Abstract
Functionally different subsets of actin filament arrays contribute to cellular organization and motility. We report the identification of a novel subset of loose actin filament arrays through regulated association with the widely expressed protein SWAP-70. These loose actin filament arrays were commonly located behind protruding lamellipodia and membrane ruffles. Visualization of these loose actin filament arrays was dependent on lamellipodial protrusion and the binding of the SWAP-70 PH-domain to a 3′-phosphoinositide. SWAP-70 with a functional pleckstrin homology-domain lacking the C-terminal 60 residues was targeted to the area of the loose actin filament arrays, but it did not associate with actin filaments. The C-terminal 60 residues were sufficient for actin filament association, but they provided no specificity for the subset of loose actin filament arrays. These results identify SWAP-70 as a phosphoinositide 3-kinase signaling-dependent marker for a distinct, hitherto unrecognized, array of actin filaments. Overexpression of SWAP-70 altered the actin organization and lamellipodial morphology. These alterations were dependent on a proper subcellular targeting of SWAP-70. We propose that SWAP-70 regulates the actincytoskeletonasaneffectororadaptorproteininresponsetoagoniststimulatedphosphatidylinositol (3,4)-bisphosphate production and cell protrusion.
INTRODUCTION
The cellular actin cytoskeleton is organized in a variety of spatially and temporally controlled assemblies of actin filaments. Actin filaments are polymerized from monomeric G-actin in lamellipodia and filopodia at the cell periphery (Wang, 1985; Machesky and Hall, 1997). These newly polymerized actin filaments are highly dynamic and are turned over rapidly (Wang, 1985). It has been proposed that the actin filaments found in the remainder of the cells have their origin in the lamellipodium (Small et al., 1998) and in small membrane ruffles occurring throughout the lamella (Schafer et al., 1998). Actin filaments are organized into various arrays such as stress fibers, lamellipodial networks, filopodial bundles, dorsal arcs, peripheral concave or convex bundles as well as geodesic arrays (Small, 1988; Heath and Holifield, 1993; Small et al., 1998). The organization of each of these assemblies is controlled and stabilized by specific sets of actin-associated proteins, conferring on them different functions. However, it is not known how many functionally different actin arrays exist in cells and how their spatial and temporal organization is regulated and coordinated. An asymmetric and polarized organization of the different actin arrays in cells is fundamental for cell migration, growth, division, differentiation, and defense (Verkhovsky et al., 1998; Weiner et al., 1999).
The induction and maintenance of a polarized distribution of different actin arrays is dependent on extracellular and intracellular signals. Leukocytes and amoebas of Dictyostelium discoideum can orient themselves in a gradient of a chemoattractant and move toward the source of the chemoattractant (Zigmond, 1981; Devreotes and Zigmond, 1988). Activation of uniformly distributed G protein-coupled receptors on the cell surface leads to the intracellular stimulation of phosphoinositide 3-kinase (PI 3-kinase) and a highly polarized accumulation of phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3] and/or phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2] (Meili et al., 1999; Rickert et al., 2000; Servant et al., 2000; Chung et al., 2001). Pleckstrin homology (PH)-domain–containing proteins have been suggested to represent targets of PI 3-kinase lipids. PH-domains are found in many proteins involved in a diverse array of physiological events, including cellular signaling and cytoskeletal organization (Haslam et al., 1993; Gibson et al., 1994). They are a structural module of ∼100–120 amino acid residues, and several of them have been demonstrated to bind with high specificity to differently phosphorylated phosphoinositides (Kavran et al., 1998; Rameh et al., 1997).
The PH-domain containing protein SWAP-70 with a molecular mass of 70 kDa was originally isolated as a B cell-specific component of an isotype switch recombination complex called SWAP (Borggrefe et al., 1998). Analysis of its PH-domain sequence reveals a good fit to a consensus motif (Isakoff et al., 1998; Ferguson et al., 2000; Thomas et al., 2001), which predicts high-affinity binding to PI(3,4)P2. Its intracellular localization has been reported to depend on cell activation. Stimulation of the B cell receptor triggers a translocation of SWAP-70 from the cytosol to the plasma membrane in B cells (Masat et al., 2000). This translocation required a functional PH-domain. Although SWAP-70 was considered to be expressed in B cells only (Borggrefe et al., 1998, 1999; Masat et al., 2000), Ishikawa et al. (1998) were able to detect RNA expression by reverse transcription-polymerase chain reaction (PCR) in a variety of different tissues. This result suggested that SWAP-70 could have a more widespread function.
In a two-hybrid screen, we identified SWAP-70 as a possible binding partner of rat myosin IXb (myr 5), a class IX myosin-RhoGAP molecule (our unpublished observation). Therefore, we sought to investigate its possible role as an effector of PI 3-kinase in the regulation of polarized actin dynamics. To this end, we used the mouse melanoma B16F1 cells that are highly motile and exhibit a high frequency of lamellipodia formation.
MATERIALS AND METHODS
Identification and Cloning of SWAP-70
SWAP-70 was identified in a two-hybrid interaction trap screen (Golemis and Khazak, 1997). A cDNA library in vector JG4-5 of serum-starved WI-38 human lung fibroblasts (a kind gift of Sardet and Gyuris, Massachusetts General Hospital, Boston, MA) was screened with bait plasmid pEG235 encoding amino acids 1440–1592 of rat myosin IXB (myr 5). The myr 5 cDNA was inserted at BamHI and NcoI sites. Plasmids were transformed into yeast strain EGY48 together with the LacZ reporter plasmid pSH18-34. Of 2 × 107 colony-forming units plated, two Leu+ and LacZ+ clones were identified. Both clones contained the identical 2.3-kb insert encoding full-length SWAP-70 as verified by analysis of cDNA and genomic DNA.
Antibodies
To raise antibodies specific for SWAP-70, rabbits were immunized with a denatured fusion protein of 6xHis-SWAP-70 (aa 89–585). An EcoRI/XhoI fragment encoding amino acids 89–585 was excised from plasmid JG4-5-SWAP-70 and subcloned into the corresponding restriction sites in pBluescriptSK (Stratagene, La Jolla, CA). Further subcloning into the BamHI/SalI sites of the pQE30 6xHis-tag expression vector (QIAGEN, Hilden, Germany) was performed by excision of a BamHI/XhoI fragment from the modified pBluescript. Fusion protein was expressed in Escherichia coli (DH5α) and purified over Ni2+-nitrilotriacetic acid resin under denaturing conditions as recommended by the manufacturer. Rabbit sera GK1 and GK2 were affinity purified by standard techniques. The purified fusion protein was coupled to CNBr-activated Sepharose-4B (Amersham BioSciences, Freiburg, Germany) in 6 M urea, 0.1 M NaHCO3, pH 8.0.
