids Res. 2004;32:D101–103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Felsenstein J. Cladistics. 1989;5:164–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.