Abstract
A genetic screen was established to clone apoptosis-inducing genes in a high-throughput format. It led to the isolation of several proapoptotic genes whose proteins are localized to mitochondria. One of the isolated genes is cytochrome bL (cybL also known as SDHC, CII-3, or QPs-1), a component of the respiratory chain complex II. It was further investigated because both cybL and another component of complex II, cybS, have recently been identified as tumor suppressor proteins, some of which act by controlling apoptosis. Our studies reveal that cell death induction by cybL expression is concomitant with a transient inhibition of complex II and the generation of reactive oxygen species. Importantly, cells that are constitutively deficient in cybL are resistant to a variety of proapoptotic cytostatic drugs and to the effects of the Fas receptor. Our results therefore identify complex II as a sensor for apoptosis induction and could explain the unexpected observation that complex II is inactivated in tumors.
INTRODUCTION
Apoptosis is the genetically determined cell death that plays a role in tissue homeostasis, development, and disease (White, 1996). Its program is initiated by the cell upon receiving defined signals. They are then converted via a signal transduction pathway into the activation of proapoptotic cysteine proteases, the so-called caspases (Salvesen and Dixit, 1997). Subsequently, these enzymes cleave specific substrates in the cell and thereby generate the typical phenotype of an apoptotic cell. To avert diseases, the induction of this destructive process and therefore the activation of proapoptotic signaling pathways must be tightly regulated.
A survey of components of proapoptotic signaling pathways reveals an intriguing correlation: Almost all genes that mediate the signal for apoptosis induction also have the dominant property to induce apoptosis upon overexpression. This capacity is even conserved across species (McCarthy and Dixit, 1998). This also holds true for adaptor proteins such as TRADD (Hsu et al., 1995) and FADD (Chinnaiyan et al., 1995) of the Fas- and the tumor necrosis factor (TNF) receptor complex or for ANT-1 of the permeability transition pore (Bauer et al., 1999). It might be explained by the observation that protein–protein interactions are in many cases responsible for the induction of cell death (Yang et al., 1998a,b). On overexpression, these interactions are generated and apoptosis is induced.
These genes therefore define protein complexes as sensors for apoptosis because they are also used by other stimuli that lead to cell death. The dominant trait of genes to induce apoptosis could therefore be used to define such sensors.
In previous work, we described a screen to isolate dominant apoptosis-inducing genes (Grimm and Leder, 1997). Herein, we used the screen in a 96-well high-throughput format by introducing single cDNAs into cells. For the first time, we were able to use this screen and one of the isolated genes to identify a novel sensor for apoptosis induction. It is complex II of the respiratory chain whose subunits cybL and cybS have recently been shown to be tumor suppressor proteins.
MATERIALS AND METHODS
Cell Culture and Transfections
The cybL-deficient cells are derivatives from Chinese hamster cells (Oostveen et al., 1995). These cells were cultivated in DMEM with 10% fetal calf serum (Sigma Chemie, Deisenhofen, Germany), 5 mM glucose, 200 mM l-glutamine, 1× minimal essential medium nonessential amino acids (both from Invitrogen, Karlsruhe, Germany), and an antibiotics solution (Sigma Chemie) with penicillin (100 U/ml) and streptomycin (0.1 mg/ml). 293T and HeLa cells were kept in DMEM supplemented with 5% fetal calf serum in a humidified 5% CO2 atmosphere at 37°C. For the generation of HeLa cells deficient in mitochondrial DNA (HeLa ρ0 cells), the cells were incubated for 2 mo in 100 μg/ml ethidium bromide in medium supplemented with 50 μg/ml uridine and 100 μg/ml pyruvate (King and Attardi, 1989). These cells are dependent on uridine addition, which is indicative for cells with an inactive respiratory chain (King and Attardi, 1989). In addition, using a RNA blot we observed that cytochrome oxidase subunit 1, which is encoded by the mitochondrial DNA, is absent in these cells (our unpublished data). HeLa wild-type (WT) cells and HeLa ρ0 cells were investigated for the presence of various apoptosis regulators: Apaf-1, cytochrome c, Smac, and caspase-3 and caspase-9. No quantitative difference in the expression levels of these proteins was found. The cybL-green fluorescent protein (GFP) fusion protein is constitutively expressed in the reconstituted cells that can grow on galactose instead of glucose, which is required for the cybL-negative cells. For transfections, the cells were split in appropriate plates and transfected with plasmid DNA by the calcium phosphate coprecipitation method as described previously (Roussel et al., 1984). For transfections in 24-well plates, 25 μl of DNA solution were mixed with 25 μl of 2× HEPES-buffered saline (HBS) (pH 6.9; 4°C) in a 96-well plate with a 12-channel pipettor (Eppendorf, Hamburg, Germany). Twenty microliters of a 0.25 M CaCl2 solution (4°C) was added and mixed, and 38 μl was distributed on cells after incubating for 25 min at room temperature. For the transfections of 293T cells, chloroquine (40 μM) was added to the medium that was changed 5 h later. The transfection efficiencies were routinely 50%.
Generation of Constructs
The isolated cybL was modified by recombinant polymerase chain reaction (PCR) with Pwo polymerase, which has proof-reading activity. Thirty-five cycles of PCR were used to amplify the mitochondrial targeting sequence of cyclophilin D (residues 1–40), and an additional 34 cycles were applied to attach it to the N terminus of cybL. The longer isoform of cybL was isolated from L929 RNA by reverse transcription-PCR by using suitable primers. The mitochondrial targeting sequence (residues 1–40) was amplified from a cyclophilin D construct by PCR. The correct sequence of all constructs was verified by sequencing. Its subcellular localization was checked by a fusion protein with cyclophilin D and yellow fluorescent protein and was found to be exclusively localized to mitochondria. The genes for the human complex II subunits of the flavin-containing protein and the iron-sulfur protein were isolated with reverse transcription-PCR and control sequenced.
Generation of a Normalized Library and cDNA Screening
The normalization and construction of a cDNA kidney library was performed as described previously (Grimm and Leder, 1997). Plating aliquots on agar revealed that the library was comprised of ∼2.5 × 105 clones. Seventy-seven percent of the clones contained inserts that comprise a medium length of 2 kb. Aliquots containing single clones were inoculated in wells of 96-well blocks (QIAGEN, Hilden, Germany) in 900 μl of LB medium and grown for 30 h at 300 rpm agitation. The plasmids were isolated as described previously (Neudecker and Grimm, 2000) and transfected into 293T cells. Subsequently, we have used visual inspection for the phenotype of apoptotic cells as the cellular read-out. When a positive clone was identified in transfected 293T cells, the DNA was again transfected to verify the result. The remaining DNA was used to transform bacteria for a large-scale plasmid isolation.
Apoptosis Quantification
Apoptosis was quantified with a fluorescence-activated cell sorting (FACS) analysis as described previously (Bauer et al., 1999). A cotransfected GFP expression plasmid was used to assess the transfection efficiency. The apoptotic cell population with subdiploid DNA content was normalized to the percentage of GFP-positive cells. Each condition was tested in at least three independent experiments. For the apoptosis determination of the different apoptosis inducers (Table 1), 2.0 μg of expression plasmids (1 μg in the case of ANT-1 and globin alpha combined with 1 μg of luciferase plasmid) together with 0.8 μg of a GFP plasmid were transfected into 293T cells, which were harvested at the indicated time points. Cytostatic drugs were used with cybL-negative or reconstituted cells at the following concentrations: doxorubicin (4 μM), paclitaxel (4 μM), menadione (40 μM), etoposide (4 μM), cisplatin (400 nM), and arsenic trioxide (30 μM). The cells were harvested 18 h after addition of the drugs. HeLa and HeLa ρ0 cells were treated with the following concentrations: doxorubicin (20 μM), paclitaxel (40 μM), menadione (40 μM), etoposide (400 μM), cisplatin (500 μM), and arsenic trioxide (10 μM). For the induction with biological apoptosis inducers, a monoclonal antibody (mAb) against the Fas receptor (100 ng/ml; Kamiya Biomedical, Thousand Oaks, CA) or TNF (50 ng/ml; BIOMOL Research Laboratories, Plymouth Meeting, PA) together with interferon-γ (100 U/ml, BIOMOL Research Laboratories) were used. For the complex I and complex II activity measurements, HeLa cells were treated with arsenic trioxide (2 μM), doxorubicin (7.5 μM), paclitaxel (4 μM), etoposide (200 μM), menadione (12.5 μM), cisplatin (200 μM), TNF (2.5 ng/ml) with cycloheximide (1.0 μg/ml), or the antibody against the Fas receptor (150 ng/ml) with interferon-γ (150 U/ml). When apoptosis was measured based on the phenotypic alterations, the ratio of transfected (GFP positive) and morphologically apoptotic cells was determined in relation to all GFP-positive and therefore transfected cells (Li and Horwitz, 1997). At least 250 cells were counted in each independent experiment. Specific apoptosis was calculated as the percentage of apoptotic cells minus the percentage of apoptotic cells in control-transfected cells (Yang et al., 1997).
Table 1.