The green fluorescent protein (GFP) antibody was a kind gift of Prof. Markus Maniak (Kassel, Germany). A rabbit polyclonal antibody directed against nonmuscle myosin (BT-561) was from Biomedical Technologies (Stoughton, MA).
Plasmids for Cell Transfections
SWAP-70 with an N-terminal hemagglutinin (HA)-tag followed by a short 19 residue linker sequence was cloned into pUHD10-3 (Resnitzky et al., 1994). Two annealed complementary oligonucleotides encoding SpeI, HA-tag and NotI sequences, a NotI/blunted Bsu 36I SWAP-70 fragment, and pUHD 10-3 previously modified by insertion of SpeI/EcoRV sites between EcoRI and BamHI sites were ligated. This resulted in a plasmid encoding amino acids MYPYDVPDYACGRRGWLGRRGWRGSRAAA followed by the complete SWAP-70 coding sequence.
For construction of the GFP-SWAP-70 plasmid, the 5′ region of SWAP-70 was amplified by PCR with the primers MB411 (GGCTCGAGGGAGCTTGAAGGAGGAGCTG) and MB412 (CACAGAGGGTCCAACACATCC). The PCR fragment was cloned into the XhoI and EcoRI restriction sites of the pEGFP-C1 vector (BD Biosciences Clontech, Palo Alto, CA). The missing region of SWAP-70 was excised with EcoRI/BamHI from pUHD-HA-SWAP-70 and ligated into the corresponding sites of the pEGFP-C1 vector.
SWAP-70 fragments encompassing amino acids 1–313 or 205–585 were amplified by PCR to construct GFP-SWAP(1–313) and GFP-SWAP(205–585). The primers used were MB411 and MB420 (GGGGATCCTCAAGGGCTGCCCAGCTTCAACAG) for SWAP(1-313) and primers MB421 (GGCTCGAGGGATGGCAATTAATGAAGTCTTTAATG) and MB422 (GGGGATCCTCACTCCGTGGTCTTTTTCTCTT) for SWAP(205-585). The PCR fragments were cloned into the XhoI and BamHI restriction sites of the pEGFP-C1 vector.
The PH(aa205-313)-GFP plasmid was constructed by PCR with primers MB433 (GGGAATTCATGGCAATTAATGAAGTCTTTAATG) and MB434 (GGGGATCCCCAGGGCTGCCCAGCTTCAACAGA). The PCR fragment was inserted into the EcoRI and BamHI restriction sites of the pEGFP-N1 vector (BD Biosciences Clontech).
The mutations R230C and RR223,224EE in GFP-SWAP-70 were introduced by the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Primers encoding the R230C mutation were MB459 (CCACAGACGGAAAAACTGGACTGAATGCTGGTTTGTACTAAAACCC) and MB460 (GGGTTTTAGTACAAACCAG-CATTCAGTCCAGTTTTTCCGTCTGTGG) and primers encoding the RR223,224EE mutations were MB457 (GCAGGGTTACATGATGAAAAAGGGCCACGAAGAGAAAAACTGGACTG) and MB458 (CAGTCCAGTTTTTCTCTTCGTGGCCCTTTTTCATCATGTAACCCTGC). All PCR-products were verified by sequencing. The SWAP-70 cDNAs were transferred from the GFP-SWAP-70 plasmids into the pECFP-C1 and the pEYFP-C1 vectors (BD Biosciences Clontech) by using the XhoI and BamHI restriction sites. YFP-SWAP(1-525) was constructed by insertion of a stop codon with PCR. The SWAP(526-585) fragment was amplified by PCR by using the primers 5′-GGCTCGAGGGATGGCAACTAATAAGACC and 5′-GGGGATCCTCACTCCGTGGTCTTTTTCTCTT and subcloned into the XhoI and BamHI sites of pEGFP-C1.
Cell Culture
Swiss 3T3 fibroblasts, HtTa-1 HeLa cells (a gift of H. Bujard, Heidelberg, Germany; Gossen and Bujard, 1992), REF52 cells, and B16F1 mouse melanoma cells (American Type Culture Collection, Manassas, VA) were cultured in DMEM (Invitrogen, Karlsruhe, Germany)/10% fetal calf serum (FCS) (Invitrogen)/2 mM l-glutamine (Invitrogen)/0.1 mg/ml penicillin/streptomycin (Roche Diagnostics, Mannheim, Germany) at 37°C in a humidified atmosphere with 5% CO2. B16F1 cells used for videomicroscopy were maintained in high glucose (4500 mg/l) DMEM (Sigma-Aldrich, St. Louis, MO) supplemented with 10% FCS (PAA Laboratories, Linz, Austria). For observation by video microscopy B16F1 cells were transferred to Ham's F12 medium (Sigma-Aldrich) containing 10% FCS.
Cell and Tissue Extract Preparations and Immunoblotting
Cells were plated on Ø 60-mm culture plates, transfected (see below), and grown until they were 80–90% confluent. Cells were washed three times with phosphate-buffered saline (PBS) (150 mM NaCl, 3 mM Na2HPO4, 1.5 mM KH2PO4, pH 7.4), harvested by scraping, and collected by centrifugation. Cells were lysed in 5 volumes of lysis buffer (100 mM NaCl, 1 mM MgCl2, 0.5 mM EGTA, 1 mM β-mercaptoethanol, 0.5% NP-40, 20 mM HEPES, pH 7.4). Tissue samples were prepared as described previously (Chieregatti et al., 1998). Immunoblotting was performed according to Towbin et al. (1979), and signals were detected using either the enhanced chemiluminescence system (Amersham Biosciences) or SuperSignal West Pico chemiluminescent substrate reagent (Pierce Chemical, Rockford, IL).
