Abstract
In budding yeast, HXT genes encoding hexose permeases are induced by glucose via a mechanism in which the F box protein Grr1 antagonizes activity of the transcriptional repressor Rgt1. Neither the mechanism of Rgt1 inactivation nor the role of Grr1 in that process has been understood. We show that glucose promotes phosphorylation of Rgt1 and its dissociation from HXT gene promoters. This cascade of events is dependent upon the F-box protein Grr1. Inactivation of Rgt1 is sufficient to explain the requirement for Grr1 but does not involve Rgt1 proteolysis or ubiquitination. We show that inactivation of Mth1 and Std1, known negative regulators of HXT gene expression, leads to the hyperphosphorylation of Rgt1 and its dissociation from HXT promoters even in the absence of glucose. Furthermore, inactivation of Mth1 and Std1 bypasses the requirement for Grr1 for induction of these events, suggesting they are targets for inactivation by Grr1. Consistent with that proposal, Mth1 is rapidly eliminated in response to glucose via a mechanism that requires Grr1. Based upon these data, we propose that glucose acts via Grr1 to promote the degradation of Mth1. Degradation of Mth1 leads to phosphorylation and dissociation of Rgt1 from HXT promoters, thereby activating HXT gene expression.
INTRODUCTION
The malleability of gene expression is a primary determinant of adaptability. All organisms can adapt to both internal and environmental changes via alterations in the pattern of gene expression. One of the primary manifestations of that capacity is the ability of cells to use different carbon sources. This is, in part, a consequence of the output of a complex network of sensors and signaling pathways that leads to the remodeling of the expression of transporters and metabolic enzymes.
The preferred carbon source for yeast, as for most cells, is glucose. Introduction of glucose to growth medium leads to the rapid repression of genes that are nonessential for its utilization and the induction of genes that facilitate its uptake and metabolism. Among the many genes induced by glucose is a family of hexose transporters encoded by the HXT genes (Gancedo, 1998; Özcan and Johnston, 1999; Van Belle and André, 2001). The HXT family consists of 17 genes encoding proteins that are closely related but subject to distinct patterns of regulation by glucose. The best-characterized members of the family include HXT1, which is induced in high but not low glucose; HXT2 and HXT4, which are induced in low but not high glucose; and HXT3, which is induced in both low and high glucose. These patterns of expression correlate roughly with the affinity of the specific transporter for glucose (Özcan and Johnston, 1999).
Glucose regulation of HXT gene expression is mediated via signals emanating from the low- and high-affinity glucose receptors Snf3 and Rgt2, respectively, both of which are closely related to members of the hexose transporter family but have extended carboxyterminal cytoplasmic domains that are required for signal transduction (Özcan et al., 1996a; Özcan et al., 1998). Although relatively little is known about the arrangement of downstream elements of that pathway, several elements required for signaling have been characterized sufficiently to predict their function.
First, repression of HXT gene expresssion in the absence of glucose is known to require RGT1, which encodes a DNA binding protein that recognizes elements in the HXT promoters (Özcan et al., 1996b). RGT1 is required for transcriptional repression of HXT1-HXT4 in the absence of glucose (Vallier et al., 1994; Özcan et al., 1996b). In contrast, the transcriptional repression of HXT2 and HXT4 observed in high glucose is apparently mediated via a separate mechanism involving MIG1 (Gancedo, 1998). In addition to its capacity to act as a transcriptional repressor, there is evidence that RGT1 can act as a transcriptional activator (Özcan and Johnston, 1995; Özcan et al., 1996b). Whether those effects are all mediated at the level of the HXT promoters is not known.
Derepression of HXT gene expression in the presence of glucose requires the F-box protein Grr1 (Özcan et al., 1993; Vallier et al., 1994; Özcan and Johnston, 1995). Inactivation of RGT1 bypasses the requirement for GRR1 to induce HXT gene expression, thereby placing it upstream of RGT1 in the glucose-signaling pathway. Because Grr1 is an established component of a Skp1/Cullin/F-box protein (SCF) E3 ubiquitin ligase complex and mediates the ubiquitination of proteins destined for proteolysis via the proteasome (Skowyra et al., 1997; Patton et al., 1998), it has been proposed that Grr1 antagonizes Rgt1 by targeting it for degradation (Özcan and Johnston, 1999). Consistent with that hypothesis, two other components of that complex, Skp1 and Cdc53, have been shown to be required for transcriptional activation in response to glucose (Li and Johnston, 1997). However, Cdc34, the E2 ubiquitin-conjugating enzyme required for the SCF-mediated ubiquitination of established SCFGrr1 targets, seems to be dispensable for HXT gene induction (Li and Johnston, 1997). Surprisingly, the protein motifs of Grr1 required for recognition of established ubiquitination targets seem to be distinct from those required for regulation of HXT gene expression, suggesting that the properties of the targets involved in those two processes are distinct (Hsiung et al., 2001).
Two other genes, MTH1 and STD1, have been shown to be important for maintenance of HXT gene repression (Schmidt et al., 1999; Schulte et al., 2000). Whereas inactivation of either gene alone results in limited defects in regulation of HXT gene expression, inactivation of both genes results in derepression in the absence of glucose, suggesting a partial functional overlap. This is consistent with the high degree of sequence homology between the encoded proteins (Std1 and Mth1 are 61% identical) (Hubbard et al., 1994). Although reports are conflicting, Mth1 has been reported to interact with the cytoplasmic domains of either one or both of the membrane-bound glucose sensors, Snf3 and Rgt2 (Schmidt et al., 1999; Lafuente et al., 2000). The inference that MTH1 can interact with HXT promoters has been derived from the analysis of mutant alleles (Özcan et al., 1993). These interactions are consistent with its reported localization to both the cytoplasmic membrane and the nucleus (Schmidt et al., 1999), although it should be noted that the retention of those proteins at the plasma membrane does not require Rgt2 or Snf3. Std1 has been reported to interact biochemically with Rgt1 (Tomas-Cobos and Sanz, 2002) as well as with Snf1, a protein kinase involved in global regulation of gene expression in response to glucose (Hubbard et al., 1994; Tomas-Cobos and Sanz, 2002; Kuchin et al., 2003). However, despite the relatively extensive analysis of Mth1 and Std1, neither their cellular functions nor their role in this pathway is understood.
To elucidate the mechanism of glucose regulation of HXT gene expression, we investigated the role of Grr1 in that process. This study confirms a recent report (Mosley et al., 2003) that Rgt1 binds to HXT1-HXT4 promoters in vivo under repressing conditions but dissociates from those promoters in the presence of glucose. Dissociation of Rgt1 from these promoters is associated with its hyperphosphorylation. Grr1 is required for both the hyperphosphorylation of Rgt1 and its dissociation from promoters. However, we show that Rgt1 is not a direct target for ubiquitination by SCFGrr1. Instead, Grr1 is required to inactivate Mth1 and Std1 in response to glucose. Mth1 inactivation seems to occur at the level of degradation. Based upon these data, we conclude that glucose acts via Grr1 to regulate the abundance of Mth1. Inactivation of Mth1 leads to hyperphosphorylation of Rgt1 and dissociation from HXT promoters.
EXPERIMENTAL PROCEDURES
Yeast Strains, Culture, and Plasmids
The relevant genotypes of the yeast strains used in this study are listed in Table 1. All strains are isogenic with W303a. All strains were grown in standard culture media and standard yeast genetic methods were used.
