Abstract
Cardiac myosin-binding protein-C (MyBP-C), also known as C-protein, is one of the major myosin-binding proteins localizing at A-bands. MyBP-C has three isoforms encoded by three distinct genes: fast-skeletal, slow-skeletal, and cardiac type. Herein, we are reporting a novel alternative spliced form of cardiac MyBP-C, MyBP-C(+), which includes an extra 30 nucleotides, encoding 10 amino acids in the carboxyl-terminal connectin/titin binding region. This alternative spliced form of MyBP-C(+) has a markedly decreased binding affinity to myosin filaments and connectin/titin in vitro and does not localize to A-bands in cardiac myocytes. When MyBP-C(+) was expressed in chicken cardiac myocytes, sarcomere structure was markedly disorganized, suggesting it has possible dominant negative effects on sarcomere organization. Expression of MyBP-C(+) is hardly detected in ventricles through cardiac development, but its expression gradually increases in atria and becomes the dominant form after 6 mo of age. The present study demonstrates an age-induced new isoform of cardiac MyBP-C harboring possible dominant negative effects on sarcomere assembly.
INTRODUCTION
The muscle sarcomere is a complex and highly ordered array of numerous proteins and the precise organization and alignment of these proteins are essential for proper muscle function. Interaction between thick and thin filaments causes sliding between these two filaments, resulting in muscle contraction. Thick filaments consist predominantly of myosin with several myosin-associated proteins. These myosin-associated proteins are important for integration and maintenance of sarcomeres.
Cardiac myosin-binding protein-C (MyBP-C), also known as C-protein, is one of the major myosin-binding proteins in vertebrate striated muscles with an approximate molecular mass of 140 kDa (Offer et al., 1973; Pepe and Drucker, 1975). MyBP-C is localized to the central cross-bridge zone in each half of the A-band region of myofibrils (Pepe and Drucker, 1975; Craig and Offer, 1976). MyBP-C modulates myosin assembly (Offer et al., 1973), actin–myosin interaction in sarcomeres (Moos et al., 1978; Hartzell, 1985), and stabilizes thick filaments (Moos et al., 1978). In addition, MyBP-C interacts with connectin/titin, which is important for the precise arrangement of actin-myosin filaments in sarcomeres (Maruyama, 1986; Fürst et al., 1992; Koretz et al., 1993).
MyBP-C, a member of the intracellular immunoglobulin (Ig) superfamily, has three isoforms, two skeletal (fast- and slow-) and one cardiac type encoded by three distinct genes. The two skeletal types of MyBP-C are composed of a total of 10 domains: seven IgI domains and three fibronectin (FN) type III repeats (Einheber and Fischman, 1990; Fürst et al., 1992; Okagaki et al., 1993; Weber et al., 1993; Harpaz and Chothia, 1994; Kurasawa et al., 1999), whereas, cardiac MyBP-C has 11 domains (C0–C10): eight IgI domains (instead of seven in the skeletal types) and three FN type III repeats (Figure 1A). In addition, cardiac MyBP-C has a unique phosphorylation domain between the C1 and C2 domain (Kasahara et al., 1994; Gautel et al., 1995; Yasuda et al., 1995; Yang et al., 1998). Through these domains, MyBP-C interacts with other sarcomeric proteins; C1–C2 domains interact with the S2 segment of myosin, C10 domain with light meromyosin (LMM), and C8–C10 with connectin/titin.
Figure 1.
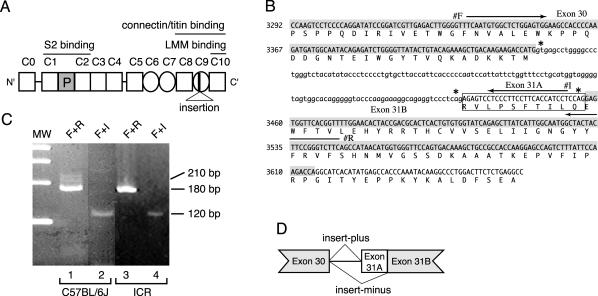
(A) Structure of mouse cardiac MyBP-C protein (cMyBP-C). The numbers above cMyBP-C indicate the position of each IgI domain (open square) and FN type III repeat (circle). The cardiac-specific phosphorylation domain is shown as a shaded square (P). Position of 10 amino acids insertion is indicated in C9 domain. (B) Genomic sequence and deduced amino acid sequence encoding the C9 IgI domain of mouse cardiac MyBP-C. The nucleotide sequence of the coding region and the deduced amino acid sequence of MyBP-C(+) are represented in capital letters. Exon 30 and 31B are highlighted by hatch. MyBP-C(+)-specific insert sequence (exon 31A) is indicated by a box. Arrows indicate the location of primer F (#F), primer I (#I) and primer R (#R). Asterisks indicate a 5′ splice donor sequence, gt, and 3′ splice acceptor sequence, CAG. (C) Identification of MyBP-C(+) in C57BL/6J and ICR mice. cDNA libraries prepared from 8-wk-old C57BL/6J hearts (lane 1 and 2) or from ICR hearts (lane 3 and 4) were subjected to PCR by using a primer F + R set (lane 1 and 3) or using a primer F + I set (lane 2 and 4). Primer F + R set amplifies 212-base pair and 182-base pair DNA fragments which are generated from MyBP-C(+) and MyBP-C(–) cDNA, respectively. Primer F + I set specifically detects MyBP-C(+) cDNA as a 118-base pairs product. A molecular weight marker (MW) indicates 400, 300, 200, and 100 base pairs, top to bottom. (D) Schematic genomic structure of mouse cardiac MyBP-C. Both MyBP-C(–) and MyBP-C(+) mRNAs are encoded by a single gene and generated by an alternative mRNA splicing.
All isoforms of MyBP-C are specific for different muscle fiber types and different developmental stages (Obinata et al., 1984; Obinata, 1985; Bähler et al., 1985; Kawashima et al., 1986; Fougerousse et al., 1998; Gautel et al., 1998; Kurasawa et al., 1999). In humans and mice, expression of the cardiac-type isoform is restricted to the heart through development, and no splicing variants of the cardiac isoform have been reported (Fougerousse et al., 1998; Gautel et al., 1998; Kurasawa et al., 1999). On the other hand, the chicken cardiac isoform containing the specific phosphorylation domain is abundantly expressed in cardiac muscle through development; however, an alternative spliced variant lacking the phosphorylation domain is expressed dominantly but transiently in embryonic skeletal muscle (Bähler et al., 1985; Yasuda et al., 1995; Mohamed et al., 1998).
Mutations in the gene MYBPC3, encoding human cardiac-type MyBP-C, have been identified to cause modest and late-onset familial hypertrophic cardiomyopathy (Bonne et al., 1995; Watkins et al., 1995; Carrier et al., 1997; Rottbauer et al., 1997; Yu et al., 1998). Familial hypertrophic cardiomyopathy is an autosomal dominant disease with typical morphological changes, including cardiomyocyte hypertrophy and/or myofibrillar disarray with fibrosis (Maron et al., 1987a,b; Seidman and Seidman, 2001). Most of the mutations found in the MYBPC3 are predicted to lead to the altered mRNA sequence and to produce the carboxyl-terminal truncation of the cardiac MyBP-C polypeptides lacking the C10 myosin binding site, or in some cases, lacking the C8–C10 connectin/titin binding site (Bonne et al., 1995; Watkins et al., 1995; Carrier et al., 1997; Kimura et al., 1997; Rottbauer et al., 1997; Yu et al., 1998). These findings demonstrate that the expression of mutant sarcomeric proteins, such as the truncated cardiac MyBP-C, induce cardiac dysfunction and alteration of myofibrils in cardiomyocytes. However, the precise mechanism for this negative effect has not yet been determined.
Herein, we identified a novel isoform of mouse cardiac MyBP-C mRNA, namely, MyBP-C(+), that has an additional 30 nucleotides by alternative splicing and encodes exactly the same amino acid sequence as previously reported, but contains an additional 10 amino acids in the connectin/titin binding region. MyBP-C(+) decreased binding to myosin filaments and connectin/titin in vitro. Green fluorescent protein (GFP)-tagged MyBP-C(+) did not correctly localize to sarcomeres, and it disrupted the A-band formation in chicken cardiomyocytes. Reverse transcription-polymerase chain reaction (RT-PCR) analysis showed that mRNA encoding MyBP-C(+) was dominantly expressed in the aged mouse atrium.
