Abstract
Telomeres are specialized functional complexes that ensure chromosome stability by protecting chromosome ends from fusions and degradation and avoiding chromosomal termini from being sensed as DNA breaks. Budding yeast Tel1 is required both for telomere metabolism and for a Rad53-dependent checkpoint responding to unprocessed double-strand breaks. We show that overexpression of a GAL1-TEL1 fusion causes transient telomere lengthening and activation of a Rad53-dependent G2/M checkpoint in cells whose telomeres are short due to the lack of either Tel1 or Yku70. Sudden telomere elongation and checkpoint-mediated cell cycle arrest are also triggered in wild-type cells by overproducing a protein fusion between the telomeric binding protein Cdc13 and the telomerase-associated protein Est1. Checkpoint activation by GAL1-TEL1 requires ongoing telomere elongation. In fact, it is turned off concomitantly with telomeres reaching a new stable length and is partially suppressed by deletion of the telomerase EST2 gene. Moreover, both telomere length rebalancing and checkpoint inactivation under galactose-induced conditions are accelerated by high levels of either the Sae2 protein, involved in double-strand breaks processing, or the negative telomere length regulator Rif2. These data suggest that sudden telomere lengthening elicits a checkpoint response that inhibits the G2/M transition.
INTRODUCTION
The ends of eukaryotic chromosomes are protected by specialized functional complexes called telomeres from being recognized and processed as double-strand breaks (DSBs) (McClintock, 1941, 1942). The critical distinction between telomeres and DSBs is that the natural ends of linear chromosomes do not undergo end-to-end fusions and other processing events normally required to promote repair of DNA breaks. In addition, the presence of DSBs triggers the activation of surveillance mechanisms, known as DNA damage checkpoints, which ensure that damaged DNA molecules are neither replicated nor segregated to daughter cells until repaired (reviewed in Longhese et al., 1998; Zhou and Elledge, 2000; Kolodner et al., 2002; Nyberg et al., 2002; Rouse and Jackson, 2002).
In most organisms, such as yeast and vertebrates, the ends of linear chromosomes contain telomeric DNA repeats of 5–25 base pairs that vary in sequence among species and terminate in a 3′ single-stranded overhang (Wellinger and Sen, 1997), which is likely important for both recruitment of the telomerase enzyme and for efficient telomeric sequence addition. Telomeric DNA is usually maintained by the action of telomerase, a specialized reverse transcriptase that uses its RNA component as a template to elongate the TG-rich strand of telomeric DNA (reviewed in Greider, 1996; Zakian, 1996). Telomerase-mediated lengthening of chromosomal ends in budding yeast involves a number of factors. In fact, the action of the Est2 catalytic reverse transcriptase protein subunit and of the TLC1 telomerase RNA is modulated by both the TLC1-binding proteins Est1 and Est3 and the single-stranded TG1–3 DNA binding protein Cdc13 (reviewed in Evans et al., 1999). Moreover, once localized at telomeres, telomerase activity has to be regulated to avoid unlimited addition of TG1-3 repeats. In budding yeast, the Rap1 protein negatively regulates telomere length, and the Rap1-binding proteins Rif1 and Rif2 are important for this negative regulation. In fact, both RIF1 and RIF2 deletions cause telomere lengthening (Hardy et al., 1992; Wotton and Shore, 1997) in a telomerase-dependent manner (Teng et al., 2000; Chan et al., 2001), and mutations in the Rif-interacting domain of Rap1 increase telomere length and accelerate telomerase action (Kyrion et al., 1992). It has been suggested that the Rap1/Rif complex may modulate telomerase activity by acting in a length-sensing mechanism based on the binding of one Rap1 protein per 15–18 base pairs of TG repeats (Conrad et al., 1990; Gilson et al., 1993; Marcand et al., 1997).
Despite the functional differences between telomeres and DSBs, a subset of the proteins required for DSB repair also have distinct roles in telomere maintenance. Among them, the RAD50, MRE11, and XRS2 gene products form a highly conserved complex called MRX, which is involved in recombinational DNA repair, DSB end resection, telomere function, and checkpoint mechanisms in Saccharomyces cerevisiae (reviewed in Haber, 1998; D'Amours and Jackson, 2002). A chromosome suffering a double-strand break can be repaired either through homologous recombination or by nonhomologous end joining (NHEJ) (reviewed in Paques and Haber, 1999), and the MRX complex controls both processes (Haber, 1998). Also the DNA-dependent protein kinase DNA-PK (Wood et al., 2001), as well as DNA ligase IV and its associated factors, XRCC4 in mammals, Lif1 and Lif2 in S. cerevisiae, are essential for NHEJ (reviewed in Paques and Haber, 1999). In particular, the MRX complex interacts with DNA ligase IV/Lif1, thereby promoting intermolecular DNA joining (Chen et al., 2001).
Besides MRX, the Yku70/Yku80 protein complex is required for both NHEJ and telomere maintenance (reviewed in Lundblad, 2000). This complex is physically associated with yeast telomeres (Gravel et al., 1998), and its lack results in a substantial reduction of telomere length (Porter et al., 1996; Boulton and Jackson, 1996, 1998). Moreover, shift to 37°C causes ykuΔ mutants to arrest cell cycle progression and accumulate single-stranded DNA in subtelomeric Y′ regions (Maringele and Lydall, 2002). The mismatch repair-associated EXO1 gene is required for both single-stranded DNA (ssDNA) generation and cell cycle arrest of yku70 mutants, suggesting that ssDNA is an important component of the arrest signal in the absence of Yku (Maringele and Lydall, 2002).
Evolutionarily conserved protein kinases, such as S. cerevisiae Mec1 and Tel1, Schizosaccharomyces pombe Rad3, and human ATM and ATR, are also key players in both telomere maintenance and checkpoint networks (reviewed in Abraham, 2001; Shiloh, 2001). In budding yeast, a Tel1-dependent checkpoint, which becomes apparent in the absence of Mec1 and converges on Rad9 and Rad53 like the canonical Rad24-Mec1 pathway (Usui et al., 2001), requires the MRX complex and responds to unprocessed DSBs (D'Amours and Jackson, 2001; Grenon et al., 2001; Usui et al., 2001). The Tel1-dependent checkpoint is also triggered by the loss of Sae2, a 345 amino acid protein, whose lack seems to cause accumulation of unprocessed DSBs (McKee and Kleckner, 1997; Rattray et al., 2001; Usui et al., 2001). The phenotype of sae2Δ mutants is very similar to that of nonnull rad50 and mre11 mutants. In fact, they accumulate meiotic unresected DSBs, in which 5′-3′ resection is blocked by the presence of the topoisomerase II-like protein Spo11 at DNA ends (McKee and Kleckner, 1997; Rattray et al., 2001). Thus, Sae2 deficiency may cause accumulation of DNA alterations that are specifically sensed by the Tel1-dependent checkpoint.
Both the lack of any of the three MRX proteins and TEL1 deletion cause marked telomere shortening (Greenwell et al., 1995; Morrow et al., 1995; Nugent et al., 1998; Tsukamoto et al., 2001), and the phenotypes of tel1Δ cells are not exacerbated by the lack of MRX proteins. This suggests that Tel1 and MRX act in the same pathway (Ritchie and Petes, 2000). The ATR-like protein Mec1, which has a central role in the major DNA damage checkpoint pathway, also contributes to telomere length maintenance, but independently of Tel1/MRX. In fact, mec1 tel1 cells, as well as mec1 cells lacking any of the MRX proteins, show more dramatic telomere shortening than each single mutant, similar to that seen in cells lacking active telomerase (Ritchie et al., 1999; Ritchie and Petes, 2000). Thus, Tel1/MRX and Mec1 are both required for telomerase-mediated replication of telomeric DNA. The absence of Tel1 and MRX proteins does not affect either telomerase catalytic activity or Cdc13 binding to telomeric DNA (Chan et al., 2001; Tsukamoto et al., 2001). However, when telomerase is artificially targeted to telomeres, their lengthening is at least as effective in mec1 mrx and mec1 tel1 as in wild-type cells (Tsukamoto et al., 2001), suggesting that Mec1, Tel1, and MRX may act by recruiting to telomeres either telomerase or telomerase-activating factors.
We have recently shown that yeast strains expressing a galactose-inducible GAL1-TEL1 fusion as the sole Tel1 source undergo a transient G2/M cell cycle arrest in the absence of exogenous DNA damage (Clerici et al., 2001). This arrest seems to be due to checkpoint activation, because it requires both the Rad53 and Chk1 kinases (Clerici et al., 2001), known to cooperate in the G2/M DNA damage checkpoint (Sanchez et al., 1999), and Rad9, whose phosphorylation and interaction with Rad53 leads to Rad53 activation (Gilbert et al., 2001). Conversely, it does not seem to involve Ddc1, a protein presumably acting in the DNA damage-sensing steps of the canonical Mec1 pathway (Clerici et al., 2001).
Herein, we show that overexpression of TEL1 in cells with short telomeres causes telomere lengthening and the activation of a Rad53-dependent checkpoint response. Sudden telomere elongation and checkpoint-mediated cell cycle arrest are also triggered in wild-type cells by overproducing a protein fusion between the telomeric binding protein Cdc13 and the telomerase-associated protein Est1. Both TEL1 overexpression-dependent telomere elongation and checkpoint activation can be partially suppressed by preventing telomerase action. Moreover, accelerating rebalancing of telomeric ends also accelerates checkpoint inactivation. Thus, as proposed for telomere shortening (Enomoto et al., 2002; Ijpma and Greider, 2003), also sudden telomere lengthening by telomerase seems to trigger a DNA damage checkpoint response.
MATERIALS AND METHODS
Plasmids
To obtain plasmid pML411, containing a SAE2 fragment spanning from 324 base pairs upstream of the coding region start codon to 225 base pairs downstream of the stop codon, a 1587-base pair SAE2 fragment was amplified by polymerase chain reaction (PCR) with yeast genomic DNA as template, the oligonucleotides PRP322 (5′-CGG AAT TCG TCT GAG TTA GCG TCT GAT TTT G-3′) and PRP323 (5′-CGG AAT TCC CTG GTA AGT TAG GTG TCA TTT G-3′) as primers, and Pfu polymerase (Stratagene, La Jolla, CA). The PCR amplification product was then cloned into the EcoRI-EcoRI sites of plasmid YEplac195 (Gietz and Sugino, 1988).
