Abstract
Mammalian LGN/AGS3 proteins and their Drosophila Pins orthologue are cytoplasmic regulators of G-protein signaling. In Drosophila, Pins localizes to the lateral cortex of polarized epithelial cells and to the apical cortex of neuroblasts where it plays important roles in their asymmetric division. Using overexpression studies in different cell line systems, we demonstrate here that, like Drosophila Pins, LGN can exhibit enriched localization at the cell cortex, depending on the cell cycle and the culture system used. We find that in WISH, PC12, and NRK but not COS cells, LGN is largely directed to the cell cortex during mitosis. Overexpression of truncated protein domains further identified the Gα-binding C-terminal portion of LGN as a sufficient domain for cortical localization in cell culture. In mitotic COS cells that normally do not exhibit cortical LGN localization, LGN is redirected to the cell cortex upon overexpression of Gα subunits of heterotrimeric G-proteins. The results also show that the cortical localization of LGN is dependent on microfilaments and that interfering with LGN function in cultured cell lines causes early disruption to cell cycle progression.
INTRODUCTION
In mammals, two proteins that are related to Drosophila Partner of Inscuteable (Pins) have been identified (Mochizuki et al., 1996; Takesono et al., 1999). They define a class of cytoplasmic nonreceptor-linked regulators of G-protein signaling. Members of this class of proteins generally contain two types of repeats: seven tetratricopeptide repeats (TPR) at the amino-terminus and three Gαi/o-Loco (GoLoco) repeats at the carboxy-terminus. TPR motifs usually mediate protein-protein interactions (Blatch and Lassle, 1999), whereas GoLoco motifs are responsible for association with Gα subunits of heterotrimeric G-proteins (Siderovski et al., 1999; Natochin et al., 2000; Bernard et al., 2001). These cytoplasmic signaling regulators behave as guanine dissociation inhibitors (GDI), preventing the exchange of GDP-bound for GTP-bound Gα (DeVries et al., 2000; Peterson et al., 2000).
In Drosophila, Pins was originally discovered as a protein that interacts with Inscuteable (Insc) in dividing neuroblasts (NBs; Schaefer et al., 2000; Yu et al., 2000) and that both Pins and Insc (Kraut et al., 1996) are asymmetrically localized to the apical cortex of NBs during mitosis and play important roles in the localization of basal cell fate determinants and mediate correct spindle orientation in dividing NBs (Kraut et al., 1996; Schaefer et al., 2000; Yu et al., 2000). The apical cortical localization of Pins in dividing Drosophila NBs depends not only on the N-terminal sequences that interact with Insc but also on the C-terminal region that binds to Gα subunit of heterotrimeric G-proteins. However, in epithelial cells, which lack Insc expression, Pins associates with the lateral cortex instead (Schaefer et al., 2000; Yu et al., 2000).
The subcellular localization of the mammalian proteins related to Drosophila Pins, AGS3 and LGN, has also been recently reported. As for AGS3, the protein is reported to be primarily cytoplasmic throughout the cell cycle (Blumer et al., 2002). A truncated version of AGS3 (AGS3-Short) lacking the N-terminal TPR repeats has also been identified (Pizzinat et al., 2001). It is enriched in the heart and shows a subcellular cytoplasmic distribution that is different from AGS3. The localization data suggested that the TPR domains might account for the differences between a homogeneous cytoplasmic staining for AGS3-Short and a punctate cytoplasmic distribution for AGS3. As for LGN, its subcellular localization has been reported by two separate studies using reagents based on the human LGN sequence. In one study, Blumer et al. (2002) used PC12 cell to show that LGN exhibits dramatic differences in its localization at specific stages of the cell cycle. The authors have shown that LGN moves from the nucleus to the midbody structure separating daughter cells during the later stages of mitosis, suggesting a role in cytokinesis. In another study, Du et al. (2001) have shown that LGN, unlike Drosophila Pins, accumulates at the spindle poles of dividing polarized MDCK cells (Du et al., 2001). The authors have also shown that LGN plays essential roles in the assembly and organization of the mitotic spindle via binding to the nuclear mitotic apparatus protein NuMA, which tethers spindles at the poles (Du et al., 2001, 2002).
The mammalian LGN has so far been shown to assume various subcellular localizations that are different than that reported for fly Pins. Using reagents generated based on the LGN sequence from mouse, we have further characterized LGN localization profile in various cell lines. We show here that similar to fly Pins, transfected tagged versions of mouse LGN as well as endogenous LGN are found enriched at the cortex of some, but not all cell lines tested in a cell cycle–dependent manner. Furthermore, we show that the C-terminal GoLoco-containing domain of LGN is sufficient for cortical localization. We also report that factors affecting the cortical localization of LGN include microfilaments and the Gα subunits of heterotrimeric G-proteins. Our data show that overexpression of the mouse LGN or prevention of LGN translation in cell lines results in cell cycle arrest. We discuss possibilities underlying the different localization data observed for LGN.
MATERIALS AND METHODS
Plasmid Constructs and Cell Transfection
The generation of full-length Pins-related LGN sequence from mouse, m-LGN (or m-Pins) cDNA (accession number AY081187), was initially based on a partial EST sequence (AA543923; IMAGE: 949074) and is described elsewhere (Yu et al., 2003). Another EST (BC021308; IMAGE: 5007832) encoding the full-length mouse LGN protein homologue is also found in the database. FLAG-tagged versions of full-length, N-terminal (aa1–384) and C-terminal (aa385–650) mouse LGN were generated by cloning into the BamHI/XhoI sites of pXJ40 (Manser et al., 1997) vector and expressing them under CMV promoter. The His-Gαi3 fusion construct (in pQE60) was a kind gift from Chen Canhe (Chen et al., 1997, 2001). The mouse Gαo transfection construct was kindly provided by Graeme Milligan (Hoffmann et al., 2001).
For transfection, Lipofectamine from Life Technologies-BRL (Life Technologies, Rockville, MD) was used according to manufacturer's instructions. Cells were seeded at ∼1–3 × 105 per 35-mm tissue culture plates in 2 ml of appropriate medium and kept at 37°C in a CO2 incubator until a confluency of 50–80% was reached, typically ∼18–24 h. For each transfection, 1–2 μg of DNA was diluted in 100 μl OptiMEM and mixed with solution B containing 30 μl of Lipofectamine in 100 μl OptiMEM. The mixture was incubated at room temperature for 45 min to form DNA-liposome complex. OptiMEM, 0.8 ml, was then added to the complex and the diluted complex was overlayed on the rinsed cells. The cells were incubated with the complex for 6 h at 37°C in a CO2 incubator after which the fresh serum-containing medium was added to the cells. The cells were typically recovered for fixing and further analysis after ∼20 h. Typically, ∼20–40% of the cells showed expression of the transfected plasmid, depending on the cell line used. The transfection experiments were repeated three times. From each experiment, ∼100 interphasic and ∼25 mitotic transfected cells were counted. The percentage of cells showing a particular subcellular localization of the transfected protein was calculated based on the data from all three experiments.