Table 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| K699 | a ade2-1 can 1-100 his3-1.15 leu2-3,112 trp1-1 ura3 | Willems et al., 1996 |
| CWY1121 | a RGT1-3HA::KanMX2 | This study |
| CWY1128 | a grr1::LEU2 RGT1-3HA::KanMX2 | This study |
| KFY907 | a mth1::ZEO RGT1-3HA::KanMX2 | This study |
| KFY913 | a std1::HphMX4 RGT1-3HA::KanMX2 | This study |
| KFY916 | a mth1::ZEO std1::HphMX4 RGT1-3HA::KanMX2 | This study |
| KFY903 | a grr1::LEU2 mth1::ZEO RGT1-3HA::KanMX2 | This study |
| KFY906 | a grr1::LEU2 std1::HphMX4 RGT1-3HA::KanMX2 | This study |
| KFY917 | a grr1::LEU2 mth1::ZEO std1::HphMX4 RGT1-3HA::KanMX2 | This study |
| CWY1341 | a rgt1::KanMX2 | This study |
| YHY452 | a grr1::LEU2 rgt1::KanMX2 | This study |
| CWY1310 | a MTH1-6xmyc::KanMX2 | This study |
| CWY1308 | a STD1-6xmyc::KanMX2 | This study |
Deletion mutants were constructed using polymerase chain reaction (PCR)-based methods as described previously (Wach et al., 1994; Longtine et al., 1998; Goldstein and McCusker, 1999). Deletion mutants were confirmed by PCR. To generate chromosomally carboxy-terminal epitope-tagged proteins, the 3′ end was amplified by PCR and cloned into the pKAN vector or one of its derivates (Haase, Wolff, and Reed, unpublished data) for targeted integration or by the PCR-based method as described previously (Longtine et al., 1998). DNA fragments were sequenced to ensure fidelity. Sequences of all oligonucleotides used for these manipulations are available upon request.
Protein and RNA Analysis
If not stated otherwise, cells were grown in 2% galactose to early log-phase, half of the cells were then shifted to 4% glucose for 120 min, after which cells were harvested by filtration. Cells were then washed in ice-cold water and pellets were stored at [–80°C].
For reverse transcription (RT) analysis, RNA isolation, cDNA synthesis, and PCR were performed as described previously (Hsiung et al., 2001). Sequences of primers are available upon request.
For Western blot analysis, protein extracts were prepared either by lyses in urea-buffer or a trichloroacetic acid (TCA)/urea extraction method. Urea-buffer extracts were prepared in urea-buffer (8 M urea, 200 mM NaCl, 10 mM Tris pH 7.5, 0.2% SDS, protease inhibitors [10 mM phenylmethylsulfonyl fluoride (PMSF), 2 mg/ml aprotenin, leupetin, and pepstatin A], phosphatase inhibitors [10 mM sodium pyrophosphate, 5 mM EDTA, 5 mM EGTA, 50 mM NaF, and 0.1 mM orthovanadate]). Cells were broken with glass beads 4×20 s in a FastPrep FP120 (BIO 101, Vista, CA/Savant Instruments, Holbrook, NY). Samples were diluted to 4 M urea before loading onto SDS-gels. For TCA-urea extraction cells were broken in 5–10 volumes of 20% TCA with glass beads 4 times 40 s in a FastPrep FP120. The TCA pellet was washed twice in acetone and then resuspended in extraction buffer (8 M urea, 4% SDS, protease inhibitors [10 mM PMSF, 2g/ml aprotenin, leupetin, and pepstatin A], phosphatase inhibitors [10 mM sodium pyrophosphate, 5 mM EDTA, 5 mM EGTA, 50 mM NaF, and 0.1 mM orthovanadate]). Samples were diluted to 4 M urea and 2% SDS before loading onto SDS-gels.
Chromatin Immunoprecipitation Assay
Cells were grown as described above. Chromatin immunoprecipitation assays were performed by a protocol based on Tanaka et al. (1997). Briefly, DNA-protein cross-links were induced in vivo by incubation of cells with formaldehyde (final concentration 1%) for 20 min at room temperature, followed by the addition of glycine to a final concentration of 125 mM for 5 min at room temperature. Cells were washed three times with ice-cold Tris-buffered saline and cell pellets were frozen. Frozen cell pellets were resuspended to 1.5 × 109 cells/ml in lysis buffer (50 mM HEPES pH 7.5, 140 mM NaCl, 1% Triton X-100, 0.1% Na deoxycholate, 50 μM PMSF, 2 μg/ml aprotinin, leupeptin, and pepstatin A). Cells were broken with glass beads 4 × 20 s at setting 4.5 in a FastPrep FP120 (BIO 101/Savant). After a 15-min centrifugation the supernatant was discarded and the pellet (chromatin fraction) was resuspended in the initial volume of lysis buffer. The DNA was fragmented to ∼500–1000 base pairs by sonication at half maximum (low setting) with a Braun Sonic 2000. After clarification, Rgt1–3HA was immunoprecipitated from an equivalent of 6.75 × 108 cells with the monoclonal anti-HA antibody 12CA5 (ascites fluid) (a generous gift from Ian Wilson, The Scripps Research Institute, La Jolla, CA) and protein A beads for 4 h at 4°C. Immune complexes were washed twice with 1 ml of lysis buffer, twice with 1 ml of lysis buffer with 250 mM NaCl, twice with 1 ml of wash buffer (10 mM Tris pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% Na deoxycholate, 1 mM EDTA), and twice with 1 ml of Tris-EDTA. Protein–DNA complexes were eluted with 50 μl of elution buffer (50 mM Tris pH 8.0, 10 mM EDTA, 1% SDS) and DNA-protein cross-linking was reversed in 1% SDS/Tris-EDTA at 65°C overnight. DNA was purified on QIAquick PCR columns (QIAGEN, Valencia, CA) according to the manufacturer's instructions. PCR reactions (15 min 94°C, 27 times [50 s 94°C, 1 min 30 s 50°C, 2 min 72°C], 10 min 72°C) were performed using HotStartTaq Master Mix kit (QIAGEN) on 1/6000 of the input (pellet fraction) and 1/60 of the immunoprecipitation. Sequences of the primers are available upon request. PCR fragments were separated on a 2.5% agarose gel and visualized by ethidium bromide.
Analysis of Rgt1 Phosphorylation
To analyze the phosphorylation status of Rgt1 protein extracts were prepared in radioimmunoprecipitation assay (RIPA) buffer (1% deoxycholic acid, 1% Triton X-100, 0.1% SDS, 250 mM NaCl, 50 mM Tris-HCl pH.7.5, 10 mM sodium pyrophosphate, 5 mM EDTA, 5 mM EGTA, 50 mM NaF, 0.1 mM orthovanadate, 1 mM PMSF, 2 μg/ml aprotinin, leupeptin, and pepstatin A), and Rgt-3HA was immunoprecipitated from 2.5 mg of protein with 12CA5 ascites fluid. Immune complexes were washed twice with 1 ml of RIPA buffer, twice with 1 ml of RIPA buffer without phosphatase inhibitors, and once in 1 ml of 50 mM Tris-HCl pH 7.5, 5 mM dithiothreitol, 0.1 mg/ml bovine serum albumin. Immunopurified Rgt1 was split into three equal parts. One part was incubated in 100 μl of phosphatase reaction mix (50 mM Tris-HCl pH 7.5, 5 mM dithiothreitol, 0.1 mM EDTA, 0.01% Brij35, 2 mM MnCl2) without phosphatase. The two other parts were incubated with phosphatase reaction mix and 1200 U of lambda-phosphatase (New England Biolabs, Beverly, MA), but to one of them a phosphatase inhibitor cocktail (final concentration: 10 mM sodium pyrophosphate, 5 mM EDTA, 5 mM EGTA, 50 mM NaF, 0.1 mM orthovanadate) was added. The reactions were incubated at 30°C for 60 min, the immuncomplexes washed with 300 μl of RIPA buffer and analyzed by Western blotting with monoclonal antibodies directed against the hemagglutinin (HA)-epitope (12CA5 or 16B12) or the myc-epitope (9E10).