Isolated genes and their characterization
|
Specific apoptosis
|
||||
|---|---|---|---|---|
| Gene | Sublocalization | Known function | 28 h | 42 h |
| Cytochrome bL (CybL, CII-3, QPs-1, SDHC) | Mitochondria, inner membrane | e--Transport in complex II of the respiratory chain | 57.7% ± 8.6 | 59.7% ± 5.3 |
| Cytochrome bS (CybS, CII-4, QPs-3, SDHD) | Mitochondria, inner membrane | e--Transport in complex II of the respiratory chain | 18.4% ± 5.4 | 45.7% ± 0.5 |
| Adenine-nucleotide-translocator (ANT-1) | Mitochondria, inner membrane | ADP/ATP exchange in the PT-pore | 32.4% ± 3.5 | 94.0% ± 0.8 |
| Voltage-dependent anion channel-2 (VDAC-2, Porin) | Mitochondria, outer membrane | Pore formation in the PT-pore | 7.0% ± 3.8 | 55.9% ± 4.6 |
| Uncoupling protein-2 (UCP-2) | Mitochondria, inner membrane | Heat generation | 16.8% ± 4.7 | 22.2% ± 6.4 |
| Medium-chain acyl-CoA dehydrogenase | Mitochondria, matrix | β-Oxidation of fatty acids | 20.7% ± 1.1 | 37.6% ± 3.0 |
| Kynurenine 3-monooxygenase | Mitochondria, outer membrane | Hydroxylation of kynurenine | 16.1% ± 1.2 | 60.7% ± 5.0 |
| Very-long-chain acyl-CoA synthetase | Membranes of peroxisomes, endoplasmic reticulum | Metabolism of very-long-chain fatty acids | 31.4% ± 0.5 | 55.3% ± 6.8 |
| Cytochrome b-558 | Plasma membrane | H2O2 generation by NADPH oxidation | 32.9% ± 4.2 | 51.5% ± 6.0 |
| Hemoglobin alpha (Hba-α1) | Cytosol | O2 Transport | 22.1% ± 4.8 | 52.7% ± 12.9 |
| Serine palmitoyltransferase, subunit 2 | Endoplasmic reticulum | Sphingolipid synthesis | 64.3% ± 4.8 | 90.7% ± 1.4 |
Measurements of Respiratory Chain Complexes
The activity of complex II was measured by a spectrophotometric test assay (Veitch et al., 1992). For this, succinate (20 mM) was added to isolated mitochondria, which is oxidized to fumarate by complex II. The electrons from this reaction are used for the ubiquinone-dependent reduction of dichlorophenylindophenol whose extinction was detected at 610 nm at equal time intervals. Antimycin was used as an inhibitor of complex III in this reaction. Succinate dehydrogenase (SDH) activity was quantified according to an established protocol (Hatefi, 1978) that uses the reduction of 2,6-dichloroindophenol coupled to succinate oxidation with the use of phenazine methosulfate as mediator. When cytochrome c was used as an electron acceptor to measure complex II activity, a protocol was used that detects the succinate-dependent, malonate-sensitive reduction of cytochrome c (Schmidt et al., 1992). Complex I activity was assessed by a spectrophotometric assay as described previously (Veitch et al., 1992).
Detection of Reactive Oxygen Intermediates (ROIs)
The production of ROIs was determined essentially as described by Li et al. (1999) by using dihydroethidine (HE) (Molecular Probes, Eugene, OR). Superoxide anions are able to oxidize HE to ethidium, which intercalated in the DNA. Briefly, cells were harvested, washed with phosphate-buffered saline, and incubated with 3.5 μM HE for 30 min (5% CO2) and analyzed by flow cytometry (FACS-Calibur; BD Biosciences, San Jose, CA). Lucigenin, a specific probe for superoxide anions, was used as described previously (Pervaiz and Clement, 2002) after pooling three 10-cm plates transfected with the indicated plasmids. Its luminescence was detected with a luminometer (Berthold, Wildbad, Germany). Under these conditions, we have not found a different oxygen consumption that is taken as an indicator for artificial superoxide production by redox cycling of lucigenin.
Immunoblotting for the Quantification of cybL and Apoptosis Regulators
After transfecting cybL into 293T cells cellular extracts were isolated, separated in a SDS-PAGE, and blotted onto polyvinylidene membranes (Amersham Biosciences, Piscataway, NJ). The membranes were analyzed by an antiserum against cybL (a gift of Brian Ackrell, University of California, San Francisco, San Francisco, CA). The signals were scanned using the Eagleye (Stratagene, La Jolla, CA), normalized to the transfection efficiency and compared. Two parallels were performed. Apoptosis regulators in HeLa and HeLa ρ0 cells were determined by an immunoblot by using antibodies against cytochrome c (BD PharMingen, San Diego, CA), Smac (BIOMOL Research Laboratories), Apaf-1 (BIOMOL Research Laboratories), and caspases-3 and caspase-9 (both from CN Bioscience, San Diego, CA).
Metabolite Analysis
Citrate, succinate, and glutamate were measured with an enzymatic bioanalysis kit (r-biopharm, Darmstadt, Germany) according to the recommendations of the manufacturer.
RESULTS
Screen for Apoptosis-inducing Genes
First, we tested the suitability of 293T cells to serve as an indicator cell line for isolating apoptosis-inducing genes. The rational was that 293T cells could have accumulated mutations during their passage in cell culture that repress apoptosis as shown for Hsp70 (Beere et al., 2000). In addition, 293T cells harbor the viral proteins E1A, E1B, and the large T antigen (Graham et al., 1977; DuBridge et al., 1987), which can modulate apoptosis (White et al., 1992; McCarthy et al., 1994). Overload of the endoplasmic reticulum (ER) is known to induce an unspecific stress response that can result in cell death (Welihinda et al., 1999). We therefore overexpressed a number of membrane proteins such as the platelet-derived growth factor-receptor and integrins that are processed in the ER. None of them could lead to cell death in 293T cells. In addition, we transfected several constitutively active oncogenes (such as H-ras and v-src) and 17 dominant-negative mutants of known genes (such as STAT-2-dn and Ubiquitin-dn) into these cells. Our intention was to induce a cellular signal that the cells normally cannot generate. However, none of these genes could lead to the phenotypic changes of apoptosis induction. We also tested a number of different drugs that have been shown to induce cell death in certain cells. Using titration curves of 29 tested substances, we observed apoptotic cell death with only four reagents (our unpublished data). The rest elicited only an unspecific toxicity effect. Furthermore, after performing the screen, we have isolated the gene ANT-1. The almost identical gene ANT-2 (90% identity), however, was unable to induce a proapoptotic signal (Bauer et al., 1999).
From all these experiments, we concluded that a defined signal is needed for apoptosis induction in 293T cells and that an unspecific damage of these cells is not sufficient for cell death induction.
After screening 40% of a cDNA library (100,000 clones), we isolated 72 different cDNAs (∼0.07%) that could induce cell death based on two morphological criteria typical for apoptotic cells: membrane blebbing (Kerr et al., 1994) and volume loss (Bortner and Cidlowski, 1998). Several genes among them are known as apoptosis inducers, and therefore serve as internal positive controls for the screen: ZIP kinase (Kawai et al., 1998), NIP3 (Chen et al., 1997), FADD (Chinnaiyan et al., 1995), PERP (Attardi et al., 2000), and CIDE-A and CIDE-B (Inohara et al., 1998).
Several Apoptosis-inducing Gene Products from the Screen Are Localized to Mitochondria
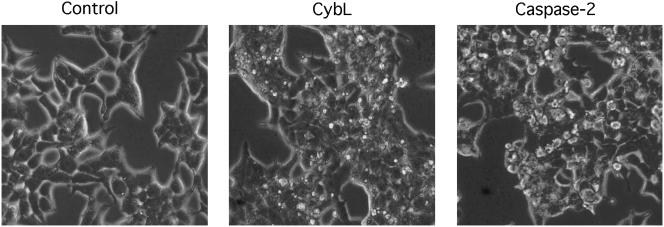
We observed that besides the canonical phenotype of apoptosis some genes generated a phenotype different from the one caused by all other proapoptotic genes, such as the cysteine protease caspase-2. These genes led to a more pronounced constriction of the cytoplasm, leading to small, rounded 293T cells. Membrane blebbing as evident in caspase-2–transfected cells, which was used as the second criterion for the isolation of proapoptotic genes, was only rarely observed (Figure 1). However, these genes produced the canonical internucleosomal cleavage of the DNA (our unpublished data). All genes were also successfully tested for their proapoptotic activity in HeLa cells. In those cases in which partial cDNAs were obtained, the complete open reading frames of the genes were isolated and apoptosis induction was verified. Sequencing revealed that most of these genes encode proteins localized in mitochondria. These organelles have recently been identified as important mediators of the apoptotic signal (Green and Reed, 1998). Table 1 lists the isolated genes with their subcellular localization, complex association and known function. Their proapoptotic effect on 293T cells is presented for two time points.
Figure 1.
Morphological differences between apoptosis induction by caspase-2 and several genes that were isolated with the screen as exemplified by cybL. Expression vectors for cybL and for caspase-2 (2 μg each) were transfected into 293T cells. Phase contrast pictures were taken after 16 h at a 320-fold magnification.