To obtain plasmid pML415, containing the RIF2 open reading frame flanked by 521 base pairs upstream of the start codon and 383 base pairs downstream of the stop codon, a 2092-base pair RIF2 fragment was amplified by PCR with yeast genomic DNA as template, the oligonucleotides PRP324 (5′-CGG AAT TCC CGA AGC TAC TTA TGC CAG AA-3′) and PRP325 (5′-CGG AAT TCA GTG CGT GCA AAA CCT AGA AG-3′) as primers, and Pfu polymerase (Stratagene). The PCR amplification product was then cloned into the EcoRI-EcoRI sites of plasmid YEplac195.
To obtain plasmid pML460, carrying the GAL1-CDC13 EST1 fusion, a 1357-base pair CDC13 fragment was amplified by PCR with plasmid PVL1091, kindly provided by V. Lundblad (Houston, TX), as template, the oligonucleotides PRP505 (5′-CGC GGA TCC ATA TGG ATA CCT TAG AAG AGC CTG AG-3′) and PRP508 (5′-TTG TAC ATG ATG GCT TTG AAG CC-3′) as primers, and Pfu as polymerase. The amplification products was then cloned into the BamH1-BamH1 sites of plasmid PVL1091, in which a 931-base pair SpeI-BamH1 fragment carrying the GAL1 promoter was inserted into the XbaI-BamH1 sites.
Yeast Strains and Media
The genotypes of all the yeast strains used in this study are listed in Table 1. All yeast strains were derivatives of W303 (MATa or MATα, ade2-1, can1-100, his3-11,15, leu2-3,112, trp1-1, ura3).
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference/Source |
|---|---|---|
| K699 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 | Longhese et al. (1997) |
| K700 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 | Longhese et al. (1997) |
| Y300 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 tel1Δ::HIS3 | Longhese et al. (2000) |
| YLL941 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 yku70Δ::HIS3 | This study |
| YLL1133 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3::TEL1::URA3 tell::GAL1-TEL1::LEU2 | This study |
| YLL1156 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 est2Δ::KANMX4 | This study |
| YLL1157 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 est2Δ::KANMX4 tell::GAL1-TEL1::LEU2 | This study |
| YLL1169 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 rad51Δ::HIS3 | This study |
| YLL1171 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 rad51Δ::HIS3 tell::GAL1-TEL1::LEU2 | This study |
| YLL1230 | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3::TEL1::URA3 tell::GAL1-TEL1::LEU2 yku70Δ::HIS3 | This study |
| DMP3469/1A | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 tell::GAL1-TEL1::LEU2 | This study |
| DMP3539/10D | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 tel1::GAL1-TEL1::LEU2 | Clerici et al. (2001) |
| DMP4086 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3/ura3 tel1::GAL1-TEL1::LEU2/TEL1 est2Δ::KANMX4/EST2 | This study |
Strain DMP3539/10D, carrying one copy of the GAL1-TEL1 fusion disrupting the TEL1 chromosomal gene, was obtained as described previously (Clerici et al., 2001). Strain DMP3469/1A was a meiotic segregant from a cross between strains DMP3539/10D and K700. Strain YLL1133, carrying the TEL1 gene integrated at the URA3 locus, was obtained by transforming strain DMP3539/10D with an NcoI-digested pSP21 (kindly provided by T. Petes, Chapel Hill, NC) derivative plasmid, in which the AatII-AatII fragment, carrying the ADE3 gene and the 2 μ origin of replication, had been deleted.
To generate the YKU70 chromosomal deletion, a yku70Δ::HIS3 cassette was constructed by PCR with pFA6a-HIS3 plasmid (Wach et al., 1994) as template, and oligonucleotides PRP171 (5′-CAA TAG TGG AGA ACT TAA CGA TCA AGT GGA TGA AAC AGG TTA TAC GTA CGC TGC AGG TCG AC-3′) and PRP172 (5′-ATT TAG CCT TTG GAT GAT TGG ATC TTC TGA CTT CTC AGA TTC TAA TCG ATG AAT TCG AGC TCG-3′) as primers, followed by transformation of strains K699 and YLL1133 with the PCR product, giving rise to strains YLL941 and YLL1230, respectively, where 1587 base pairs of the YKU70 coding region were replaced by the Kluyveromyces lactis HIS3 gene.
To generate the RAD51 chromosomal deletion, a rad51Δ::HIS3 cassette was constructed by PCR with pFA6a-HIS3 plasmid (Wach et al., 1994) as template, and oligonucleotides PRP410 (5′-AGG CCT ACT AAT TTG TTA TCG TCA TAT GTC TCA AGT TCA AGA ACA CGT ACG CTG CAG GTC GAC-3′) and PRP411 (5′-AAT AGA GAC AAG AGA CCA AAT ACC TAC TCG TCT TCT TCT CTG GGG ATC GAT GAA TTC GAG CTC G-3′) as primers, followed by transformation of strains K699 and DMP3539/10D with the PCR product, giving rise to strains YLL1169 and YLL1171, respectively, where 1161 base pairs of the RAD51 coding region were replaced by the Kluyveromyces lactis HIS3 gene.
To generate the EST2 chromosomal deletion, an est2Δ::KANMX4 cassette was constructed by PCR with pFA6a-KANMX4 plasmid as template, and oligonucleotides PRP406 (5′-GAT TTA TAC TCA TGA AAA TCT TAT TCG AGT TCA TTC AAG ACA AGC CGT ACG CTG CAG GTC GAC-3′) and PRP407 (5′-TAT ATA TGC TTG CAA GTG TTG AAT TTC CTT TCT CAA AAG AAT GAT ATC GAT GAA TTC GAG CTC G-3′) as primers, followed by transformation of strains K699 and DMP3539/10D with the PCR product, giving rise to strains YLL1156 and YLL1157, respectively, where 2549 base pairs of the EST2 coding region were replaced by the KANMX4 gene. After transformation, clones were picked directly from the G418-selective plates (∼25 generations) to perform the experiment described in Figure 3, B and C, and to verify concomitantly the accuracy of the disruption (see below).
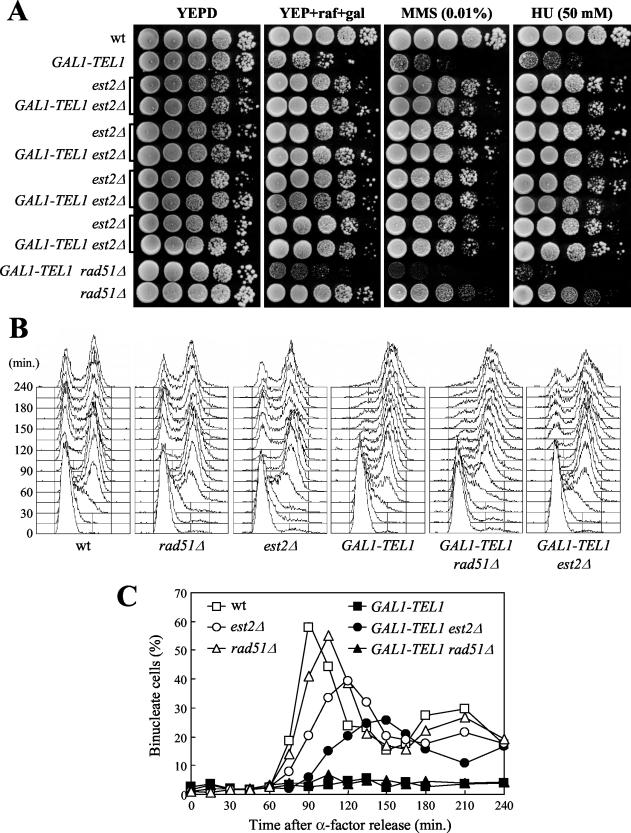
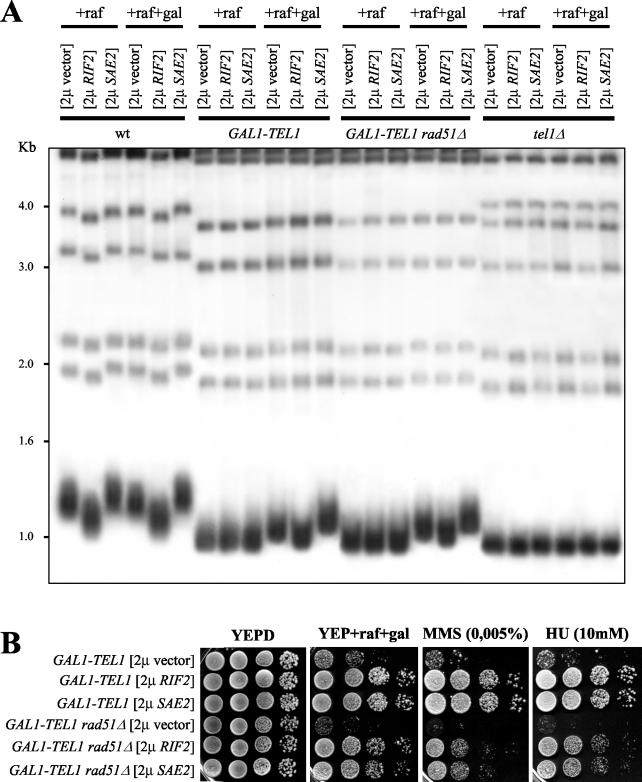
Figure 3.
Effects of EST2 and RAD51 deletions on galactose-induced GAL1-TEL1 cells. (A) A diploid strain heterozygous for the GAL1-TEL1 and est2Δ alleles (DMP4086; see MATERIALS AND METHODS) was allowed to sporulate, and tetrads were dissected on YEPD plates. Forty-eight hours after tetrad dissection (25 generations), est2Δ and GAL1-TEL1 est2Δ clones from spores of the same tetrad (indicated by square parenthesis) were inoculated from the dissection plate into YEP + raf and grown for eight further generations. Serial dilutions of these cultures and of wild-type (K699), rad51Δ (YLL1169), GAL1-TEL1 (DMP3539/10D), and GAL1-TEL1 rad51Δ (YLL1171) cell cultures logarithmically growing in YEP + raf were spotted on YEPD and on YEP + raf + gal plates with or without MMS and HU at the indicated concentrations. (B and C) Wild-type (K699), rad51Δ (YLL1169), est2Δ (YLL1156), GAL1-TEL1 (DMP3539/10D), GAL1-TEL1 rad51Δ (YLL1171), and GAL1-TEL1 est2Δ (YLL1157) cell cultures growing logarithmically in YEP + raf were synchronized in G1 with α-factor and released from G1 block at time zero in YEP + raf + gal. For the est2Δ and GAL1-TEL1 est2Δ cell cultures, est2Δ disruptant clones (see MATERIALS AND METHODS) grown on selective medium (∼25 generations) were inoculated in YEP + raf and grown further for not >15 generations (40 generations combined). Galactose was added 30 min before the release. Samples were collected at the indicated times after α-factor release to analyze DNA content by fluorescence-activated cell sorting analysis (B) and the percentage of binucleate cells by propidium iodide staining (C).