RESULTS
Glucose Induction of HXT Gene Expression Is Associated with the Loss of Rgt1 from HXT Gene Promoters
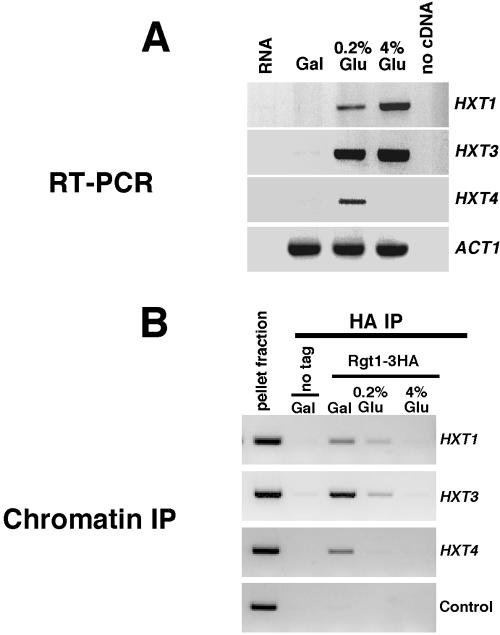
Several lines of evidence suggest that Rgt1 is a transcriptional repressor acting at the promoters of multiple HXT genes. However, when this study was initiated Rgt1 had not been shown to bind to promoter DNA in vivo nor was it known whether binding is regulated in response to glucose. To evaluate whether Rgt1 binds to HXT promoters in vivo, an HA epitope was introduced into the chromosomal allele of RGT1. We analyzed Rgt1 binding to the promoters of HXT1, HXT3, and HXT4. These genes were selected because they had been previously shown to be differently regulated in response to glucose (Özcan and Johnston, 1999). HXT1 is maximally induced at high glucose and only modestly induced in low glucose, whereas HXT3 is induced efficiently at both low glucose concentrations. HXT4 induced in low glucose but repressed in high glucose (Figure 1A).
Figure 1.
Induction of HXT gene expression is associated with dissociation of Rgt1 from HXT gene promoters. Cells expressing Rgt1-HA were grown in medium containing 2% galactose. The culture was split and either left untreated or adjusted to a final concentration of 0.2% (wt/vol) or 4% (wt/vol) glucose and then maintained for an additional 2 h. (A) RNA was isolated and analyzed by RT-PCR for HXT1, HXT3, and HXT4 along with ACT1 as a control. (B) Binding of Rgt1-HA to HXT1, HXT3, and HXT4 promoters was analyzed in cells from the same culture by chromatin immunoprecipitation. A primer pair for HFI1 and a strain expressing untagged Rgt1 were included as controls for nonspecific immunoprecipitation.
Association of Rgt1-HA with chromosomal DNA was analyzed in vivo by chromatin immunoprecipitation. Cells were either grown in 2% galactose or shifted from galactose to low glucose (0.2%) or high glucose (4%). Consistent with a role as a transcriptional repressor, Rgt1-HA was found to be associated with all three promoters in galactose-grown cells (Figure 1B). No association with any of the three HXT promoters was observed in cells growing on 4% glucose. Although, HXT4 is repressed under those conditions, that repression is known to be independent of RGT1 (Özcan and Johnston, 1995; Özcan, 2002). Finally, Rgt1 was associated with both the HXT1 and HXT3 promoters in 0.2% glucose, which partially induces their expression, but was undetectable at the HXT4 promoter in 0.2% glucose where expression is fully induced. This suggests that Rgt1 is differentially regulated at these promoters. Although it is possible that Rgt1 remains associated with the promoters in a form that is undetectable by chromatin immunoprecipitation, the simplest interpretation of these results is that Rgt1 dissociates from the HXT promoters under conditions where transcription is induced. These data are most consistent with Rgt1 acting as a transcriptional repressor at HXT promoters in the absence of glucose and induction of HXT gene expression by glucose occurring as a consequence of dissociation of Rgt1 from those promoters. Furthermore, the finding that Rgt1 is absent from these promoters under inducing conditions seems inconsistent with the proposal, based upon studies with HXT promoter-reporter fusions and one hybrid analysis (Özcan et al., 1996b), that Rgt1 acts as a transcriptional activator at HXT promoters (see DISCUSSION). Additional support for this contention comes from our observation that HXT gene expression is unaffected by inactivation of Rgt1 (Figure 2A).
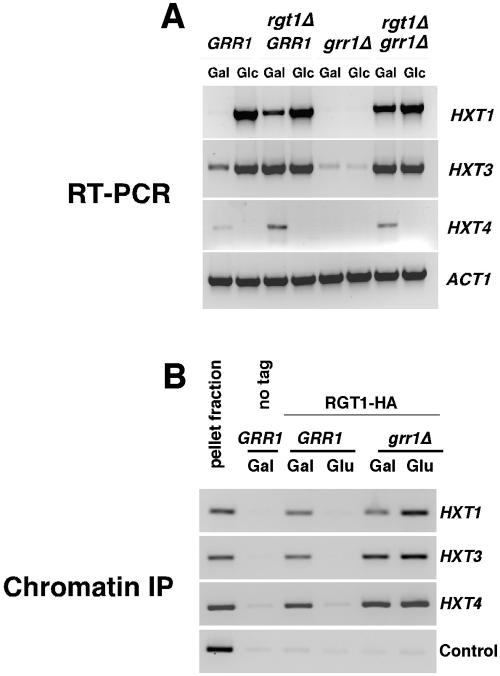
Figure 2.
Grr1 is required for regulation of Rgt1 binding to HXT promoters. (A) Grr1 acts via Rgt1 to induce HXT gene expression in response to glucose. Wild-type cells or cells carrying inactivated chromosomal alleles of either RGT1, GRR1, or both, were grown on galactose and then either adjusted to 4% glucose or left unchanged and cultured for an additional 2 h. RNA was then isolated and analyzed by RT-PCR for HXT1, HXT3, and HXT4 along with ACT1 as a control. (B) Glucose acts via Grr1 to induce dissociation of Rgt1 from HXT gene promoters. Wild-type and grr1Δ mutant cells carrying either Rgt1-HA or untagged Rgt1 were cultured as described in A and then binding of Rgt1-HA to HXT gene promoters analyzed by chromatin immunoprecipitation. A primer pair for HFI1 and a strain expressing untagged Rgt1 were included as controls for nonspecific immunoprecipitation.
Grr1 Is Required for Phosphorylation of Rgt1 and Dissociation from HXT Gene Promoters
Induction of HXT gene expression requires the F-box protein Grr1. The defect in induction of HXT gene expression resulting from inactivation of GRR1 is suppressed by inactivation of RGT1, suggesting that Grr1 antagonizes the activity of Rgt1 as a repressor at HXT gene promoters (Marshall-Carlson et al., 1991; Özcan and Johnston, 1995) (Figure 2A). To evaluate whether Grr1 is required for the dissociation of Rgt1 from HXT promoters in response to glucose, we performed chromatin immunoprecipitation of Rgt1 in a grr1Δ mutant by using the protocol outlined above. Consistent with previously published observations, cells deficient in GRR1 failed to induce HXT1, HXT3, or HXT4 in response to glucose (Figure 2A). Chromatin immunoprecipitation of Rgt1 from strains growing in galactose or shifted to 4% glucose revealed that Rgt1 remains associated with promoter DNA of all three HXT genes in the grr1Δ mutant cells regardless of the presence or absence of glucose (Figure 2B). Glucose induces the dissociation of Rgt1 from those promoters in wild-type cells. Based upon these observations, we conclude that Grr1 is required for dissociation of Rgt1 from HXT promoters.