Selecting genes from the apoptosis-inducing cDNAs requires additional information of a gene to integrate it into a physiological or pathological context. We concentrated on cytochrome bL (cybL, also known as QPs-1, SDHC, and CII-3), a protein serving as a membrane anchor for other components of complex II in the respiratory chain (Scheffler, 1998). Complex II is also a component of the Krebs cycle. Rather than translocating protons, like other constituents of the respiratory chain, it serves to feed electrons from the Krebs cycle to the respiratory chain (Saraste, 1999). cybL was chosen for further investigation, because cytochrome bS (cybS), another membrane component of complex II, was also discovered by the screen. Most importantly, mutations in cybS and cybL have recently been shown to be responsible for hereditary paragangliomas (Baysal et al., 2000; Niemann and Muller, 2000). Furthermore, the chromosomal region of cybS is frequently deleted in many solid tumors (Koreth et al., 1999). Because apoptosis sensitivity and tumorigenesis are often inversely correlated, these findings indicated that complex II might be a sensor for apoptosis induction.
cybL Expression Leads to a Transient Inhibition of Complex II Activity and to the Production of Reactive Oxygen Intermediates
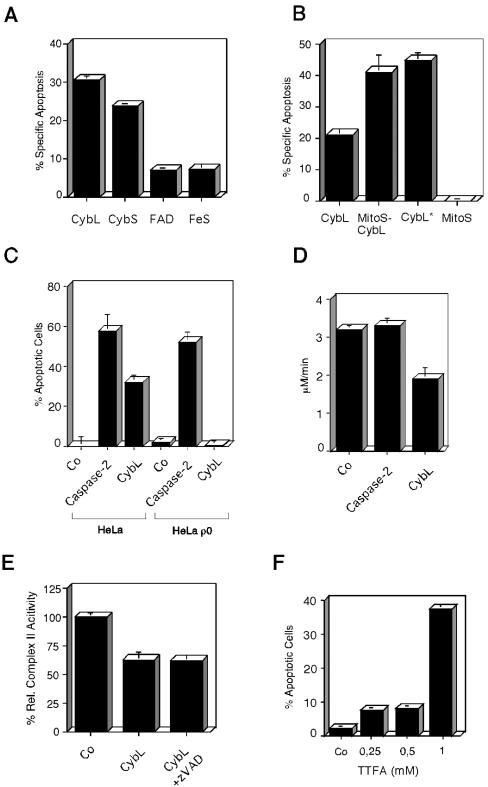
In keeping with our previous results that a specific signal is required for apoptosis induction, we wanted to establish the specificity of the signal for apoptosis induction. To this end, we transfected all four components of complex II of the respiratory chain into 293T cells and assessed cell death by FACS analysis. Figure 2A reveals that the flavin-containing protein (FAD) and the iron-sulfur protein (FeS) that comprise the catalytic center for the succinate oxidation did not lead to cell death. In contrast, the two membrane-anchoring proteins cybL and cybS were able to induce apoptosis. An immunoblot analysis showed that a 3.8-fold induction of cybL over its endogenous protein level already suffices to induce apoptosis after only 16 h (our unpublished data).
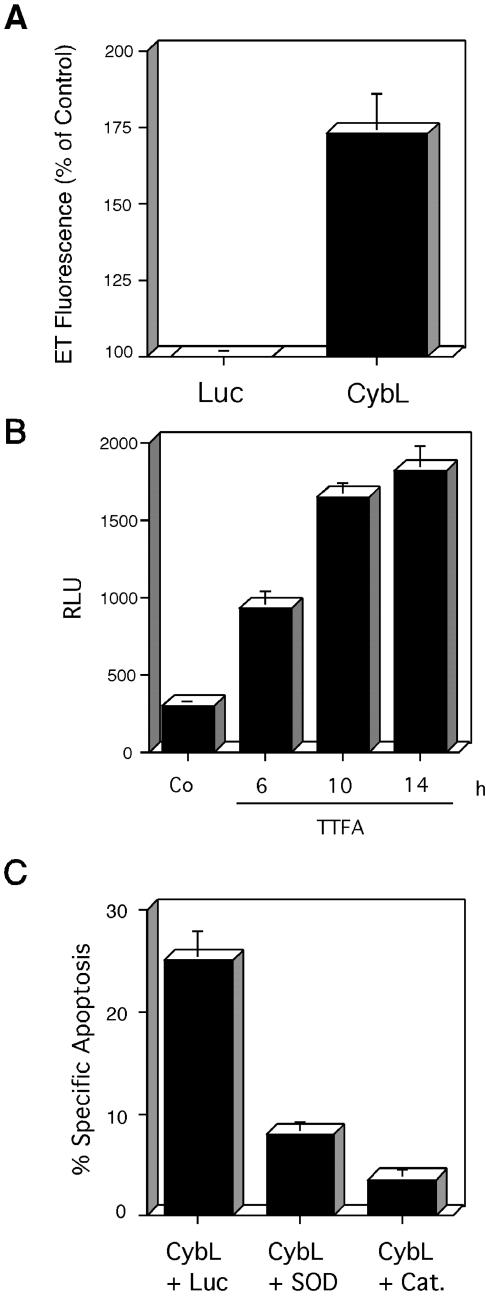
Figure 2.
Characterization of the proapoptotic stimulus exerted by cybL expression. (A) Test of all components of complex II of the respiratory chain for apoptosis induction. Expression plasmids of cybL and cybS as well as the FAD and the FeS were transfected into 293T cells. Apoptosis was quantified 26 h later by using FACS analysis. (B) Importance of mitochondria for cell death induced by cybL. The isolated cybL isoform was fused at the N terminus to the coding sequence of the mitochondrial import signal of cyclophilin D. Plasmids coding for this fusion protein (MitoS-CybL), the import sequence alone (MitoS), the isolated cybL (CybL) and a longer splice-isoform of cybL (CybL*) were transfected into 293T cells together with a GFP-expression construct. Apoptosis was measured 26 h after starting the transfection by using FACS. Shown are the means and the standard deviations of apoptosis induction of three independent experiments with each construct. (C) Role of the mitochondrial respiratory chain for apoptosis induction by cybL. Wild-type HeLa cells and HeLa ρ0 cells, which harbor mitochondria without mitochondrial DNA and therefore have a defective respiratory chain, were transfected with a control plasmid, an expression vector for cybL or caspase-2, and a GFP construct. Twenty-two hours after starting the transfection the extent of apoptosis in the GFP-positive cells was determined by counting morphologically apoptotic cells. The mean and the SD of three independent results are given for each construct. (D) Effect of cybL expression on the activity of complex II of the respiratory chain. Ten micrograms of an expression plasmid for cybL, for caspase-2, or a control vector was transfected into 10-cm dishes with 293T cells. Thirteen hours after starting the transfection, the mitochondria were isolated and complex II activity of the respiratory chain was assessed with succinate, which reduces dichlorophenylindophenol whose concentration was measured photometrically. For each transfection, a data point was taken every 10 s. Shown are the activities in total cell extracts as the means and SD of three independent experiments for each construct. (E) Effect of cybL expression on complex II activity as measured by an assay with cytochrome c as electron acceptor. Eighteen hours after the transfection of cybL into 293T cells (with and without caspase inhibitor zVAD), mitochondrial extracts were prepared and tested for complex II activity. These measurements were normalized to the transfection efficiency as determined by cotransfected GFP expression vectors. (F) Apoptosis induction by the complex II inhibitor TTFA in HeLa cells. Increasing concentrations of TTFA were added to HeLa cells and apoptosis was detected by FACS analysis.
Subsequently, we wanted to test whether cybL exerts its proapoptotic effect in mitochondria. The isolated cybL is a splice-isoform and does not contain the second described exon. It therefore lacks part of the sequence (residues 8–26) that was proposed to act as a mitochondrial import sequence (Elbehti-Green et al., 1998). Nevertheless, a hemagglutinin-tagged protein of this cybL isoform was found to colocalize to the fluorescence signal emitted by the mitochondria-specific dye Mitotracker (our unpublished data). We speculated that the import of overexpressed cybL into this organelle might be a limiting step for its proapoptotic activity. Consequently, we fused the import sequence of cyclophilin D to the N terminus of cybL. Figure 2B shows that this cyclophilin d-cybL fusion construct and the longer isoform of cybL led to an equal increase in the apoptosis-inducing activity compared with the shorter cybL isoform when 293T cells were transfected. We also saw a similar increase of cell death in HeLa cells (our unpublished data).