The above-mentioned est2Δ::KANMX4 cassette was also used to transform a TEL1/GAL1-TEL1 diploid strain, where one TEL1 copy was substituted with the GAL1-TEL1 fusion. The resulting diploid strain TEL1/GAL1-TEL1 EST2/est2Δ (DMP4086) was allowed to sporulate and tetrads were dissected on YEPD plates. Forty-eight hours after tetrad dissection (25 generations), segregant clones were used for the experiments described in Figures 3A and 4, C and D, and concomitantly analyzed for the presence of the est2Δ and GAL1-TEL1 markers.
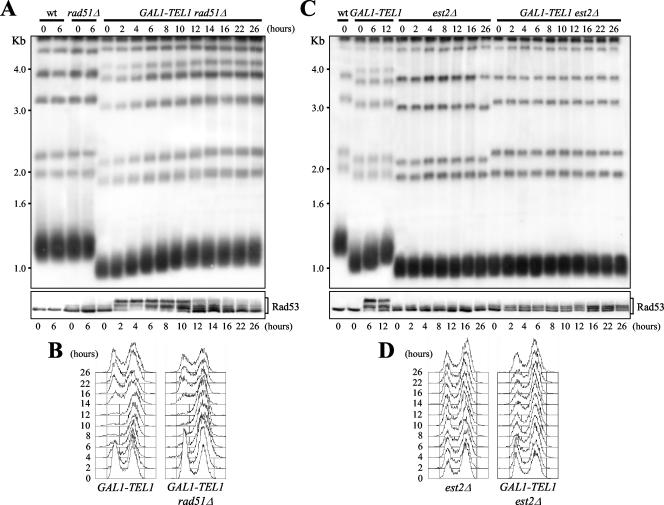
Figure 4.
Kinetics of telomere elongation of galactose-induced GAL1-TEL1 cells in the absence of EST2 or RAD51. Strains were as follows: wt (K699), GAL1-TEL1 (DMP3539/10D), rad51Δ (YLL1169), GAL1-TEL1 rad51Δ (YLL1171), est2Δ, and GAL1-TEL1 est2Δ. est2Δ and GAL1-TEL1 est2Δ cell cultures were from spores obtained and treated as in Figure 3. Cell cultures exponentially growing in YEP + raf (time 0) were shifted to YEP + raf + gal and samples were taken at the indicated times to analyze telomere length (A and C, top), the pattern of Rad53 phoshorylation by Western blot by using anti-Rad53 antibodies (A and C, bottom) and DNA content by fluorescence-activated cell sorting analysis (B and D). (A and C) Genomic DNA was digested with XhoI and hybridized with a poly(GT) telomere-specific probe. Similar results were obtained by analyzing four independent est2Δ and GAL1-TEL1 est2Δ spores.
Strains YLL848 and YLL932, carrying the TEL1-HA3 and GAL1-TEL1-HA3 alleles at the TEL1 chromosomal locus were constructed as described previously (Clerici et al., 2001).
Wild-type, GAL1-TEL1, tel1Δ and rad51Δ strains carrying either the 2 μ vector, or 2 μ RIF2 or 2 μ SAE2 plasmids were constructed by transforming strains K699, DMP3539/10D, Y300 and YLL1169 with plasmids YEplac195 (2 μ URA3), pML415 (2 μ RIF2 URA3) and pML411 (2 μ SAE2 URA3), respectively. Wild-type strains carrying either the empty vector or a derivative plasmid with the GAL1-CDC13-EST1 fusion were constructed by transforming strain K699 with plasmids YCplac111 (ARS1 CEN4 LEU2) and pML460 (ARS1 CEN4 LEU2 GAL1-CDC13-EST1), respectively.
The accuracy of all gene replacements and integrations was verified by Southern blot analysis or PCR. Standard yeast genetic techniques and media were according to Rose et al. (1990). Cells were grown in YEP medium (1% yeast extract, 2% bactopeptone, 50 mg/l adenine) supplemented with 2% glucose (YEPD) or 2% raffinose (YEP + raf) or 2% raffinose and 1% galactose (YEP + raf + gal). Transformants carrying the KANMX4 cassette were selected on YEPD plates containing 400 μg/ml G418 (US Biochemical Corp., Cleveland, OH).
Search for High Copy Number Suppressors
To search for multicopy suppressors of the hydroxyurea (HU)-sensitivity caused by GAL1-TEL1 overexpression, the GAL1-TEL1 strain DMP3539/10D was transformed with a S. cerevisiae genomic library based on the multicopy 2 μ vector YEp24 (Carlson and Botstein, 1982). Ura+ transformants were tested for their ability to grow at 25°C on YEP + raf + gal plates in the presence of 150 mM HU, which inhibits the untransformed strain. Plasmids from transformants showing cosegregation of HU-resistance with the URA3 vector marker were recovered and introduced again into the DMP3539/10D strain, to confirm their ability to suppress GAL1-TEL1 HU-hypersensitivity. Restriction analysis allowed us to identify several classes of plasmids containing different yeast genomic fragments (to be published elsewhere). The nucleotide sequences of both ends of the smallest DNA insert of each plasmid class were determined and compared with the whole S. cerevisiae genomic sequence in the Saccharomyces Genome Database. Because most inserts contained several open reading frames, the suppressor genes were identified by cloning subfragments of the inserts into the 2 μ plasmid YEplac195 and testing the derivative plasmids for their ability to suppress the HU-hypersensitivity of the galactose-induced GAL1-TEL1 strain.
Southern Blot Analysis of Telomere Length
Yeast DNA was prepared according to standard methods and digested with the XhoI enzyme. The resulting DNA fragments were separated by gel electrophoresis in 0.8% agarose gel and transferred to a GeneScreen nylon membrane (PerkinElmer Life Sciences, Boston, MA), followed by hybridization with a 32P-labeled poly(GT) probe and exposure to x-ray–sensitive films. Standard hybridization conditions were used.
Other Techniques
Synchronization experiments, total protein extract preparation, and Western blot analysis were performed as described in Paciotti et al., 2000.
RESULTS
Checkpoint Activation by Ectopic TEL1 Expression Correlates with Telomere Lengthening
Strains carrying a GAL1-TEL1 fusion at the TEL1 chromosomal locus as the sole Tel1 source (tel1::GAL1-TEL1, from now ahead simply indicated as GAL1-TEL1) undergo a transient Rad53-dependent G2/M cell cycle arrest with concomitant phosphorylation of Rad9, Chk1, and Rad53 after shift to galactose-induced conditions. This cell cycle arrest also leads to growth defects and sensitivity to DNA-damaging agents (Clerici et al., 2001; Figure 1A).
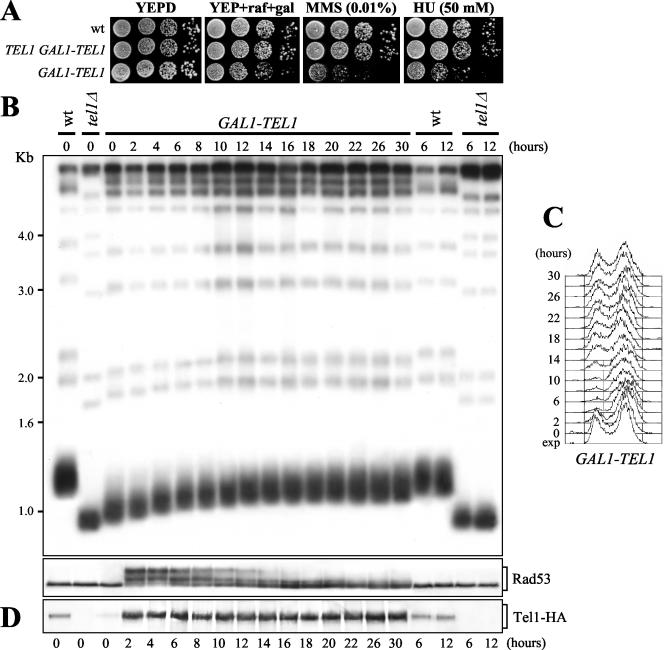
Figure 1.
Ectopic TEL1 overexpression leads to telomere elongation. Isogenic strains were as follows: wild-type (K699), tel1Δ (Y300), GAL1-TEL1 (DMP3539/10D), and TEL1 GAL1-TEL1 (YLL1133). (A) Serial dilutions of YEP + raf exponentially growing cell cultures were spotted on YEPD and on YEP + raf + gal plates with or without MMS and HU at the indicated concentrations. (B and C) Wild-type, tel1Δ, and GAL1-TEL1 cell cultures exponentially growing in YEP + raf for >80 generations (time 0) were shifted to YEP + raf + gal and samples were taken at the indicated times to analyze telomere length (B, top), the pattern of Rad53 phosphorylation by Western blot by using anti-Rad53 antibodies (B, bottom), and DNA content by fluorescence-activated cell sorting analysis (C). Genomic DNA in B was digested with XhoI and hybridized with a poly(GT) telomere-specific probe. (D) TEL1-HA3 (YLL848), tel1Δ (Y300), and GAL1-TEL1-HA3 (YLL932) cell cultures exponentially growing in YEP + raf for >80 generations (time 0) were shifted to YEP + raf + gal, and samples were taken at the indicated times to analyze the amount of Tel1-HA3–tagged protein by Western blot by using anti-hemagglutinin (HA) antibodies. Kinetics of Rad53 phosphorylation and cell cycle progression were also determined (our unpublished data) and were identical to those shown in B and C. exp, exponentially growing cells.