MATERIALS AND METHODS
cDNA and Sequencing
A cDNA clone encoding “insert-plus” mouse cardiac MyBP-C [MyBP-C(+)] was isolated from a 12-wk-old mouse BALB/c heart cDNA library (BD Biosciences Clontech, Palo Alto, CA) among several “insert-minus” mouse cardiac MyBP-C [MyBP-C(–)] cDNAs reported previously (Kasahara et al., 1994). The newly isolated clone and previously isolated clone were sequenced with a SequiTherm EXCEL2 Long-Read DNA sequencing kit-LC (Epicentr, Madison, WI) or a Thermo Sequenase Cy5.5 dye terminator cycle sequencing kit (Amersham Biosciences UK, Little Chalfont, Buckinghamshire, United Kingdom). Analysis of DNA sequence data was carried out with the GENETYX program (SDC Software, Tokyo, Japan). Consequently, we found some errors on the nucleotide and amino acid sequences encoding MyBP-C(–) reported previously and then corrected them here; T441 was revised to C (no change in the encoded amino acid sequence), a G was inserted at 2638, and a C was deleted at 2700; therefore, the amino acid sequence QSMPWECPGPALPLSPSCLL (844–863) was replaced with AVNAVGMSRPSPASQPFMPI.
PCR and RT-PCR Samples and Primers
In this study, we used two cDNA libraries: one was prepared from total RNA isolated from 8-wk-old ICR mouse hearts by using reverse-transcription with oligo-dT primers (Chomczynski and Sacchi, 1987) and the other was prepared from 8-wk-old C57BL/6J (SuperScript mouse heart cDNA library; Invitrogen, Carlsbad, CA).
Messenger RNAs were extracted using a QuickPrep MicromRNA purification kit (Amersham Biosciences UK) from ventricles and atria of newborn, 4-wk-, 8-wk-, and >6-mo-old ICR mice. These mRNAs were reverse-transcripted with oligo-dT primers. The following primers were used for PCR reactions: primer F, 5′-TCAATGTGGCTCTGGAGTGG-3′ (3335–3354); primer R, 5′-GAAGACCCGGAAGTAGTAGC-3′ (3527–3546); and primer I, 5′-AGGATGGTGAAGGAAGGGAG-3′ (3433–3452).
Genomic tail DNA was prepared from ICR mice according to the method described by Blin and Stafford (1976). By a PCR reaction with a primer F + R set, the genomic DNA fragment was isolated, sequenced, and a comparison with human cardiac MyBP-C genomic DNA sequence (accession no. U91629, Entrez) was made to define the exon-intron structure.
Generation of Recombinant MyBP-Cs by Using a Baculovirus Expression System and the Binding Assay
Both of the XhoI-EcoRI fragments encoding full-length MyBP-C(–) and MyBP-C(+) were subcloned into pBlueBacHis2A (Invitrogen). Preparation of recombinant viral DNA was performed using a Bac-N-Blue transfection kit (Invitrogen) according to the technical bulletin. Consequent recombinant MyBP-Cs with histidine tags were purified from lysates of sf9 cells by using a HiTrap affinity column (Amersham Biosciences UK), which was saturated with nickel ions, and then dialyzed against binding buffer (0.1 M KCl, 20 mM imidazole-HCl, pH 7.0). The supernatant separated by centrifugation at 50,000 rpm for 30 min (TLA-100; Beckman Coulter, Fullerton, CA) was used for the binding reactions.
Crude myosin was prepared from rabbit ventricles as described previously (Perry, 1955), and the myosin was purified using DEAE-Sephadex A-50 (Richards et al., 1967). Reconstitution of myosin filaments was performed during a dialysis against the binding buffer. β-Connectin prepared from chicken pectoralis muscle (Kimura and Maruyama, 1983), which was kindly provided by Dr. S. Kimura (Chiba University, Chiba, Japan), was dialyzed against the binding buffer. Quantitations of the MyBP-Cs, reconstituted myosin filaments, and β-connectin were performed by a bicinchoninic acid protein assay reagent kit (Pierce Chemical, Rockfold, IL).
For binding assays, recombinant MyBP-C(–) or MyBP-C(+) at varying concentrations was incubated with reconstituted myosin filaments (0.5 μM) or β-connectin (0.04 μM) for 30 min at 4°C, and then centrifuged at 50,000 rpm for 30 min. Supernatants and pellets were subjected to SDS-PAGE, stained with Coomassie Blue, scanned with a densitograph (Atto, Tokyo, Japan), and analyzed using NIH Image.
Vector Constructs
Using a Transformer site-directed mutagenesis kit (BD Biosciences Clontech), each A106 in the cDNA encoding mouse cardiac MyBP-C(–) and MyBP-C(+) was substituted to G. The XhoI-EcoRI fragment encoding the full length of MyBP-C(–) open reading frame and that of MyBP-C(+) open reading frame were subcloned into pEGFP-C3 (BD Biosciences Clontech), and the vector was named pGMC(–) or pGMC(+), respectively. To construct pGMCΔ7-10, the pGMC(–) was partially digested with ApaI and then ligated. As for the pGMCΔ9-10, the pGMC(–) was digested at the EcoRV site and the BamHI site and then blunted and ligated by a Blunting Kit (Takara Bio., Kyoto, Japan). To construct pGMCΔ10(–) and pGMCΔ10(+), the pGMC(–), and the pGMC(+) were digested, respectively, with HindIII and BamHI, and then blunted and ligated by the blunting kit.
Cell Culture and Transfection
Cardiac myocytes were prepared from chicken embryos at embryonic day 9–10, according to Lin et al. (1989). Chick ventricles were incubated in 4 ml of trypsin solution (0.125% trypsin in phosphate-buffered saline (PBS) containing 1 mM EGTA) for 5 min at room temperature. The supernatant containing dispersed cells was placed into 25 ml of growth medium (10% fetal calf serum, 10 U/ml penicillin, 10 μg/ml streptomycin in minimal essential medium with l-glutamine and Earle's salts; Invitrogen). The remaining tissue fragments were incubated in 4 ml of the fresh trypsin solution as described above, and the dispersed cells were harvested for an additional two times. The cell suspension was filtered to remove large clumps of cells and then centrifuged for 5 min. Cells were plated for 1 h at 37°C in a humidified 5% CO2 incubator (Hyde et al., 1969). The supernatant containing floating myocytes was collected and centrifuged for 5 min. The cells were resuspended in growth medium, grown on glass coverslips at an initial density of 1–2 × 105 cells/35-mm dish. The following day, cells were transfected with the expression vector using Effectene transfection reagent (QIAGEN, Tokyo, Japan).
Indirect Immunofluorescence Microscopy and Immunoblotting
Three days after transfection, cells were rinsed with PBS and fixed with 10% formalin in PBS for 15 min. The cells were then permeabilized with 0.2% Triton X-100 in PBS for 10 min. Atrial and ventricular tissues isolated from ICR mice >20 mo of age were prefixed with 4% paraformaldehyde in PBS for 30 min, followed by immersion in liquid nitrogen-cooled isopentane. The longitudinal frozen sections (3 μm) were fixed with 4% paraformaldehyde in PBS for 10 min. After rinsing repeatedly with PBS, the cells and the sections were blocked with 1% bovine serum albumin in PBS and incubated with primary antibodies for 1 h. After repeated washing with 1% bovine serum albumin in PBS, they were incubated with rhodamine-conjugated goat anti-rabbit IgG or anti-mouse IgG as secondary antibodies for 1 h. After a final washing with PBS, the specimens were mounted in 50% glycerol in PBS. All steps were carried out at room temperature. Samples were examined with an Axioskop (Carl Zeiss, Jena, Germany) with a cooled charge-coupled device camera, CoolSNAP (Nippon ROPER, Chiba, Japan,) or with an Axiovert 135 TV (Carl Zeiss) with a cooled charge-coupled device camera, PXL37 (Photometrics, Tucson, AZ).
For immunoblotting, proteins were transferred to nitrocellulose membranes (Towbin et al., 1979). Membranes were treated with 5% skimmed milk in tris-buffered saline for 1 h and then incubated with primary antibody followed by treatment with alkaline phosphatase-conjugated secondary antibody. The membrane was washed with Tris-buffered saline and stained with nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate p-toluidine salt in + phosphatase buffer (0.1 M NaCl, 2 mM MgCl2, 0.1 M Tris-HCl, pH 9.5).