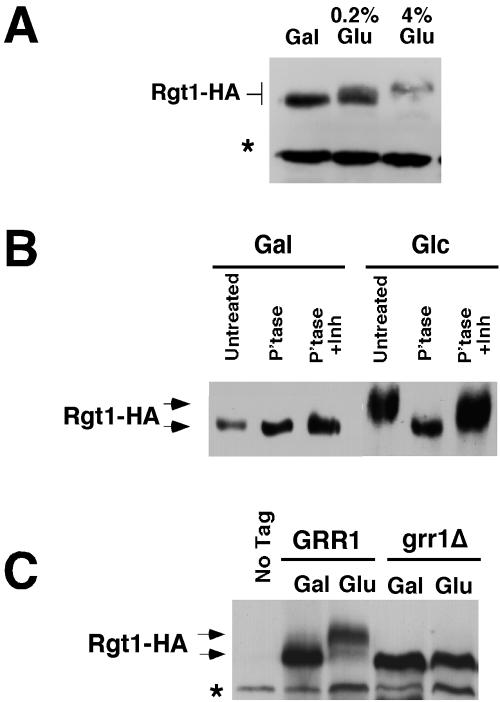
Grr1 has an established role in ubiquitin-mediated proteolysis and is also required for the glucose-induced loss of Rgt1 from HXT gene promoters. Therefore, we evaluated the status of the Rgt1 protein in cells growing on galactose and after induction with 0.2 or 4% glucose. Rgt1-HA is detected as a polypeptide with apparent molecular mass of 160 kDa based upon its migration of SDS-polyacrylamide gels, significantly greater than its predicted molecular mass of 132 kDa (Figure 3A). Furthermore, the Rgt1 polypeptide migrates with a progressively lower mobility when cells are grown on increasing concentrations of glucose, suggesting that Rgt1 becomes covalently modified upon exposure to glucose. Further analysis of Rgt1 provided no evidence that the protein was ubiquitinated (our unpublished data). In contrast, Rgt1 was phosphorylated in response to glucose. When Rgt1-HA immunoprecipitated from glucose-grown cells was treated with lambda phosphatase a substantial increase in mobility was observed. The mobility of the phosphatase-treated protein was similar to untreated Rgt1-HA from galactose-grown cells (Figure 3B). An increase in mobility was not observed if phosphatase inhibitors were added along with the phosphatase. Phosphatase treatment of Rgt1 from galactose-grown cells resulted in a small, but reproducible, increase in mobility. We conclude that Rgt1 is phosphorylated under both inducing and noninducing conditions. However, the phosphorylation observed in the presence of glucose is more extensive. Thus, hyperphosphorylation of Rgt1 is associated with induction of HXT gene expression and dissociation of Rgt1 from HXT gene promoters.
Figure 3.
Rgt1 is hyperphosphorylated in response glucose via a pathway that requires Grr1. (A) Rgt1 is progressively modified in response to glucose. Cells expressing Rgt1-HA were grown in galactose and then adjusted to either 0.2% (wt/vol) or 4% (wt/vol) glucose and grown for an additional 2 h. Rgt1-HA was then analyzed in cell extracts by SDS-PAGE followed by immunoblotting with anti-HA antibody. The asterisk identifies a 130-kDa background band that can be used as a loading control. (B) Rgt1 is hyperphosphorylated in glucose-grown cells. Rgt1-HA was immunoprecipitated from cells grown on galactose or 4% glucose as described previously. Rgt1-HA immune complexes were then treated with lambda protein phosphatase, either without or with a phosphatase inhibitor cocktail, or left untreated and analyzed by SDS-PAGE followed by immunoblotting with anti-HA antibody. (C) Hyperphosphorylation of Rgt1 in response to glucose requires Grr1. Wild-type cells or grr1Δ mutants expressing Rgt1-HA were grown on galactose or adjusted to 4% glucose for 2 h. Protein extracts were analyzed by SDS-PAGE and immunoblotting with anti-HA antibody. Wild-type cells expressing untagged Rgt1 were used as a control. The asterisk identifies a 130-kDa background band that can be used as a loading control.
Grr1 has no known role in protein phosphorylation. Analysis of Rgt1 phosphorylation in grr1Δ mutants revealed that GRR1 is required for induction of hyperphosphorylation of Rgt1 in the presence of glucose (Figure 3C) and for dissociation from HXT gene promoters. Although it is not clear whether Rgt1-HA from grr1Δ mutants is phosphorylated, its mobility is equivalent to phosphatase-treated Rgt1-HA. Thus, it seems that Grr1 is required not only for the hyperphosphorylation of Rgt1 associated with its dissociation from HXT promoters but also for the phosphorylation observed under noninducing conditions.
Grr1 Regulation of Rgt1 Phosphorylation and Promoter Binding and of HXT Gene Induction Is Mediated via Mth1 and Std1
The role of Grr1 in the regulation of HXT gene expression seems to be mediated via phosphorylation of Rgt1 and dissociation from HXT promoters. In the interest of identifying the direct target of Grr1 in this process, we have evaluated the regulation of Rgt1 phosphorylation in cells carrying deficiencies in protein kinases with known roles in the specific regulation of HXT gene expression or with general roles in the regulation of carbohydrate metabolism. These included, but were not limited to SKS1 (Yang and Bisson, 1996; Vagnoli and Bisson, 1998), its close homolog VHS1 (YDR247W), SNF1 (Hardie et al., 1998), and TPK1, TPK2 and TPK3 (Toda and Sass, 1988). None of the kinases analyzed exhibited a substantial defect in the glucose-induced phosphorylation of Rgt1 (our unpublished data). Inactivation of Reg1, a regulatory subunit of the Glc7 protein phosphatase implicated in the regulation of HXT gene expression (Özcan and Johnston, 1995), resulted in a modest effect on Rgt1 phosphorylation. However, the magnitude of its effect on phosphorylation or HXT gene expression seemed insufficient to explain the defect observed in grr1Δ mutants (our unpublished data). Thus, it is unlikely that Reg1 is the primary target of Grr1 for this regulation. However, it is possible that Grr1 simultaneously targets more than one of these proteins for inactivation in response to glucose.
A number of observations implicate two closely related proteins, Mth1 and Std1, in the glucose-dependent regulation of HXT gene expression. First, in agreement with previous studies with HXT promoter-lacZ reporter fusions (Schmidt et al., 1999), we find that inactivation of either MTH1 or STD1 alone causes defects in repression of HXT gene expression on nonglucose carbon sources (Figure 4A). This is most noticeable in the analysis of HXT3, which is fully derepressed in mth1Δ mutants and partially derepressed in std1Δ mutants. HXT4 is modestly affected by mth1Δ and HXT1 is largely unaffected by either of the single mutations. However, inactivation of both MTH1 and STD1 results in a dramatic derepression of all three genes under the same conditions (Schmidt et al., 1999; Figure 4A). Next, the Mth1 protein is absent from cells growing on glucose. Finally, both Mth1 and Std1 can interact with Rgt2/Snf3 (Schmidt et al., 1999) and Std1 can bind to Rgt1 (Tomas-Cobos and Sanz, 2002). Together, these observations led us to evaluate the role of Mth1 and Std1 in regulation of Rgt1 phosphorylation and promoter binding and as potential targets of Grr1.
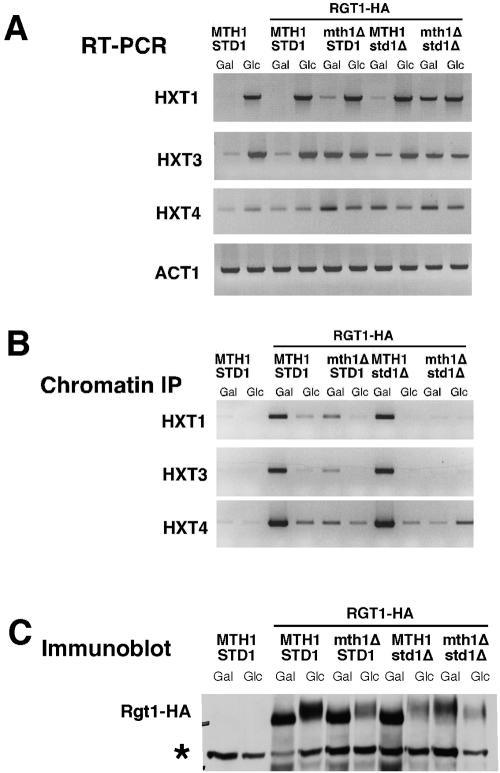
Figure 4.