Cell Cultures and Immunoblotting
The human amniotic-derived cell line WISH (ATCC CCL 25), monkey kidney COS-1 and normal rat kidney NRK cells were grown in DMEM medium supplemented with 2 mM glutamine, 5% FCS, 100 U/ml penicillin, and 10 μg/ml streptomycin. The mouse neuronal-derived PC12 cell line was grown as described by Greene and Tischler (1976). For immunofluorescence microscopy, coverslips were coated with 20 μg/ml laminin (Upstate Biotechnology, Lake Placid, NY) for 1 h at 37°C before seeding the cells. MDCK cells (strain II), a polarized epithelial cell line derived from dog kidney, were cultured on polycarbonate filters (transwell clear, Costar, Cambridge, MA) in DMEM medium supplemented with 10% fetal bovine serum and 1% penicillin (100 U/ml)/10 μg/ml streptomycin. The cells were plated at 2.5 × 106 cells/filter for 2 d before fixing and processing for immunofluorescence.
About 100 μg of each total cell lysate was separated on 10 or 12% SDS-PAGE and then transferred onto nitrocellulose membrane using wet transfer system from Bio-Rad (Richmond, CA). The membrane was then blocked with 5% milk at 4°C overnight after which primary antibody incubation was carried out for 2 h at room temperature. The dilution for primary antibody was typically 1 μg/ml. Secondary anti-HRP antibody from rabbit (Vector Laboratories, Burlingame, CA) was used at 1:10,000 dilution and followed by detection of bands using ECL detection kit from Amersham (Amersham, Buckinghamshire, UK).
Fixation and Immunofluorescence
Cells were washed in PBS and fixed in either 3.7% paraformaldehyde for 20 min at room temperature or kept in chilled methanol for 10 min at 4°C. They were then permeabilized with PBS containing 0.1% Triton X-100 for 5 min and blocked in 10% FBS for 1 h at room temperature. Fixed cells were incubated with primary antibody at 4°C overnight, washed with PBS, and then treated with fluorescent secondary antibody for 2 h at room temperature. To stain DNA, TOPRO-3 dye from Molecular Probes (Eugene, OR) was used. Coverslips were mounted with Vectashield (Vector Laboratories) and examined under confocal microscopy (Bio-Rad, MRC1024).
For drug treatments, WISH cells were plated on the previous day on 35-mm dishes and allowed to grow till 80% confluent. For microfilaments depolymerization, cells were washed with PBS and incubated with serum-free medium (SFM) with or without 1 μM latrunculin A (Molecular Probes) for 60 min in a CO2 incubator (Rosin-Arbesfeld et al., 2001). For microtubules depolymerization, cells were washed with PBS and then overlaid with or without 0.5 μg/ml colchicine (Sigma, St. Louis, MO) in complete culture medium. The cells were then cultured for 24 h in the CO2 incubator. The treated cells were rinsed twice with PBS and then fixed for 20 min in 4% paraformaldehyde or for 10 min with chilled methanol at –20°C before proceeding with the immunofluorescence analysis.
Antibodies
Polyclonal anti-LGN antibodies were produced in rabbits by injecting a GST-fusion product containing 194 amino acids from the C-terminal domain of mouse LGN (aa478–672). The anti-LGN antibodies were affinity purified. For this purpose, ∼100 μg of the GST-fusion protein was run on a 12% SDS-PAGE, transferred onto a nitrocellulose membrane, and stained with Ponceau red for 5 min. The membrane strip containing the fusion protein was cut, washed with PBS, and incubated with 10 ml serum at 4°C for 12–14 h. After rinsing with PBS, the antigen-bound antibody was eluted in 200 μl elution buffer (Pierce, Rockford, IL; Cat. No. 21009) and the eluted product was immediately neutralized with 1 M Tris, pH 9.5. Finally, the eluted antibody was recovered in the supernatant after spinning at 14,000 rpm for 10 min. The affinity-purified anti-LGN antibody was used in immunofluorescence and on Western blots at final concentrations of 2 μg/100 μl and 1 μg/μl, respectively.
Rabbit polyclonal (Affinity Bioreagents, Cambridge, UK; Cat. No. PA1–984) and mouse monoclonal (M2 from Sigma, St. Louis, MO) anti-FLAG antibodies were used at 2 μg/100 μl and 1 μg/100 μl, respectively. Other antibodies used were Vti1α at 1 μg/100 μl (BD Transduction Laboratories, Lexington, KY; Cat. No. V85620–050), ZO-1 at 1 μg/100 μl (Zymed, South San Francisco, CA; Cat. No. 61–7300), β-Catenin at 1 μg/100 μl (Transduction Labs; Cat. No. C19220–050), β-tubulin at 1:5 dilution (E7, hybridoma bank), rabbit Gαi3 (Upstate Biotechnology; Cat. No. 06–270) and rabbit Gαi1–2 at 1:50 dilution (Upstate Biotechnology; Cat. No. 06–236) and monoclonal mouse Gαo MAB 0371 at 1:50 dilution (Chemicon, Temecula, CA).
Rabbit (Cat. No. 711–095-152 and 711–165-152) and mouse (Cat. No. 715–095-150 and 715–165-150) FITC- and Cy3-conjugated goat IgG (AffiniPure, H+L) secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). TRITC-Phalloidin at 0.1 mg/ml (Sigma; Cat. No. P1951) and goat polyclonal antiactin antibody (I-19) at 1 μg/ml (Santa Cruz Biotechnology, Santa Cruz, CA; Ca. No. sc-1616) were used to detect actin. A secondary bovine anti-goat IgG HRP antibody (Santa Cruz; Cat. No. sc-2350) was also used at 0.4 μg/ml for immunoblots.
Protein Binding and GDI Assay
Direct binding between mouse LGN and various Gα subunits was assayed using His columns. Various 35S-labeled LGN protein products were generated using the TnT-coupled transcription/translation kit from Promega (Madison, WI) and incubated with 1 μg His-Gαi2/3/o protein and His beads in 250 μl binding buffer (20 mM Tris 7.5, 70 mM NaCl, 1 mM DTT, 0.6 mM EDTA, 0.01% Triton X, 20 μM GDP) for 40 min at 25°C. The reaction mixtures were washed at room temperature, and protein complexes were analyzed on acrylamide gels. Protein gels were then dried under vacuum and autoradiographed.
To assay the GDI activity of mouse LGN, [35S]GTPγ binding experiments were performed with some modification to the protocol described by DeVries et al. (2000). Reaction mixtures containing 50 nM His-Gαi3/o-GDP, and 1 μM GST-LGN (aa385–672) or control GST were incubated in buffer A (50 mM Tris, pH 8.0, 1 mM DTT, 1 mM EDTA, 10 mM MgSO4). Experiments were started by adding 2 μM [35S]GTPγ in 50-μl reaction volume and incubated at 30°C for different time periods. The reactions were terminated by washes with ice-cold buffer before measuring the scintillation counts.
BrdU Labeling and Morpholino Treatment
For BrdU labeling, cells were transfected with various mouse LGN constructs (FL-FLAG, N-FLAG, or C-FLAG). Thirty-six hours after transfection, the cells were incubated for 60 min in 1 mM BrdU, fixed, and then stained with anti-FLAG (Affinity Bioreagents) and anti-BrdU (Boehringer Mannheim, Indianapolis, IN) antibodies according to manufacturer's instructions. Transfected cells positive for FLAG were scored for BrdU staining under confocal microscopy. Typically, ∼100 cells were counted for each experiment. The experiments were repeated three times to obtain the percentage of FLAG-positive cells that are labeled with BrdU.