Consequently, we focused our subsequent experiments on mitochondria to define the relevant sensor for apoptosis. The most prominent function of mitochondria is the generation of ATP through the respiratory chain. We therefore explored its importance for apoptosis induction by using HeLa ρ0 cells. These cells have been depleted of mitochondrial DNA by prolonged incubation with ethidium bromide and thereby harbor an inactive respiratory chain. Figure 2C shows that cybL was unable to induce apoptosis in HeLa ρ0 cells, whereas caspase-2 was active in the parental cells as well as in the mutated cell line. Because CybL is a component of complex II of the respiratory chain, we speculated that it might influence the catalytic activity of this protein complex. To test this, we isolated mitochondria from control and cybL-transfected cells and used succinate as a specific substrate for complex II activity. In this assay, complex II activity was measured 13 h after transfection before any signs of apoptosis were manifest. Furthermore, caspase-2–transfected cells were used to determine whether the effect on complex II activity was specific for CybL or a common event in apoptosis induction. Figure 2D reveals that cybL transfection can repress this enzymatic activity by 41%, whereas caspase-2–transfected cells did not display a reduced complex II performance. We also used an assay for complex II activity that uses cytochrome c as a nonartificial electron acceptor. This experiment likewise revealed a reduction of complex II after transfection of cybL, which remained unchanged when zVAD, a pan-caspase-inhibitor, was added to the cells (Figure 2E).
The drug thenoyltrifluoroacetone (TTFA), an inhibitor of complex II (Suno and Nagaoka, 1984), was used to support the correlation between the transient inhibition of complex II and apoptosis induction. Figure 2F shows that TTFA, like cybL, could induce apoptosis in almost 40% of HeLa cells as assessed by DNA degradation.
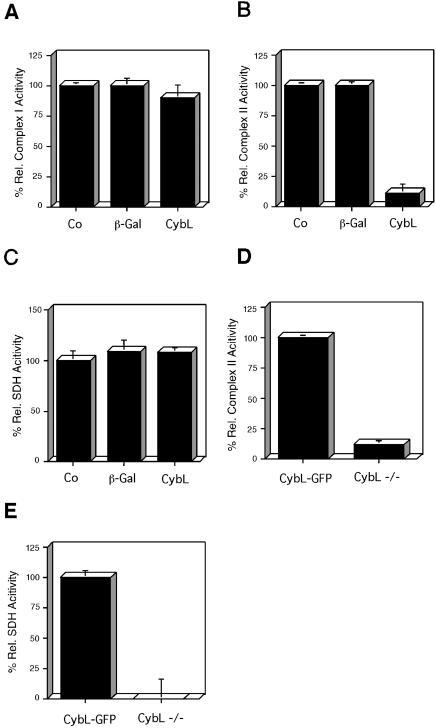
Because our starting hypothesis about the screen assumed specific proapoptotic signals, we wanted to test how specifically overexpressed cybL represses complex II of the respiratory chain. We therefore transfected cybL into cells and measured the activities of complex I and complex II. Figure 3, A and B, show that cybL expression affected only complex II, whereas complex I was not compromised in its function. The enzymatic activities of complex II include both the SDH reaction that oxidizes succinate as well as the transfer of the electrons generated thereby to ubiquinone (coenzyme Q reductase activity). We detected that cybL expression only reduced complex II activity, whereas the succinate dehydrogenase reaction was unaltered (Figure 3, B and C). It can therefore be concluded that the iron-sulfur protein and the flavin-containing protein that constitute the SDH activity remain structurally intact and that overexpressed cybL uncouples the transfer of electrons to ubiquinone. A similar situation has been described with a mutation of cybL in Caenorhabditis elegans (Ishii et al., 1998; Senoo-Matsuda et al., 2001). However, although the SDH activity is evident in the enzymatic tests that use artificial electron acceptors, in intact cells the electrons cannot be transferred to downstream components, and SDH is likewise inactive. In cells constitutively deficient in cybL (Oostveen et al., 1995), we have found that both the SDH activity and the complex II activity were reduced compared with cells in which cybL was reconstituted with a cybL-GFP fusion construct (Figure 3, D and E).
Figure 3.
Effect of cybL expression on the activity of different complexes of the respiratory chain. (A–C) The same extracts of untreated 293T cells (Co) or cells transfected with the indicated expression plasmids for β-gal or cybL were investigated for their complex I activity (A), complex II function (B), and SDH (C). Shown are the activities normalized to the transfection efficiencies and relative to the untreated control as means and SD of three independent experiments. (D and E) Complex II activity and succinate dehydrogenase in cybL-negative and -positive cells. (D) Complex II activity was assessed in cybL-negative (cybL –/–) and -reconstituted (cybL-GFP) cells. (E) Succinate dehydrogenase activity was investigated in cybL-negative and -positive cells. Shown are the activities relative to the cybL-reconstituted cells as means and SD of three independent experiments.
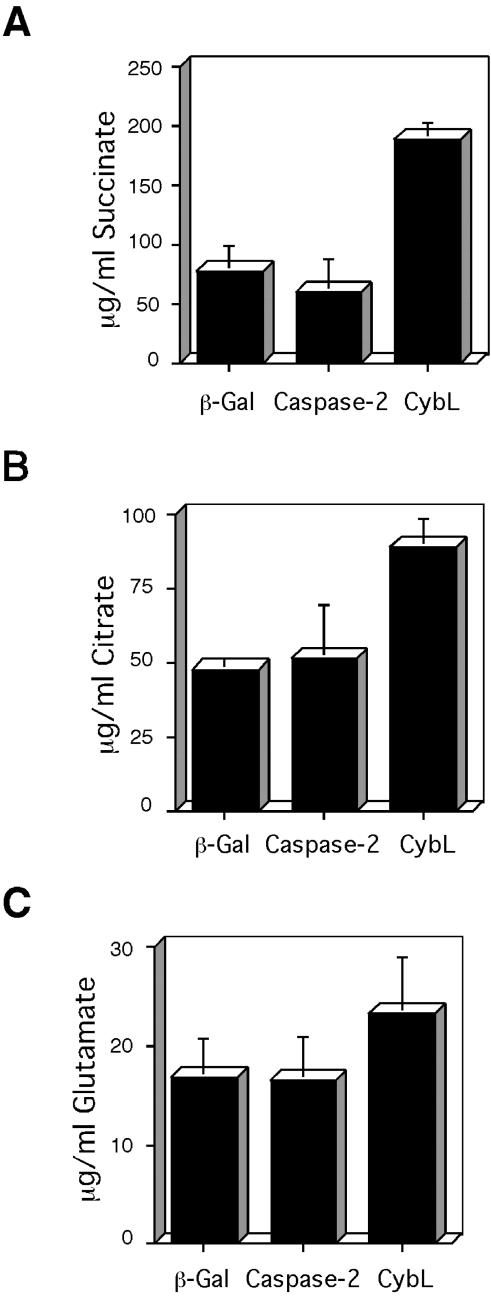
A block of complex II is expected to stop the Krebs cycle because it supplies the substrates for complex II in the form of succinate. Hence, we tested whether cybL expression has an effect on the concentrations of Krebs cycle metabolites. Figure 4 shows that two components of the Krebs cycle, succinate (Figure 4A) and citrate (Figure 4B), were found to be increased. Glutamate, an amino acid that is only indirectly derived from the Krebs cycle metabolite α-ketoglutarate, was not significantly up-regulated (Figure 4C). The concentrations of these metabolites in the supernatant of transfected cells did not change (our unpublished data).
Figure 4.
Consequences of cybL expression on metabolites. The concentrations of succinate (A), citrate (B), and glutamate (C) in the same extracts of 293T cells were determined after transfection of expression plasmids for β-gal, caspase-2, or cybL. Shown are the concentrations normalized to the transfection efficiencies as the means and SD of three independent experiments.
A transient inhibition of various complexes of the respiratory chain was implicated in the generation of ROIs. In the case of complex II, the semichinone Qs·– was suggested to be a major contributor to ROI production (McLennan and Degli Esposti, 2000). We therefore tested the generation of ROIs. Figure 5A demonstrates that before appreciable apoptosis was generated, cybL led to the generation of oxygen radicals as measured by HE. ROI detection was also successful using the superoxide anion-specific reagent lucigenin (our unpublished data). Because TTFA likewise induces apoptosis (Figure 2F), it was investigated whether it also generates ROIs. Figure 5B reveals that this reagent led to an increase of ROIs even at a point in time when no signs of apoptosis were detectable (after 6 h). To find out whether ROIs are necessary for cybL-induced cell death, we cotransfected cybL and two different ROI-scavenging enzymes, Cu/Zn superoxide dismutase and catalase. Both enzymes could lead to a reduction of apoptosis with catalase being the more efficient construct (Figure 5C).
Figure 5.
Role of reactive oxygen species in apoptosis induced by cybL. (A) Expression of cybL generates ROIs. 293T cells were transfected with a control vector (Luc) or a vector encoding CybL. ROI generation was measured after 14 h by staining with hydroethidine, which is oxidized to ethidium (ET) by superoxide radicals in cells. The results are expressed as percentage of cells with high fluorescence intensity compared with the control-transfected cells. The means and the standard deviations of three independent experiments are given. (B) Generation of ROIs by the complex II inhibitor TTFA. TTFA was applied to HeLa cells and ROI formation was detected at the indicated time points using lucigenin. (C) Catalase and superoxide dismutase can reduce apoptosis induced by cybL. An expression vector for cybL was cotransfected with a control vector encoding luciferase or with expression vectors for superoxide dismutase (SOD) or for catalase (Cat.) at a ratio of 1:1. The means and the standard deviations of three independent experiments are given as quantified by FACS analysis.