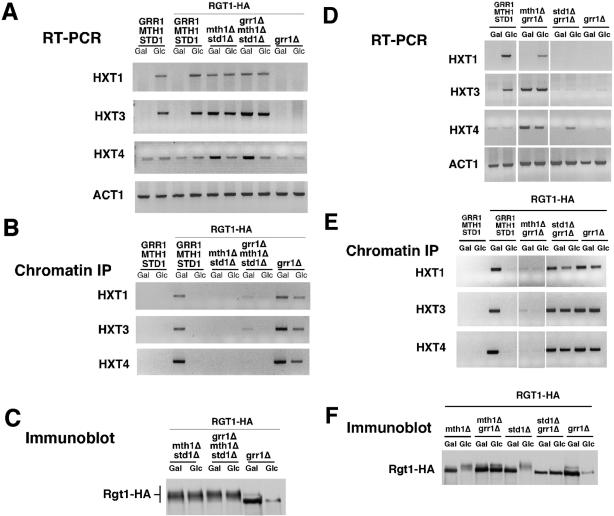
Maintenance of repression of HXT gene expression by Rgt1 under noninducing conditions is dependent upon MTH1 and STD1. Wild-type cells and cells in which MTH1, STD1, or both genes were inactivated expressing RGT1-HA were grown in galactose medium or adjusted to 4% glucose for 2 h. (A) RNA was isolated and analyzed by RT-PCR for HXT transcript abundance. ACT1 is presented as a control. (B) Rgt1-HA binding to the HXT promoters was analyzed by chromatin immunoprecipitation. Wild-type cells without Rgt1-HA are presented as a control. (C) Rgt1-HA was analyzed by SDS-PAGE and immunoblotting with anti-HA antibody. The asterisk indicates a 130-kDa background band recognized by anti-HA and serves as a loading control.
To determine whether Mth1 and Std1 play a role in the regulation of HXT gene expression at the level of Rgt1, we evaluated Rgt1-HA binding to the HXT promoters in both single mth1Δ and std1Δ mutants and in cells carrying disruptions of both genes (Figure 4B). Rgt1 was found to associate at wild-type levels with all three HXT promoters in std1Δ mutants growing in galactose. In contrast, mth1Δ mutants exhibited substantially reduced binding at all three promoters. Binding to HXT3 was most dramatically affected consistent with the substantial defect in regulation of that gene in mth1Δ mutants. However, glucose was found to reduce the association of Rgt1 with all three promoters, indicating that binding was still subject to regulation in the absence of Mth1. Strikingly, binding of Rgt1 to each of the three promoters was fully disrupted in the mth1Δ std1Δ mutants. Thus, the derepression of HXT gene expression observed in those mutants is associated with a defect in promoter binding by Rgt1.
Analysis of the Rgt1 protein in these mutants revealed that the protein seems hypophosphorylated on galactose and hyperphosphorylated on glucose in both mth1Δ and std1Δ single mutants (Figure 5C). Consistent with that observation Rgt1 retains the capacity to interact with each of those promoters when hypophosphorylated. However, the extent of that binding is decreased in the mth1Δ consistent with the derepression of HXT3 observed under those conditions (Figure 4A). Furthermore, Rgt1 is lost from all of those promoters in the presence of glucose, which induces expression of the respective genes. Finally, consistent with the induction of expression observed in galactose-grown cells, the Rgt1 protein was found to be hyperphosphorylated in mth1Δ std1Δ mutants under both repressing and nonrepressing conditions (Figure 4C). Together, these data suggest that Std1 is more important for repressing HXT1 expression, whereas Mth1 is a more important for HXT3 and, perhaps, HXT4, a finding in general agreement with previously reported observations (Tomas-Cobos and Sanz, 2002).
Figure 5.
The failure of grr1Δ mutants to derepress HXT gene expression is bypassed by inactivation of MTH1 and STD1. The effect of inactivation of GRR1 on HXT gene expression was examined in wild-type cells and cells in which MTH1 and STD1 (A–C) or each gene alone (D–F) were inactivated. Cells were grown in galactose medium or adjusted to 4% glucose for 2 h. (A and D) RNA was isolated and analyzed by RT-PCR for the abundance of HXT transcripts. ACT1 is presented as a control. (B and E) Rgt1-HA binding to HXT promoters was analyzed by chromatin immunoprecipitation. Wild-type cells with untagged Rgt1 are presented as a control. (C and F) Rgt1-HA was analyzed by SDS-PAGE and immunoblotting with anti-HA antibody. Differences in abundance in the grr1Δ mutant on glucose and galactose are due to differences in the amount of protein loaded for these samples.
We reasoned that if Mth1 and Std1 were targets for inactivation by Grr1, then inactivation of those genes should bypass the requirement for Grr1 for induction of HXT gene expression. We therefore performed an analysis of HXT gene expression, Rgt1 promoter binding, and Rgt1 phosphorylation status with mth1Δ and std1Δ mutant cells in which GRR1 had also been inactivated (Figure 5). That analysis demonstrates that the defect in glucose induction of HXT gene expression caused by grr1Δ is fully suppressed in the mth1Δ std1Δ double mutant (Figure 5A). That suppression is associated with the persistent hyperphosphorylation of Rgt1 (Figure 5C) and its failure to associate with HXT promoters (Figure 5B). Furthermore, inactivation of Grr1 fails to restore Rgt1 binding to HXT promoters in mth1Δ std1Δ mutants (Figure 5B). Based upon these observations, we conclude that Grr1 is required for inactivation of Mth1 and perhaps Std1. It is the inactivation of those proteins that leads to the phosphorylation of Rgt1, its dissociation from HXT promoters, and, as a consequence, HXT gene expression.
To achieve a more precise understanding of the role of Grr1 in the regulation of individual targets, we evaluated HXT expression and Rgt1 in cells in which either Mth1 or Std1 was inactivated along with Grr1. Inactivation of Grr1 in the std1Δ mutant enhanced Rgt1 binding to HXT promoters, blocked phosphorylation of Rgt1, and suppressed HXT gene expression (Figure 5, D–F). In contrast, inactivated of Grr1 in mth1Δ mutants failed to block the dissociation of Rgt1 from HXT promoters or to suppress Rgt1 phosphorylation (Figure 5, E and F). Furthermore, grr1Δ had little effect on the derepression of HXT3 and HXT4 gene expression caused by mth1Δ (Figure 5D). We suspect that HXT1 is regulated by STD1 in the absence of Mth1. This leads us to conclude that Grr1 is essential for the inactivation of Mth1 but that Std1 can be inactivated by an independent mechanism.
Loss of Mth1 in Response to Glucose Requires Grr1
The finding that inactivation of MTH1 and STD1 bypassed the requirement for GRR1 in the regulation of HXT gene expression prompted us to evaluate the role of Grr1 in the regulation of those genes and their protein products. We first determined the relative abundance of MTH1 and STD1 mRNA and protein in cells growing in galactose or shifted into glucose. In agreement with published observations (Schmidt et al., 1999), STD1 mRNA (Figure 6A) and protein (Figure 6B) were found to be unaffected by carbon source. Mth1 protein, which was detectable in galactose-grown cells, was lost when cells were analyzed after addition of glucose (Figure 6B). However, we found that MTH1 mRNA was only modestly affected by glucose (Figure 6A; our unpublished data), suggesting that the loss of the Mth1 protein is posttranslational. MTH1 RNA was previously shown to be undetectable during continuous growth on glucose (Schmidt et al., 1999). Because it was possible that this difference was a consequence of differences in the culture conditions, we analyzed the accumulation of MTH1 RNA during continuous growth on glucose or galactose (Figure 6A, bottom). Our data show that although MTH1 RNA accumulation is diminished on glucose it remains detectable.