Morpholino treatment of cells was performed as described by Gene-Tools (Eugene, OR). The antisense morpholino sequence was 5′ GAATGGTCTTCCCTCATGCTTATCA-3′ (overlapping the ATG start codon, underlined) and was traced within the cell with its 3′ carboxy-fluorescein modification. Typically, morpholino phosphorodiamidate oligonucleotides (MOs) contain ∼25 bps overlapping with the first AUG translational start site. They have a high affinity for RNA, although they do not recruit RNAseH but exhibit high efficacy through nonclassical antisense approach (Summerton, 1999; Larson and Ekker, 2001). Morpholino oligos can block translation of mRNA by steric blocking, preventing assembly of a functional ribosome complex. The delivery of morpholino to cells was performed as per manufacturer's protocol. Briefly, the special delivery solution was mixed with morpholino for 15 min at room temperature and then added to the cells for ∼3 h, after which the cells were allowed to recover for ∼20 h before being harvested for further analyses. Harvested cells were fixed and analyzed by FACScan using WinMDI3.5 software. A Becton Dickinson (Lincoln Park, NJ) FACScan machine was used to acquire and analyze 21,000 events (using Cellquest and Modfit). DNA analysis was by propidium iodide staining (50 μg/ml at 37°C for 30 min). The morpholino experiment was repeated three times, and each time it was done in triplicates.
Morpholino-treated cells were also analyzed by immunostaining and immunoblotting using affinity-purified anti-LGN antibodies to determine the effect of morpholino on LGN protein levels. For the immunoblots, the band intensity was quantified using the Bio-Rad multianalyst version-1 software and Bio-Rad (model GS-700) imaging densitometer. For the immunostaining, ∼100 mitotic morpholino-treated cells were counted in each experiment in order to determine the percentage of morpholino-treated cells showing loss of LGN staining.
RESULTS
LGN-FLAG Is Enriched at the Cell Cortex in Mitotic WISH, PC12, and NRK But Not in COS Cell Cultures
Fly Pins displays cortical localization in dividing cells (Schaefer et al., 2000; Yu et al., 2000), whereas the mammalian LGN homologue localizes to spindle poles (Du et al., 2001) or midbody structures (Blumer et al., 2002), depending on the cell lines used. These findings prompted us to further characterize the subcellular localization profile of LGN in various cell culture systems. As a first step, we carried out an overexpression study of an LGN-FLAG construct in various cell line systems and followed the localization of exogenous LGN during mitosis using anti-FLAG antibody. Depending on the cell line used, the overexpression experiments revealed a cortical localization profile for LGN during mitosis that has not been reported before (Figures 1 and 2).
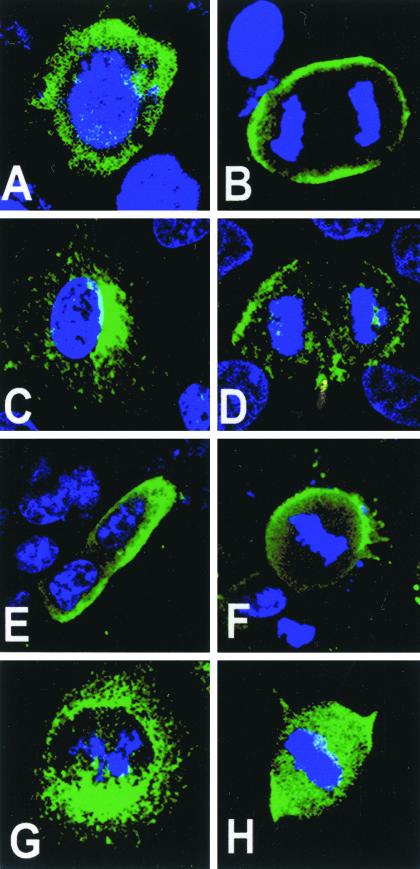
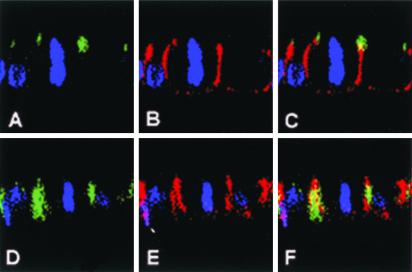
Figure 1.
Overexpression and localization of exogenous LGN-FLAG in cell lines. LGN-FLAG was overexpressed in WISH (A and B), NRK (C and D), PC12 (E and F), and COS (G and H) cells. LGN localization was detected with anti-FLAG (green) and DNA was stained with TOPRO3 (blue). (A, C, E, and G) Interphase cells; (B, D, F, and H) mitotic cells. WISH, NRK, and PC12 cells localize LGN-FLAG to the perinucleus/cytoplasm in interphase and to the cell cortex during mitosis. In mitotic COS, LGN remains cytoplasmic (H).
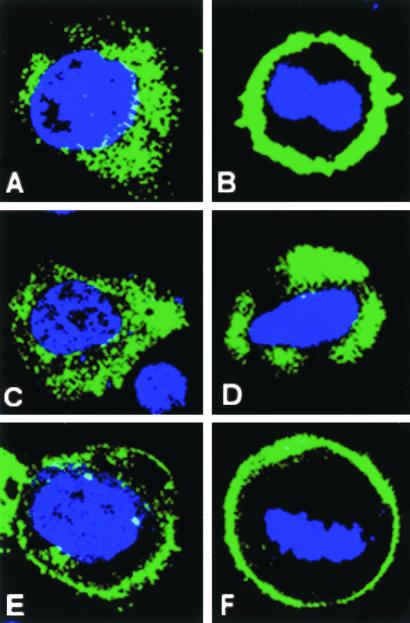
Figure 2.
Domain dissection of LGN. FLAG-tagged LGN constructs were transfected into WISH cells, and their subcellular localization was detected using anti-FLAG antibody (green). (A) Perinuclear/cytoplasmic staining for FL-LGN at interphase; (B) a cortical localization for FL-LGN at metaphase. N-FLAG (amino acids 1–384) remains mainly in the cytoplasm during interphase (C) and metaphase (D). C-FLAG (amino acids 385–672) is enriched at the cell cortex during both interphase (E) and metaphase (F). Residual cytoplasmic/perinuclear staining for C-FLAG can also be seen at interphase (E). The cell cycle stage was determined by DNA staining (blue).
In WISH cells, the full-length LGN-FLAG (FL-FLAG) localized to the perinuclear region/cytoplasm during interphase (in 100% of transfected cells) and was mainly redirected to the cortex (in 90% of the transfected cells) during mitosis (Figures 1, A and B, and 2, A and B). In NRK cells, LGN-FLAG was also largely perinuclear during interphase (Figure 1C) and enriched at the cell cortex during mitosis (Figure 1D). A similar cell cycle–dependent localization profile for LGN-FLAG was also observed in PC12 cells (Figure 1, E and F). COS cells, on the other hand, did not show cortical localization for LGN-FLAG as they enter mitosis. In these cells, LGN-FLAG was perinuclear during interphase (Figure 1G) and remained in the cytoplasm of all transfected cells during mitosis (Figure 1H). In these experiments, higher amounts of cortical LGN accumulated at the cortex of dividing WISH cells compared with other cell lines used.