Primary Antibodies
The following primary antibodies were used: C315, a monoclonal antibody specific for chicken cardiac MyBP-C (Kawashima et al., 1986); αccp, a polyclonal antibody that recognizes both chicken and mouse cardiac MyBP-C and reacts with both MyBP-C(–) and MyBP-C(+) (Yasuda et al., 1995); A4.1045, a monoclonal antibody against the sarcomeric myosin heavy chain developed by Helen M. Blau (obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA); and Pc72C, a polyclonal antibody against the carboxyl-terminal (M-line) region of connectin/titin was kindly provided from Dr. S. Kimura (Chiba University) (Soeno et al., 1999).
Statistical Analysis
To study the influence of the expression of MyBP-C variants on the endogenous cross-striated sarcomeres in cardiomyocytes, cells were transfected with plasmids and 3 d later, processed for immunostaining as described above. The percentage of cells expressing the MyBP-Cs and exhibiting alteration of sarcomeres as observed after staining with A4.1045 was determined for each cover glass. The data obtained from three independent transfections were analyzed. For each construct, three coverglasses were examined and between 240 and 394 transfected cells were scored. The data were analyzed by one-way analysis of variance followed by the multiple comparison analysis with the test of Ryan (Ryan, 1959, 1960).
RESULTS
Isolation of a Variant of Cardiac MyBP-C
One of the major myosin binding proteins, cardiac MyBP-C/C-protein localizing at A-bands consists of eight IgI domains (Figure 1A, open square) and three FN type III repeats (Figure 1A, circle). We isolated a novel variant of cardiac MyBP-C cDNA from a 12-wk-old BALB/c mouse heart cDNA library, which we previously used for isolating a full-length cDNA (Kasahara et al., 1994). The novel cDNA of ∼4.2 kb, encodes identical eight IgI and three FN type III repeats, except for an additional 30 bases insertion located at the 5′ end of the exon 31, which was previously reported as an intron sequence between exon 30 and exon 31 (Figure 1B, exon 31A, sequence in the box). The insertion does not result in a frame-shift mutation, nor does it contain a termination codon. Therefore, this cDNA encodes the entire amino acids of the mouse cardiac-type MyBP-C with an extra10 amino acids in the C9 domain (Figure 1A, insertion), which is important for connectin/titin binding (Freiburg and Gautel, 1996) and incorporation into sarcomeres (Gilbert et al., 1996). We term the previously reported cardiac MyBP-C as insert-minus or MyBP-C(–) and the novel variant MyBP-C containing the extra amino acids as insert-plus or MyBP-C(+).
To confirm that the MyBP-C(+) cDNA was not an artifact of the cDNA library used, two additional cDNA libraries prepared from whole hearts of 8-wk-old C57BL/6J mice or ICR mice were screened by PCR. As shown in Figure 1B, a combination of primer F and R was designed to recognize both the insert-plus (212-base pair products) and the insert–minus cDNA (182-base pair products) and a combination of primer F and I was designed only for amplifying the insert-plus cDNA (118 base pairs). As shown in Figure 1C (lane 1), the PCR reaction with an F + R primer set amplified a 182-base pair PCR product representing MyBP-C(–) with an additional few bands with higher molecular weight, including a 212-base pair product in the C57BL/6J cDNA library. The nucleotide sequence confirmed that the 212-base pair PCR product contains exon 31A. When we used an F + I primer set, a 118-base pair PCR product from MyBP-C(+) was clearly amplified in the same cDNA library (Figure 1C, lane 2). Although the 212-base pair MyBP(+) products were hardly detected in the ICR cDNA library (Figure 1C, lane 3), the 118-base pair products were clearly detected (Figure 1C, lane 4). These results demonstrate the expression of cardiac-type MyBP-C(+) in all the cDNA libraries examined and its expression is significantly lower than that of MyBP-C(–).
Genomic Southern blotting demonstrated that a single gene encodes mouse cardiac MyBP-C (Harris et al., 2002). Therefore, MyBP-C(+) and MyBP-C(–) are expected be generated from a single gene, with an identical sequence except for the 30-base pair nucleotides insertion in MyBP-C(+), which is coded in the genomic region previously reported as an intron. In addition, two consensus splice acceptor sequences CAG (Figure 1B, *) were located next to the newly identified exon 31A and the known exon 31B with a consensus 5′ splice donor sequence gt (Figure 1B, *) located at the 5′ end of intron next to the coding sequence (exon 30), which is identical to the sequence of MyBP-C(–) as we described previously (Kasahara et al., 1994). Therefore, we concluded that both MyBP-C(+) and MyBP-C(–) mRNAs were encoded by a single gene and generated by an alternative splicing (Figure 1D).
MyBP-C(+) Reduced the Binding Affinity to Myosin Filaments and Connectin/Titin
The 10 amino acid insert encoded by exon 31A is located at the C9 domain, a domain that interacts with connectin/titin (Figure 1A). Therefore, we examined whether this insert affects its binding to connectin/titin. Full-length MyBP-C(–) or MyBP-C(+) protein were produced in sf9 cells by using a baculovirus expression system and were used for a cosedimentation assay with reconstituted myosin filaments as well as β-connectin.
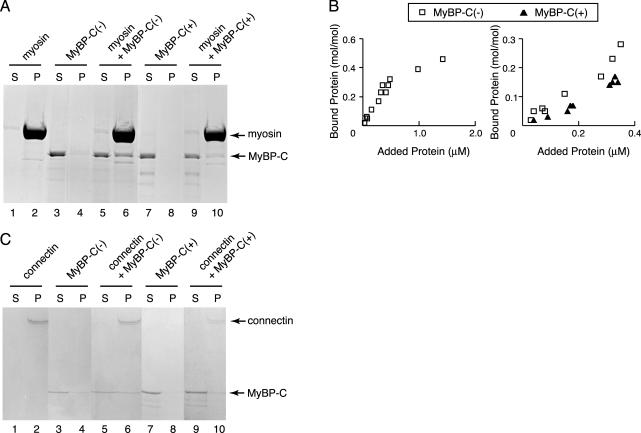
MyBP-C(–) or MyBP-C(+) was mixed with reconstituted myosin filaments prepared from rabbit cardiac muscles for 30 min at 4°C. Representative results obtained using 0.5 μM cardiac myosin and 0.15 μM recombinant MyBP-C(–) or 0.17 μM MyBP-C(+) are shown in Figure 2A. Before mixing, the majority of myosin fractionated to pellets (P) (lane 2), whereas MyBP-C fractionated to supernatants (S) (lanes 3 and 7). When MyBP-C was mixed with reconstituted myosin, MyBP-C bound to myosin fractionated in pellets (lanes 6 and 10) and unbound MyBP-C stayed in supernatants (lanes 5 and 9). Densitometric analysis showed that 40% of MyBP-C(–) or 20% of MyBP-C(+) was associated with the myosin filaments under this experimental condition.
Figure 2.
Binding of recombinant MyBP-C to myosin filaments or connectin/titin. (A) Cosedimentation assays were performed using 0.5 μM of cardiac myosin filaments and 0.15 μM MyBP-C(–) or 0.17 μM MyBP-C(+) at 0.1 M KCl. Supernatant (S) and pellet (P) were separated by centrifugation, followed by SDS-PAGE and Coomassie Brilliant Blue (CBB) staining. Arrow indicates position of myosin or MyBP-C. (B) Left, equilibrium binding reactions between MyBP-C(–) and cardiac myosin filaments. Different concentrations of MyBP-C (0–1.5 μM) were incubated with a constant concentration (0.5 μM) of cardiac myosin filaments. Samples were separated by centrifugation and dissolved by SDS-PAGE. CBB-stained gels were densitometrically scanned, and the concentrations of bound protein were normalized to the myosin concentrations. Right, binding reactions of MyBP-C (0–0.4 μM) and cardiac myosin (0.5 μM) below the saturated concentration. Open square, binding of MyBP-C(–) to the myosin filaments; closed triangle, binding of MyBP-C(+) to the myosin filaments. (C) Binding assays were performed at 0.04 μM β-connectin and 0.1 μM MyBP-C(–) or 0.13 μM MyBP-C(+) at 0.1 M KCl. (S) and (P) on the SDS-gels stained with CBB represent supernatant and pellet fractions after the centrifugation, respectively. Arrow indicates position of β-connectin or MyBP-C.