We further studied this phenomenon by asking whether checkpoint activation in galactose-induced GAL1-TEL1 cells might be related to alterations in telomere metabolism. In fact, the lack of Tel1 results in short but stable telomeres (Greenwell et al., 1995; Morrow et al., 1995). Because Tel1 levels in the GAL1-TEL1 strain depend on the expression of the GAL1-TEL1 fusion, this might in turn influence telomere length in uninduced versus galactose-induced conditions. As shown in Figure 1B, lane 0, telomeres of GAL1-TEL1 cells exponentially growing in raffinose (GAL1 promoter off) were shorter than those of wild-type cells and only slightly longer than those of tel1Δ cells. Thus, the amount of Tel1 protein resulting from GAL1-TEL1 basal expression, which is lower than that produced by endogenous TEL1 under the same conditions (Figure 1D, lane 0), is insufficient to maintain wild-type telomere length. However, although telomere length was affected, the checkpoint was not activated under these conditions. In fact, neither tel1Δ nor GAL1-TEL1 cells grown in raffinose contained phosphorylated Rad53 (Figure 1B bottom, lane 0), which normally occurs upon activation of the known DNA damage checkpoint pathways and is detectable as changes of Rad53 electrophoretic mobility (Sanchez et al., 1996). Moreover, telomere length did not change in wild-type and tel1Δ cells shifted to galactose (Figure 1B, top, lanes 6 and 12), which normally progressed through cell cycle (our unpublished data). Conversely, GAL1-TEL1 cells shifted to galactose arrested with 2C DNA content (Figure 1C) and phosphorylated Rad53 (Figure 1B, bottom), while gradually lengthened telomeres (Figure 1B, top). This telomere lengthening paralleled cell cycle arrest and checkpoint activation. In fact, at the same time telomere length reached a new equilibrium (10/12 h after the shift) (Figure 1B, top), GAL1-TEL1 cells divided, as indicated by the appearance of cells with 1C DNA content (Figure 1C). Accordingly, the amount of phosphorylated Rad53 started to decrease (Figure 1B, bottom), although Tel1 levels remained high throughout the experiment (Figure 1D). Therefore, activation of the Rad53-dependent checkpoint by ectopic Tel1 overexpression correlates with changes in telomere length.
Telomere Lengthening and Checkpoint Activation Is Triggered by TEL1 Overexpression Only in Cells with Short Telomeres
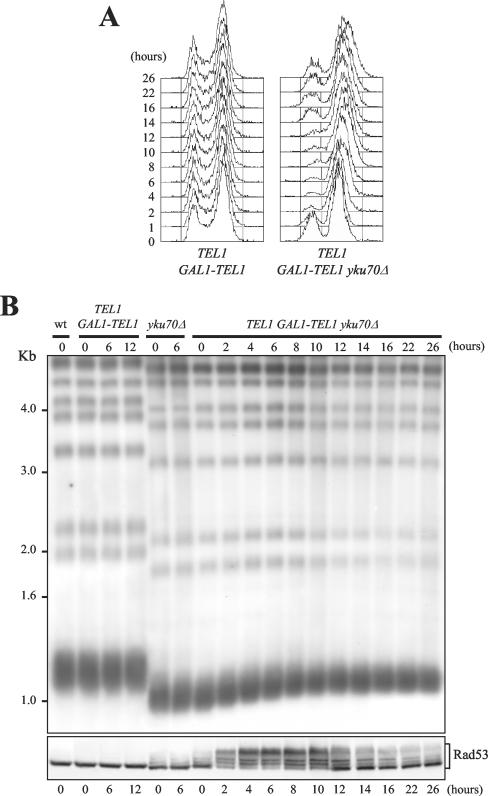
We next asked whether GAL1-TEL1 overexpression might activate the Rad53-dependent response also in the presence of wild-type TEL1. To this end, we integrated a fully functional TEL1 gene at the URA3 locus in the GAL1-TEL1 strain. The derivative TEL1::URA3 tel1::GAL1-TEL1 strain (from now ahead simply indicated as TEL1 GAL1-TEL1) did not show any growth defect, was as sensitive as wild type to HU and methyl methanesulfenate (MMS) (Figure 1A) and did not undergo any cell cycle arrest after shift to galactose-containing medium (Figure 2A). Moreover, Rad53 phosphorylation was not detectable by Western blot analysis of galactose-induced TEL1 GAL1-TEL1 cell extracts (Figure 2B, bottom). Finally, telomere length in TEL1 GAL1-TEL1 cells did not change after shift to galactose and was similar to that of wild-type cells (Figure 2B, top, lanes 6 and 12). Thus, increasing Tel1 levels in TEL1 cells with normal telomere length neither causes telomere lengthening nor activates the checkpoint. On the other hand, both telomere elongation and checkpoint activation are triggered by Tel1 overproduction in GAL1-TEL1 cells with short telomeres. Thus, it is possible that TEL1 overexpression may elicit these responses only when it is induced in cells with short telomeres that might be more accessible to telomerase action. If these were the case, GAL1-TEL1 induction might cause telomere lengthening and activate the checkpoint also in TEL1 cells whose telomeres are short due to deletion of the YKU70 gene. As shown in Figure 2B, lane 0, telomeres of both yku70Δ and TEL1 GAL1-TEL1 yku70Δ cells exponentially growing at 23°C in raffinose were shorter than those of TEL1 GAL1-TEL1 cells under the same conditions. After shift to galactose, TEL1 GAL1-TEL1 yku70Δ cells gradually lengthened telomeres (Figure 2B, top) and arrested with 2C DNA content (Figure 2A), undivided nuclei (our unpublished data), and high levels of phosphorylated Rad53 (Figure 2B, bottom), until telomeres reached a new equilibrium length (10/12 h). At this time, cells started also to resume cell cycle progression and the amount of phosphorylated Rad53 began to decrease. Conversely, telomere length did not change in galactose-induced TEL1 GAL1-TEL1 and yku70Δ cells (Figure 2B, top), which progressed through the cell cycle (Figure 2A; our unpublished data), although the lack of Yku70 caused some Rad53 phosphorylation (Figure 2B, bottom). Therefore, TEL1 overexpression seems to trigger telomere elongation and Rad53-dependent checkpoint activation even in the presence of endogenously expressed TEL1, as far as it is turned on in cells with short telomeres.
Figure 2.
GAL1-TEL1 induction triggers telomere lengthening and checkpoint activation in cells with short telomeres. Wild-type (K699), TEL1 GAL1-TEL1 (YLL1133), TEL1 GAL1-TEL1 yku70Δ (YLL1230), and yku70Δ (YLL941) cell cultures exponentially growing in YEP + raf for >80 generations (time 0) were shifted to YEP + raf + gal and samples were taken at the indicated times to analyze DNA content by FACS analysis (A), telomere length (B, top), and the pattern of Rad53 phosphorylation by Western blot by using anti-Rad53 antibodies (B, bottom). Genomic DNA in B was digested with XhoI and hybridized with a poly(GT) telomere-specific probe.
Est2 Is Required for the Tel1-induced Cell Cycle Arrest and Telomere Elongation
Based on the above-mentioned data, checkpoint activation by TEL1 overexpression might be due to ongoing telomere lengthening and, in this case, it should likely be suppressed by preventing telomere elongation. We therefore analyzed the kinetics of cell cycle progression and telomere elongation of galactose-induced GAL1-TEL1 cells lacking the RAD51 gene, which is necessary for recombinational repair and telomere elongation in the absence of telomerase (Le et al., 1999), or the EST2 gene, which encodes the catalytic subunit of telomerase (Lingner et al., 1997). In addition, we analyzed the ability of the same strains to form colonies on YEP + raf + gal plates in the absence or in the presence of HU and MMS. In fact, GAL1-TEL1 induction also causes growth defects and hypersensitivity to genotoxic drugs, the latter likely due to additive effects on checkpoint activation of TEL1 overexpression and DNA damage caused by the drugs (Figure 1A; Clerici et al., 2001).
As shown in Figure 3, deletion of RAD51 did not suppress the effects of GAL1-TEL1 induction. In fact, GAL1-TEL1 rad51Δ cells showed more severe growth defects compared with GAL1-TEL1 cells, as well as higher hypersensitivity to HU and MMS than both GAL1-TEL1 and rad51Δ cells on galactose-containing plates (Figure 3A). Moreover, both GAL1-TEL1 rad51Δ and GAL1-TEL1 cells released from the G1 block in galactose-containing medium remained arrested with undivided nuclei until the end of the experiment (Figure 3, B and C). Finally, when GAL1-TEL1 rad51Δ cells exponentially growing in raffinose were shifted to galactose, their kinetics of telomere lengthening and reequilibration (Figure 4A, top), Rad53 phosphorylation (Figure 4A, bottom), and cell cycle arrest (Figure 4B) were comparable with those observed in similarly treated GAL1-TEL1 cell cultures (Figure 1 and 4). Some Rad53 phosphorylation was detected in rad51Δ cells in all the analyzed conditions (Figure 4A, bottom), suggesting that the lack of recombinational repair might cause accumulation of unrepaired DNA lesions, which might activate the checkpoint even without external DNA damage.
On the contrary, all the effects of GAL1-TEL1 expression were partially counteracted by deletion of EST2. In fact, all the analyzed GAL1-TEL1 est2Δ meiotic segregants from a GAL1-TEL1/TEL1 est2Δ/EST2 heterozygous strain showed higher ability than GAL1-TEL1 cells to form colonies on galactose-containing plates and behaved similarly to est2Δ segregants both in the presence and in the absence of genotoxic drugs (Figure 3A). Moreover, when GAL1-TEL1 est2Δ cell cultures were released from α-factor G1 arrest into galactose-containing medium, they began to divide nuclei (Figure 3C) and to undergo cell division (Figure 3B) 105 and 120 min after release, respectively, whereas similarly treated GAL1-TEL1 cells remained arrested with 2C DNA contents and undivided nuclei throughout the experiment (Figure 3, B and C). It is worth noting that the suppression of the G2/M cell cycle arrest caused by deletion of EST2 was apparent despite entry into S phase and nuclear division were slower in est2Δ cells than in wild-type (Figure 3, B and C).
These results prompted us to verify whether telomere elongation was abrogated in galactose-induced GAL1-TEL1 est2Δ cells. Indeed, shift to galactose of GAL1-TEL1 est2Δ cells did not cause any change in their telomere length, which was similar to that of est2Δ mutants (Figure 4C, top). Moreover, galactose-induced GAL1-TEL1 est2Δ cells did not arrest cell cycle progression (Figure 4D) and showed a reduction of Rad53 phosphorylation compared with similarly treated GAL1-TEL1 cells (Figure 4C, bottom). Interestingly, est2Δ cells showed some Rad53 phosphorylation (Figure 4C, bottom) that might depend on unprotected telomeric ends due to telomere erosion.