The C Terminus of Mouse LGN Is Sufficient for Cortical Localization
Pins and Pins-related proteins contain N-terminal TPR repeats and C-terminal GoLoco repeats. Their N-terminal TPR repeats have been shown to be involved in the interaction with other proteins such as Insc and NuMA (Schaefer et al., 2000; Yu et al., 2000; Du et al., 2001) whereas the C-terminal GoLoco repeats mediate binding to Gα subunits of heterotrimeric G-proteins (DeVries et al., 2000). In flies, The C termini of Pins have been shown to be required for cortical localization of the protein (Yu et al., 2002).
To dissect the domains responsible for directing LGN to different localization sites within the cell, we generated and expressed constructs containing various FLAG-tagged fragments of the mouse LGN protein in WISH cells and assayed their localization during the cell cycle using anti-FLAG antibody (Figure 2). Transfected cells expressing the N-terminus (N-FLAG; aa1–384) or the C termini (C-FLAG; amino acids 385–672) of LGN showed different localization profiles. For N-FLAG, ∼95% of transfected interphasic cells showed predominantly cytoplasmic staining (Figure 2C), whereas the remainder ∼5% exhibited nuclear localization. However, ∼99% of transfected mitotic cells failed to localize N-FLAG to their cortex (Figure 2D). In contrast for C-FLAG, ∼99% of transfected cells showed predominantly cortical staining throughout the cell cycle from interphase to telophase (Figure 2, E and F). These experiments indicate that the C termini of LGN contains a cortical localization signal, similar to that reported for fly Pins (Yu et al., 2002).
Endogenous LGN Also Localizes to the Cortex of Mitotic PC12 and WISH But Not COS Cells
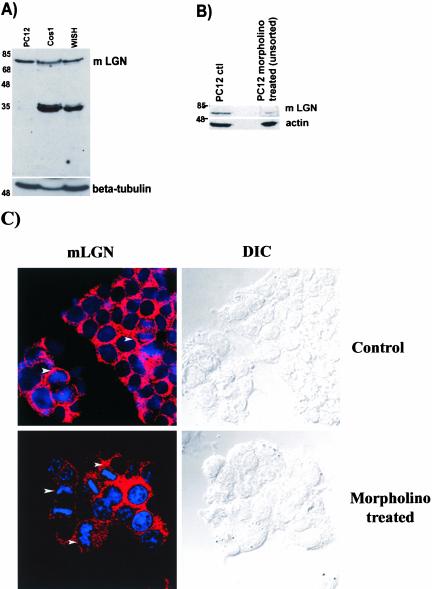
The overexpression studies showed preferential cortical localization for LGN-FLAG during mitosis in WISH, PC12, and NRK but not COS cell lines. In order examine the localization profile of endogenous LGN in these cell lines, anti-LGN antibodies were raised against mouse LGN in rabbits and used for immunostaining. As a first approach to determine the specificity of the antibody, affinity-purified anti-LGN antisera were used on immunoblots to probe total protein extracts from PC12, WISH, and COS cells. In this experiment, a single band of the expected LGN size was detected in the PC12 cell extract (Figure 3A). The intensity of the single LGN band obtained in the PC12 cell extract was reduced fivefold upon treatment of PC12 cells with an LGN-specific morpholino before blotting, suggesting that the anti-LGN antibody is specific (Figure 3B). In addition, treatment with the LGN-specific morpholino caused loss of cortical staining detected with anti-LGN in ∼26% of the mitotic PC12 cells (Figure 3C), further suggesting that the antibody is LGN specific. WISH and COS cell extracts, on the other hand, showed in addition to the LGN band other lower size bands (Figure 3A), similar to the previously reported data by Blumer et al. (2002). No protein bands were detected in PC12, WISH, or COS cells extracts on the immunoblot when a preimmune serum was used or when the cell extracts were subjected to antigen absorption treatment before immunoblotting (our unpublished results), again indicating the LGN antibody is specific. Additional lower molecular weight bands for LGN were also reported by other researchers (Blumer et al., 2002), and they are either LGN degradation products or possibly LGN isoforms produced by alternative splicing events. Nevertheless, the single LGN band obtained in PC12 cells, the drastic reduction of its intensity upon treatment of PC12 cells with an LGN-specific morpholino and the loss of cortical LGN staining in the treated cells suggest that the anti-LGN antibody is specific.
Figure 3.
Immunoblot analysis. Total protein extracts, 100 μg, were loaded in each lane and immunoblotted. (A) Protein extracts from PC12, COS, and WISH cell lines were probed with anti-LGN antibody at a concentration of 1 μg/ml. Anti-LGN detects a prominent band of the expected LGN molecular weight in the PC12 extract. COS and WISH cell extracts show the LGN band and other additional lower size bands. β-Tubulin is used as a protein-loading marker. (B) An immunoblot of PC12 cell extracts before and after morpholino treatment. The intensity of the LGN band is reduced fivefold upon morpholino treatment compared with that of the control untreated PC12 cells. In this immunoblot, actin was used as a protein-loading marker. (C) Confocal and DIC images of control and morpholino-treated PC12 cells stained with anti-LGN (red) and DNA (blue). Arrowheads indicate cells in mitosis showing cortical LGN staining in control cells but not in the morpholino-treated cells.
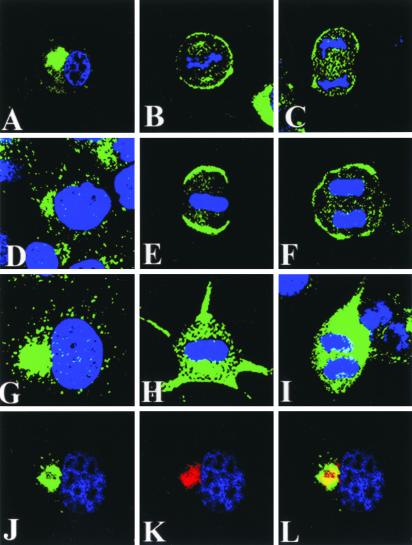
Staining of WISH and PC12 cell lines with anti-LGN antibody revealed a cortical localization profile for the endogenous LGN protein (Figure 4) that is similar to that observed when a FLAG-tagged version of mouse LGN was transfected into these cells and detected with anti-FLAG antibody (see Figures 1 and 2). The cortical staining detected in mitotic PC12 cells with anti-LGN was also sensitive to treatment with LGN-specific morpholino (Figure 3C; showing loss of staining in ∼26% of the treated cells), further supporting the observation of mitotic-dependent cortical LGN localization. In PC12 cells, endogenous LGN was predominantly perinuclear during interphase and partially accumulated at the cell cortex during mitosis (Figure 4, A–C). Some LGN staining associated with the cytoplasm and spindle apparatus could also be seen in metaphase cells (Figure 4B). A similar localization profile was also demonstrated for endogenous LGN in WISH cells (Figure 4, D–F). In these cells, LGN cortical staining appeared to be strongest at the two opposite poles of metaphase cells and was more intense compared with LGN staining in other cell lines. A cortical localization for LGN in dividing WISH cells was also observed when affinity-purified LGN antibodies generated independently by other laboratories (Blumer et al., 2002) were used. Interestingly, these antibodies also stained midbody and spindle poles in WISH cells (our unpublished results). Similar to the LGN-FLAG localization data obtained in the overexpression studies, a cortical localization for endogenous LGN was not detected in mitotic COS cells, and the LGN protein remained cytoplasmic (Figure 4, H and I). In interphase COS cells, LGN was found in the perinuclear region that is usually occupied by Golgi and other membrane compartments (Figure 4G) and colocalized with Vti1α (Figure 4, J–L). Vti1α is a SNARE protein that colocalizes with Golgi markers in various cell lines (Xu et al., 1998; Antonin et al., 2000).