To analyze the binding of MyBP-C proteins more precisely, various concentrations of MyBP-C proteins were mixed with a constant concentration of myosin (0.5 μM). Baculovirus derived MyBP-C(–) was found to bind to myosin with a molar ratio of ∼0.5:1 at the saturated concentration which is similar to that of native MyBP-C (0.6:1) as reported previously (Alyonycheva et al., 1997) (Figure 2B, left). At a concentration between 0 and 0.4 μM, which is below the saturating concentration, MyBP-C(–) binds myosin with higher affinity than that of MyBP-C(+) (Figure 2B, right).
Similar results were obtained using β-connectin purified from chicken pectoralis muscle. Using 0.04 μM β-connectin mixed with 0.1 μM MyBP-C(–) or 0.13 μM MyBP-C(+) demonstrated that MyBP-C(+) (Figure 2C, lane 10) had a weaker interaction with β-connectin than that of MyBP-C(–) (Figure 2C, lane 6). These results demonstrate that an insertion of 10 amino acids into the connectin/titin binding domain of MyBP-C resulted in decreased affinity to connectin/titin as well as myosin.
MyBP-C(+) Does Not Effectively Assemble into Sarcomeres
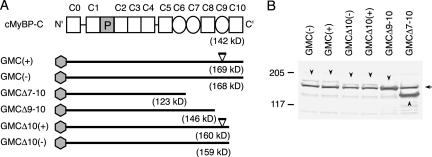
Multiple reports described that carboxyl-terminus deletion mutants of cardiac as well as skeletal MyBP-C, either deleting the myosin binding domain (C10) or the connectin/titin binding domain (C8–C10), resulted in diffusely localized MyBP-C protein impairing the endogenous sarcomeric structure in transfected cells (Gilbert et al., 1996; McConnell et al., 1999, 2001; Yang et al., 1998, 1999; Flavigny et al., 1999). Therefore, we asked whether this 10 amino acid insertion in C9 domain modifies its assembly into sarcomeres. GFP-tagged expression vectors encoding MyBP-C(+) or MyBP-C(–), as well as a series of carboxyl-terminus deletion mutants were generated (Figure 3A): GMC(+) represents GFP-tagged full-length MyBP-C(+), GMC(–), GFP-tagged full-length MyBP-C(–), GMCΔ7–10, GMC(–) with deletion of C7 to C10, GMCΔ9-10, a deletion mutant without C9 and C10, GMCΔ10(+), GMC(+) with a deletion of C10 and GMCΔ10(–), GMC(–) with a deletion of C10.
Figure 3.
(A) Structure of mouse cardiac MyBP-C protein (cMyBP-C) and construct of expression vectors. As shown in Figure 1A, the numbers above cMyBP-C indicate the position of each IgI domain (open square) and FN type III repeat (circle). Shaded hexagons indicate a GFP-tag. Estimated molecular weights are shown under each construct. The positions of the 10 amino acid insert at the C9 domain of MyBP-C are indicated by the open triangles in GMC(+) and GMCΔ10(+). GMC(+) stands for GFP-tagged full-length MyBP-C(+), GMC(–) for GFP-tagged full-length MyBP-C(–), GMCΔ7-10 for GMC(–) with deletion of C7 to C10, GMCΔ9-10 for a deletion mutant without C9 and C10, GMCΔ10(+) for GMC(+) with a deletion of C10, and GMCΔ10(–) for GMC(–) with a deletion of C10. (B) Western blot analysis of the recombinant proteins using anti-MyBP-C antibody (αccp). Molecular masses (in kilodaltons) are shown. An arrow indicates the position of endogenous chicken cardiac MyBP-C, and arrowheads indicate the product from each construct. Note that the relative mobility of GMCΔ9-10 is virtually the same as that of the endogenous MyBP-C.
These vectors were transfected into chicken cardiomyocytes and protein expression was confirmed by Western blotting by using anti-MyBP-C antibody (αccp). GFP-tagged full-length MyBP-C(–) [GMC(–)] or MyBP-C(+) [GMC(+)] proteins migrated at a slightly higher molecular weight (arrowheads) than that of endogenous chicken cardiac MyBP-C (arrow). A series of carboxyl-terminus deletion mutants were also detected with anti-MyBP-C antibody (αccp) (Figure 3B, arrowheads) and anti-GFP antibody (our unpublished data).
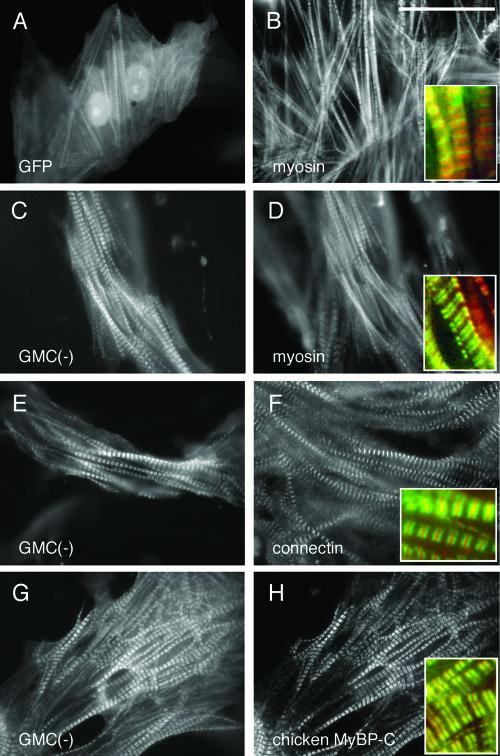
After confirming the molecular size of the GFP-fused various MyBP-C protein constructs, we examined the intracellular localization of these proteins. First, we examined the localization of nonfused GFP proteins. The majority of GFP proteins were diffusely localized in the cytoplasm (Figure 4A). Some nonfused GFP proteins were broadly incorporated into the sarcomere structure and showed overlapping staining with myosin (Figure 4B, red) that was observed in 51% of the total of 240 transfected cells (Figure 4, A and B and Table 1). However, GFP signals were not completely colocalized with myosin (red color), and the strong green GFP signals were easily detected at the gap between the A-bands (M lines) (enlarged image in Figure 4B). When GFP proteins were detected in myofibrils, 93% of cells showed organized sarcomere structure (Figure 4, A and B, and Table 1). This indicates that GFP proteins nonspecifically associate with myofibrils, but these proteins do not have deleterious effects on sarcomere organization.
Figure 4.
Incorporation of GFP-MyBP-C(–) into the A-band of sarcomeres. Chicken cardiomyocytes were transfected with pGMC(–) or pEGFP-C3 (nonfused GFP vector). Three days after the transfection, cells were fixed and immunostained with A4.1045, specific for sarcomeric myosin heavy chain (B and D); Pc72C, specific for M-line connectin/titin (F); or C315, specific for chicken cardiac MyBP-C (H). Distribution of GFP-alone (A) and localization of GMC(–) (C, E, and G) were observed using its green fluorescence. The inserts show the enlarged image (3.5×) of GFP or GMC(–) (green) and the corresponding sarcomere protein (red). Bar, 40 μm.
Table 1.
Distribution of GFP or MyBP-Cs and the A-band pattern in transfected cells
| GFP | GMC(-) | GMC(+) | GMC Δ7-10 | GMC Δ9-10 | GMC Δ10(+) | GMC Δ10(-) | |
|---|---|---|---|---|---|---|---|
| Cells with diffuse distribution of GFP or MyBP-Cs, % | 49 ± 12 | 27 ± 3 | 73 ± 1 | 61 ± 18 | 67 ± 7 | 73 ± 6 | 74 ± 15 |
| Cells with organized A-bands, % | 73 ± 4 | 76 ± 3 | 47 ± 5 | 47 ± 11 | 43 ± 11 | 35 ± 6 | 23 ± 4 |
| Number of the cells examined | 240 | 331 | 394 | 357 | 363 | 341 | 342 |
| Cells with organized A-bands in cells with myofibril localization of GFP or MyBP-Cs, % | 93 (114/122)* | 99 (241/244)* | 21 (22/107)* | 39 (56/143)* | 35 (42/120)* | 18 (16/90)* | 18 (16/91)* |
, Asterisks denote ratio of cells with organized A-bands to cells with myofibril localization of GFP or MyBP-Cs.