Cell Transfections and Fluorescence Microscopy
For immunofluorescence microscopy, Swiss3T3 and HtTA-1 HeLa cells were plated on sterile coverslips and transfected with FuGENE reagent (Roche Diagnostics). Cells were treated with pharmacological agents 36–48 h after transfection. B16F1 cells were cultured on Ø30-mm dishes and transfected by using Superfect reagent (QIAGEN). Transfected B16F1 cells were replated on laminin coated coverslips 4–6 h before analysis or pharmacological treatments. Coverslips were coated for 1 h with laminin (25 μg/ml) (Sigma-Aldrich) in coating buffer (150 mM NaCl, 50 mM Tris, pH 7.5) and rinsed with PBS immediately before plating of cells. AlF4– (50 μM AlCl2 [Merck, Darmstadt, Germany] and 30 mM NaF [Merck] final concentration) was diluted from stock solutions into complete medium and then added to the cells by replacing the growth medium. Cells were normally fixed in 4% paraformaldehyde (PFA; Merck) for 30 min. Free aldehyde groups were blocked with 0.1 M glycine (Roth, Karlsruhe, Germany) and then cells were permeabilized in 0.05% saponin (Sigma-Aldrich) for 20 min. Unspecific binding sites were blocked with 5% normal goat serum (Dianova, Hamburg, Germany). To remove cytosolic proteins cells were treated with 0.3% Triton X-100 in the presence of 4% PFA for 3 min followed by 4% PFA for 30 min. All reagents were buffered with PBS. After incubation with primary antibody directed against SWAP-70 (GK2), cells were stained with fluorescein isothiocyanate- or rhodamineconjugated goat anti-rabbit IgG (Dianova). Filamentous actin was stained with fluorescein isothiocyanate- or Alexa 594-conjugated phalloidin (Molecular Probes, Eugene, OR). Some cells were fixed with glutaraldehyde (Sigma-Aldrich) to achieve better actin cytoskeleton preservation. Cells were first extracted with 0.25% Triton X-100 in the presence of 0.5% glutaraldehyde for 1 min and then fixed further with 1% glutaraldehyde for 15 min. Free aldehyde groups were blocked with 0.5 mg/ml sodium borohydride and unspecific binding sites with 5% normal goat serum in PBS. Triton X-100, glutaraldehyde, and borohydride were buffered with CB-buffer (150 mM NaCl, 5 mM EGTA, 5 mM glucose, 5 mM MgCl2, 10 mM 2-(N-morpholino)ethanesulfonic acid, pH 6.1). Antibody stainings were produced as described for paraformaldehyde-fixed cells. The cells were analyzed with an Axiophot fluorescence microscope (Carl Zeiss, Jena, Germany), by using 63× or 100× objectives. Images were acquired with a dual mode cooled charge-coupled device camera C 4880 (Hamamatsu, Bridgewater, NJ) controlled by High Performance Image Control (Hipic) software (Hamamatsu). Images were imported and processed in Adobe Photoshop 5.5. Videomicroscopy of B16F1 cells was performed as described previously (Hahne et al., 2001).
Generation of GST-SWAP-70 Fusion Proteins
The cDNA coding for full-length SWAP-70 or just the PH domain (aa 205–313) was amplified by PCR by using primers MB432 (GGGGATCCGGGAGCTTGAAGGAGGAGCTG), MB433B (GGCTCGAGTCACTCCGTGGTCTTTTTCTCTT) and MB418 (GGGGATCCATGGCAATTAATGAAGTCTTTAATG), MB419 (GGCTCGAGTCAAGGGCTGCCCAGCTTCAACAG), respectively. The PCR fragments were cloned into the BamHI and XhoI restriction sites of the pGEX4T-1 vector (Pharmacia), yielding plasmids GST-SWAP-70 and GST-PH(205-313). GST-SWAP-R230C and GST-SWAPRR223,224EE plasmids were obtained by QuikChange site-directed mutagenesis as described above for the GFP plasmids. For cloning of the GST-PH-R230C and GST-PH-RR223,224EE plasmids the mutated SWAP-70 cDNAs served as templates for the initial PCR. All PCR-products were verified by sequencing.
GST-SWAP-70 and GST-PH fusion proteins were expressed in E. coli (DH5α or XL1Blue for GST-PH domains) and purified over glutathione-Sepharose 4B as recommended by the manufacturer (Amersham Biosciences).
Phospholipid Binding Assay
Nitrocellulose strips containing spots of equal amounts (100 pmol) of different phospholipids (PIP-strips) were purchased from Echelon Biosciences (Salt Lake City, UT). Phospholipid-binding assays were performed as recommended by the manufacturer. Briefly, the PIP-strips were blocked with 3% fatty acid-free bovine serum albumin (Sigma-Aldrich) and incubated with the GST-SWAP fusion proteins overnight at 4°C. Bound protein was visualized by standard immunodetection. GST-SWAP-70 fusion proteins were detected by SWAP-70 antibody GK2 and GST-PH fusion proteins were detected by anti-glutathione S-transferase (GST) antibody (Amersham Biosciences). Secondary antibodies were either coupled to peroxidase and detected by chemiluminescence (Pierce Chemical) or to alkaline phosphatase and detected by 2,2′-di-p-nitrophenyl-5,5′-diphenyl-3,3′-[3,3′-dimethoxy-4,4′-diphenylene]-ditetrazolium chloride)/(5-bromo-4-chloro-3-indolyl-phosphate) color reaction (Promega).
Online Supplemental Material
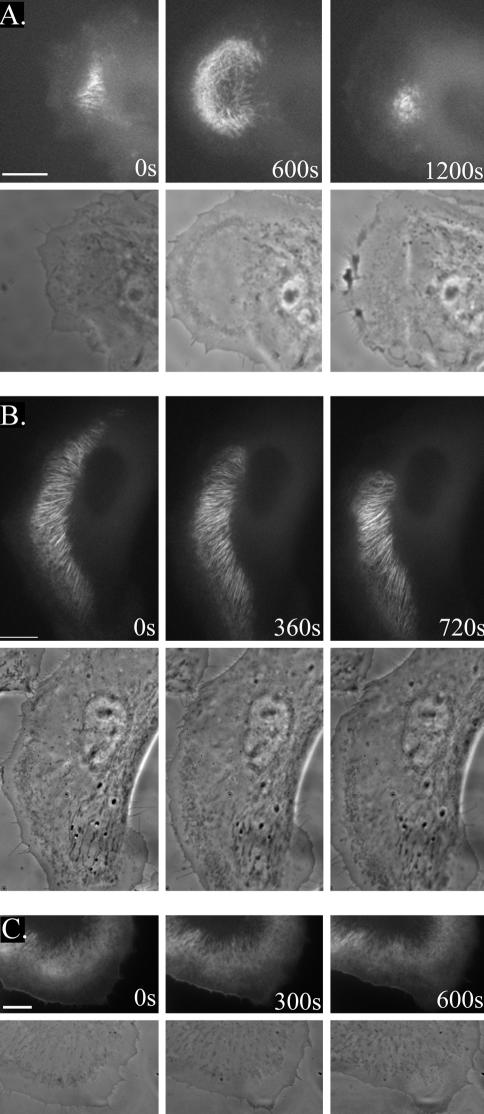
Time-lapse images are included as online videos, supplementing Figure 5 (Videos 1, 2, and 3). The videos 1, 2, and 3 contain the selected time-lapse frames shown in Figure 5A, B, and C, respectively. The time intervals of successive frames for Video 1 are 20 s and for Videos 2 and 3 are 30 s, respectively. All cells were monitored simultaneously by fluorescence and phase contrast microscopy. All videos are available at www.molbiolcell.org.