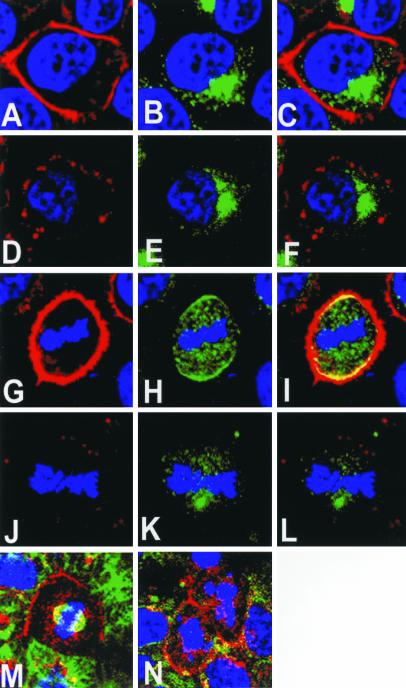
Figure 4.
Subcellular localization of endogenous LGN in cell lines. Confocal images of PC12 (A–C), WISH (d–F), and COS cells (G–L) stained for LGN (green) and DNA (blue) are shown. In all cell lines, LGN localizes to a perinuclear space during interphase (A, D, G, and J). Colocalization in interphase COS cells between LGN (J; green) and Vti1α Golgi marker (K; red) is shown in L (yellow). Note the cortical localization of LGN during mitosis is only observed in PC12 (B and C) and WISH (E and F) but not COS (H and I) cells. In metaphase (H) and anaphase (I) COS cells, LGN remains in the cytoplasm. Some cytoplasmic staining of LGN can also be seen in mitotic PC12 (B and C) and WISH (E and F) cells.
To determine which membrane subdomain LGN is associated with during mitosis, polarized epithelial MDCK cells were stained with anti-LGN antibody and analyzed using confocal microscopy (Figure 5). Analysis of the images showed that LGN is not distributed randomly over the cortex (Figure 5, D and F); staining was generally absent from the basal and apical membrane and was largely confined to the lateral cell membrane during mitosis. The localization of LGN was compared with that of two other membrane proteins, ZO-1 (Figure 5, A and C) and β-catenin (Figure 5, B and C, and E and F). The ZO-1 protein is localized in tight junctions, and β-catenin is a basolateral membrane protein in polarized epithelial cells. In vertical optical sections, LGN staining was absent from the apical and basal membrane but was present on the lateral membrane where its localization overlaps with β-catenin (Figure 5, D–F).
Figure 5.
Cortical subdomain localization of LGN in polarized MDCK cells. Confocal vertical optical sections (X-Z axis) of polarized MDCK cells at metaphase are shown.(A–C) A metaphase MDCK cell double stained for apical membrane marker ZO-1 (A; green) and β-catenin basolateral membrane marker (B; red). There is little overlap between the two membrane markers (C; yellow). (d--F) A metaphase MDCK cell double stained for LGN (D; green) and β-catenin (E; red). LGN and β-catenin show colocalization at the lateral membrane subdomain (F; yellow). The cell cycle stage was determined by DNA staining (blue).
LGN Cortical Localization Is Dependent on Microfilaments But Not Microtubules
To assess the role of microfilaments and microtubules on LGN cortical localization, WISH cells were subjected to treatments with latrunculin B and colchicine (Figure 6). Cells treated with the microfilaments-destabilizing drug, latrunculin B, were double-stained with anti-LGN antibody and phalloidin. In interphase cells, latrunculin treatment did not affect the perinuclear localization of LGN (Figure 6, E and F). On the other hand, the cortical LGN localization that is normally seen in control cells during mitosis (Figure 6, H and I) was abolished upon latrunculin treatment (Figure 6, K and L). As expected, the cortical microfilament staining was also abolished in treated cells (Figure 6, D and J). In contrast, treatment with colchicine, the microtubule-destabilizing drug, disrupted the mitotic spindle but did not affect LGN cortical localization at mitosis (Figure 6, M and N). The data indicates that LGN cortical localization during mitosis is dependent on microfilaments but not microtubules.
Figure 6.
Effects of cytoskeleton on LGN cortical localization. (A–L) Confocal images of cycling WISH cells stained with phalloidin (red), LGN (green), and DNA (blue); (A–C) a control interphase cell with LGN localizing at the perinuclear region (B, green) and actin microfilaments at the cell cortex (A, red); (C) a merged image of A and B; (D) the loss of cortical actin microfilaments upon latrunculin treatment, but the perinuclear LGN staining (E) remains unaffected in the drug-treated cell. (F) A merged image of D and E; (I) a merged image of G (phalloidin) and H (LGN) showing areas of overlap (yellow) between cortical actin and LGN in a control metaphase cell; note that some LGN staining in the cytoplasm is also visible at this stage. (J–L) A metaphase cell treated with latrunculin B; note the cytoplasmic staining of LGN (K) and the absence of phalloidin staining in the treated cell (J); (L) a merged image of J and K. (M and N) Metaphase WISH cells without (M) or with (N) colchicine treatment and stained for LGN (red), tubulin (green), and DNA (blue); note the absence of spindle staining in the treated cell, but no effect on LGN cortical staining (N).
LGN Cortical Localization Can Be Influenced by Gα Subunits of Heterotrimeric G-proteins
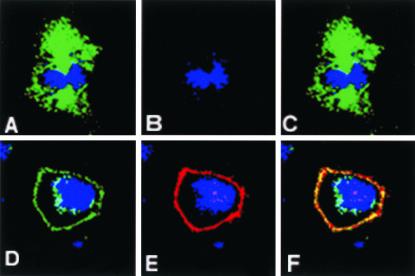
The identification of a cortical localization signal for LGN in its C-terminal region implicated a role for Gα subunits of heterotrimeric G-proteins in the localization of LGN. To investigate the role of G-proteins in LGN cortical localization, we transfected COS cells with various Gαi/o constructs (Figure 7). We used COS cells because our data showed that they do not localize LGN cortically during mitosis (Figure 7, A and C). COS cells also lack the expression of some G-protein members including Gαo (Luo and Denker, 1999; Figure 7B). Ectopic expression of Gαo in COS cells redirected most of LGN to the cell cortex (Figure 7, D and F), indicating that the G-proteins can localize LGN to the cortex in these cells. The cortical localization of LGN was observed in all Gαo-transfected COS cells. COS cells also directed the ectopically expressed Gαo protein to their cell cortex (Figure 7, E and F). In this system, the overexpression of Gαo was associated with abnormal rounded-cell morphology.
Figure 7.