In contrast, in 73% of the cells expressing GFP-MyBP-C(–) protein, GFP signals were colocalized with sarcomeric myosin at A-bands; therefore, the gap between the A-bands (M-lines) were clearly observed as fluorescent negative areas (Figure 4, C and D versus A and B). Coimmunostaining with anti-connectin/titin antibody (red), a marker for the M-line, showed a narrow red line (M-line), indicating MyBP-C(–) is localized exclusive from the M-line. Of note, fluorescent signals from GFP and connectin/titin overlapped besides the M-line (yellow). However, we assume this is an artifact from the strong fluorescent signals. Indeed, anti-MyBP-C antibody recognized endogenous MyBP-C proteins exclusive from the M-line, colocalizing with GMC(–) proteins (Figure 4, G and H).
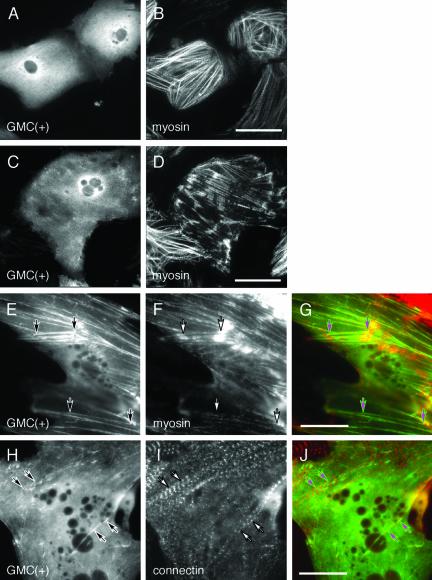
Interestingly, in the 73% of cells expressing GFP-MyBP-C(+) proteins, they diffusely localized in the cytoplasm without forming striated sarcomere structures (Figure 5, A and C, and Table 1). However, GFP-MyBP-C(+) proteins were detected as filamentous structure and were not concentrated at the M-line (Figure 4, A and B versus Figure 5, E and J). In rest of the cells (27%), GFP-MyBP-C(+) proteins colocalized with myofibrils (Figure 5, E and H).
Figure 5.
Localization of GFP-MyBP-C(+) in cardiomyocytes. Chicken cardiomyocytes were transfected with pGMC(+). Three days after the transfection, cells were fixed and immunostained with A4.1045 (B, D, F, and G) or Pc72C (I and J). GMC(+) was visualized using its green fluorescence (A, C, E, G, H, and J). Myosin filaments and connectin/titin were observed using a rhodamine channel. (G and J) Composite images GMC(+) (green) and the corresponding sarcomere protein (red). Arrows indicate colocalization of GMC(+) with myosin filaments (E–G) or with connectin/titin (H–J). Bars, 40 μm (B and D) or 30 μm (G and J).
When GFP-MyBP-C(+) proteins were diffusely localized, 57% of the cells showed well-organized sarcomeric myosin staining (Figure 5B) and 43% of cells did not (Figure 5D). When GFP-MyBP-C(+) were detected in the myofibrils, only 21% of cells showed organized sarcomere structure and 79% of these cells displayed somewhat disorganized sarcomere structure detected with anti-myosin antibody (Figure 5, E–G) and anti-connectin/titin antibody (Figure 5, H–J). In addition, we sometimes detected condensed GFP signals in the cytoplasm (our unpublished data). These data are summarized in Table 1.
These results indicate that GFP-MyBP-C(–) was effectively incorporated into sarcomere structures and colocalized with the endogenous MyBP-C protein; however, GFP-MyBP-C(+) was not effectively targeted to A-bands. When GFP-MyBP-C(+) proteins were incorporated myofibrils along with myosin filaments or connectin/titin (Figure 5, E–J, arrow), sarcomere structures were significantly disturbed (p = 0.0004, multiple analyses using the test of Ryan; Table 1). This suggests that MyBP-C(+) has dominant negative effects on sarcomere organization.
Alteration of Sarcomere Structure by MyBP-C(+) or Carboxyl-Terminus Deletion Mutants
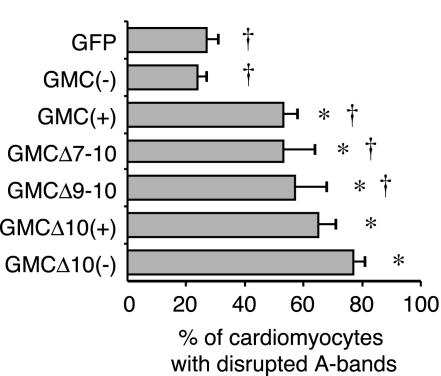
To further examine the effects of MyBP-C(+) on sarcomere structure, we compared it to several carboxyl-terminus deletion mutants that have been shown to disorganize sarcomere structure (Figure 3A) (Gilbert et al., 1996; Flavigny et al., 1999). Consistent with previous reports, 60–74% of transfected cells showed diffusely localized carboxyl-terminus deletion mutants (Table 1). Altered sarcomere structure was also detected in 53% of the cells expressing GMCΔ7-10 mutants, 57% of GMCΔ9-10, 65% of GMCΔ10(+), and 77% of GMCΔ10(–) (Figure 6, *p < 0.001). Accordingly GMCΔ10(–) showed the most severe effects on sarcomere disorganization, which was significantly higher than GMCΔ9–10, GMCΔ7–10, and GFP-MyBP-C(+) (Figure 6, †p < 0.01).
Figure 6.
Alteration of sarcomere structure by MyBP-C(+) or the carboxy-terminus deletion mutants. Three days after transfection, chicken cardiomyocytes were fixed and immunostained with the antibody for sarcomeric myosin heavy chain. GFP-positive cardiomyocytes were scored and the percentage of cardiomyocytes exhibiting an A-band disruption was determined. Average values and standard deviations are indicated. *p < 0.001 compared with GMC(–); †p < 0.01 compared with GMCΔ10(–) (test of Ryan).
MyBP-C(+) mRNA Is Dominantly Expressed in the Atria of Aged Mice
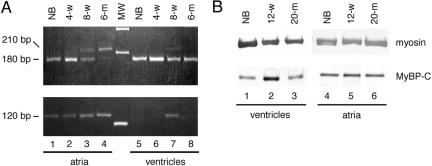
According to PCR amplification from an 8-wk-old whole heart cDNA library, expression levels of mRNA of MyBP-C(+) is lower compared with MyBP-C(–) (Figure 1C). To further examine MyBP-C(+) expression levels during different stages of heart development, we performed RT-PCR with RNA isolated from ICR mouse atria or ventricles at the newborn stage, 4 wk, 8 wk, and >6 mo of age (Figure 7A). In atria, the level of the expression of MyBP-C(–) mRNA detected as a 182-base pair product was dominant at the newborn stage and 4 wk of age, and then it gradually decreased at 8 wk and >6 mo of age. In contrast, the expression of the 212-base pair products representing MyBP-C(+) gradually increased and was dominant >6 mo of age. In ventricles, 182-base pair product of MyBP-C(–) mRNA was dominantly expressed at all stages examined and 212-base pair product of MyBP-C(+) was detected only at 8 wk of age (Figure 7A, top). This was confirmed by another primer set amplifying MyBP-C(+) cDNA as a 118-base pair product (Figure 7A, bottom). Although, the proportion of the expression of MyBP-C(–) versus MyBP-C(+) is different among individual animals examined, MyBP-C(–) is dominantly expressed in ventricles in all postnatal developmental stages, whereas MyBP-C(+) is dominantly expressed in the atria of older mice.
Figure 7.
Expression of MyBP-C during the aging of the heart. (A) RT-PCR analysis with primer F + R sets (top) or with primer F + I sets (bottom). Lanes 1–4, mRNAs from atria; lanes 5–8, mRNAs from ventricles of ICR mice, newborn (NB), 4 wk (4-w), 8 wk (8-w) and >6 mo of age (6-m). MW represents from top to bottom, 300, 200, 100 base pairs. (B) Western blotting by using anti-sarcomeric myosin antibody (top) or anti-MyBP-C antibody (αccp) which reacts to both MyBP-C(–) and MyBP-C(+) (bottom). Lanes 1–3, proteins from ventricles; lanes 4–6, proteins from atria of ICR mice, newborn (NB), 12 wk (12-w) and >20 mo of age (20-m).