Figure 5.
SWAP-70 localizes to loose actin filament arrays located behind actively extending lamellipodia. B16F1 cells were transfected with GFP-SWAP-70, replated on laminin-coated coverslips and images were acquired 1 h (A) or 4 h (B and C) after replating. Corresponding fluorescence and phase contrast images are shown. (A) Spreading cell. The lamellipodium protrudes, achieves its maximal size at the 600-s time point and then collapses. (B) Slowly migrating cell, which is changing the direction of migration. (C) Ruffling cell with SWAP-70 localizing to the cell edge of retracting area. Bars, 10 μm. See also Videos 1, 2, and 3.
RESULTS
SWAP-70 Is Widely Expressed
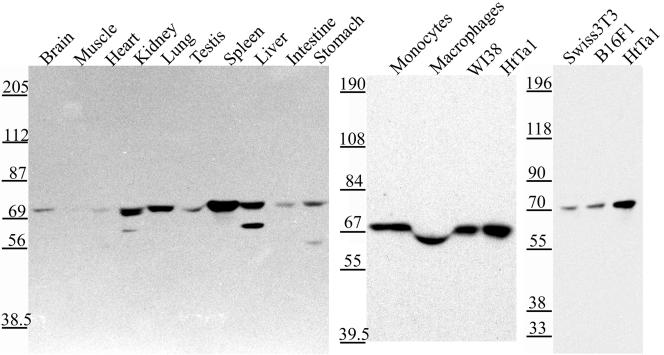
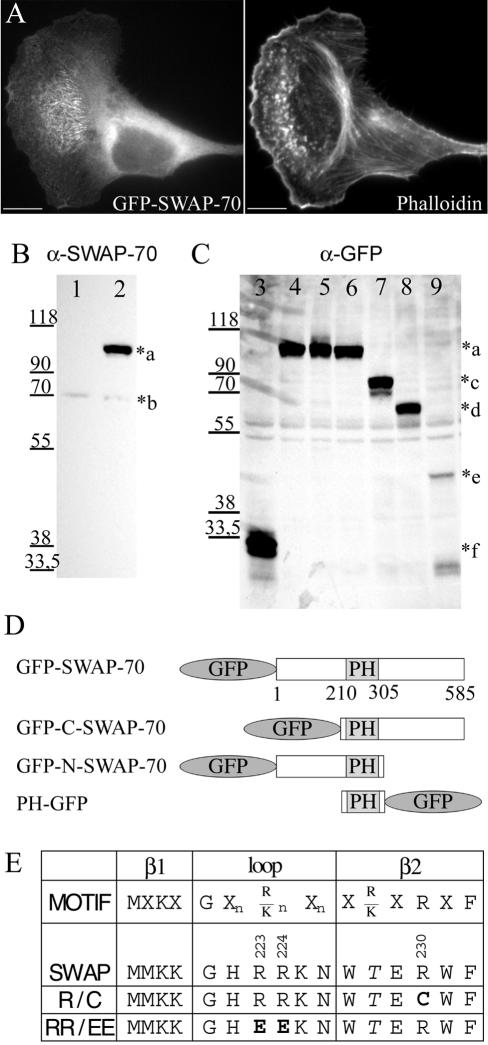
Human full-length SWAP-70 was identified in a two-hybrid screen by using a part of the tail region (amino acids 1440–1592) of rat myosin IXb as a bait. The physiological signifi-cance of this interaction remains to be determined. SWAP-70 has been reported to be a B cell-specific protein (Borggrefe et al., 1998, 1999; Masat et al., 2000). However, Ishikawa et al. (1998) had reported the cloning of SWAP-70 (KIAA0640) from a human brain cDNA library. Furthermore, reverse transcription-PCR experiments had detected SWAP-70 mRNA expression in all of the 10 different tissues analyzed (Ishikawa et al., 1998). In addition, our SWAP-70 cDNA was derived from a WI-38 lung fibroblast cDNA library. To assess the expression of SWAP-70 protein in various rat tissues and cell lines, we raised polyclonal antibodies specific for SWAP-70. Affinity-purified antibodies recognized on immunoblots a single protein band of the expected molecular mass of 70 kDa (Figure 1). SWAP-70 was detected in every tissue tested, being most abundant in spleen and fairly abundant in kidney, lung, and liver. SWAP-70 was also expressed in human monocytes and macrophages, the human cell lines WI-38 and HtTA-1 HeLa, and the mouse cell lines Swiss3T3 and B16F1 (Figure 1). Northern blot analysis revealed a similar tissue distribution of mRNA expression (our unpublished data).
Figure 1.
SWAP-70 is widely expressed. Equal amounts of protein from different adult rat tissues and human and mouse cell homogenates were immunoblotted with the affinity-purified SWAP-70 antibody GK2. Tissues and cells are indicated as described above each lane. Molecular mass standards in kilodaltons are shown on the left.