Effects of Gαo on LGN cortical localization. (A–F) Confocal images of COS cells stained for LGN (green) and Gao (red). (A–C) A control metaphase COS cell with cytoplasmic LGN (A) and no Gαo expression (B); (C) a merged image of A and B. (d--F) A COS cell transfected with Gαo; LGN (D) and exogenous Gαo (E) are directed to the cell cortex of the transfected COS cell; note that a residual perinuclear staining for LGN can still be seen in this experiment; (F) a merged image of D and E showing areas of overlap (yellow) between Gαo and LGN. In all images, DNA staining is in blue.
LGN Interacts Directly with Gα Subunits of Heterotrimeric G-proteins and Acts as a GDI
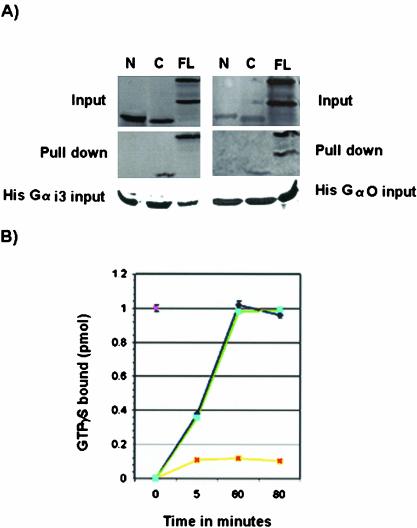
Pins and its related proteins are characterized by the presence of C-terminal GoLoco repeats, which bind Gα proteins (Siderovski et al., 1999; DeVries et al., 2000; Natochin et al., 2000; Bernard et al., 2001). Like its mammalian homologues, mouse LGN was able to bind Gαi3-GDP in vitro via its C termini (Figure 8A). No binding to Gαi3 was observed with mouse LGN constructs lacking the C-terminal GoLoco repeats (Figure 8A). As expected, binding of mouse LGN to Gαi3 inhibited its rate of exchange of GDP for GTP (Figure 8B). Mouse LGN was also able to bind Gαo (Figure 8A) but no GDI activity was observed with Gαo (Figure 8B).
Figure 8.
GDI activity of mouse LGN. (A) Binding of mouse LGN to Gα subunits of heterotrimeric G-proteins. In vitro–translated LGN constructs containing amino acids 1–384 from the N-terminus (N), amino acids 385–670 from the C termini (C), or full length (FL) were incubated with His-Gαi3 or His-Gαo bound to His columns. Only FL and C products copurify with His-Gαi3/o. No binding is detected between construct N and Gαi3 or Gαo. In the FL lane, two bands are detected, an upper band corresponding to LGN and a lower band being a byproduct of the translation reaction. (B) GDI activity of mouse LGN on Gαi3. Time course experiments showing the rate of [35S]GTPγ binding by Gαi3 were carried out in the presence of 1 μM GST-LGN (yellow). Control experiments using GST alone are shown in blue. GST-LGN inhibits ∼90% of [35S]GTPγ binding to Gαi3 but the GST control has no effect. The effect of LGN on Gαi3 activity is observed as early as 5 min and lasts >80-min time period. GST-LGN effect on Gαo (green) is similar to the GST control (blue), indicating mouse LGN has no GDI activity on Gαo.
Loss/Ectopic Expression of LGN Causes Cell Cycle Defects
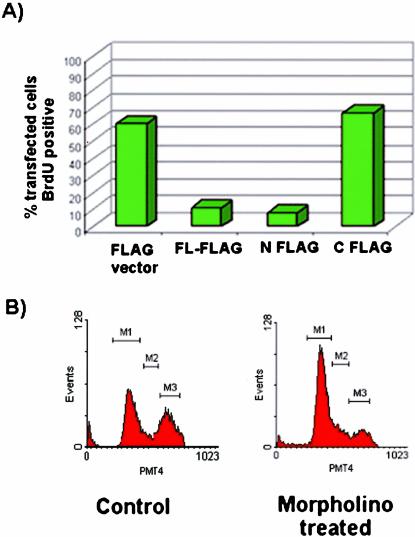
To study the function of LGN in cell cycle progression, we carried out BrdU labeling experiments in PC12 cells ectopically expressing three LGN constructs, FL-FLAG, N-FLAG, and C-FLAG (Figure 9A). In these cells, ectopic expression of FL-FLAG or N-FLAG prevented incorporation of BrdU in transfected cells (Figure 9A), indicating cell cycle arrest at the G1/S transition stage. In contrast, ectopic expression of the C-FLAG construct or the FLAG vector alone (control) did not affect cell cycle progression as indicated by BrdU incorporation in transfected cells (Figure 9A).
Figure 9.
Effects of LGN on cell cycle progression. (A) Effect of ectopic expression of various mouse LGN constructs, FL-FLAG, N-FLAG, and C-FLAG on cell cycle in PC12 cells. Anti-BrdU and anti-FLAG antibodies were used for detection purposes. The bars represent percentage of BrdU-positive cells. The majority of FL-FLAG– or N-FLAG–transfected cells were BrdU-negative, whereas C-FLAG– or FLAG-vector–transfected cells were BrdU-positive, indicating that overexpression of either FL or N-FLAG but not C-FLAG results in cell cycle arrest. (B) Effect of LGN removal by morpholino treatment on cell cycle in PC12 cells. For control cells, the percentage of cells in G1 phase (M1) was ∼52%, S phase (M2) ∼11%, and G2 phase (M3) ∼37%. For morpholino-treated cells, the scores were ∼70% for M1, ∼10% for M2, and ∼21% for M3, indicating that the loss of LGN causes partial delay/disruption of the cell cycle progression at the G1/S phase boundary.
The effect of LGN removal on cell cycle progression was assayed by treating cycling PC12 cells with the LGN-specific morpholino and carrying out FACScan analysis. In this experiment, a higher percentage of cells accumulated at G1 than that with control untreated cells, indicating a delay/disruption of the cell cycle progression at the G1/S boundary (Figure 9B).
DISCUSSION
In this report, we have characterized a novel dynamic cortical localization pattern of mammalian LGN that partly resembles that of fly Pins. We have mapped the cortical localization signal of LGN to its C-terminal Gα-interacting domain and shown that LGN function is required for cell cycle progression. The cell cycle–dependent cortical localization of LGN requires microfilaments and can be influenced by the Gα subunits of heterotrimeric G-proteins.