Thus, both the G2/M cell cycle arrest and Rad53 phosphorylation caused by GAL1-TEL1 overexpression are at least partially dependent on telomerase action, whereas they do not require Rad51-dependent homologous recombination. Moreover, the reduced checkpoint activation in galactose-induced GAL1-TEL1 est2Δ cells correlates with their inability to elongate telomeres.
Overexpression of RIF2 or SAE2 Can Suppress G2/M Cell Cycle Arrest and Accelerate Telomere Length Reequilibration in Galactose-induced GAL1-TEL1 Cells
To better understand the molecular mechanisms underlying GAL1-TEL1 effects, we searched for multicopy suppressors of the HU hypersensitivity of galactose-induced GAL1-TEL1 cells, which correlates with checkpoint activation. Among the several plasmids identified in a S. cerevisiae genomic library in the multicopy 2 μ vector YEp24 (see MATERIALS AND METHODS), 14 plasmids carried the SAE2 gene and 2 plasmids carried the RIF2 gene. As shown in Figure 5A, both SAE2 and RIF2 on 2 μ plasmids were able to suppress the hypersensitivity to genotoxic agents of galactose-induced GAL1-TEL1 cells. We decided to analyze in more detail the basis of suppression, because both genes are functionally related to TEL1, although in different ways. In fact, the Rif2 protein, together with Rif1 and Rap1, is required to avoid unlimited addition of TG1–3 repeats by telomerase, possibly acting as a negative regulator of telomerase activity (Wotton and Shore, 1997; Teng et al., 2000; Chan et al., 2001). On the other hand, the lack of SAE2, which is likely involved in processing of DSBs, triggers the activation of the Tel1-dependent checkpoint (Usui et al., 2001).
Figure 5.
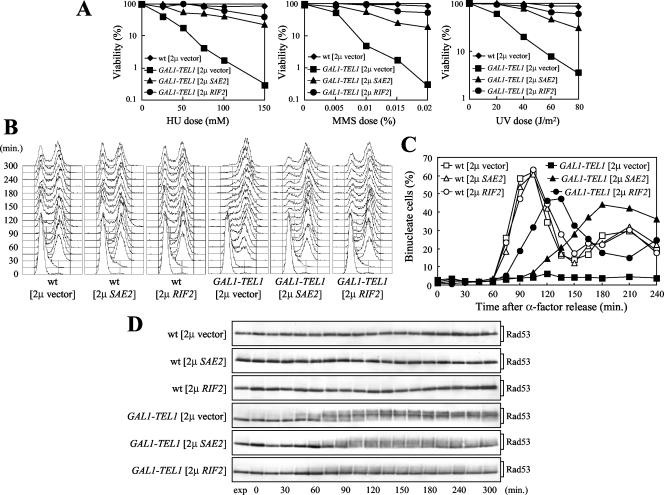
Effects of high copy number SAE2 and RIF2 on galactose-induced GAL1-TEL1 cells. Strains were as follows: wild-type [2 μ vector], wild-type [2 μ SAE2], wild-type [2 μ RIF2], GAL1-TEL1 [2 μ vector], GAL1-TEL1 [2 μ SAE2], and GAL1-TEL1 [2 μ RIF2] (see MATERIALS AND METHODS). (A) Dose-response killing curves were determined by plating serial dilutions of SC-ura + raf exponentially growing cell cultures on YEP + raf + gal plates with or without MMS or HU at the indicated concentrations. One set of YEP + raf + gal plates was exposed to the indicated UV doses. Plates were incubated at 25°C and colony-forming units were counted after 3 d. (B–D) Cell cultures growing logarithmically in SC-ura + raf were synchronized in G1 with α-factor and released from the G1 block at time 0 in YEP + raf + gal. Galactose was added 30 min before the release. Samples were collected at the indicated times after α-factor release to analyze DNA content (B), the percentage of binucleate cells (C), and the pattern of Rad53 phosphorylation by Western blot by using anti-Rad53 antibodies (D). exp, exponentially growing cells.
Further analysis clearly showed that, although to different extents, high copy number SAE2 and RIF2 suppress the G2/M cell cycle arrest of galactose-induced GAL1-TEL1 cells. In fact, when cells were released from a G1 arrest in galactose-containing medium, GAL1-TEL1 cells containing SAE2 or RIF2 on 2 μ plasmids initiated nuclear division at 120 and 90 min after release from α-factor, respectively (Figure 5, B and C). These cells also showed a reduction in the amount of Rad53 phosphorylation, compared with GAL1-TEL1 cells carrying the empty 2 μ vector (Figure 5D), which remained arrested with 2C DNA contents and undivided nuclei until the end of the experiment (Figure 5, B and C).
In agreement with previous data (Wotton and Shore, 1997) and with its role in negative regulation of telomerase, the RIF2 gene on a 2 μ plasmid caused telomere shortening (Figure 6A). Conversely, high levels of Rif2 did not cause any detectable alteration of telomere length in tel1Δ or GAL1-TEL1 cells grown in raffinose (Figure 6A), suggesting that either telomeres in these cells are already too short to be targeted by Rif2, or Tel1 is required for Rif2-dependent inhibition of telomerase. On the other hand, an excess of Rif2 partially counteracted the Tel1-dependent telomere elongation in galactose-induced GAL1-TEL1 cells (Figure 6A). In fact, telomeres of galactose-induced GAL1-TEL1 cells carrying the RIF2 gene on 2 μ plasmids were shorter than those of galactose-induced GAL1-TEL1 cells. GAL1-TEL1 [2 μ RIF2] telomeres were slightly longer than those of the same cells under uninduced conditions, likely due to minimal perturbation of the balance between Tel1-dependent elongation and Rif2-dependent shortening. Thus, Rif2 might suppress TEL1 overexpression-dependent checkpoint activation by counteracting Tel1 access to telomeric ends and/or its functions in promoting telomere elongation. Consistent with a role of Rif2 in negatively regulating telomerase activity (Teng et al., 2000; Chan et al., 2001), these Rif2-dependent suppression events do not require Rad51, which collaborates in maintaining telomere ends in the absence of telomerase independently of RAD50 (Le et al., 1999). In fact, an excess of Rif2 similarly promoted telomere lengthening (Figure 6A) and was able to suppress the hypersensitivity to genotoxic agents (Figure 6B) of galactose-induced GAL1-TEL1 cells both in the absence and in the presence of RAD51.
Figure 6.
Effects of high copy number SAE2 and RIF2 on telomere length. Isogenic strains were as follows: wild-type [2 μ vector], wild-type [2 μ RIF2], wild-type [2 μ SAE2], GAL1-TEL1 [2 μ vector], GAL1-TEL1 [2 μ RIF2], GAL1-TEL1 [2 μ SAE2], GAL1-TEL1 rad51Δ [2 μ vector], GAL1-TEL1 rad51Δ [2 μ RIF2], GAL1-TEL1 rad51Δ [2 μ SAE2], tel1Δ [2 μ vector], tel1Δ [2 μ RIF2], and tel1Δ [2 μ SAE2] (see MATERIALS AND METHODS). (A) Cell cultures exponentially growing in SC-ura + raf for >80 generations were shifted to SC-ura + raf + gal and samples were taken 14 h later to analyze telomere length. (B) Serial dilutions of SC-ura + raf exponentially growing cell cultures were spotted on YEPD and on YEP + raf + gal plates with or without MMS and HU at the indicated concentrations.
In contrast to what was observed with multicopy RIF2, 2 μ plasmids carrying the SAE2 gene caused a slight telomere elongation in wild-type cells both in raffinose and in galactose, as well as in galactose-induced GAL1-TEL1 cells (Figure 6A). Tel1 was likely required for Sae2-induced telomere lengthening, because telomere length was not influenced by multicopy SAE2 in tel1Δ cells or in GAL1-TEL1 cells grown in raffinose (Figure 6A). The effects of Sae2 high levels on telomere length (Figure 6A), hypersensitivity to DNA-damaging agents and growth rate (Figure 6B) were not abrogated in GAL1-TEL1 rad51Δ cells, indicating that Rad51-dependent homologous recombination was not required.
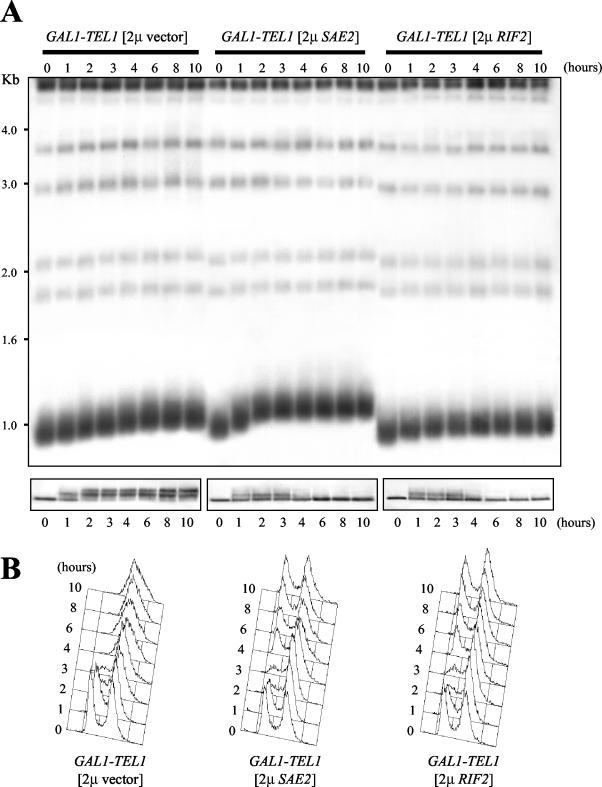
Thus, high levels of Sae2 and Rif2 have opposite effects on steady-state telomere length of galactose-induced GAL1-TEL1 cells despite they both suppress the Tel1-dependent cell cycle arrest. This suggests that the suppression mechanism might involve changes in the kinetics of the Tel1-induced telomere lengthening. Indeed, as shown in Figure 7, shift to galactose caused telomere elongation in both GAL1-TEL1 [2 μ SAE2] and GAL1-TEL1 [2 μ RIF2] cells, but a stable length was reached in these cells within 2 h, whereas GAL1-TEL1 cells carrying the empty vector were still elongating telomeres 8 h after the shift. Telomere ends in galactose-induced cells resuming cell cycle progression were stable but shorter in GAL1-TEL1 [2 μ RIF2] cells compared with GAL1-TEL1 [2 μ vector] cells, which in turn showed slightly shorter telomeres than GAL1-TEL1 [2 μ SAE2] cells under the same conditions. Although the shift to galactose activated the checkpoint in both GAL1-TEL1 [2 μ SAE2] and GAL1-TEL1 [2 μ RIF2] cells, the faster telomere length equilibration induced by Sae2 and Rif2 overexpression paralleled both faster checkpoint inactivation and resumption of cell cycle progression, compared with GAL1-TEL1 cells containing the empty 2 μ vector. In fact, Rad53 was phosphorylated immediately after shift to galactose in GAL1-TEL1 [2 μ SAE2] and GAL1-TEL1 [2 μ RIF2] cells, but the amount of phosphorylated Rad53 started to decrease (Figure 7A, bottom) and cells reentered cell cycle (Figure 7B) concomitantly with telomere length reaching a steady state.