SWAP-70 Associates with Filament Arrays Generated behind Actively Protruding Lamellipodia That Lack Myosin II
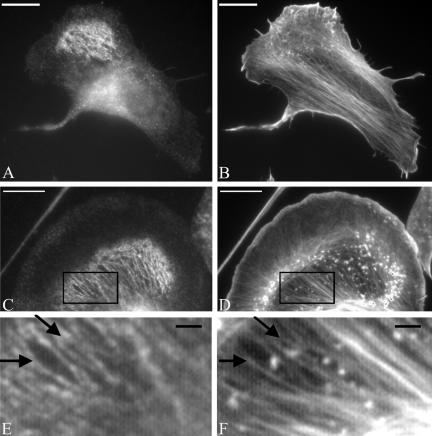
In B16F1 mouse melanoma cells, SWAP-70 exhibited in many cells a diffuse localization. However, in cells with circular membrane ruffles (actin clouds; Ballestrem et al., 1998), SWAP-70 was localized specifically on filaments immediately adjacent to the actin clouds (Figure 2). By using glutaraldehyde to preserve actin filaments and double labeling with phalloidin, it could be demonstrated that SWAP-70 colocalized with very fine, loose actin filament arrays (Figure 2, C–F) that were easily overlooked against the more prominent actin networks of lamellipodia and stress fiber bundles. Therefore, SWAP-70 highlighted a subset of actin filament arrays that were not assembled into distinct filament bundles. Significantly, the same filament arrays were found to lack nonmuscle myosin II (Figure 3). The filament arrays labeled by SWAP-70 were neither colocalizing with microtubules nor intermediate filaments, nor the actin-binding proteins tropomyosin, α-actinin, and vinculin (our unpublished data).
Figure 2.
Endogenous SWAP-70 associates with a subset of loose actin filament arrays. B16F1 cells were plated on 25 μg/ml laminin for 5 h before paraformaldehyde (A and B) or glutaraldehyde (C and D) fixation and double labeling with SWAP-70 antibody GK2 (A and C) and phalloidin (B and D). Areas boxed in C and D are shown at higher magnification in E and F, respectively. Arrows in E and F show the colocalization of SWAP-70 (E) with fine actin filament bundles (F). Bars, A–D, 10 μm; E and F, 1.5 μm.
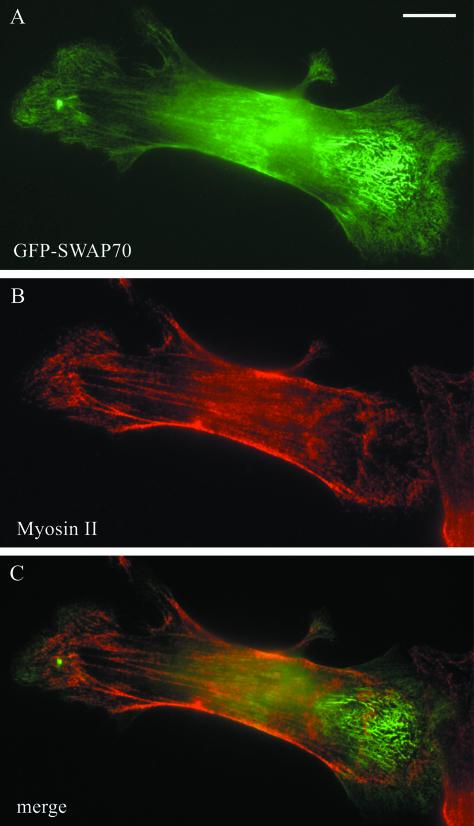
Figure 3.
SWAP-70 does not localize on myosin II-containing actin fibers. B16F1 cells were transfected with GFP-SWAP-70 as described in MATERIALS AND METHODS and fixed for 3 min in 4% PFA, 0.1% Triton X-100 followed by fixation in 4% PFA for 30 min. Cells were labeled for myosin II by indirect immunofluorescence by using a secondary antibody coupled to Alexa 568. GFP-SWAP-70 (A), myosin II (B), and merge of the two stainings (C). Bar, 10 μm.
To investigate the SWAP-70 localization in live cells, we expressed a fusion protein of SWAP-70 with GFP at its N or C termini. Both constructs localized in the same way as endogenous SWAP-70, when expressed at low levels (Figure 4A), and exhibited the expected molecular weight when probed by immunoblotting with antibodies directed against SWAP-70 or GFP (Figure 4, B and C; only data from the N-terminal fusion are shown).
Figure 4.
GFP-constructs. (A) Thirty hours after transfection with GFP-SWAP-70, B16F1 cells were replated on laminin-coated coverslips and after an additional 5 h fixed with paraformaldehyde and stained with Alexa 594-phalloidin. Bars, 10 μm. (B and C) B16F1 cells were transfected with different GFP-SWAP-70-constructs for 30 h and then analyzed by Western blotting with polyclonal affinitypurified SWAP-70 antibody GK2 (B) or monoclonal GFP antibody (C). Equal amounts of protein (B, 10 μg; C, 40 μg) from cell homogenates were loaded. Cells were transfected with the following constructs: lanes 1 and 3: pEGFP-C1; lanes 2 and 4: GFP-SWAP-70; lane 5: GFP-SWAP-R230C; lane 6: GFP-SWAP-RR223,224EE; lane 7: GFP-SWAP(205-585); lane 8: GFP-SWAP(1-313) and lane 9: PH(205-313)GFP. The *a–f indicates the size of the different constructs: a (∼100 kDa): GFP-SWAP-70; b (70 kDa): endogenous SWAP-70; c (∼75 kDa): GFP-SWAP(205-585); d (∼67 kDa): GFP-SWAP(1-313); e (∼43 kDa): PH(205-313)-GFP; and f (∼30 kDa): GFP. (D) Schematic representation of SWAP-70 GFP-fusion proteins. (E) Residues conserved in PH domains binding to PI(3,4)P2 and PI(3,4,5)P3 (Isakoff et al., 1998) are aligned to the SWAP-70 sequence. Mutated residues in constructs R230C and RR223,224EE are indicated in bold.
In resting, nonmotile cells, GFP-SWAP-70 was distributed diffusely. However, when cells started to extend lamellipodia, GFP-SWAP-70 became concentrated in fine, loose actin filament arrays located in the lamellae regions (Figure 5A; Video 1). The GFP-SWAP-70–labeled filaments followed the direction of extension, but they did not extend into lamellipodia (Figure 5B; Video 2). In cells switching from protrusion to retraction of lamellipodia, GFP-SWAP-70 became frequently localized to the cell edge, whereby the filamentous localization was gradually lost (Figure 5C; Video 3). In summary, the filamentous GFP-SWAP-70 localization was strictly coupled to lamellipodia extension and as such was highly dynamic. In accordance with this notion, an increase in number of cells exhibiting active lamellipodial extension or rosettes of dynamic actin “spots” by the application of aluminum fluoride and vanadate, respectively, correlated with an increase in number of cells with fine, loose actin filament arrays labeled by SWAP-70 (our unpublished data). The localization of SWAP-70 to loose actin filament arrays coupled to lamellipodial extension was not restricted to B16F1 cells, but was also observed in other cells such as Swiss 3T3, REF52, and HeLa (supplemental Figure 1).