Evidence for a cortical localization of LGN during mitosis is supported by several observations. First, ectopically expressed full-length tagged-LGN protein can localize at the cell cortex in various mitotic cell lines. Second, the overexpression studies show that the LGN C terminus alone is able to accumulate at the cell cortex during mitosis. Third, COS cells, which normally do not localize LGN, redirect this protein to their cortex upon overexpression of heterotrimeric G-proteins. Fourth, an LGN-specific morpholino causes loss of cortical LGN staining in 26% of treated mitotic cells. The mitotic cortical localization of LGN described in this article has not been previously reported. Du et al. (2001) have shown that LGN associates with the spindle poles during mitosis, and its function is required to regulate mitotic spindle organization. They have also shown that the N-terminus of human LGN binds the nuclear mitotic apparatus protein NuMA, which tethers spindles at the poles, and that this interaction is required for the LGN phenotype. In a separate study, LGN was reported to be nuclear and to move to midbody domain in late mitotic phases (Blumer et al., 2002). The different LGN localization data shown in these studies are surprising because they all describe the same LGN protein but use different reagents. The various localization data suggest an ability of LGN to localize to different subcellular compartments, depending on the cell cycle stage and the cellular context. Interestingly, a partial cortical staining for LGN in WISH cells is also observed with LGN antibodies (personal observation) obtained from other laboratories (Blumer et al., 2002). Mouse LGN and human LGN share a high degree of identity at the amino acid level. Mouse LGN shares 92% identity with human LGN, 60% with AGS3, and 49% with fly Pins (Yu et al., 2003). The difference in LGN localization data between these studies may reflect variations in the multiple LGN isoforms produced in the cell and/or their posttranslational modification status. It has been previously reported the LGN locus produces several LGN peptides (Blumer et al., 2002), suggesting the existence of splice variants or alternative promoters as reported for AGS3 (Pizzinat et al., 2001). The human LGN gene contains 14 exons and exon 1 encodes an amino-terminal 12aa that is not found in all ESTs for LGN (Blumer et al., 2002). Therefore, it is possible that the different antibodies generated for the human and mouse LGN proteins may recognize distinct protein epitopes that are present on the various LGN isoforms produced in the cell. In this scenario, it could be envisaged that, because different LGN isoforms might be localized differently to various subcellular sites, our reagents perhaps preferentially recognize the LGN isoforms that localize to the cortex during mitosis. Cell lines may also localize proteins differently. Indeed, no cortical localization for LGN could be detected in COS cells, but WISH cells accumulate higher levels of LGN at their cell cortex compared with other cell lines (this article) and this may facilitate the detection of the protein in these cells.
Domain dissection analysis suggests that the region containing the C-terminal GoLoco motifs of LGN is sufficient for membrane targeting and that the cortical localization of this domain is cell cycle independent. This implies roles for heterotrimeric G-proteins in LGN cortical localization, which would be consistent with the observations that LGN can directly bind Gαi2/3/o subunits. A function for heterotrimeric G-proteins in LGN cortical localization is also supported by evidence from ectopic expression studies in COS cells, which cannot normally localize endogenous LGN to the cortex during mitosis. The ectopic expression studies in COS cells indicate a possible role for Gαo in directing the cortical localization of LGN during mitosis in these cells. Because LGN does not act as a GDI for Gαo, its Gαo-driven cortical localization in COS cells may be independent of its GDI activity. LGN can bind directly to Gαo in vitro and this lends support to the notion that LGN cortical localization by Gαo may be a direct event. This notion can also be supported by the cortical localization data of Gαo in transfected COS cells.
Work on the Pins-related protein from rat, AGS3, has also shown that in crude fractionation assays overexpression of G-proteins can direct AGS3-Short to the membrane (Pizzinat et al., 2001). Interestingly, fly Pins also contains a cortical localization domain in its C termini (Yu et al., 2002) and the C termini of mouse LGN exhibit similar cortical localization when expressed in fly neuroblasts (Yu et al., 2003). Gα subunits have been found segregated onto the plasma membrane and membranes of several organelles such as the endoplasmic reticulum, Golgi complex, and the nucleus (Ercolani et al., 1990; Stow et al., 1991; deAlmeida et al., 1994; Hamilton and Nathanson, 1997). Based on the localization data and interaction of LGN with heterotrimeric G-proteins, it is plausible to suggest that during mitosis, LGN is released from the perinuclear domain and becomes accessible to binding by plasma membrane-associated heterotrimeric G-proteins.
The functional significance of the cell cycle–dependent cortical localization of LGN is not yet clear. It is interesting to note here that LGN localization to the cell cortex during mitosis is somewhat similar to what is described for fly Pins. In Drosophila, Pins is normally found in the lateral cortex of epithelial cells and only become asymmetrically localized upon the expression of inscuteable in neuroblasts, for which a mammalian homologue has not been found so far (Schaefer et al., 2000; Yu et al., 2000). Pins is also dependent on heterotrimeric G-proteins activity for its localization (Schaefer et al., 2001). Furthermore, Pins plays important roles in neuroblast asymmetric cell divisions and the available data suggest that its interaction with Gα facilitates receptor-independent G-protein signaling (Scheafer et al., 2000, 2001). Interestingly, the mouse LGN gene reported in this study can also bind fly Inscuteable and rescue defects associated with Pins mutations in the fly (Yu et al., 2003), showing functional conservation. In conclusion, LGN and Pins share the domains required for cortical localization, and both proteins can assume this dynamic localization depending on the presence of a suitable partner.
LGN, like other Pins-related proteins, complexes with Gα subunits of heterotrimeric G-proteins and inhibits dissociation of GDP from Gαi (Mochizuki et al., 1996; DeVries et al., 2000; Natochin et al., 2000; Peterson et al., 2000; Scheafer et al., 2000; Bernard et al., 2001). Whether LGN interferes with G-proteins activity at the plasma membrane remains to be determined. Interestingly, interfering with LGN expression causes cell cycle arrest. However, the cell cycle arrest at the G1/S stage is incomplete because we were still able to find some treated cells in the mitotic phase. The role of LGN in cell cycle progression may be due to interference with G-proteins activity or the function of some other protein(s) at different cellular sites. Interestingly, a nuclear localization for LGN has also been reported (Blumer et al., 2002). Whether LGN is required at the nucleus or other subcellular sites for the cell cycle progression is still not clear, and further investigations are required to address these issues.
Acknowledgments
We thank Hing Fook Siong for technical assistance and Xavier Morin (King's College London) for valuable discussions. We also thank researchers at IMCB (A-STAR, Singapore), especially from Wan-Jin Hong and Walter Hunziker's laboratories for their generous supply of reagents.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-04-0212. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-04-0212.