Expression of Cardiac MyBP-C Protein in the Aged Mouse Heart
To further examine the expression of MyBP-C(+) protein in the aged mouse atria, we tried to raise a specific antibody for MyBP-C(+) recognizing the unique 10 amino acid insertion but we were unsuccessful after several immunizations. Western blotting using an anti-MyBP-C antibody (αccp), which recognizes both of MyBP-C(–) and MyBP-C(+) shows that the total amount of ventricular MyBP-C expression is increased at 12 wk of age and then decreased at >20 mo of age. In contrast, the expression of the total amount of atrial MyBP-C, MyBP-C(–) plus MyBP-C(+), was constant at all three stages examined (Figure 7B).
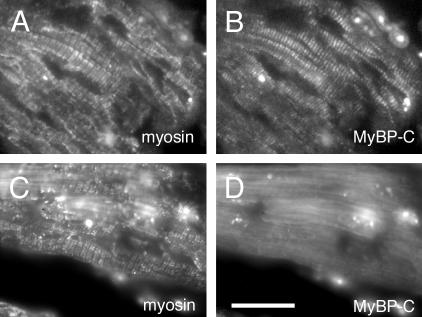
In aged mouse hearts (> 20 mo of age), ventricular MyBP-C exhibited striated sarcomeric pattern colocalizing with myosin (Figure 8, A and B); however, atrial MyBP-C was diffusely localized in the cytoplasm despite the striated sarcomere formation stained with anti-myosin antibody (Figure 8, C and D). These results indicate that the localization of MyBP-C is specifically stage and tissue (aria versus ventricles) regulated. According to the dominant expression of MyBP-C(+) in aged mouse demonstrated by RT-PCR, diffusely localized MyBP-C proteins in aged atria are likely the MyBP-C(+) form.
Figure 8.
Typical distribution of MyBP-C in the aged mouse heart. Cryosections (3 μm) of ventricles from >20-mo-old mice (A and B) and atria from >20-mo-old mice (C and D) were immunostained with anti-sarcomeric myosin antibody (A and C) or anti-MyBP-C antibody (αccp). Ventricular MyBP-C showed striated sarcomere structure (B), whereas atrial MyBP-C were diffusely localized in the cytoplasm without assembling into organized myosin filaments (C and D). Bar, 20 μm.
DISCUSSION
We found a novel splicing variant of MyBP-C/C-protein mRNA that has an additional 30 nucleotide sequence, resulting in a 10 amino acid insertion in the C9 domain, which is known as the connectin/titin binding region. We term this insertion-plus variant MyBP-C(+) and the previously described insertion-minus MyBP-C MyBP-C(–). A 30-base pair insertion sequence (exon 31A) was found in the cardiac MyBP-C genomic sequence previously reported as an intron sequence between exon 30 and 31B. A consensus splice donor site, gt, was found at the 5′ end of this intron, whereas two consensus acceptor sites were found with 30-base pair distance; one followed by exon 31A and the other followed by exon 31B. Cardiac MyBP-C is coded by a single gene. MyBP-C(+) contains the identical coding sequence with a 30-base pair insertion; therefore, the mRNAs encoding these two isoforms of mouse cardiac MyBP-C are generated by an alternative splicing mechanism using the two acceptor sites regulated in a spatial and temporal manner. Similar internal splicings have been found in other genes to produce multiple isoforms at different developmental stages and in different tissues (Paul et al., 1986; reviewed in Breitbart et al., 1987; Magnuson et al., 1991), including human and mouse fibronectin (Tamkun et al., 1984; Kornblihtt et al., 1985; reviewed in Schwarzbauer, 1991).
Biochemical analyses demonstrated several unique characteristics of MyBP-C(+) compared with MyBP-C(–). In a cosedimentation assay, baculovirus-derived MyBP-C(+) decreased binding affinity to connectin/titin and myosin filaments, whereas MyBP-C(–) showed a very similar binding affinity as native MyBP-C(–) described previously (Alyonycheva et al., 1997). These results indicate the baculovirus derived MyBP-C(–) and the native MyBP-C(–) have very similar protein structures, but the 10 amino acid insertion at C9 domain changes its structure.
Six of 10 amino acid residues are hydrophobic, and these hydrophobic residues likely disrupt the α-helical stretch of MyBP-C(–) and replace it with beta-sheets at the C9 domain according to the Chou-Fasman protein secondary analyses of a GENETYX program. Because C8, C9, and C10 domains of MyBP-C are known to directly bind to a subset of Ig domains of connectin/titin (Freiburg and Gautel, 1996), conformational changes due to the 10 amino acid insertion will decrease the association between MyBP-C and connectin/titin. In addition, our experiments showed that the insertion into the C9 domain significantly decreased the binding to myosin filaments. The major myosin-binding domain of cardiac and skeletal MyBP-C was mapped at the carboxyl-terminal C10 domain (Okagaki et al., 1993; Alyonycheva et al., 1997), but a recent report from Welikson and Fischman demonstrated that C7–C9 domains of the chicken skeletal MyBP-C also have an ability to bind with myosin (Welikson and Fischman, 2002). Therefore, we assume that the inserted 10 amino acids in the C9 domain modify the structure of MyBP-C, resulting in alteration of conformation or direction of the C10 domain, which is important for binding with myosin, or the inserted 10 amino acids directly reduced binding to myosin filaments.
Supporting these in vitro binding assays, transfection experiments showed that GFP-MyBP-C(+) was not effectively incorporated into sarcomere structures compared with GFP-MyBP-C(–). Because MyBP-C(+) could still bind to myosin filaments and connectin/titin with lower affinity, it sometimes associated with myofibrils, but it could not fully compete with and replace the endogenous chick MyBP-C protein, which does not include the 10 amino acid insertion (Yasuda et al., 1995). We could not fully explain why MyBP-C(+) is sometimes incorporated into myofibrils and sometimes diffusely distributed in the cytoplasm. It may be due to the different accumulation level of the MyBP-C(+) in different cells or the timing of association of MyBP-C(+) with sarcomeric components during sarcomere assembly.
The carboxyl-terminus end of MyBP-C is known to be important for sarcomere assembly, and expression of the carboxyl-terminus deletion mutants impaired the endogenous sarcomeric structure in the transfected myocytes (Gilbert et al., 1996; McConnell et al., 1999, 2001; Flavigny et al., 1999; Yang et al., 1998, 1999). We confirmed these results in >50% of the cells transfected with the several carboxyl-terminus deletion mutants. In our hands, GMCΔ10(–) showed the strongest effect on sarcomere disorganization in chicken cardiomyocytes followed by GMCΔ10(+), GMCΔ9-10, and then GMCΔ7-10, indicating that the C10 deletion mutants have the strongest effects and that further truncation decreases the effects. Because MyBP-C binds to connectin/titin through C8–C10 domains, the C10 deletion mutant is expected to bind connectin/titin with higher affinity than other deletion mutants.
We hypothesized that the lack of myosin binding but the preserved connectin/titin binding may cause the severe effects on sarcomere disorganization. These mutants could be replaced with the endogenous MyBP-C more effectively because of partially reserved binding to connectin/titin. Accordingly, GMCΔ7-10, which lacks both connectin/titin and myosin binding, showed the mildest effects on sarcomere disorganization, but still >50% of the transfected cells with GMCΔ7-10 showed disorganized sarcomere structure. We assume that interaction with other sarcomeric proteins through domain C0–C6 of GMCΔ7-10 is responsible for the effects, including the N-terminal myosin S2 binding site (Gruen and Gautel, 1999).
Surprisingly, MyBP-C(+) disorganizes sarcomere structure as effectively as carboxyl-terminus deletion mutant GMCΔ7-10, showing a similar deleterious effect on sarcomeres. As assessed by mRNA levels at least, MyBP-C(+) is dominantly expressed in aged mouse atria; therefore, it may act as a disruptive polypeptide via a dominant-negative effect on remaining MyBP-C(–) or it may partially compensate for the decreased expression of MyBP-C(–) in aged atria. It is generally accepted that the aged heart is associated with a number of characteristic morphological, histological, biochemical, and functional changes, including a reduction in the number of myocytes, the development of cardiac fibrosis, diastolic dysfunction, and a decrease in the intracellular response to β-adrenergic stimulation (reviewed in Roffe, 1998). These changes are associated with hypertrophy as a result of a net decrease in the number of cardiomyocytes (Fleg, 1986) and isoform transition of myosin heavy chain (MHC) from α-MHC to β-MHC in rat ventricles (Lompre et al., 1984; Schuyler and Yarbrough, 1990; Wahr et al., 2000). We have not examined the molecular mechanisms explaining the switch from MyBP-C(–) to MyBP-C(+) and its effects on aged atrium nor have we investigated whether any novel cardiac MyBP-C isoform is expressed in other animals besides the mouse. But interestingly, it has been demonstrated that senescent rat atrial myocytes displayed abnormal myofilament arrays (Feldman and Navaratnam, 1981).