The PH-Domain of SWAP-70 Binds Phosphoinositides
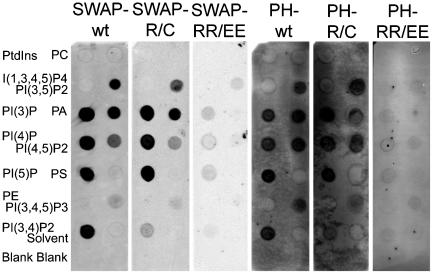
As a first step in addressing the mechanism of SWAP-70 targeting to the loose actin filament arrays, we sought to investigate the involvement of the SWAP-70 PH-domain. In vitro phospholipid binding studies were performed with purified GST-SWAP-70 and GST-PH fusion proteins (Figure 6). PIP-strips containing equal amounts of different phospholipids immobilized on nitrocellulose were incubated with the fusion proteins. Both fusion proteins bound specifically to a number of phosphorylated phosphoinositides, namely, phosphatidylinositol (3)-phosphate, phosphatidylinositol (4)-phosphate, phosphatidylinositol (5)-phosphate, phosphatidylinositol (3,4)-bisphosphate, phosphatidylinositol(3,5)bisphosphate, and phosphatidylinositol(4,5)-bisphosphate. A weaker binding of GST-PH and only a faint binding of GST-SWAP-70 was observed to PI(3,4,5)P3. No binding was observed to the head group I(1,3,4,5)P4, phosphatidylinositol, phosphatidylethanolamine, phosphatidylcholine, and phosphatidylserine. Interestingly, the fusion proteins also interacted with phosphatidic acid. Control experiments with GST alone detected no binding (our unpublished data).
Figure 6.
The PH domain of SWAP-70 binds phosphoinositides in vitro. GST-SWAP-70 constructs were expressed in E. coli and purified. The PIP-strips were incubated with different fusion proteins indicated on top of each PIP-strip at concentrations of 0.5 μg/ml (SWAP-wt, SWAP-R/C, SWAP-RR/EE, and PH-RR/EE) or 0.01 μg/ml (PH-wt and PH-R/C). Bound fusion proteins were detected by indirect immunostaining. The GST-SWAP-70 fusion proteins were detected by GK2 antibody and the GST-PH fusion proteins with GST antibody. The phospholipids (100 pmol) spotted on PIP-strips were as follows: phosphatidylinositol (PtdIns), phosphatidylcholine (PC), inositol(1,3,4,5)-tetraphosphate [I(1,3,4,5)P4], phosphatidylinositol(3,5)-bisphosphate [PI(3,5)P2], phosphatidylinositol (3)-phosphate [PI(3)P], phosphatidic acid (PA), phosphatidylinositol (4)-phosphate [PI(4)P], phosphatidylinositol(4,5)-bisphosphate [PI(4,5)P2], phosphatidylinositol (5)-phosphate [PI(5)P], phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylinositol(3,4,5)-trisphosphate [PI(3,4,5)P3], and phosphatidylinositol(3,4)-bisphosphate [PI(3,4)P2].
To modify or delete the phosphoinositide binding, we constructed two PH-domain mutants. Based on previous studies with PH-domains, we speculated that R230 might be important for binding to the 3-phosphate. Therefore, we mutated this residue to a cysteine as in a mutant of Btk that causes agammaglobulinemia (deWeers et al., 1994). Introduction of this mutation caused a specific reduction of the binding of GST-SWAP-70 and GST-PH to PI(3,4)P2 (Figure 6). To abolish the binding of the SWAP-70 PH-domain to phosphoinositides, we mutated the two positively charged arginine residues 223 and 224 in the β1/β2-loop to negatively charged glutamic acid residues. As predicted, the introduction of these two point mutations abolished phosphoinositide and also phosphatidic acid binding (Figure 6).
The Binding of a 3′-Phosphoinositide by the PH-Domain Is Necessary for Localization of SWAP-70 to the Loose Actin Filament Arrays
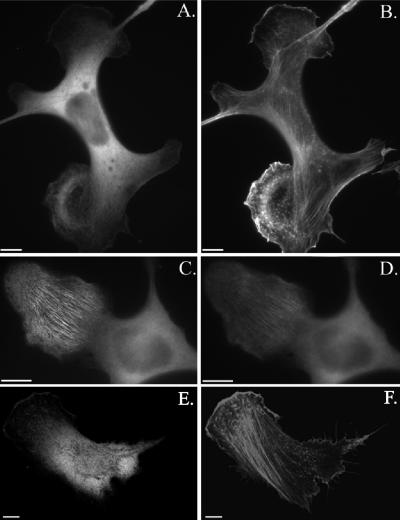
Next, we investigated how the altered phospholipid binding by the mutant PH-domains affected the cellular localization of SWAP-70. GFP-SWAP-70-RR/EE no longer localized to the fine, loose actin filament arrays, demonstrating that phospholipid binding to the PH-domain is necessary for the specific subcellular localization of SWAP-70 (Figure 7A). Occasionally, GFP-SWAP-70-RR/EE exhibited a faint colocalization with the phalloidin staining. Introduction of the PH-domain R/C mutation into GFP-SWAP-70 seemed to reduce the SWAP-70 actin filament association. To compare the localization of wild-type SWAP-70 and SWAP-70-R/C directly in the same cells, we cotransfected cells with CFP-SWAP-70 and YFP-SWAP-70-R/C. As shown in Figure 7, C and D, YFP-SWAP-70-R/C exhibited an equal intensity of cytoplasmic staining to CFP-SWAP-70, but a much fainter labeling of the fine actin filament arrays. This result suggests that a 3′-phosphoinositide could be the physiological ligand of the SWAP-70 PH-domain, a notion also supported by the finding that the SWAP-70 localization was abolished by the specific PI 3-kinase inhibitor wortmannin (our unpublished data). Although the PH-domain of SWAP-70 is necessary for proper SWAP-70 localization, it is not sufficient. The isolated PH-domain of SWAP-70 was not targeted to the fine loose actin filament arrays and its localization seemed to be mostly cytosolic (Figure 7, E and F)
Figure 7.