References
- Antonin, W., Riedel, D., and von Mollard, G.F. (2000). The SNARE Vti1a-β is localized to small synaptic vesicles and participates in a novel SNARE complex. J. Neurosci. 20, 5724–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, M.L., Peterson, Y.K., Chung, P., Jourdan, J., and Lanier, S.M. (2001). Selective interaction of AGS3 with G-proteins and the influence of AGS3 on the activation state of G-proteins. J. Biol. Chem. 276, 1585–1593. [DOI] [PubMed] [Google Scholar]
- Blatch, G.L., and Lassle, M. (1999). The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21, 932–939. [DOI] [PubMed] [Google Scholar]
- Blumer, J.B., Chandler, L.J., and Lanier, S.M. (2002). Expression analysis and subcellular distribution of the two G-protein regulators AGS3 and LGN indicate distinct functionality. Localization of LGN to the midbody during cytokinesis. J. Biol. Chem. 277, 15897–15903. [DOI] [PubMed] [Google Scholar]
- Chen, C., Zheng, B., Han, J., and Lin, S.C. (1997). Characterization of novel RGS proteins that bind to Gα proteins and inhibits pheromone signalling in yeast. J. Biol. Chem. 272, 8679–8685. [DOI] [PubMed] [Google Scholar]
- Chen, C., Wang, H., Fong, C.W., and Lin, S.C. (2001). Multiple phosphorylation sites in RGS16 differently modulate its GAP activity. FEBS Lett. 504, 16–22. [DOI] [PubMed] [Google Scholar]
- deAlmeida, J.B., Holtzman, E.J., Peters, P., Ercolani, L., Ausiello, D.A., and Stow, J.L. (1994). Targeting of chimeric G alpha i proteins to specific membrane domains. J. Cell Sci. 107, 507–515. [DOI] [PubMed] [Google Scholar]
- DeVries, L., Fischer, T., Tronchere, H., Brothers, G.M., Strockbine, B., Siderovski, D.P., and Farquhar, M.G. (2000). Activator of G protein signaling 3 is a guanine dissociation inhibitor for Galpha i subunits. Proc. Natl. Acad. Sci. USA 97, 14364–14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Q., Stukenberg, T., and Macara, I.G. (2001). A mammalian partner of inscuteable binds NuMA and regulates mitotic spindle organization. Nat. Cell Biol. 3, 1069–1075. [DOI] [PubMed] [Google Scholar]
- Du, Q., Taylor, L., Compton, D.A., and Macara, I.G. (2002). LGN blocks the ability of NuMA to bind and stabilize microtubules. A mechanism for mitotic spindle assembly regulation. Curr. Biol. 12, 1928–1933. [DOI] [PubMed] [Google Scholar]
- Ercolani, L., Stow, J.L., Boyle, J.F., Holtzman, E.J., Lin, H., Grove, J.R., and Ausiello, D.A. (1990). Membrane localization of the pertussis toxin-sensitive G-protein subunits alpha i-2 and alpha i-3 and expression of a metallothionein-alpha i-2 fusion gene in LLC-PK1 cells. Proc. Natl. Acad. Sci. USA 87, 4635–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, L.A., and Tischler, A.S. (1976). Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. USA 73, 2424–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, S.E., and Nathanson, N.M. (1997). Differential localization of G-proteins, Gαo and Gαi-1, -2, and -3, in polarized epithelial MDCK cells. Biochem. Biophys. Res. Commun. 234, 1–7. [DOI] [PubMed] [Google Scholar]
- Hoffmann, M., Ward, R.J., Cavalli, A., Carr, I.C., and Milligan, G. (2001). Differential capacities of the RGS1, RGS16 and RGS-GAIP regulators of G protein signaling to enhance alpha2A-adrenoreceptor agonist-stimulated GTPase activity of G(o1)alpha. J. Neurochem. 78, 797–806. [DOI] [PubMed] [Google Scholar]
- Kraut, R., Chia, W., Jan, L.Y., Jan, Y.N., and Knoblich, J.A. (1996). Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature 383, 50–55. [DOI] [PubMed] [Google Scholar]
- Larson, J.D., and Ekker, S.C. (2001). Morphant technology in model developmental systems. Genesis 30, 89–93. [DOI] [PubMed] [Google Scholar]
- Luo, Y., and Denker, B.M. (1999). Interaction of heterotrimeric G protein Gαo with Purkinje cell protein-2. Evidence for a novel nucleotide exchange factor. J. Biol. Chem. 274, 10685–10688. [DOI] [PubMed] [Google Scholar]
- Manser, E., Huang, H.Y., Loo, T.H., Chen, X.Q., Dong, J.M., Leung, T., and Lim, L. (1997). Expression of constitutively active α-PAK reveals effects of the kinase on actin and focal complexes. Mol. Cell. Biol. 17, 1129–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki, N., Cho, G., Wen, B., and Insel, P.A. (1996). Identification and cDNA cloning of a novel human mosaic protein, LGN, based on interaction with Gαi2. Gene 181, 39–43. [DOI] [PubMed] [Google Scholar]
- Natochin, M., Lester, B., Peterson, Y.K., Bernard, M.L., Lanier, S.M., and Artemyev, N.O. (2000). AGS3 inhibits GDP dissociation from galpha subunits of the Gi family and rhodopsin-dependent activation of transducin. J. Biol. Chem. 275, 40981–40985. [DOI] [PubMed] [Google Scholar]
- Peterson, Y.K., Hazard, S., 3rd, Graber, S.G., and Lanier, S.M. (2000). Stabilization of the GDP-bound conformation of Gialpha by a peptide derived from the G-protein regulatory motif of AGS3. J. Biol. Chem. 275, 33193–33196. [DOI] [PubMed] [Google Scholar]
- Pizzinat, N., Takesono, A., and Lanier, S.M. (2001). Identification of a truncated form of the G-protein regulator AGS3 in heart that lacks the tetratricopeptide repeat domains. J. Biol. Chem. 276, 16601–16610. [DOI] [PubMed] [Google Scholar]
- Rosin-Arbesfeld, R., Ihruke, G., and Beinz, M. (2001). Actin-dependent membrane association of the APC tumour suppressor in polarized mammalian epithelial cells. EMBO J. 20(21), 5929–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, M., Shevchenko, A., Shevchenko, A., and Knoblich, J.A. (2000). A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr. Biol. 10, 353–362. [DOI] [PubMed] [Google Scholar]
- Schaefer, M., Petronczki, M., Dorner, D., Forte, M., and Knoblich, J.A. (2001). Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell 107, 183–194. [DOI] [PubMed] [Google Scholar]
- Siderovski, D.P., Diverse-Pierluissi, M., and De Vries, L. (1999). The GoLoco motif: a Galphai/o binding motif and potential guaninenucleotide exchange factor. Trends Biochem. Sci. 24, 340–341. [DOI] [PubMed] [Google Scholar]
- Stow, J.L., de Almeida, J.B., Narula, N., Holtzman, E.J., Ercolani, L., and Ausiello, D.A. (1991). A heterotrimeric G protein, G alpha i-3, on Golgi membranes regulates the secretion of a heparan sulfate proteoglycan in LLC-PK1 epithelial cells. J. Cell Biol. 114, 1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerton, J. (1999). Morpholino antisense oligomers: the case for Rnase H-independent structural type. Biochem. Biophys. Acta 1489, 141–158. [DOI] [PubMed] [Google Scholar]
- Takesono, A., Cismowski, M.J., Ribas, C., Bernard, M., Chung, P., Hazard, S., 3rd., Duzic, E., and Lanier, S.M. (1999). Receptor-independent activators of heterotrimeric G-protein signaling pathways. J. Biol. Chem. 274, 33202–33205. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Wong, S.H., Tang, B.L., Subramaniam, V.N., Zhang, T., and Hong, W. (1998). A 29-kilodalton Golgi soluble N-ethylmaleimide-sensitive factor attachment protein receptor (Vti1-rp2) implicated in protein trafficking in the secretory pathway. J. Biol. Chem. 273, 21783–21789. [DOI] [PubMed] [Google Scholar]
- Yu, F., Morin, X., Cai, Y., Yang, X., and Chia, W. (2000). Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100, 399–409. [DOI] [PubMed] [Google Scholar]
- Yu, F., Ong, C.T., Chia, W., and Yang, X. (2002). Membrane targeting and asymmetric localization of Drosophila partner of inscuteable are discrete steps controlled by distinct regions of the protein. Mol. Cell. Biol. 22, 4230–42340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F., Morin, X., Kaushik, R., Bahri, S., Yang, X., and Chia, W. (2003). A mouse homologue of Drosophila pins can asymmetrically localize and substitute for pins function in Drosophila neuroblasts. J. Cell Sci. 116, 887–896. [DOI] [PubMed] [Google Scholar]