In summary, we found a novel alternatively spliced form of cardiac MyBP-C that is abundantly expressed in aged mouse atria. This isoform, MyBP-C(+), shows a decreased binding to myosin filaments and connectin/titin and a decreased incorporation into sarcomere structure, suggesting it is involved in the morphological and functional changes of cardiac muscle during aging.
Acknowledgments
We thank Ellen O. Weinberg and Julie R. McMullen for critical reading of the manuscript; Hiroshi Abe, Nozomi Kunugihara, Kentaro Kayukawa, Donald A. Fischman, and Robert E. Welikson for valuable suggestions; and Sumiko Kimura for β-connectin and Pc72C. This research was supported by research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the National Center of Neurology and Psychiatry of the Ministry of Health, Labour and Welfare of Japan.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-10-0685. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-10-0685.
References
- Alyonycheva, T.N., Mikawa, T., Reinach, F.C., and Fischman, D.A. (1997). Isoform-specific interaction of the myosin-binding proteins (MyBPs) with skeletal and cardiac myosin is a property of the C-terminal immunoglobulin domain. J. Biol. Chem. 272, 20866–20872. [DOI] [PubMed] [Google Scholar]
- Bähler, M., Moser, H., Eppenberger, H.M., and Wallimann, T. (1985). Heart C-protein is transiently expressed during skeletal muscle development in the embryo, but persists in cultured myogenic cells. Dev. Biol. 112, 345–352. [DOI] [PubMed] [Google Scholar]
- Blin, N., and Stafford, D. W. (1976). A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 3, 2303–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonne, G., Carrier, L., Bercovici, J., Cruaud, C., Richard, P., Hainque, B., Gautel, M., Labeit, S., James, M., and Beckmann, J. (1995). Cardiac myosin binding protein-C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nat. Genet. 11, 438–440. [DOI] [PubMed] [Google Scholar]
- Breitbart, R.E., Andreadis, A., and Nadal-Ginard, B. (1987). Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu. Rev. Biochem. 56, 467–495. [DOI] [PubMed] [Google Scholar]
- Carrier, L., et al. (1997). Organization and sequence of human cardiac myosin binding protein C gene (MYBPC3) and identification of mutations predicted to produce truncated proteins in familial hypertrophic cardiomyopathy. Circ. Res. 80, 427–434. [PubMed] [Google Scholar]
- Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Craig, R., and Offer, G. (1976). The location of C-protein in rabbit skeletal muscle. Proc. R. Soc. Lond. B. Biol. Sci. 192, 451–461. [DOI] [PubMed] [Google Scholar]
- Einheber, S., and Fischman, D.A. (1990). Isolation and characterization of a cDNA clone encoding avian skeletal muscle C-protein: an intracellular member of the immunoglobulin superfamily. Proc. Natl. Acad. Sci. USA 87, 2157–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, M.L., and Navaratnam, V. (1981). Ultrastructural changes in atrial myocardium of the ageing rat. J. Anat. 133, 7–17. [PMC free article] [PubMed] [Google Scholar]
- Flavigny, J., Souchet, M., Sebillon, P., Berrebi-Bertrand, I., Hainque, B., Mallet, A., Bril, A., Schwartz, K., and Carrier, L. (1999). COOH-terminal truncated cardiac myosin-binding protein C mutants resulting from familial hypertrophic cardiomyopathy mutations exhibit altered expression and/or incorporation in fetal rat cardiomyocytes. J. Mol. Biol. 294, 443–456. [DOI] [PubMed] [Google Scholar]
- Fleg, J.L. (1986). Alterations in cardiovascular structure and function with advancing age. Am. J. Cardiol. 57, 33C–44C. [DOI] [PubMed] [Google Scholar]
- Fougerousse, F., Delezoide, A.L., Fiszman, M.Y., Schwartz, K., Beckmann, J.S., and Carrier, L. (1998). Cardiac myosin binding protein C gene is specifically expressed in heart during murine and human development. Circ. Res. 82, 130–133. [DOI] [PubMed] [Google Scholar]
- Freiburg, A., and Gautel, M. (1996). A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur. J. Biochem. 235, 317–323. [DOI] [PubMed] [Google Scholar]
- Fürst, D.O., Vinkemeier, U., and Weber, K. (1992). Mammalian skeletal muscle C-protein: purification from bovine muscle, binding to titin and the characterization of a full-length human cDNA. J. Cell Sci. 102, 769–778. [DOI] [PubMed] [Google Scholar]
- Gautel, M., Furst, D.O., Cocco, A., and Schiaffino, S. (1998). Isoform transitions of the myosin binding protein C family in developing human and mouse muscles: lack of isoform transcomplementation in cardiac muscle. Circ. Res. 82, 124–129. [DOI] [PubMed] [Google Scholar]
- Gautel, M., Zuffardi, O., Freiburg, A., and Labeit, S. (1995). Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? EMBO J. 14, 1952–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, R., Kelly, M.G., Mikawa, T., and Fischman, D.A. (1996). The carboxyl terminus of myosin binding protein C (MyBP-C, C-protein) specifies incorporation into the A-band of striated muscle. J. Cell Sci. 109, 101–111. [DOI] [PubMed] [Google Scholar]
- Gruen, M., and Gautel, M. (1999). Mutations in beta-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. J. Mol. Biol. 286, 933–949. [DOI] [PubMed] [Google Scholar]
- Harpaz, Y., and Chothia, C. (1994). Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J. Mol. Biol. 238, 528–539. [DOI] [PubMed] [Google Scholar]
- Harris, S.P., Bartley, C.R., Hacker, T.A., McDonald, K.S., Douglas, P.S., Greaser, M.L., Powers, P.A., and Moss, R.L. (2002). Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ. Res. 90, 594–601. [DOI] [PubMed] [Google Scholar]
- Hartzell, H.C. (1985). Effects of phosphorylated and unphosphorylated C-protein on cardiac actomyosin ATPase. J. Mol. Biol. 186, 185–195. [DOI] [PubMed] [Google Scholar]
- Hyde, A., Blondel, B., Matter, A., Cheneval, J.P., Filloux, B., and Girardier, L. (1969). Homo- and heterocellular junctions in cell cultures: an electrophysiological and morphological study. Prog. Brain Res. 31, 283–311. [DOI] [PubMed] [Google Scholar]
- Kasahara, H., Itoh, M., Sugiyama, T., Kido, N., Hayashi, H., Saito, H., Tsukita, S., and Kato, N. (1994). Autoimmune myocarditis induced in mice by cardiac C-protein. Cloning of complementary DNA encoding murine cardiac C-protein and partial characterization of the antigenic peptides. J. Clin. Investig. 94, 1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima, M., Kitani, S., Tanaka, T., and Obinata, T. (1986). The earliest form of C-protein expressed during striated muscle development is immunologically the same as cardiac-type C-protein. J. Biochem. 99, 1037–1047. [DOI] [PubMed] [Google Scholar]
- Kimura, S., and Maruyama, K. (1983). Preparation of native connectin from chicken breast muscle. J Biochem 94, 2083–2085. [DOI] [PubMed] [Google Scholar]
- Kimura, A., et al. (1997). Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat. Genet. 16, 379–382. [DOI] [PubMed] [Google Scholar]
- Koretz, J.F., Irving, T.C., and Wang, K. (1993). Filamentous aggregates of native titin and binding of C-protein and AMP-deaminase. Arch. Biochem. Biophys. 304, 305–309. [DOI] [PubMed] [Google Scholar]
- Kornblihtt, A.R., Umezawa, K., Vibe-Pedersen, K., and Baralle, F.E. (1985). Primary structure of human fibronectin: differential splicing may generate at least 10 polypeptides from a single gene. EMBO J. 4, 1755–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasawa, M., Sato, N., Matsuda, A., Koshida, S., Totsuka, T., and Obinata, T. (1999). Differential expression of C-protein isoforms in developing and degenerating mouse striated muscles. Muscle Nerve 22, 196–207. [DOI] [PubMed] [Google Scholar]