Figure 7.
Kinetics of telomere elongation of galactose-induced GAL1-TEL1 cells in the presence of high copy number SAE2 and RIF2. GAL1-TEL1 [2 μ vector], GAL1-TEL1 [2 μ SAE2], and GAL1-TEL1 [2 μ RIF2] cell cultures exponentially growing in SC-ura + raf for >80 generations (time 0) were shifted to SC-ura + raf + gal and samples were taken at the indicated times to analyze telomere length (A, top), the pattern of Rad53 phoshorylation by Western blot by using anti-Rad53 antibodies (A, bottom) and DNA content by fluorescence-activated cell sorting analysis (B). In A, genomic DNA was digested with XhoI and hybridized with a poly(GT) telomere-specific probe.
Thus, reentry into the cell cycle of GAL1-TEL1 cells overexpressing SAE2 and RIF2 coincides with telomere length reequilibration.
Overexpression of a CDC13-EST1 Fusion Causes Telomere Lengthening and a Checkpoint-induced Cell Cycle Arrest
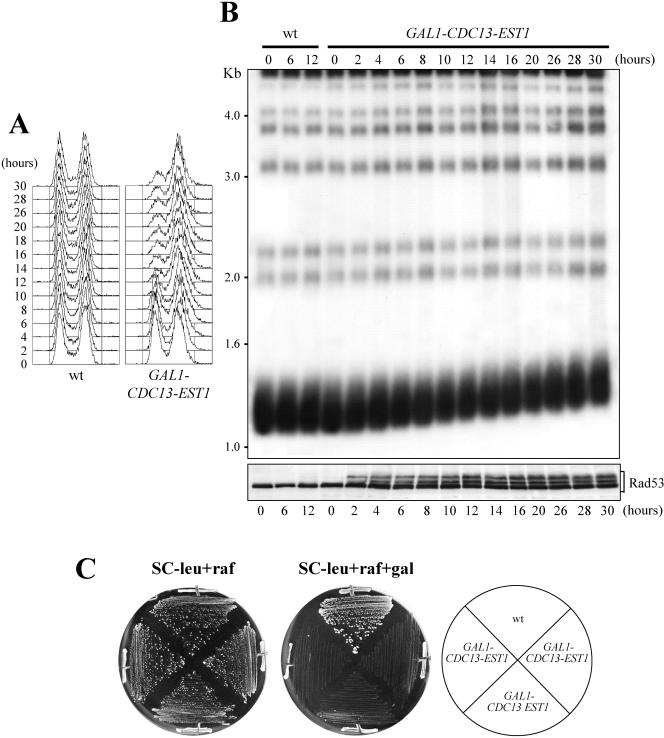
Based on the data described above, we hypothesized that sudden increase in telomere length might cause checkpoint-mediated cell cycle arrest. To gain independent supports to this hypothesis, we asked whether other conditions, besides GAL1-TEL1 overexpression in cells with short telomeres, could cause telomere lengthening and concomitant activation of a checkpoint response. Because it was shown that overexpression of EST2 and EST3 has no effect on telomere length, whereas high levels of the telomerase-associated Est1 protein caused slight telomere elongation after 75 generations (Friedman et al., 2003; Seto et al., 2003), we first analyzed galactose-induced GAL1-EST1 cells. These cells did not show any change in telomere length and did not arrest cell cycle progression within 36 h after shift to galactose (our unpublished data), suggesting that other telomeric proteins may be rate limiting for telomere elongation under these conditions. Because telomere length homeostasis can be maintained by restricting access of telomerase to chromosomal termini, this situation might be overcome by directly tethering telomerase to the ends. In S. cerevisiae, recruitment of telomerase to the chromosome termini depends on the telomere-binding protein Cdc13 (Nugent et al., 1996; Evans and Lundblad, 1999). It has been shown that fusing Cdc13 directly to the Est1 protein results in greatly elongated telomeres, consistent with a role for Est1 in mediating telomerase access to telomeres (Evans and Lundblad, 1999). We therefore analyzed the effects of overexpressing a GAL1-CDC13-EST1 fusion in otherwise wild-type cells. As shown in Figure 8, when cultures of these cells exponentially growing in raffinose were shifted to galactose-containing medium, most cells arrested with 2C DNA content (Figure 8A) and undivided nuclei (our unpublished data). This cell cycle arrest paralleled telomere lengthening (Figure 8B, top) and Rad53 phosphorylation (Figure 8B, bottom), indicating that the checkpoint was activated under these conditions. Thus, overexpression of CDC13-EST1 causes both telomere lengthening and activation of a Rad53-dependent checkpoint when induced in cells with full-length telomeres. Importantly, GAL1-CDC13-EST1 cells could not form colonies on galactose-containing plates (Figure 8C) and were still elongating telomeres and arrested with high levels of phosphorylated Rad53 even 30 h after galactose addition. This indicates that checkpoint activation persisted much longer in these cells than in GAL1-TEL1 overexpressing cells and may be due to the more direct effects of the Cdc13-Est1 fusion protein on telomerase activity compared with Tel1.
Figure 8.
Effects of CDC13-EST1 overexpression on telomere length and cell cycle progression. Strains were as follows: wild-type [ARS1 CEN4 vector], wild-type [ARS1 CEN4 GAL1-CDC13-EST1] (see MATERIALS AND METHODS). (A and B) Cell cultures exponentially growing in SC-leu + raf for >80 generations (time 0) were shifted to SC-leu + raf + gal and samples were collected at the indicated times to analyze DNA content by fluorescence-activated cell sorting analysis (A), telomere length (B, top), and the pattern of Rad53 phosphorylation by Western blot by using anti-Rad53 antibodies (B, bottom). In B, genomic DNA was digested with XhoI and hybridized with a poly(GT) telomere-specific probe. (C) Cells growing in SC-leu + glu for >80 generations were isolated on SC-leu + raf and on SC-leu + raf + gal plates that were incubated at 25°C for 4 d. Three independent GAL1-CDC13-EST1 transformants are shown.
DISCUSSION
Telomere length is maintained as a result of a dynamic equilibrium between lengthening and shortening. Telomere shortening can arise from either incomplete replication of the ends or telomere-specific processing activities such as active degradation. Sequence loss is counterbalanced by the addition of telomeric sequences by telomerase. Because telomeres are stable and not apparently detected as damaged DNA, whereas DNA strand breaks are signals for checkpoint-induced cell cycle arrest and/or repair, these natural chromosomal termini must have properties that functionally and structurally distinguish them from broken ends.
It has been proposed that telomeres normally interconvert between two states, uncapped and capped (Blackburn, 2000, 2001), in which the structure can become competent or inaccessible, respectively, to the action of telomerase or other enzymes. Telomere capping has been suggested to protect the physical integrity of the chromosomal termini and to avoid the telomeres from being recognized as broken ends. When telomere capping fails, the telomeres can be sensed as irreparable DNA double-strand breaks and may be the substrates for the actions of several enzymes that can lead to end-to-end fusion, degradation, and elongation by telomerase (reviewed in Dubrana et al., 2001; Kolodner et al., 2002). A detailed knowledge of the factors and the molecular mechanisms involved in end protection is crucial for understanding how cells distinguish between a double-strand break and a normal telomere end.
Telomere Lengthening and Checkpoint Activation
It has been suggested that telomere length, telomere-specific proteins, and/or the higher order telomeric DNA-protein complex may contribute to telomere capping and confer properties that keep these ends from being detected as DSBs (reviewed in Lundblad, 2000; Blackburn, 2001). Uncoupling telomere lengthening from capping would make these chromosome ends susceptible to DNA repair activities and suitable to elicit a DNA damage checkpoint response. For example, the lack of telomerase may cause uncapping, and the unprotected telomeres may signal to arrest cell cycle progression. In fact, as telomeres shorten in telomerase-deficient yeast cells, a DNA damage response leads to G2/M cell cycle arrest that is mediated by Mec1, whereas it does not seem to involve Tel1 (Enomoto et al., 2002; Ijpma and Greider, 2003). In agreement with these data, we observed that est2Δ cell cultures from spore clones progressively accumulate G2-arrested cells (our unpublished data) and show Rad53 phosphorylation (Figure 4).
In this study, we propose that a checkpoint response can be triggered also by sudden telomere lengthening. We show that induction of TEL1 overexpression in tel1Δ or yku70Δ cells with short telomeres causes telomere lengthening, which correlates with the activation of a Rad53-dependent checkpoint leading to G2/M cell cycle arrest. Moreover, both telomere elongation and checkpoint activation require telomerase, because they are partially relieved by deletion of the EST2 gene, encoding the catalytic telomerase subunit. Finally, checkpoint-induced cell cycle arrest and sudden telomere elongation can be triggered also by overexpression of a CDC13-EST1 fusion in otherwise wild-type cells. The telomere lengthening property of this fusion is likely a consequence of telomerase delivery to telomeres rather than of perturbation of Cdc13 function. In fact, it has been shown that the same extensive telomere lengthening is conferred also when only the DNA binding domain of Cdc13 was fused to Est1, whereas leaving the full-length Cdc13 intact (Evans and Lundblad, 1999).