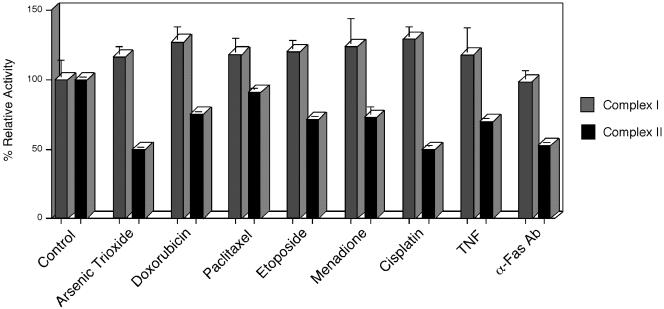
Because a transient reduction of complex II activity by TTFA or by cybL transfection correlated with apoptosis induction (Figure 2), we were interested whether apoptosis-inducing reagents would also lead to such a reduction. Consequently, we applied different apoptosis stimuli to HeLa cells and measured complex II activity before any signs of apoptosis induction were evident. Figure 6 shows that all inducers led to a reduction of complex II activity, whereas the enzymatic activity of complex I was not significantly changed.
Figure 6.
Specific reduction of complex II activity in HeLa cells after apoptosis induction. HeLa cells were treated with the indicated apoptosis stimuli. Before appreciable signs of apoptosis could be detected, the activities of complex I and complex II (ubiquinone-dependent DCIP reduction) were assessed by spectrophotometric test assays. Shown are the activities in the same cell extracts relative to the untreated control as means and SD of three independent experiments for each reagent.
Cells Deficient in cybL are Resistant to Distinct Apoptosis Inducers
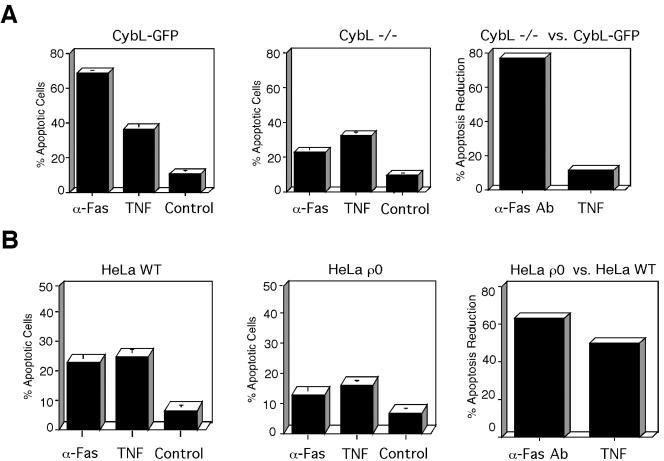
The above-mentioned data implicated complex II to be a sensor for apoptosis induction. To further evaluate the contribution of complex II for apoptosis induction we used recently described cybL-deficient cells (Oostveen et al., 1995). In contrast to the experiments described above, cybL (and therefore complex II) is constitutively inactivated in these cells. As a control, we took cells that harbor a cybL-GFP fusion protein that restores complex II activity. A number of cytostatic drugs therapeutically used for apoptosis induction were applied and apoptosis was quantified. Figure 7A shows that in cybL-deficient cells as opposed to cybL-reconstituted cells, a diverse range of cytostatic drugs such as etoposide (a topoisomerase II inhibitor), paclitaxel (an inhibitor of tubulin disassembly), or doxorubicin (a DNA-binding molecule) were significantly less active for apoptosis induction. The repression of apoptosis induction ranged from 61% with paclitaxel to ∼90% with etoposide and doxorubicin. Only arsenic trioxide (an activator of proapoptotic nuclear bodies; Zhu et al., 1997) did not show a significantly different apoptosis induction. A comparable difference in cell death was observed with doxorubicin when apoptosis was quantified by a caspase activity assay (our unpublished data). To check the specificity of the effect, we investigated the activity of the same cytostatic drugs on cells deficient in the complete respiratory chain. Even though cell-specific differences can influence the response, Figure 7B shows that, compared with cybL-deficient and -reconstituted cells, HeLa ρ0 cells are much less resistant to apoptosis induction than WT HeLa cells (Figure 7A). With the exception of menadione, whose apoptosis induction was completely reduced, none of the activities was repressed by >42%. Cell death by etoposide was even more pronounced in HeLa ρ0 cells than in WT cells. Because cytostatic drugs often use signal transduction pathways that are also used by physiological apoptosis inducers, we wanted to test the activation of two cellular membrane receptors that mediate apoptosis. To this end, we used an antibody against the Fas receptor (Nagata and Golstein, 1995) and the ligand for the TNF receptor (Wallach et al., 1997). Figure 8A shows that in complex II-deficient cells the activation of the Fas receptor was repressed by 77% compared with cells in which complex II is reconstituted. TNF, on the other hand, was equally active for apoptosis induction in these two cell lines (33 vs. 37%). HeLa and HeLa ρ0 cells were again investigated as a control. In contrast to complex II-deficient cells, we saw an equal reduction (50 and 63%) in apoptosis induction compared with the WT cells with both TNF and anti-Fas antibody (Figure 8B).
Figure 7.
cybL-deficient cells are specifically resistant to cytostatic drug-induced apoptosis. (A) Comparison of apoptosis in cells lacking cybL or reconstituted with cybL. CybL-deficient (CybL –/–) as well as cells that express a cybL construct (CybL-GFP) were treated with the indicated cytostatic drugs for 18 h after which the extent of apoptosis was quantified by FACS analysis. The means and the SD of three independent measurements are given. The right hand panel shows the comparison of the percentage of apoptosis reduction between complex II-deficient versus -reconstituted cells. The means of the apoptosis inductions for each inducer (minus control) were put in relation and its percentage was subtracted from 100%. (B) Comparison of the apoptosis potential of cells with and without an intact respiratory chain. HeLa and HeLa ρ0 cells were incubated with the same drugs used in (A) and apoptosis was quantified by FACS. The right panel states the percentage of apoptosis reduction between HeLa ρ0 versus WT HeLa cells that was calculated as in A.
Figure 8.
Quantification of apoptosis induction by activation of the TNF- and the Fas-receptor. (A) Apoptosis in cells with (CybL-GFP) and without (CybL –/–) an intact complex II after stimulation with TNF or an anti-Fas receptor mAb. The induced cell death was quantified by FACS analysis 14 h after application of the stimuli. In the right panel, the percentage of apoptosis reduction between complex II-deficient versus -reconstituted cells is shown. The means of the apoptosis inductions for each inducer were compared, and the percentage of repression was calculated as in Figure 7. (B) Comparison of apoptosis induction by TNF and anti-Fas mAb in WT HeLa and HeLa ρ0 cells. Apoptosis was induced and quantified as in A. The percentage of apoptosis repression of HeLa ρ0 versus WT HeLa cell was calculated as in A.
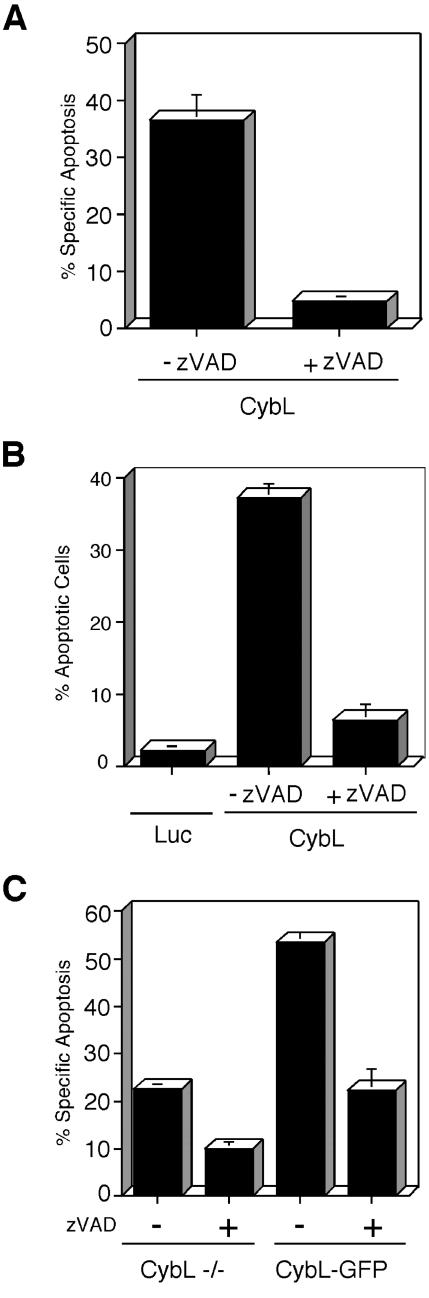
Recent reports have supported the notion that not all apoptotic cell death is mediated by caspases (McCarthy et al., 1997; Foghsgaard et al., 2001). We therefore tested whether these cysteine proteases are downstream components of the proapoptotic signal induced by cybL. Figure 9A shows that the proapoptotic effect of transfected cybL could be suppressed by zVAD, a pan-specific inhibitor of caspases. This was also apparent when apoptosis was quantified by morphological criteria (Figure 9B). In addition, zVAD also reduced doxorubicin-stimulated apoptosis in both cybL-positive and -negative cells (Figure 9C).