- Lin, Z.X., Holtzer, S., Schultheiss, T., Murray, J., Masaki, T., Fischman, D.A., and Holtzer, H. (1989). Polygons and adhesion plaques and the disassembly and assembly of myofibrils in cardiac myocytes. J. Cell Biol. 108, 2355–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lompre, A.M., Nadal-Ginard, B., and Mahdavi, V. (1984). Expression of the cardiac ventricular alpha- and beta-myosin heavy chain genes is developmentally and hormonally regulated. J. Biol. Chem. 259, 6437–6446. [PubMed] [Google Scholar]
- Magnuson, V.L., Young, M., Schattenberg, D.G., Mancini, M.A., Chen, D.L., Steffensen, B., and Klebe, R.J. (1991). The alternative splicing of fibronectin pre-mRNA is altered during aging and in response to growth factors. J. Biol. Chem. 266, 14654–14662. [PubMed] [Google Scholar]
- Maron, B.J., Bonow, R.O., Cannon, R.O., Leon, M.B., and Epstein, S.E. (1987a). Hypertrophic cardiomyopathy. Interrelations of clinical manifestations, pathophysiology, and therapy (1). N. Engl. J. Med. 316, 780–789. [DOI] [PubMed] [Google Scholar]
- Maron, B.J., Bonow, R.O., Cannon, R.O., Leon, M.B., and Epstein, S.E. (1987b). Hypertrophic cardiomyopathy. Interrelations of clinical manifestations, pathophysiology, and therapy (2). N. Engl. J. Med. 316, 844–852. [DOI] [PubMed] [Google Scholar]
- Maruyama, K. (1986). Connectin, an elastic filamentous protein of striated muscle. Int. Rev. Cytol. 104, 81–114. [DOI] [PubMed] [Google Scholar]
- McConnell, B.K., et al. (1999). Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J. Clin. Investig. 104, 1235–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, B.K., et al. (2001). Comparison of two murine models of familial hypertrophic cardiomyopathy. Circ. Res. 88, 383–389. [DOI] [PubMed] [Google Scholar]
- Mohamed, A.S., Dignam, J.D., and Schlender, K.K. (1998). Cardiac myosin-binding protein C (MyBP-C): identification of protein kinase A and protein kinase C phosphorylation sites. Arch. Biochem. Biophys. 358, 313–319. [DOI] [PubMed] [Google Scholar]
- Moos, C., Mason, C.M., Besterman, J.M., Feng, I.N., and Dubin, J.H. (1978). The binding of skeletal muscle C-protein to F-actin, and its relation to the interaction of actin with myosin subfragment-1. J. Mol. Biol. 124, 571–586. [DOI] [PubMed] [Google Scholar]
- Obinata, T. (1985). Changes in myofibrillar protein isoform expression during chicken skeletal muscle development. Zool. Sci. 2, 833–847. [Google Scholar]
- Obinata, T., Reinach, F.C., Bader, D.M., Masaki, T., Kitani, S., and Fischman, D.A. (1984). Immunochemical analysis of C-protein isoform transitions during the development of chicken skeletal muscle. Dev. Biol. 101, 116–124. [DOI] [PubMed] [Google Scholar]
- Offer, G., Moos, C., and Starr, R. (1973). A new protein of the thick filaments of vertebrate skeletal myofibrils. Extraction, purification and characterization. J. Mol. Biol. 74, 653–676. [DOI] [PubMed] [Google Scholar]
- Okagaki, T., Weber, F.E., Fischman, D.A., Vaughan, K.T., Mikawa, T., and Reinach, F.C. (1993). The major myosin-binding domain of skeletal muscle MyBP-C (C protein) resides in the COOH-terminal, immunoglobulin C2 motif. J. Cell Biol. 123, 619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, J.I., Schwarzbauer, J.E., Tamkun, J.W., and Hynes, R.O. (1986). Cell-type-specific fibronectin subunits generated by alternative splicing. J. Biol. Chem. 261, 12258–12265. [PubMed] [Google Scholar]
- Pepe, F.A., and Drucker, B. (1975). The myosin filament. III. C-protein. J. Mol. Biol. 99, 609–617. [DOI] [PubMed] [Google Scholar]
- Perry, S.V. (1955). Myosin adenosine triphosphatase. Methods Enzymol. 2, 582–588. [Google Scholar]
- Richards, E.G., Chung, C.S., Menzel, D.B., and Olcott, H.S. (1967). Chromatography of myosin on diethylaminoethyl-Sephadex A-50. Biochemistry 6, 528–540. [DOI] [PubMed] [Google Scholar]
- Roffe, C. (1998). Ageing of the heart. Br. J. Biomed. Sci. 55, 136–148. [PubMed] [Google Scholar]
- Rottbauer, W., et al. (1997). Novel splice donor site mutation in the cardiac myosin-binding protein-C gene in familial hypertrophic cardiomyopathy. Characterization Of cardiac transcript and protein. J. Clin. Investig. 100, 475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, T.A. (1959). Multiple comparisons in psychologic research. Psychol. Bull. 56, 26–47. [DOI] [PubMed] [Google Scholar]
- Ryan, T.A. (1960). Significance tests for multiple comparisons of proportions, variances, and other statistics. Psychol. Bull. 57, 318–328. [DOI] [PubMed] [Google Scholar]
- Schuyler, G.T., and Yarbrough, L.R. (1990). Effects of age on myosin and creatine kinase isoforms in left ventricles of Fischer 344 rats. Mech. Ageing Dev. 56, 23–38. [DOI] [PubMed] [Google Scholar]
- Schwarzbauer, J.E. (1991). Alternative splicing of fibronectin: three variants, three functions. Bioessays 13, 527–533. [DOI] [PubMed] [Google Scholar]
- Seidman, J.G., and Seidman, C. (2001). The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell 104, 557–567. [DOI] [PubMed] [Google Scholar]
- Soeno, Y., Yajima, H., Kawamura, Y., Kimura, S., Maruyama, K., and Obinata, T. (1999). Organization of connectin/titin filaments in sarcomeres of differentiating chicken skeletal muscle cells. Mol. Cell Biochem. 190, 125–131. [PubMed] [Google Scholar]
- Tamkun, J.W., Schwarzbauer, J.E., and Hynes, R.O. (1984). A single rat fibronectin gene generates three different mRNAs by alternative splicing of a complex exon. Proc. Natl. Acad. Sci. USA 81, 5140–5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahr, P.A., Michele, D.E., and Metzger, J.M. (2000). Effects of aging on single cardiac myocyte function in Fischer 344 x Brown Norway rats. Am. J. Physiol. 279, H559–H565. [DOI] [PubMed] [Google Scholar]
- Watkins, H., Conner, D., Thierfelder, L., Jarcho, J. A., MacRae, C., McKenna, W. J., Maron, B.J., Seidman, J.G. and Seidman, C.E. (1995). Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat. Genet. 11, 434–437. [DOI] [PubMed] [Google Scholar]
- Weber, F.E., Vaughan, K.T., Reinach, F.C., and Fischman, D.A. (1993). Complete sequence of human fast-type and slow-type muscle myosin-binding-protein C (MyBP-C). Differential expression, conserved domain structure and chromosome assignment. Eur. J. Biochem. 216, 661–669. [DOI] [PubMed] [Google Scholar]
- Welikson, R.E., and Fischman, D.A. (2002). The C-terminal IgI domains of myosin-binding proteins C and H (MyBP-C and MyBP-H) are both necessary and sufficient for the intracellular crosslinking of sarcomeric myosin in transfected non-muscle cells. J. Cell Sci. 115, 3517–3526. [DOI] [PubMed] [Google Scholar]
- Yang, Q., Sanbe, A., Osinska, H., Hewett, T.E., Klevitsky, R., and Robbins, J. (1998). A mouse model of myosin binding protein C human familial hypertrophic cardiomyopathy. J. Clin. Investig. 102, 1292–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q., Sanbe, A., Osinska, H., Hewett, T.E., Klevitsky, R., and Robbins, J. (1999). In vivo modeling of myosin binding protein C familial hypertrophic cardiomyopathy. Circ. Res. 85, 841–847. [DOI] [PubMed] [Google Scholar]
- Yasuda, M., Koshida, S., Sato, N., and Obinata, T. (1995). Complete primary structure of chicken cardiac C-protein (MyBP-C) and its expression in developing striated muscles. J. Mol. Cell Cardiol. 27, 2275–2286. [DOI] [PubMed] [Google Scholar]
- Yu, B., et al. (1998). Molecular pathology of familial hypertrophic cardiomyopathy caused by mutations in the cardiac myosin binding protein C gene. J. Med. Genet. 35, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]