Checkpoint-induced cell cycle arrest caused by telomere shortening requires Mec1 (Ijpma and Greider, 2003), whereas Mec1 does not seem to be involved in the TEL1 overexpression-dependent cell cycle arrest (Clerici et al., 2001). Because it is known that Mec1 and Tel1 have partially overlapping functions (Ritchie et al., 1999), increased Tel1 levels in our system might totally substitute for Mec1, thus explaining why Mec1 is not required for checkpoint activation by GAL1-TEL1.
Together, our data suggest that sudden telomerase-dependent increase in telomere length may cause disequilibrium between telomere sequence addition and the amount of telomere binding proteins available. Because telomerase-mediated extension of the TG1–3 strand is tightly coupled with the synthesis of the complementary C1–3A strand (Diede and Gottschling, 1999), uncontrolled telomerase action could produce long single-stranded DNA regions. These unprotected 3′ overhangs may be detected as DNA damage and trigger a checkpoint response (Maringele and Lydall, 2002).
TEL1 Overexpression and Telomere Elongation
The telomerase-mediated telomere lengthening caused by Tel1 overproduction suggests that Tel1 may activate the telomerase pathway, for example by increasing the ability of telomerase to access the telomere. In agreement with this hypothesis, artificial telomerase targeting to chromosomal ends causes telomere lengthening in the absence of Tel1 and Mec1, whereas both telomerase catalytic activity and Cdc13 binding to telomeres are unchanged (Chan et al., 2001; Tsukamoto et al., 2001). Together, these data suggest that Tel1 allows telomerase action in vivo by modulating its interaction with telomere substrate, rather than altering its catalytic properties.
Indeed, the MRX complex, which is required for Tel1-dependent checkpoint response to unprocessed DSBs (Usui et al., 2001) and acts in the same pathway as Tel1 to maintain telomere length (Ritchie and Petes, 2000), is required both to activate the checkpoint and to promote telomere elongation in galactose-induced GAL1-TEL1 cells (our unpublished data). Because the MRX complex has been proposed to prepare a 3′ single-stranded tail at the end of a telomere to be used as a telomerase substrate (Nugent et al., 1998; Diede and Gottschling, 2001; Lobachev et al., 2002), processing of the ends by MRX complex may be essential to elongate telomeres in the presence of high levels of Tel1.
On the other hand, an unprotected chromosome end can be channeled into recombination pathways (DuBois et al., 2002) that may extinct the damage signals arising from the dysfunctional ends and allow cells to overcome checkpoint arrest. Deletions of the LIF1 and LIF2 genes, which are necessary for nonhomologous end joining pathway (Herrmann et al., 1998; Vaillant and Marcand, 2001), neither suppress nor exacerbate the growth defects and hypersensitivity to DNA-damaging agents of galactose-induced GAL1-TEL1 cells (our unpublished data). This indicates that the NHEJ pathway may not be involved in cellular response to ectopic TEL1 overexpression. Conversely, we found that deletion of RAD51, which does not reduce telomere lengthening and checkpoint activation in galactose-induced GAL1-TEL1 cells, also enhances their cell lethality, suggesting that the homologous recombination repair pathway can protect cells from the effects caused by turning on GAL1-TEL1.
We observed telomere elongation and checkpoint activation only when TEL1 expression is turned on in cells with short telomeres. This suggests that short telomeres, although stable under unperturbed conditions, might be more susceptible to the action of telomerase, once TEL1 is overexpressed, than full-length telomeres. In fact, it has been proposed that, when telomeres shorten below a critical level, Rap1/Rif molecules are lost from the telomeres, which then shift the equilibrium at the terminus toward a structure permissive to telomeric sequence addition (reviewed in Lundblad, 2000; Blackburn, 2001).
Telomere Length Reequilibration and Checkpoint Inactivation
Tel1-dependent telomere lengthening is transient. When telomeres reach an equilibrium length, although Tel1 is still present at high levels, the checkpoint is inactivated and cell cycle progression resumes. This suggests that, once telomeres in galactose-induced GAL1-TEL1 cells reach a certain length, they might fold into a higher order structure. This configuration may limit telomere accessibility to telomerase or to telomere binding proteins required for telomerase action.
The mean length and length distribution of telomeres are determined by a balance between lengthening and shortening activities. It has been proposed that telomeres themselves control elongation by telomerase, when their length exceeds a certain threshold. In fact, telomere elongation is restricted to a few base pairs per generation and its rate decreases progressively with increasing telomere length (Marcand et al., 1999). The Rap1, Rif1, and Rif2 proteins, together with the gradual folding of the telomere into a restrictive higher order configuration, may regulate the gradual decline of telomerase activity. We propose that a sudden telomerase-dependent telomere lengthening can uncouple telomeric sequence addition from the binding of telomeric proteins that provide the capping functions necessary to protect telomere termini. Unprotection of the ends in turn elicits a checkpoint response. In this case, it should be possible to suppress checkpoint activation in galactose-induced GAL1-TEL1 cells by increasing the levels of telomeric proteins with capping functions. Indeed, we found that overproduction of Rif2 accelerates both reentry into the cell cycle and telomere length reequilibration in galactose-induced GAL1-TEL1 cells. We suggest that an excess of Rif2 may accelerate recapping of telomeric ends and/or prevent telomerase activity, either directly or through Tel1/MRX inhibition, thus blocking further telomere elongation and shutting off the checkpoint. Consistent with this, deletion of either RIF1 or RIF2 causes a telomerase-mediated lengthening of telomeric ends (Hardy et al., 1992; Wotton and Shore, 1997; Teng et al., 2000; Chan et al., 2001), which may occur because the lack of Rif proteins either relieves telomerase from an inhibitory effect or affects telomere capping. Capping properties of Rif proteins may be related to their suggested structural role in establishing a chromatin structure that limits telomerase access (Diede and Gottschling, 1999; Teng et al., 2000). Tel1 in turn may be required for the Rif-dependent inhibition of telomerase. In fact, telomere elongation in rif2Δ mutants is largely dependent on Tel1 (Craven and Petes, 1999; Chan et al., 2001), and the addition of synthetic Rap1-binding sites, which results in telomere shortening in wild-type cells, fails to cause the same effect in a tel1Δ strain (Ray and Runge, 1999).
High levels of the Sae2 protein, likely involved in processing DSBs (McKee and Kleckner, 1997; Rattray et al., 2001), concomitantly accelerate reentry into the cell cycle and resetting of stable telomere length in GAL1-TEL1 cells. Telomere length is increased in Sae2-overproducing cells compared with wild-type, suggesting that this protein might facilitate telomere length reequilibration and subsequent checkpoint switch off in galactose-induced GAL1-TEL1 cells by generating structures at telomeres that enhance Tel1/MRX action. It is worth mentioning that SAE2 deletion enhances the ability of DNA damage to activate Tel1 in the absence of Mec1 (Usui et al., 2001) and causes accumulation of unresected DSBs (McKee and Kleckner, 1997; Rattray et al., 2001). Based on the possible role of Sae2 in processing DSBs, it is tempting to speculate that an excess of Sae2 might promote processing of the chromosomal termini, thus facilitating the generation of telomeric ssDNA that triggers telomerase recruitment and action (Nugent et al., 1998; Diede and Gottschling, 2001).
Thus, high levels of Sae2 and Rif2 have opposite effects on telomere length, but they both facilitate telomere length reequilibration and checkpoint inactivation, indicating that not telomere length per se, but the protection of telomeric ends from further elongation is critical to allow checkpoint switch-off.
The finding that sudden telomerase-mediated telomere elongation can trigger a Rad53-dependent cell cycle arrest implies that cells must tightly control telomerase action during telomere replication. In vivo polymerization by telomerase occurs in the late S and G2/M cell cycle phases in cycling cells (McCarroll and Fangman, 1988). Accidental de novo telomeric DNA addition that may cause overelongation could be prevented by dynamic remodeling of telomeric proteins at telomeres (Smith et al., 2003). In this view, the checkpoint pathway might provide a protective mechanism against unregulated telomerase action leading to formation of unprotected telomere ends, which in turn may favor chromosome rearrangements.
Acknowledgments
We thank J. Diffley for antibodies against Rad53, T. Petes for the pSP21 plasmid, and V. Lundblad for pVL1091 plasmid and for helpful insights. Special thanks to S. Piatti and D. Lydall for critical reading of the manuscript to V. Geli and to all the members of our laboratory for useful discussions and criticisms. This work was supported by grants from Telethon-Italy (E.1247) and Associazione Italiana per la Ricerca sul Cancro to M.P.L. and from Consorzio Interuniversitario Biotecnologie and Fondo per gli Investimenti della Ricerca di Base to G.L.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-11-0719. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-11-0719.