Figure 9.
Involvement of caspases in CybL-mediated cell death. (A) The caspase inhibitor zVAD reduces apoptosis after cybL transfection. CybL was transfected into 293T cells that were treated with or without 70 μM zVAD. The induced cell death was quantified by FACS analysis 26 h after transfection by FACS analysis. Shown are the means and the SD of three independent experiments. (B) zVAD reduces the morphological changes of apoptosis induced by cybL. HeLa cells were transfected with cybL together with an expression plasmid for GFP and treated with zVAD or DMSO. Apoptosis was quantified after 24 h by counting transfected (GFP-positive) and morphologically apoptotic cells. (C) zVAD can attenuate cell death both in cybL-positive and -negative cells. Doxorubicin was used to induce apoptosis in cells expressing or lacking cybL. zVAD (70 μM) was added to repress caspase activity and cell death was quantified after 16 h.
DISCUSSION
Genes isolated with this screen can be used to define apoptosis sensors in the cell. Based on our data, we propose complex II of the mitochondrial respiratory chain to be such a sensor. Up to now the only indication for this came from the recent discovery that it harbors the tumor suppressor proteins cybL (Niemann and Muller, 2000) and cybS (Baysal et al., 2000). This finding was very unexpected and extended the role of mitochondria for pathological conditions in an important way. Because some tumor suppressor proteins act by regulating apoptosis, this study could explain their role as tumor suppressors. Our experiments show that cybL has both the dominant activity to induce apoptosis upon overexpression (Figure 2) and can reduce cell death when inactivated (Figures 7 and 8).
Unexpectedly, many different proapoptotic drugs seem to use complex II for apoptosis induction. However, we have also found distinct cytostatic drugs such as arsenic trioxide that retain their activity in complex II-deficient cells (Figure 7). In HeLa cells, on the other hand, arsenic trioxide led to a reduction of complex II activity. This could indicate that apoptosis induction via a reduction of complex II activity is cell-specific. Our findings could therefore become relevant for the therapeutical treatment of tumors in which complex II is inactivated. Because therapeutic drugs such as etoposide use endogenous apoptosis mediators (Bennett et al., 1998; Lowe et al., 1993), we also tested TNF and Fas, two biological cell death inducers. The equal apoptosis induction that we observed after TNF treatment in complex II-deficient and -reconstituted cells (Figure 8A) argues that TNF can still recruit complex I for apoptosis induction in these cells (Higuchi et al., 1998). In contrast, the potent reduction of the Fas-mediated apoptosis response could contribute to tumorigenesis in complex II deficiency. In line with this, the Fas receptor or its ligand are mutated or repressed in many different tumors and are therefore supposed to act as tumor suppressors (Muschen et al., 2000).
It is of note that cybL and cybS are binding a heme that seems not to be involved in the respiratory chain. Because it is exhibiting a redox potential of around –200 mV (Crouse et al., 1995), it could serve to sense the redox potential in healthy cells that lies around the same value and is disturbed in many pathways leading to apoptosis. This could provide an answer as to why so many different apoptosis inducers activate complex II for cell death.
Cell death is correlated with a transient inhibition of complex II by various apoptosis inducers (Figure 6), TTFA, or by cybL (Figure 2). For that, cybL overexpression could act through inhibiting complex assembly or by titrating out other components of this complex. This would lead to a block of the electron flow, the generation of oxygen radicals and apoptosis induction. This is in line with a recent report that proposed complex II of the respiratory chain to be a major generator of ROIs, possibly by the semichinone Qs·– (McLennan and Degli Esposti, 2000). Likewise, complex I and III of the respiratory chain are inhibited for apoptosis induction by TNF (Higuchi et al., 1998) and ceramide (Gudz et al., 1997), respectively. However, if complex II is constitutively nonfunctional (as in cybL-deficient cells), it cannot be transiently inhibited and used for apoptosis induction. Hence, the insensitivity to apoptosis induction observed in complex II-negative cells (Figures 7 and 8). However, we cannot exclude that additional downstream consequences of complex II inactivation contribute to the insensitivity for apoptosis induction.
If some proapoptotic stimuli use other complexes of the respiratory chain, what could be the advantage of using complex II for cell death induction and what could explain the presence of mutations in cybS and cybL in tumors (Rustin and Rotig, 2002)? The reason might be that complex II, which is the only respiratory complex fully encoded by nuclear genes, is not an obligatory part of the respiratory chain for electrons from complex I. They are transferred to complex III, bypassing complex II. Therefore, its transient inhibition still allows the functioning of the remaining respiratory chain and the generation of ATP (Wiegand et al., 1999). In contrast, the inhibition of all other complexes of the respiratory chain would result in a complete block of the respiratory chain and a reduction of the ATP level. However, a sustained concentration of ATP is crucial for the cell to induce the active cell suicide program of apoptosis (Leist et al., 1997). On the other hand, in tumor cells with an inactive complex II the remaining respiratory chain could suffice to produce enough ATP for the proliferation of these cells. Apoptosis induction in HeLa ρ0 cells in which the complete respiratory chain is inactivated supports this view. These cells are more resistant to apoptosis induction than their WT HeLa cells (Gamen et al., 1995; Higuchi et al., 1997) (Figures 6B and 7B). But, with the exception of TNF, they are more sensitive for apoptosis induction than their complex II-deficient counterparts in relation to their WT cells.
It is of particular note that cybL, the gene that we found in this work to be proapoptotic, is polycistronic with ced-9, the homolog of mammalian Bcl-2 and a major death regulator in C. elegans (Hengartner and Horvitz, 1994). Our work could therefore help to understand the connection between these two genes.
Interestingly, all proteins of the genes from the screen are components of enzymatic complexes or transport proteins, which reflects both the specificity of the proapoptotic signal as well as the active processes required for apoptosis induction. A substantial number of proteins encoded by the proapoptotic genes are localized to mitochondria (Table 1). This organelle has been implicated in the generation of proapoptotic signals (Green and Reed, 1998). We therefore assume that the distinct apoptotic phenotype induced in 293T cells (Figure 1) is generated when mitochondria are directly activated for apoptosis.
As in the case of cybL, it will be important to determine the exact nature of the apoptosis sensors defined by the other genes and their role in physiological or pathological apoptosis induction. For example, two components (VDAC and ANT-1) of the permeability transition (PT-) pore have been isolated. This protein complex spans the inner and the outer mitochondrial membrane (Zoratti and Szabo, 1995) and is activated by many proapoptotic stimuli (Zamzami et al., 1996). ANT-1 was recently described by us to activate the PT-pore, apparently by rendering inactive endogenous inhibitors (Bauer et al., 1999). Similar to the PT-pore (Deigner et al., 2000) or complex II, many other apoptosis sensors can be expected to be involved in pathological conditions: The first anabolic enzyme of ceramide synthesis, serine palmitoyltransferase, was isolated by the screen (Table 1). Dominant alleles of this enzyme have recently been shown to be responsible for hereditary sensory neuropathy type I, a degenerative disease of peripheral sensory neurons (Bejaoui et al., 2001; Dawkins et al., 2001). In addition, the enzyme kynurenine 3-monoxygenase leads to the generation of 3-hydroxykynurenine, a metabolite that is elevated in Huntington's disease (Pearson and Reynolds, 1992), Parkinson's disease (Ogawa et al., 1992), hepatic encephalopathy (Pearson and Reynolds, 1991) and human immunodeficiency virus infection (Sardar et al., 1995), and it can contribute to the cell death observed in these diseases (Okuda et al., 1996). Two genes identified by the screen (VDAC and UCP-2) have recently been shown to be up-regulated in irradiation-induced apoptosis of B cell lymphomas by using gene microarrays (Voehringer et al., 2000). Unexpectedly, we isolated hemoglobin alpha, a gene previously implicated primarily in the transport of oxygen (Leder et al., 1982). However, an excess of α-hemoglobin is used as a model for β-thalassemia (Scott et al., 1993), which can lead to cell death of bone marrow cells (Schrier, 1997).
It is to be noted that several of the isolated genes are, directly or indirectly, involved in lipid metabolism. Lipids, especially ceramides, have been known for some time to play an important role in apoptosis (Perry, 1999). This is underscored by our isolation of serine palmitoyltransferase. Interestingly, we also detected two enzymes of fatty acid metabolism, indicating that apoptosis by a dysregulation of fatty acids as observed in obesity (Shimabukuro et al., 1998) or in human myopathies (Listenberger and Schaffer, 2002) might be controlled by such genes. In addition, acyl-CoA dehydrogenase feeds electrons to complex II of the respiratory chain, possibly linking lipid metabolism with the proapoptotic activity of complex II as described in this work. Consequently, the screen opens up many avenues for investigations, and we expect this assay to allow the definition of additional sensors for apoptosis induction in the future.