The PH-domain is necessary for proper localization of SWAP-70 to loose actin filament arrays, but it is not sufficient. B16F1 cells were transfected with GFP-SWAP-RR/EE (A and B) and PHGFP (E and F) or cotransfected with CFP-SWAP-70 (C) and YFP-SWAP-70R230C (D). GFP-SWAP-70-RR/EE– and PH-GFP–transfected cells were stained with Alexa 594-phalloidin (B and F). The cell shown in C and D was treated with AlF4– for 20 min. before fixation. Fluorescence images of representative cells are shown. In A–D, epifluorescence images are shown and in E, F a 0.4-μm confocal section. Bars, 10 μm.
The Very C-Terminal Region of SWAP-70 Contains Actin Filament-targeting Information
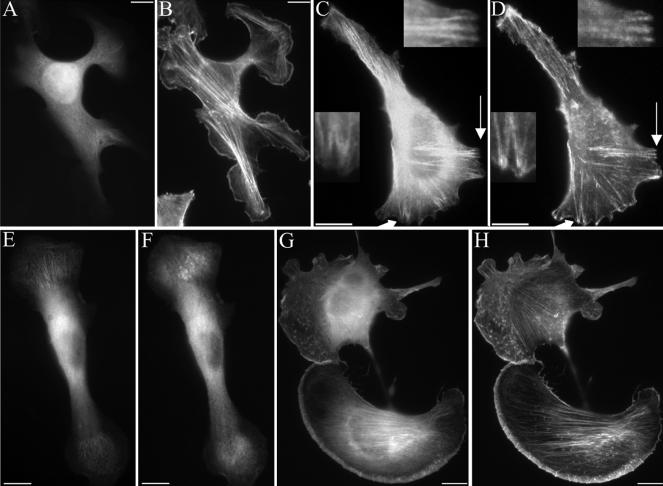
To identify the region of SWAP-70 that associates with actin filament arrays, we truncated SWAP-70 after the PH-domain and expressed it as GFP-SWAP(1-313) fusion protein. GFP-SWAP(1-313) failed to associate with actin filaments and was diffusely distributed (Figure 8, A and B). Truncation of the N-terminal region yielded the GFP-SWAP(205-585) fusion protein. In contrast to GFP-SWAP(1-313), this fusion protein containing the PH-domain and the C-terminal region localized to actin filaments (Figure 8, C and D). This places the actin filament-binding region to the C-terminal region of SWAP-70. Importantly, the GFP-SWAP(205-585) was not restricted to the subset of loose actin filament arrays, but it was located on actin filament bundles in general, with the exception of those in focal contacts (Figure 8, C and D, insets). Therefore, the specificity for binding to a subset of actin filaments must be provided by the N-terminal region. The very C-terminal region is involved in actin filament binding because truncation of the last 60 amino acids compromised the targeting to loose actin filament arrays (Figure 8, E and F). This construct was still enriched in areas of the loose actin filament arrays labeled by SWAP-70. However, in these areas it exhibited a diffuse or spotty and only occasionally filamentous localization. It also localized partly to the plasma membrane. To test whether the very C-terminal 60 residues contain the targeting information for actin filaments, we expressed a GFP-SWAP(526-585) fusion protein. This GFP-SWAP(526-585) protein exhibited a striking colocalization with actin filaments stained by phalloidin (Figure 8, G and H), demonstrating that it indeed exhibits F-actin targeting information. However, in vitro F-actin co-sedimentation experiments with purified GST-SWAP(526-585) or full-length GST-SWAP-70 did not reveal any detectable binding to F-actin (our unpublished data), suggesting that the association with actin filaments is dependent on intermediaries or cofactors.
Figure 8.
Truncation of both, the N-terminal region or the C-terminal region of SWAP-70 abolishes the localization to the subset of loose actin filament arrays. The C-terminal 60 residues contain F-actin–targeting information. B16F1 cells were transfected with GFP-SWAP(1-313) (A and B), GFP-SWAP(205-585) (C and D), or GFP-SWAP(526-585) (G and H) and double stained with Alexa 594-phalloidin (B, D, and H). The cell in C and D was fixed in the presence of Triton X-100 to improve visibility of colocalization of GFP-SWAP(205-585) with F-actin. The arrows in C and D point at the regions enlarged in the insets. In the insets the lack of GFP-SWAP(205-585) in focal contacts is shown. (E and F) B16F1 cells were cotransfected with CFP-SWAP-70 (E) and YFP-SWAP(1-525) missing the C-terminal 60 amino acids (F). Cells were fixed and viewed in the fluorescence microscope. Bars, 10 μm.
Overexpression of SWAP-70 Alters Actin Organization and Cellular Morphology
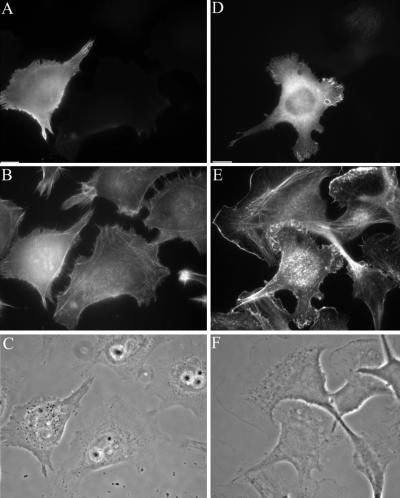
Overexpression of either HA-tagged SWAP-70 or GFP-SWAP-70 in HtTa-1 HeLa cells and B16F1 cells caused alterations in the actin cytoskeleton and cellular morphology (Figure 9). HtTa-1 HeLa cells showed a loss of lamellipodia and the acquisition of highly refractile retraction fiber-like extensions (Figure 9, A–C). The tips of these fibers were still dynamic and showed often an accumulation of SWAP-70 protein. As a consequence of SWAP-70 overexpression, the HtTa-1 HeLa cells adopted a spindle-shaped or rounded morphology. Overexpression of GFP-SWAP-70 in B16F1 cells also led to alterations in lamellipodial morphology and actin organization (Figure 9, D–F). The lamellipodia were no longer smooth and extended but instead were strongly ruffling and generally less extended. Interestingly, treatment of cells with aluminum fluoride reversed the morphological changes induced by SWAP-70 overexpression to a large extent (our unpublished data). The overexpression of SWAP-70 constructs that were not able to localize to the fine loose actin arrays, such as the SWAP-70-RR/EE PH-domain mutant and the truncated SWAP(1-313) and SWAP(205-585) constructs did not induce any obvious alterations (our unpublished data). Therefore, proper phosphoinositide-dependent subcellular targeting of overexpressed SWAP-70 constructs seemed to be a prerequisite for the induction of cellular alterations in the actin organization.