References
- Abraham, R.T. (2001). Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15, 2177–2196. [DOI] [PubMed] [Google Scholar]
- Blackburn, E.H. (2000). Telomere states and cell fates. Nature 408, 53–56. [DOI] [PubMed] [Google Scholar]
- Blackburn, E.H. (2001). Switching and signaling at the telomere. Cell 106, 661–673. [DOI] [PubMed] [Google Scholar]
- Boulton, S.J., and Jackson, S.P. (1996). Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomere maintenance. Nucleic Acid Res. 24, 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton, S.J., and Jackson, S.P. (1998). Component of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17, 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, M., and Botstein, D. (1982). Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell 28, 145–154. [DOI] [PubMed] [Google Scholar]
- Chan, S.W.L., Chang, J., Prescott, J., and Blackburn, E.H. (2001). Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr. Biol. 11, 1240–1250. [DOI] [PubMed] [Google Scholar]
- Chen, L., Trujillo, K., Ramos, W., Sung, P., and Tomkinson, A.E. (2001). Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8, 1105–1115. [DOI] [PubMed] [Google Scholar]
- Clerici, M., Paciotti, V., Baldo, V., Romano, M., Lucchini, G., and Longhese, M.P. (2001). Hyperactivation of the yeast DNA damage checkpoint by TEL1 and DDC2 overexpression. EMBO J. 20, 6485–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, M.N., Wright, J.H., Wolf, A.J., and Zakian, V. (1990). RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell 63, 739–750. [DOI] [PubMed] [Google Scholar]
- Craven, R.J., and Petes, T.D. (1999). Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast Saccharomyces cerevisiae. Genetics 152, 1531–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours, D., and Jackson, S.P. (2001). The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15, 2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours, D., and Jackson, S.P. (2002). The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Mol. Cell. Biol. 3, 317–327. [DOI] [PubMed] [Google Scholar]
- Diede, S.J., and Gottschling, D.E. (1999). Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases α and δ. Cell 99, 723–733. [DOI] [PubMed] [Google Scholar]
- Diede, S.J., and Gottschling, D.E. (2001). Exonuclease activity is required for sequence addition and Cdc13 loading at de novo telomere. Curr. Biol. 11, 1336–1340. [DOI] [PubMed] [Google Scholar]
- DuBois, M.L., Haimberger, Z.M., McIntosh, M.W., and Gottschling, D.E. (2002). A quantitative assay for telomere protection in Saccharomyces cerevisiae. Genetics 161, 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrana, K., Perrod, S., and Gasser, S.M. (2001). Turning telomeres off and on. Curr. Opin. Cell Biol. 13, 281–289. [DOI] [PubMed] [Google Scholar]
- Enomoto, S., Glowczewski, L., and Berman, J. (2002). MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 2626–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, S.K., and Lundblad, V. (1999). Est1 and Cdc13 as comediators of telomere access. Science 286, 117–120. [DOI] [PubMed] [Google Scholar]
- Evans, S.K., Bertuch, A.A., and Lundblad, V. (1999). Telomeres and telomerase: at the end, it all comes together. Trends Cell Biol. 9, 329–331. [DOI] [PubMed] [Google Scholar]
- Friedman, K.L., Heit, J.J., Morgan, D.M., and Cech, T.R. (2003). N-terminal domain of yeast telomerase reverse transcriptase: recruitment of Est3p to the telomerase complex. Mol. Biol. Cell 14, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and Sugino, A. (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six base-pair restriction sites. Gene 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Gilbert, C.S., Green, C.M., and Lowndes, N.F. (2001). Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8, 129–136. [DOI] [PubMed] [Google Scholar]
- Gilson, E., Roberge, M., Girardo, R., Rhodes, D., and Gasser, S.M. (1993). Distortion of the DNA double helix by RAP1 at silencers and multiple telomeric binding sites. J. Mol. Biol. 231, 293–310. [DOI] [PubMed] [Google Scholar]
- Gravel, S., Larrivee, M., Labrecque, P., and Wellinger, R.J. (1998). Yeast Ku as a regulator of chromosomal DNA end structure. Science 280, 741–745. [DOI] [PubMed] [Google Scholar]
- Greenwell, P.W., Kronmal, S.L., Porter, S.E., Gassenhuber, J., Obermaier, B., and Petes, T.D. (1995). TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human Ataxia Telangiectasia gene. Cell 82, 823–829. [DOI] [PubMed] [Google Scholar]
- Greider, C.W. (1996). Telomere length regulation. Annu. Rev. Biochem. 65, 337–365. [DOI] [PubMed] [Google Scholar]
- Grenon, M., Gilbert, C.S., and Lowndes, N.F. (2001). Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3, 844–847. [DOI] [PubMed] [Google Scholar]
- Haber, J.E. (1998). The many interfaces of Mre11. Cell 95, 583–586. [DOI] [PubMed] [Google Scholar]
- Hardy, C.F.J., Sussel, L., and Shore, D. (1992). A Rap1-interacting protein involved in silencing and telomere length regulation. Genes Dev. 6, 801–814. [DOI] [PubMed] [Google Scholar]
- Herrmann, G., Lindahl, T., and Schar, P. (1998). Saccharomyces cerevisiae LIF1: a function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J. 17, 4188–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijpma, A.S., and Greider, C.W. (2003). Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner, R.D., Putnam, C.D., and Myung, K. (2002). Maintenance of genome stability in Saccharomyces cerevisiae. Science 297, 552–557. [DOI] [PubMed] [Google Scholar]
- Kyrion, G., Boakye, K.A., and Lustig, A.J. (1992). C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 5159–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, S., Moore, J.K., Haber, J.E., and Greider, C.W. (1999). RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152, 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner, J., Hughes, T.R., Shevchenko, A., Mann, M., Lundblad, V., and Cech, T.R. (1997). Reverse transcriptase motifs in the catalityc subunit of telomerase. Science 276, 561–567. [DOI] [PubMed] [Google Scholar]
- Lobachev, K.S., Gordenin, D.A., and Resnick, M.A. (2002). The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangement. Cell 108, 183–193. [DOI] [PubMed] [Google Scholar]
- Longhese, M.P., Foiani, M., Muzi Falconi, M., Lucchini, G., and Plevani, P. (1998). DNA damage checkpoint in budding yeast. EMBO J. 17, 5525–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese, M.P., Paciotti, V., Fraschini, R., Zaccarini, R., Plevani, P., and Lucchini, G. (1997). The novel DNA damage checkpoint protein Ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 16, 5216–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese, M.P., Paciotti, V., Neecke, H., and Lucchini, G. (2000). Checkpoint proteins influence telomeric silencing and length maintenance in budding yeast. Genetics 155, 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad, V. (2000). DNA ends: maintenance of chromosome termini versus repair of double strand breaks. Mutat. Res. 451, 227–240. [DOI] [PubMed] [Google Scholar]
- Marcand, S., Brevet, V., and Gilson, E. (1999). Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 18, 3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand, S., Gilson, E., and Shore, D. (1997). A protein-counting mechanism for telomere length regulation in yeast. Science 275, 986–990. [DOI] [PubMed] [Google Scholar]
- Maringele, L., and Lydall, D. (2002). EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Δ mutants. Genes Dev. 16, 1919–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll, R.M., and Fangman, W.L. (1988). Time of replication of yeast centromeres and telomeres. Cell 54, 505–513. [DOI] [PubMed] [Google Scholar]
- McClintock, B. (1941). The stability of broken ends of chromosomes in Zea mays. Genetics 26, 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B. (1942). The fusion of broken ends of chromosomes following nuclear fusion. Proc. Natl. Acad. Sci. USA 28, 458–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee, A.H., and Kleckner, N. (1997). A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediates stages of meiotic prophase and characterization of a new gene. SAE2. Genetics 146, 797–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, D.M., Tagle, D.A., Shiloh, Y., Collins, F.S., and Hieter, P. (1995). TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82, 831–840. [DOI] [PubMed] [Google Scholar]
- Nugent, C.I., Bosco, G., Ross, L.O., Evans, S.K., Salinger, A.P., Moore, J.K., Haber, J.E., and Lundblad, V. (1998). Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 8, 657–660. [DOI] [PubMed] [Google Scholar]
- Nugent, C.I., Hughes, T.R., Lue, N.F., and Lundblad, V. (1996). Cdc13p: a single-strand telomeric DNA-binding protein with a dual roles in yeast telomere maintenance. Science 274, 249–251. [DOI] [PubMed] [Google Scholar]
- Nyberg, K.A., Michelson, R.J., Putnam, C.W., and Weinert, T.A. (2002). Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36, 617–656. [DOI] [PubMed] [Google Scholar]
- Paciotti, V., Clerici, M., Lucchini, G., and Longhese, M.P. (2000). The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 14, 2046–2059. [PMC free article] [PubMed] [Google Scholar]
- Paques, F., and Haber, J.E. (1999). Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, S.E., Greenwell, P.W., Ritchie, K.B., and Petes, T.D. (1996). The DNA-binding protein Hdf1 (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acid Res. 24, 582–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray, A.J., McGill, C.B., Shafer, B.K., and Strathern, J.N. (2001). Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for. SAE2/COM1. Genetics 158, 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, A., and Runge, K.W. (1999). Varying the number of telomere-bound proteins does not alter telomere length in tel1Δ cells. Proc. Natl. Acad. Sci. USA 96, 15044–15049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, K.B., and Petes, T.D. (2000). The Mre11/Rad50/Xrs2 complex and the Tel1 function in a single pathway for telomere maintenance in yeast. Genetics 155, 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, K.B., Mallory, J.C., and Petes, T.D. (1999). Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 6065–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., and Hieter, P. (1990). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Rouse, J., and Jackson, S.P. (2002). Interfaces between the detection, signaling, and repair of DNA damage. Science 297, 547–551. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., Bachant, J., Wang, H., Hu, F.H., Liu, D., Tezlaff, M., and Elledge, S.J. (1999). Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science 286, 1166–1171. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., Desany, B.A., Jones, W.J., Liu, Q., Wang, B., and Elledge, S.J. (1996). Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271, 357–360. [DOI] [PubMed] [Google Scholar]
- Seto, A.G., Livengood, A.J., Tzfati, Y., Blackburn, E.H., and Cech, T.R. (2003). A buldged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev. 16, 2800–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh, Y. (2001). ATM and ATR: networking cellular responses to DNA damage. Curr. Opin. Genet. Dev. 11, 71–77. [DOI] [PubMed] [Google Scholar]
- Smith, C.D., Smith, D.L., DeRisi, J.L., and Blackburn, E.H. (2003). Telomeric protein distributions and remodeling through the cell cycle in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 556–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, S.C., Chang, J., McCowan, B., and Zakian, V. (2000). Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6, 947–952. [DOI] [PubMed] [Google Scholar]
- Tsukamoto, Y., Taggart, A.K.P., and Zakian, V.A. (2001). The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11, 1328–1335. [DOI] [PubMed] [Google Scholar]
- Usui, T., Ogawa, H., and Petrini, J.H.J. (2001). A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7, 1255–1266. [DOI] [PubMed] [Google Scholar]
- Vaillant, M.F., and Marcand, S. (2001). NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the Ligase IV pathway. Genes Dev. 15, 3005–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Pohlmann, R., and Philippsen, P. (1994). New heterologous modules for classical or PCR-based gene disruption in Saccharomyces cerevisiae. Yeast 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wellinger, R.J., and Sen, D. (1997). The DNA structures at the ends of eukaryotic chromosomes. Eur. J. Cancer 33, 735–749. [DOI] [PubMed] [Google Scholar]
- Wood, R.D., Mitchell, M., Sgouros, J., and Lindahl, T. (2001). Human DNA repair genes. Science 291, 1284–1289. [DOI] [PubMed] [Google Scholar]
- Wotton, D., and Shore, D. (1997). A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11, 748–760. [DOI] [PubMed] [Google Scholar]
- Zakian, V.A. (1996). Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet. 30, 141–172. [DOI] [PubMed] [Google Scholar]
- Zhou, B.S., and Elledge, S.J. (2000). The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439. [DOI] [PubMed] [Google Scholar]