Acknowledgments
The superb technical assistance of F. Neudecker-Voβ is highly acknowledged. This work was supported by the Bavarian Government, Roche Diagnostics (Mannheim, Germany) and Xantos Biomedicine (Munich, Germany). T.M. is supported by the Sonderforschungsbereich 190.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-10-0631. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-10-0631.
References
- Attardi, L.D., Reczek, E.E., Cosmas, C., Demicco, E.G., McCurrach, M.E., Lowe, S.W., and Jacks, T. (2000). PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes 14, 704–718. [PMC free article] [PubMed] [Google Scholar]
- Bauer, M.K.A., Schubert, A., Rocks, O., and Grimm, S. (1999). Adenine nucleotide translocase-1, a component of the permeability transition pore, can dominantly induce apoptosis. J. Cell Biol. 147, 1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysal, B.E., et al. (2000). Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science 287, 848–851. [DOI] [PubMed] [Google Scholar]
- Beere, H.M., Wolf, B.B., Cain, K., Mosser, D.D., Mahboubi, A., Kuwana, T., Tailor, P., Morimoto, R.I., Cohen, G.M., and Green, D.R. (2000). Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat. Cell Biol. 2, 469–475. [DOI] [PubMed] [Google Scholar]
- Bejaoui, K., Wu, C., Scheffler, M.D., Haan, G., Ashby, P., Wu, L., de Jong, P., and Brown, R.H. (2001). SPTLC1 is mutated in hereditary sensory neuropathy, type 1. Nat. Genet. 27, 261–262. [DOI] [PubMed] [Google Scholar]
- Bennett, M., Macdonald, K., Chan, S.W., Luzio, J.P., Simari, R., and Weissberg, P. (1998). Cell surface trafficking of Fas: a rapid mechanism of p53-mediated apoptosis. Science 282, 290–293. [DOI] [PubMed] [Google Scholar]
- Bortner, C.D., and Cidlowski, J.A. (1998). A necessary role for cell shrinkage in apoptosis. Biochem. Pharmacol. 56, 1549–1559. [DOI] [PubMed] [Google Scholar]
- Chen, G., Ray, R., Dubik, D., Shi, L., Cizeau, J., Bleackley, R.C., Saxena, S., Gietz, R.D., and Greenberg, A.H. (1997). The E1B 19K/Bcl-2-binding protein Nip3 is a dimeric mitochondrial protein that activates apoptosis. J. Exp. Med. 186, 1975–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnaiyan, A.M., O'Rourke, K., Tewari, M., and Dixit, V.M. (1995). FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell 81, 505–512. [DOI] [PubMed] [Google Scholar]
- Crouse, B.R., Yu, C.A., Yu, L., and Johnson, M.K. (1995). Spectroscopic identification of the axial ligands of cytochrome b560 in bovine heart succinate-ubiquinone reductase. FEBS Lett. 367, 1–4. [DOI] [PubMed] [Google Scholar]
- Dawkins, J.L., Hulme, D.J., Brahmbhatt, S.B., Auer-Grumbach, M., and Nicholson, G.A. (2001). Mutations in SPTLC1, encoding serine palmitoyltransferase, long chain base subunit-1, cause hereditary sensory neuropathy type I. Nat. Genet. 27, 309–312. [DOI] [PubMed] [Google Scholar]
- Deigner, H.P., Haberkorn, U., and Kinscherf, R. (2000). Apoptosis modulators in the therapy of neurodegenerative diseases. Expert Opin. Investig. Drugs 9, 747–764. [DOI] [PubMed] [Google Scholar]
- DuBridge, R.B., Tang, P., Hsia, H.C., Leong, P.M., Miller, J.H., and Calos, M.P. (1987). Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7, 379–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbehti-Green, A., Au, H.C., Mascarello, J.T., Ream-Robinson, D., and Scheffler, I.E. (1998). Characterization of the human SDHC gene encoding of the integral membrane proteins of succinate-quinone oxidoreductase in mitochondria. Gene 213, 133–140. [DOI] [PubMed] [Google Scholar]
- Foghsgaard, L., Wissing, D., Mauch, D., Lademann, U., Bastholm, L., Boes, M., Elling, F., Leist, M., and Jaattela, M. (2001). Cathepsin B acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J. Cell Biol. 153, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamen, S., Anel, A., Montoya, J., Marzo, I., Pineiro, A., and Naval, J. (1995). mtDNA-depleted U937 cells are sensitive to TNF and Fas-mediated cytotoxicity. FEBS Lett. 376, 15–18. [DOI] [PubMed] [Google Scholar]
- Graham, F.L., Smiley, J., Russell, W.C., and Nairn, R. (1977). Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36, 59–74. [DOI] [PubMed] [Google Scholar]
- Green, D.R., and Reed, J.C. (1998). Mitochondria and apoptosis. Science 281, 1309–1312. [DOI] [PubMed] [Google Scholar]
- Grimm, S., and Leder, P. (1997). An apoptosis-inducing isoform of neu differentiation factor (NDF) identified using a novel screen for dominant, apoptosis-inducing genes. J. Exp. Med. 185, 1137–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudz, T.I., Tserng, K.Y., and Hoppel, C.L. (1997). Direct inhibition of mitochondrial respiratory chain complex III by cell-permeable ceramide. J. Biol. Chem. 272, 24154–24158. [DOI] [PubMed] [Google Scholar]
- Hatefi, Y. (1978). Resolution of complex II and isolation of succinate dehydrogenase (EC 1.3.99.1). Methods Enzymol. 53, 27–35. [DOI] [PubMed] [Google Scholar]
- Hengartner, M.O., and Horvitz, H.R. (1994). C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian protooncogene bcl-2. Cell 76, 665–676. [DOI] [PubMed] [Google Scholar]
- Higuchi, M., Aggarwal, B.B., and Yeh, E.T. (1997). Activation of CPP32-like protease in tumor necrosis factor-induced apoptosis is dependent on mitochondrial function. J. Clin. Investig. 99, 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, M., Proske, R.J., and Yeh, E.T. (1998). Inhibition of mitochondrial respiratory chain complex I by TNF results in cytochrome c release, membrane permeability transition, and apoptosis. Oncogene 17, 2515–2524. [DOI] [PubMed] [Google Scholar]
- Hsu, H., Xiong, J., and Goeddel, D.V. (1995). The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell 81, 495–504. [DOI] [PubMed] [Google Scholar]
- Inohara, N., Koseki, T., Chen, S., Wu, X., and Nunez, G. (1998). CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J. 17, 2526–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, N., Fujii, M., Hartman, P.S., Tsuda, M., Yasuda, K., Senoo-Matsuda, N., Yanase, S., Ayusawa, D., and Suzuki, K. (1998). A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature 394, 694–697. [DOI] [PubMed] [Google Scholar]
- Kawai, T., Matsumoto, M., Takeda, K., Sanjo, H., and Akira, S. (1998). ZIP kinase, a novel serine/threonine kinase which mediates apoptosis. Mol. Cell. Biol. 18, 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr, J.F., Winterford, C.M., and Harmon, B.V. (1994). Apoptosis. Its significance in cancer and cancer therapy. Cancer 73, 2013–2026. [DOI] [PubMed] [Google Scholar]
- King, M.P., and Attardi, G. (1989). Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science 246, 500–503. [DOI] [PubMed] [Google Scholar]
- Koreth, J., Bakkenist, C.J., and McGee, J.O. (1999). Chromosomes, 11Q and cancer: a review. J. Pathol. 187, 28–38. [DOI] [PubMed] [Google Scholar]
- Leder, P., Konkel, D., Leder, A., and Nishioka, Y. (1982). Globin genes: a paradigm of gene structure, function, and evolution. Natl. Cancer Inst. Monogr. 60, 49–54. [PubMed] [Google Scholar]
- Leist, M., Single, B., Castoldi, A.F., Kuhnle, S., and Nicotera, P. (1997). Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. J. Exp. Med. 185, 1481–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P.F., Dietz, R., and von Harsdorf, R. (1999). p53 regulates mitochondrial membrane potential through reactive oxygen species and induces cytochrome c-independent apoptosis blocked by Bcl-2. EMBO J. 18, 6027–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., and Horwitz, M.S. (1997). Use of green fluorescent protein in studies of apoptosis of transfected cells. Biotechniques 23, 1026–1029. [DOI] [PubMed] [Google Scholar]
- Listenberger, L.L., and Schaffer, J.E. (2002). Mechanisms of lipoapoptosis: implications for human heart disease. Trends Cardiovasc. Med. 12, 134–138. [DOI] [PubMed] [Google Scholar]
- Lowe, S.W., Ruley, H.E., Jacks, T., and Housman, D.E. (1993). p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 74, 957–967. [DOI] [PubMed] [Google Scholar]
- McCarthy, J.V., and Dixit, V.M. (1998). Apoptosis induced by Drosophila reaper and grim in a human system. Attenuation by inhibitor of apoptosis proteins (cIAPs). J. Biol. Chem. 273, 24009–24015. [DOI] [PubMed] [Google Scholar]