Figure 6.
Grr1 is required for loss of Mth1 in response to glucose. (A) Analysis of MTH1 and STD1 mRNA in wild-type cells and grr1 mutants. Top, wild-type cells and grr1Δ mutants were grown in galactose or adjusted to 4% glucose for 2 h and then the abundance of MTH1, STD1, and HXT1 RNA was analyzed by RT-PCR. ACT1 RNA is shown as a control. Bottom, wild-type cells were grown continuously on rich medium containing either 2% galactose or 4% glucose and then the abundance of MTH1 and ACT1 RNA analyzed by RT-PCR. (B) Analysis of the Std1 and Mth1 proteins in wild-type cells and grr1 mutants. Extracts from wild-type or grr1Δ mutant cells carrying myc-tagged Std1 or myc-tagged Mth1 were analyzed by SDS-PAGE and immunoblotting with anti-myc antibodies. Cdc28 is shown as a loading control. (C) Mth1 protein is lost rapidly upon addition of glucose to wild-type cells. Wild-type cells expressing myc-tagged MTH1 were grown in galactose medium and either left unchanged or adjusted to 4% glucose. Samples were taken just before addition of glucose and at the times indicated after the addition of glucose. Extracts from the cells were analyzed by SDS-PAGE and immunoblotting with anti-myc antibody. Cdc28 is shown as a loading control. (D) Loss of Mth1 protein in response to glucose is dependent upon Grr1. Wild-type cells or grr1Δ mutant cells were treated as indicated in the experiment shown in C.
Whereas inactivation of GRR1 had little effect on the abundance of MTH1 RNA (Figure 6A) in cells growing on either carbon source, it had a substantial effect on the abundance of the Mth1 protein (Figure 6B). The abundance of the Mth1 protein from galactose-grown grr1Δ mutants was significantly increased over that from wild-type cells and it persisted after a shift to glucose for 2 h. Both of these effects are consistent with a role for Grr1 in determining the stability of the Mth1 protein.
Because the known role of Grr1 in the recognition of proteins destined for degradation via the ubiquitin proteasome pathway, we evaluated the kinetics of loss of Mth1 in cells shifted from galactose into glucose. Cells growing in galactose were either maintained in that carbon source or shifted to glucose and samples taken each 5 min over a 30-min time course (Figure 6C). Mth1 was rapidly lost from the glucose-, but not the galactose-grown culture. Thus, it seems that although the MTH1 mRNA persists in the presence of glucose (Figure 6A), the protein is rapidly depleted. We repeated this experiment analyzing MTH1 RNA and protein each 15 min over a time course of 2 h (Figure 6D). Although a decrease in MTH1 RNA was observed after 30 min in that experiment, the Mth1 protein was undetectable within 15 min (and probably less). Consistent with the finding that GRR1 is required for that instability, parallel analysis of Mth1 abundance in grr1Δ mutants revealed that Mth1 persists even in the presence of glucose for the duration of the experiment (Figure 6D). The behavior of the Mth1 protein is consistent with Grr1 playing a direct role in Mth1 destabilization in response to glucose by acting as an adaptor for substrate recognition by the SCF E3 ubiquitin ligase.
In contrast to Mth1, both STD1 RNA (Figure 6A) and protein (Figure 6B) undergo a noticeable decrease in abundance under both inducing and repressing conditions in the grr1Δ cells. Although the relevance of this decrease is unclear, these data suggest that, if Std1 is regulated by glucose, the regulation occurs via a mechanism that does not involve the regulation of protein abundance. Furthermore, the mechanism is likely to be largely independent of Grr1.
DISCUSSION
GRR1 was initially identified and named based upon the finding that mutants in that gene interfere with glucose repression (Glucose Repression Resistant) (Bailey and Woodward, 1984). This study demonstrates that glucose acts via Grr1 to block the function of Mth1 and, perhaps, Std1 as negative regulators of HXT transcription, thereby inducing Rgt1 phosphorylation and dissociation from HXT promoters. One of those proteins, Mth1, is regulated at the level of its abundance via a mechanism involving Grr1.
We show herein that glucose regulates Rgt1 at the level of protein phosphorylation and promoter binding. A similar observation was recently reported (Mosley et al, 2003). Dissociation of Rgt1 from HXT promoters is tightly correlated with its hyperphosphorylation and with activation of the HXT transcription. Furthermore, glucose induction of HXT transcripts is unaffected in cells lacking Rgt1. This seems inconsistent with the proposal that Rgt1 acts as a transactivator at HXT promoters (Özcan et al., 1996b). That model is based, in part, upon the observation that Rgt1 can act as a transcriptional activator when tethered to a heterologous promoter via LexA. Our data suggests instead that Rgt1 is absent from transcriptionally active HXT promoters arguing that if Rgt1 plays a role in the activation of HXT genes, it is indirect.
It was anticipated, based upon the role of Grr1 in the ubiquitin-proteasome pathway, that Rgt1 would be a target for ubiquitin-mediated degradation. Surprisingly, inactivation of Grr1 does not substantially affect the abundance of Rgt1 nor is Rgt1 ubiquitinated in response to glucose. Instead, we find that Rgt1 is phosphorylated in response to glucose in Grr1-dependent manner consistent with the recent report by Özcan and colleagues (Mosley et al., 2003). Furthermore, our genetic and biochemical analysis places Mth1 and Std1 upstream of Rgt1 and downstream of Grr1. Consistent with its role as a negative regulator of HXT gene expression, we find that in the absence of glucose Mth1 and, to a lesser extent, Std1 are required to maintain Rgt1 in the hypophosphorylated state, and, consequently, for its association with the HXT promoters. Finally, a role for Grr1 in that process is supported by the finding that inactivation of these genes relieves the requirement for Grr1 for HXT gene expression.
The rapid Grr1-dependent loss of Mth1 suggests it as a likely target for ubiquitin-dependent degradation. Yet, there remains significant ambiguity both in the literature and in our own analysis regarding a role for ubiquitination in that context. Although Skp1 and Cdc53 are clearly important for derepression induced by glucose (Li and Johnston, 1997), studies of thermosensitive mutants of Cdc34, the E2 ubiquitin-conjugating enzyme responsible for all know ubiquitination involving SCFGrr1, suggest that it is not required (Li and Johnston, 1997; our unpublished data). Another E2 enzyme, Ubc8, has been implicated in regulation of fructose-1,6-bisphosphatase (Fbp1) stability by glucose (Schule et al., 2000). Like HXT gene expression, Fbp1 proteolysis depends upon Grr1 (Horak et al., 2002). However, the two processes are distinguishable based upon the strong dependence of Fbp1 proteolysis upon Reg1 and its lack of dependence upon Rgt2 and Snf3. Although no role for Ubc8 in SCF-dependent ubiquitination has been described, it remains a potential collaborator with SCFGrr1 in regulation of HXT gene expression and Mth1 stability. However, it remains to be established that the role of Grr1 in regulation of Mth1 abundance involves protein ubiquitination.
Std1 is both structurally and functionally related to Mth1 (Hubbard et al., 1994). Both proteins have been shown to associate with the cytoplasmic domains of one or both hexose receptors, Rgt2 or Snf3, as well as with Rgt1 (Schmidt et al., 1999; Lafuente et al., 2000). Although inactivation of Mth1 is sufficient to derepress HXT2, HXT3, and HXT4, both Mth1 and Std1 must be inactivated for full induction of Rgt1 hyperphoshorylation and HXT gene expression in the absence of glucose (Figures 4 and 5; Schmidt et al., 1999). Furthermore, both genes must be eliminated to bypass the requirement for Grr1 in those processes. Consequently, we can conclude that Grr1 is somehow involved in the regulation of Std1. However, whereas the behavior of Mth1 in response to glucose and in grr1Δ mutants argues in favor of a role for Grr1 in Mth1 turnover, Std1 is not similarly regulated. Rather, the level of the Std1 protein seems to be unaffected by glucose (Figure 6; Schmidt et al., 1999). Unlike Mth1, both STD1 RNA and protein seem to decrease in grr1 mutants. Finally, although inactivation of Grr1 has little or no effect on Rgt1 modification or HXT gene expression in cells lacking Mth1, it has a dramatic effect on those phenotypes in cells lacking Std1, suggesting that Grr1 is primarily involved in the regulation of Mth1. Together, these results argue that Mth1 is the primary target of Grr1 for regulation of HXT gene expression by glucose.