- McCarthy, N.J., Whyte, M.K., Gilbert, C.S., and Evan, G.I. (1997). Inhibition of Ced-3/ICE-related proteases does not prevent cell death induced by oncogenes, DNA damage, or the Bcl-2 homologue Bak. J. Cell Biol. 136, 215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, S.A., Symonds, H.S., and Van Dyke, T. (1994). Regulation of apoptosis in transgenic mice by simian virus 40 T antigen-mediated inactivation of p53. Proc. Natl. Acad. Sci. USA 91, 3979–3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan, H.R., and Degli Esposti, M. (2000). The contribution of mitochondrial respiratory complexes to the production of reactive oxygen species. J. Bioenerg. Biomembr. 32, 153–162. [DOI] [PubMed] [Google Scholar]
- Muschen, M., Warskulat, U., and Beckmann, M.W. (2000). Defining CD95 as a tumor suppressor gene. J. Mol. Med. 78, 312–325. [DOI] [PubMed] [Google Scholar]
- Nagata, S., and Golstein, P. (1995). The Fas death factor. Science 267, 1449–1456. [DOI] [PubMed] [Google Scholar]
- Neudecker, F., and Grimm, S. (2000). High-throughput method for isolating plasmid DNA with reduced lipopolysaccharide content. Biotechniques 28, 107–109. [PubMed] [Google Scholar]
- Niemann, S., and Muller, U. (2000). Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nat. Genet. 26, 268–270. [DOI] [PubMed] [Google Scholar]
- Ogawa, T., Matson, W.R., Beal, M.F., Myers, R.H., Bird, E.D., Milbury, P., and Saso, S. (1992). Kynurenine pathway abnormalities in Parkinson's disease. Neurology 42, 1702–1706. [DOI] [PubMed] [Google Scholar]
- Okuda, S., Nishiyama, N., Saito, H., and Katsuki, H. (1996). Hydrogen peroxide-mediated neuronal cell death induced by an endogenous neurotoxin, 3-hydroxykynurenine. Proc. Natl. Acad. Sci. USA 93, 12553–12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostveen, F.G., Au, H.C., Meijer, P.J., and Scheffler, I.E. (1995). A Chinese hamster mutant cell line with a defect in the integral membrane protein CII-3 of complex II of the mitochondrial electron transport chain. J. Biol. Chem. 270, 26104–26108. [DOI] [PubMed] [Google Scholar]
- Pearson, S.J., and Reynolds, G.P. (1991). Determination of 3-hydroxykynurenine in human brain and plasma by high-performance liquid chromatography with electrochemical detection. Increased concentrations in hepatic encephalopathy. J. Chromatogr. 565, 436–440. [DOI] [PubMed] [Google Scholar]
- Pearson, S.J., and Reynolds, G.P. (1992). Increased brain concentrations of a neurotoxin, 3-hydroxykynurenine, in Huntington's disease. Neurosci. Lett. 144, 199–201. [DOI] [PubMed] [Google Scholar]
- Perry, D.K. (1999). Ceramide and apoptosis. Biochem. Soc. Trans. 27, 399–404. [DOI] [PubMed] [Google Scholar]
- Pervaiz, S., and Clement, M.V. (2002). Hydrogen peroxide-induced apoptosis: oxidative or reductive stress? Methods Enzymol. 352, 150–159. [DOI] [PubMed] [Google Scholar]
- Roussel, M.F., Rettenmier, C.W., Look, A.T., and Sherr, C.J. (1984). Cell surface expression of v-fms-coded glycoproteins is required for transformation. Mol. Cell. Biol. 4, 1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustin, P., and Rotig, A. (2002). Inborn errors of complex II–unusual human mitochondrial diseases. Biochim. Biophys. Acta 1553, 117–122. [DOI] [PubMed] [Google Scholar]
- Salvesen, G.S., and Dixit, V.M. (1997). Caspases: intracellular signaling by proteolysis. Cell 91, 443–446. [DOI] [PubMed] [Google Scholar]
- Saraste, M. (1999). Oxidative phosphorylation at the fin de siecle. Science 283, 1488–1493. [DOI] [PubMed] [Google Scholar]
- Sardar, A.M., Bell, J.E., and Reynolds, G.P. (1995). Increased concentrations of the neurotoxin 3-hydroxykynurenine in the frontal cortex of HIV-1-positive patients. J. Neurochem. 64, 932–935. [DOI] [PubMed] [Google Scholar]
- Scheffler, I.E. (1998). Molecular genetics of succinate:quinone oxidoreductase in eukaryotes. Prog. Nucleic Acid Res. Mol. Biol. 60, 267–315. [DOI] [PubMed] [Google Scholar]
- Schmidt, D.M., Saghbini, M., and Scheffler, I.E. (1992). The C-terminus of the succinate dehydrogenase IP peptide of Saccharomyces cerevisiae is significant for assembly of complex II. Biochemistry 31, 8442–8448. [DOI] [PubMed] [Google Scholar]
- Schrier, S.L. (1997). Pathobiology of thalassemic erythrocytes. Curr. Opin. Hematol. 4, 75–78. [DOI] [PubMed] [Google Scholar]
- Scott, M.D., van den Berg, J.J., Repka, T., Rouyer-Fessard, P., Hebbel, R.P., Beuzard, Y., and Lubin, B.H. (1993). Effect of excess alpha-hemoglobin chains on cellular and membrane oxidation in model beta-thalassemic erythrocytes. J. Clin. Investig. 91, 1706–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoo-Matsuda, N., Yasuda, K., Tsuda, M., Ohkubo, T., Yoshimura, S., Nakazawa, H., Hartman, P.S., and Ishii, N. (2001). A defect in the cytochrome b large subunit in complex II causes both superoxide anion overproduction and abnormal energy metabolism in Caenorhabditis elegans. J. Biol. Chem. 276, 41553–41558. [DOI] [PubMed] [Google Scholar]
- Shimabukuro, M., Zhou, Y.T., Levi, M., and Unger, R.H. (1998). Fatty acid-induced beta cell apoptosis: a link between obesity and diabetes. Proc. Natl. Acad. Sci. USA 95, 2498–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suno, M., and Nagaoka, A. (1984). Inhibition of lipid peroxidation by a novel compound (CV-2619) in brain mitochondria and mode of action of the inhibition. Biochem. Biophys. Res. Commun. 125, 1046–1052. [DOI] [PubMed] [Google Scholar]
- Veitch, K., Hombroeckx, A., Caucheteux, D., Pouleur, H., and Hue, L. (1992). Global ischaemia induces a biphasic response of the mitochondrial respiratory chain. Anoxic pre-perfusion protects against ischaemic damage. Biochem. J. 281, 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voehringer, D.W., Hirschberg, D.L., Xiao, J., Roederer, M., Lock, C.B., Herzenberg, L.A., Steinman, L., and Herzenberg, L.A. (2000). Gene microarray identification of redox and mitochondrial elements that control resistance or sensitivity to apoptosis. Proc. Natl. Acad. Sci. USA 97, 2680–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallach, D., Boldin, M., Varfolomeev, E., Beyaert, R., Vandenabeele, P., and Fiers, W. (1997). Cell death induction by receptors of the TNF family: towards a molecular understanding. FEBS Lett. 410, 96–106. [DOI] [PubMed] [Google Scholar]
- Welihinda, A.A., Tirasophon, W., and Kaufman, R.J. (1999). The cellular response to protein misfolding in the endoplasmic reticulum. Gene Expr. 7, 293–300. [PMC free article] [PubMed] [Google Scholar]
- White, E. (1996). Life, death, and the pursuit of apoptosis. Genes Dev. 10, 1–15. [DOI] [PubMed] [Google Scholar]
- White, E., Sabbatini, P., Debbas, M., Wold, W.S., Kusher, D.I., and Gooding, L.R. (1992). The 19-kilodalton adenovirus E1B transforming protein inhibits programmed cell death and prevents cytolysis by tumor necrosis factor alpha. Mol. Cell. Biol. 12, 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand, F., et al. (1999). Respiratory chain inhibition induces tolerance to focal cerebral ischemia. J. Cereb. Blood Flow Metab. 19, 1229–1237. [DOI] [PubMed] [Google Scholar]
- Yang, X., Chang, H.Y., and Baltimore, D. (1998a). Autoproteolytic activation of pro-caspases by oligomerization. Mol. Cell 1, 319–325. [DOI] [PubMed] [Google Scholar]
- Yang, X., Chang, H.Y., and Baltimore, D. (1998b). Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science 281, 1355–1357. [DOI] [PubMed] [Google Scholar]
- Yang, X., Khosravi-Far, R., Chang, H.Y., and Baltimore, D. (1997). Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 89, 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamzami, N., Susin, S.A., Marchetti, P., Hirsch, T., Gomez-Monterrey, I., Castedo, M., and Kroemer, G. (1996). Mitochondrial control of nuclear apoptosis. J. Exp. Med. 183, 1533–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Koken, M.H., Quignon, F., Chelbi-Alix, M.K., Degos, L., Wang, Z.Y., Chen, Z., and de The, H. (1997). Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA 94, 3978–3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoratti, M., and Szabo, I. (1995). The mitochondrial permeability transition. Biochim. Biophys. Acta 1241, 139–176. [DOI] [PubMed] [Google Scholar]