Std1 associates with Snf1, a protein kinase involved in global regulation by glucose via a pathway independent of Snf3 and Rgt2 (Hubbard et al., 1994; Tomas-Cobos and Sanz, 2002; Kuchin et al., 2003). Furthermore, Std1 seems to act as a positive regulator of Snf1 (Kuchin et al., 2003). However, it is not clear whether this association is relevant to the regulation of HXT gene expression by glucose. Although Snf1 may be responsible for a portion of the regulation of Rgt1, it is not essential for glucose-inducible phosphorylation of Rgt1 (our unpublished results) nor for induction of HXT gene expression by glucose (Özcan and Johnston, 1995). Consequently, it seems likely that there are multiple signal transduction pathways involved in glucose regulation of the HXT genes that are, at least in part, independently regulated. The distinction between these pathways may account, in part, for the differences in the behavior of the HXT genes in response to different levels of glucose.
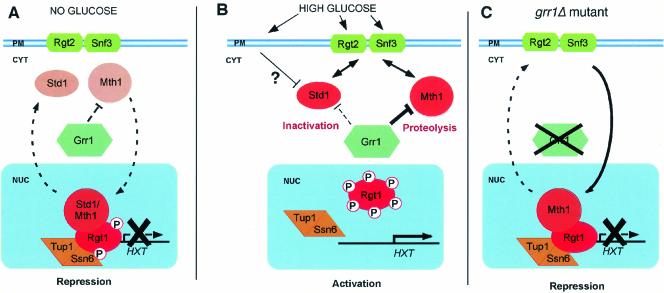
Based upon our analysis of the role of Grr1 in the regulation of HXT gene expression and observations of others, we propose the model presented in Figure 7. We suggest that repression of HXT gene expression occurs via Rgt1 binding to HXT gene promoters. However, maintenance of repression depends upon Mth1 and Std1 perhaps via a direct interaction, consistent with the observed interaction between Std1 and Rgt1 (Tomas-Cobos and Sanz, 2002). The extent to which these regulators affect expression varies between the different HXT genes, Std1 having its predominant effect on HXT1 and Mth1 having a more pronounced effect on HXT3 and HXT4 (and probably HXT2). On exposure to glucose multiple signaling pathways are activated, a primary pathway for HXT regulation involving the glucose receptors, Rgt2 and Snf3, and a second, as yet undefined, pathway. Our data suggest that Rgt2/Snf3 signaling occurs primarily by elimination of the Mth1 protein via a Grr1-dependent mechanism. Unlike Mth1, inactivation of Std1 seems to be largely Grr1-independent. That pathway may involve Snf1, a global regulator of glucose repression, consistent with the capacity of those proteins to form a complex (Hubbard et al., 1994; Tomas-Cobos and Sanz, 2002; Kuchin et al., 2003). Inactivation of Mth1 and Std1 may occur as a consequence of their retention in the cytoplasm by the Rgt2/Snf3 receptor proteins. Both proteins have been reported to interact with the cytoplasmic tails of these transmembrane receptors via two-hybrid analysis (Schmidt et al., 1999; Lafuente et al., 2000), although the specific conditions under which those interactions occur is not known. This would require cycling of these proteins between the nucleus and the cytoplasm. Finally, we propose that the inactivation of Mth1 and Std1 leads to the phosphorylation of Rgt1 by an as yet unidentified protein kinase. Phosphorylation leads to dissociation of Rgt1 from HXT promoters, thereby activating HXT gene expression. Clearly, many of the details of this model remain to be established.
Figure 7.
Regulation of HXT transcription by glucose and Grr1. This model supposes that Std1 and Mth1 cycle between the nucleus and the cytoplasm. In the nucleus, they stabilize the interaction between the transcriptional repressor Rgt1 and the HXT gene promoters and in doing so repress HXT transcription. Their association with Rgt1 suppresses phosphorylation by an unidentified protein kinase. In the cytoplasm, they interact transiently with Rgt2 and Snf3 at the plasma membrane. (A) In the absence of glucose, localization to the nucleus is favored and transcription is repressed. Mth1 is inactivated in the cytoplasm by a Grr1-dependent mechanism. Std1 is inactivated, at least in part, via a Grr1-independent mechanism. (B) In the presence of glucose, cytoplasmic localization is favored due to tethering in the cytoplasm by association with the glucose receptors Rgt2 and Snf3. Mth1 is inactivated via Grr1-dependent proteolysis, whereas inactivation of Std1 is largely independent of Grr1. Rgt1 becomes phosphorylated as a consequence of the dissociation of Mth1 and Std1 and dissociates from the HXT promoters thereby activating transcription. (C) In the absence of Grr1, Mth1 is stabilized, leading to accumulation in the nucleus. Mth1 is sufficient to explain the repression of HXT gene expression independent of the status of Std1. Std1 may still be regulated via a Grr1-independent mechanism under these conditions. Because Mth1 is constitutively associated with Rgt1, the basal phosphorylation of Rgt1 is suppressed and HXT gene transcription is repressed.
Grr1 plays an important, but as yet undescribed role in a number of nutrient-regulated transcription systems. The targets of the SPS signaling system required for the response of cells to extracellular amino acids (Forsberg and Ljungdahl, 2001) and the Rgt1/Snf3 sensor system for regulation of hexose permeases are among the best studied. There are similarities between these signal transduction systems in addition to the involvement of Grr1. Notably, both use members of a family of membrane-bound sensors related to the permeases that they regulate (Van Belle and André, 2001) and both exert transcriptional repression via the Ssn6/Tup1 transcriptional corepressor (Özcan and Johnston, 1999; Andrèasson and Ljungdahl, 2002). Like HXT gene regulation, activation of the targets of SPS (Ssy1-Ptr3-Ssy5) signaling seems to involve SCF components (Iraqui et al., 1999) but is unaffected by mutations that affect binding of targets for phosphorylation-dependent ubiquitination (Hsiung et al., 2001). However, beyond Grr1 the analogy between elements of the signal transduction systems conveying signals to the nucleus from the cell membrane remains unclear. Strikingly, the transcriptional activation in the SPS system involves proteolytic processing of two transcriptional regulators, Stp1 and Stp2 (Andrèasson and Ljungdahl, 2002). However, processing of those proteins does not seem to involve Grr1 nor have regulators analogous to Mth1 and Std1 been identified in that system (Özcan and Johnston, 1999). Consequently, it is difficult, based upon analogy to the HXT system, to predict the role of Grr1 in SPS signaling. However, it remains possible that the mechanism by which Grr1 regulates Mth1 is conserved between these pathways or that a single process mediated by Grr1 leads to the inactivation of Mth1 along with elements of those other pathways.
The diverse roles of F-box proteins in cellular regulation are only beginning to be fully appreciated. Activation of the transcriptional regulator nuclear factor-κB, which has been known to involve ubiquitination target for many years, is now known to occur via SCF-mediated phosphorylation-dependent ubiquitination (Brivanlou and Darnell, 2002). Over the past several years, a number of other ubiquitin-dependent mechanisms for gene-specific, as well as general, transcriptional regulation have begun to be elucidated (Hoppe et al., 2001; Brivanlou and Darnell, 2002). More sophisticated understanding of these regulatory networks will undoubtedly reveal a tightly regulated and highly integrated system of ubiquitination-dependent events. Grr1, as an F-box protein involved in a broad range of cellular processes including cell cycle regulation, morphogenesis, and transcriptional control provides an excellent subject for such studies.
Acknowledgments
We thank Mark Johnston, Mark Longtine, Kim Nasmyth, Steven Reed, and Dieter Wolf for strains, plasmids, and reagents. We thank Robertus deBruin and Peter Kaiser for helpful comments on the manuscript. We also acknowledge members of the Wittenberg laboratory and the TSRI Cell Cycle Group for advice and helpful discussion. K.F. was the recipient of award APART Fellowship from the Austrian Academy of Sciences. This work was supported by funding to C.W. (GM-43487 and GM-59441) from the U.S. Public Health Service.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-03-0135. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-03-0135.
References
- Andrèasson, C., and Ljungdahl, P.O. (2002). Receptor-mediated endoproteolytic activation of two transcription factors in yeast. Genes Dev. 16, 3158–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, R.B., and Woodward, A. (1984). Isolation and characterization of a pleiotropic glucose repression resistant mutant of Saccharomyces cerevisiae. Mol. Gen. Genet. 193, 507–512. [DOI] [PubMed] [Google Scholar]
- Brivanlou, A.H., and Darnell, J.E., Jr. (2002). Signal transduction and the control of gene expression. Science 295, 813–818. [DOI] [PubMed] [Google Scholar]
- Forsberg, H., and Ljungdahl, P.O. (2001). Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40, 91–109. [DOI] [PubMed] [Google Scholar]
- Gancedo, J.M. (1998). Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62, 334–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, A.L., and McCusker, J.H. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541–1553. [DOI] [PubMed] [Google Scholar]
- Hardie, D.G., Carling, D., and Carlson, M. (1998). The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67, 821–855. [DOI] [PubMed] [Google Scholar]
- Hoppe, T., Rape, M., and Jentsch, S. (2001). Membrane-bound transcription factors: regulated release by RIP or RUP. Curr. Opin. Cell Biol. 13, 344–348. [DOI] [PubMed] [Google Scholar]
- Horak, J., Regelmann, J., and Wolf, D.H. (2002). Two distinct proteolytic systems responsible for glucose-induced degradation of fructose-1,6-bisphosphatase and the Gal2p transporter in the yeast Saccharomyces cerevisiae share the same protein components of the glucose signaling pathway. J. Biol. Chem. 277, 8248–8254. [DOI] [PubMed] [Google Scholar]
- Hsiung, Y.G., Chang, H.C., Pellequer, J.L., La Valle, R., Lanker, S., and Wittenberg, C. (2001). F-box protein Grr1 interacts with phosphorylated targets via the cationic surface of its leucine-rich repeat. Mol. Cell. Biol. 21, 2506–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard, E.J., Jiang, R., and Carlson, M. (1994). Dosage-dependent modulation of glucose repression by MSN3 (STD1) in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 1972–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqui, I., Vissers, S., Bernard, F., de Craene, J.O., Boles, E., Urrestarazu, A., and André, B. (1999). Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-Box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol. 19, 989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin, S., Vyas, V.K., Kanter, E., Hong, S.P., and Carlson, M. (2003). Std1p (Msn3p) positively regulates the Snf1 kinase in Saccharomyces cerevisiae. Genetics 163, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente, M.J., Gancedo, C., Jauniaux, J.C., and Gancedo, J.M. (2000). Mth1 receives the signal given by the glucose sensors Snf3 and Rgt2 in Saccharomyces cerevisiae. Mol. Microbiol. 35, 161–172. [DOI] [PubMed] [Google Scholar]
- Li, F.N., and Johnston, M. (1997). Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp 1, coupling glucose sensing to gene expression and the cell cycle. EMBO J. 16, 5629–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A., 3rd, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Marshall-Carlson, L., Neigeborn, L., Coons, D., Bisson, L., and Carlson, M. (1991). Dominant and recessive suppressors that restore glucose transport in a yeast snf3 mutant. Genetics 128, 505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley, A.L., Lakshmanan, J., Aryal, B.K., and Özcan, S. (2003). Glucose-mediated phosphorylation converts the transcription factor Rgt1 from a repressor to an activator. J. Biol. Chem. 278, 10322–10327. [DOI] [PubMed] [Google Scholar]
- Özcan, S. (2002). Two different signals regulate repression and induction of gene expression by glucose. J. Biol. Chem. 277, 46993–46997. [DOI] [PubMed] [Google Scholar]
- Özcan, S., Dover, J., and Johnston, M. (1998). Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 17, 2566–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan, S., Dover, J., Rosenwald, A.G., Wolfl, S., and Johnston, M. (1996a). Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. USA 93, 12428–12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan, S., Freidel, K., Leuker, A., and Ciriacy, M. (1993). Glucose uptake and catabolite repression in dominant HTR1 mutants of Saccharomyces cerevisiae. J. Bacteriol. 175, 5520–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan, S., and Johnston, M. (1995). Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell. Biol. 15, 1564–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan, S., and Johnston, M. (1999). Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 63, 554–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan, S., Leong, T., and Johnston, M. (1996b). Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol. Cell. Biol. 16, 6419–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., and Tyers, M. (1998). Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 14, 236–243. [DOI] [PubMed] [Google Scholar]
- Schmidt, M.C., McCartney, R.R., Zhang, X., Tillman, T.S., Solimeo, H., Wolfl, S., Almonte, C., and Watkins, S.C. (1999). Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 4561–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schule, T., Rose, M., Entian, K.D., Thumm, M., and Wolf, D.H. (2000). Ubc8p functions in catabolite degradation of fructose-1,6-bisphosphatase in yeast. EMBO J. 19, 2161–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte, F., Wieczorke, R., Hollenberg, C.P., and Boles, E. (2000). The HTR1 gene is a dominant negative mutant allele of MTH1 and blocks Snf3- and Rgt2-dependent glucose signaling in yeast. J. Bacteriol. 182, 540–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra, D., Craig, K.L., Tyers, M., Elledge, S.J., and Harper, J.W. (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., Knapp, D., and Nasmyth, K. (1997). Loading of an MCM protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90, 649–660. [DOI] [PubMed] [Google Scholar]
- Toda, T., and Sass, P. (1988). The cAMP-dependent protein kinase genes in yeast. Oxf. Surv. Eukaryot. Genes 5, 133–161. [PubMed] [Google Scholar]
- Tomas-Cobos, L., and Sanz, P. (2002). Active Snf1 protein kinase inhibits expression of the Saccharomyces cerevisiae HXT1 glucose transporter gene. Biochem. J. 368, 657–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnoli, P., and Bisson, L.F. (1998). The SKS1 gene of Saccharomyces cerevisiae is required for long-term adaptation of snf3 null strains to low glucose. Yeast 14, 359–369. [DOI] [PubMed] [Google Scholar]
- Vallier, L.G., Coons, D., Bisson, L.F., and Carlson, M. (1994). Altered regulatory responses to glucose are associated with a glucose transport defect in grr1 mutants of Saccharomyces cerevisiae. Genetics 136, 1279–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Belle, D., and André, B. (2001). A genomic view of yeast membrane transporters. Curr. Opin. Cell Biol. 13, 389–398. [DOI] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Pohlmann, R., and Philippsen, P. (1994). New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- Willems, A.R., Lanker, S., Patton, E.E., Craig, K.L., Nason, T.F., Mathias, N., Kobayashi, R., Wittenberg, C., and M. Tyers. (1996). Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86, 453–463. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and Bisson, L.F. (1996). The SKS1 protein kinase is a multicopy suppressor of the snf3 mutation of Saccharomyces cerevisiae. Yeast 12, 1407–1419. [DOI] [PubMed] [Google Scholar]