Figure 9.
Overexpression of SWAP-70 alters the actin organization. GFP-SWAP-70 (A and D) was overexpressed in HtTa-1 HeLa cells (A–C) and B16F1 cells (D–F). Cells were fixed, permeabilized, and double stained with Alexa 594-phalloidin (B and E). Corresponding phase contrast images are shown in C and F. Bars, 10 μm. Note that GFP-SWAP-70–overexpressing cells show either a loss (HtTa-1 HeLa) of lamellipodia or an altered lamellipodial morphology (B16F1).
DISCUSSION
SWAP-70 Identifies a Transitional Subset of Actin Filaments in Motile Cells
Cell motility involves protrusion of a cell front and subsequent retraction of the rear. Protrusion is based on the forward, cyclic growth, or polymerization of actin filaments in lamellipodia and filopodia. Retraction, on the other hand, is based on the interaction of preformed actin filaments with myosin-II in contractile bundles. And the majority of these preformed filaments seem to be derived or initiated via polymerization in lamellipodia at the cell front (Small et al., 1998). There must therefore be a major switch in the proteins associated with actin from generation in lamellipodia to incorporation into other arrays in the body of the cytoskeleton. Indeed, the regulated association of different actin binding proteins with actin is implicit in general ideas about the formation and function of the actin cytoskeleton. And there are various examples of actin bundling proteins that are used in the organization of parallel actin arrays for specific functions in different cell types (Bartles, 2000). In contrast to bona fide bundling proteins, SWAP-70 was associated with a subset of actin filaments that were not in obvious bundled arrays.
Two observations suggest that SWAP-70 marks filaments in the actin cytoskeleton that are in transition between generation in the lamellipodium and incorporation into contractile bundles. First, SWAP-70 filaments were both spatially and temporarily associated with the formation of lamellipodia and membrane ruffles. Speckle microscopy has shown a continuum of retrograde flow of actin behind lamellipodia (Waterman-Storer et al., 1998), consistent with the idea that a proportion of filaments generated in the lamellipodium contribute to the network of actin that makes up the rest of the actin cytoskeleton. Second, SWAP-70–labeled filaments conspicuously lacked myosin-II. In fibroblasts, the interaction of myosin-II with actin is required for both the assembly and maintenance of actin filament bundles (Burridge and Chrzanowska-Wodnicka, 1996) that are in turn required for retraction. We therefore conclude that SWAP-70 associates with newly formed actin filaments generated in lamellipodia, that have shed lamellipodia-associated proteins (Small et al., 2002), but have yet to acquire other associated proteins, in particular myosin, to cooperate in the assembly of contractile bundles (Verkhovsky et al., 1995). Such a subset of actin filaments is likely detected only in rapidly migrating cells where significant forward movement can occur before complexing with myosin and other proteins takes place. Whether SWAP-70 plays a role in regulating myosin association remains to be established. Nevertheless, SWAP-70 seems to be the first example of a protein marking a subset of filaments in arrays distinct from lamellipodia meshwork and compact bundles.
Spatiotemporal Control of SWAP-70 Recruitment to the Subset of Loose Actin Filament Arrays
The regulated association of SWAP-70 with loose actin filament arrays was coupled to active lamellipodial extension, and it was blocked by the inhibition of PI 3-kinase. Several observations argue for a direct rather than an indirect regulation of SWAP-70 localization by 3′-phosphoinositides and in particular PI(3,4)P2. SWAP-70 with point mutations in the PH-domain was no longer found associated with the loose actin arrays. The PH-domain of SWAP-70 was demonstrated to bind monophosphorylated phosphoinositides, PI(3,4)P2, and phosphatidic acid in vitro. Replacement of two arginine residues by glutamic acids in the β1-β2 loop of the PH-domain abolished in vitro binding to the mentioned phospholipids and in vivo association with the loose actin arrays. Mutation of an arginine residue in the β2-strand important for 3-phosphate coordination (Salim et al., 1996; Franke et al., 1997; Ferguson et al., 2000) to cysteine reduced selectively the binding of SWAP-70 to PI(3,4)P2 and the association of SWAP-70 with the loose actin arrays. Oxidative stress has been reported to induce highly elevated levels of PI(3,4)P2 in cells (Van der Kaay et al., 1999). Exposure of cells to oxidative stress, a spatially unrestricted signal, caused a quantitative PI 3-kinase-dependent translocation of SWAP-70 to loose actin arrays and the plasma membrane (our unpublished data). Steep gradients of the PI 3-kinase lipid products PI(3,4)P2 and/or PI(3,4,5)P3 have been observed in agonist stimulated neutrophils and amoebas of Dictyostelium discoideum. High concentrations of these phosphoinositides were detected at actin-rich leading edges of cells (Meili et al., 1999; Rickert et al., 2000; Servant et al., 2000; Chung et al., 2001). Therefore, generation of agonist stimulated PI(3,4)P2 in extending lamellipodia could serve as a spatiotemporal signal for recruitment of SWAP-70 via its PH-domain. Other phosphoinositides cannot be excluded as SWAP-70 PH-domain ligands, because the SWAP-70 PH domain bound several phosphoinositides in vitro. The binding of SWAP-70 to PI(3,4,5)P3, the main product of PI 3-kinase, was very weak. This is in accordance with predictions based on structural data (Thomas et al., 2001) that the D-5 inositol-phosphate–binding site might be blocked in the SWAP-70 PH-domain.
Phosphoinositide binding of the PH-domain may be followed by the association of SWAP-70 with actin filaments. The actin association domain was demonstrated to reside in the C-terminal 60 residues of SWAP-70. A SWAP-70 fragment lacking the N-terminal region, but still containing the PH-domain, no longer localized to the specific subset of loose actin arrays. It associated with the bulk of the actin filaments present in cells, but was excluded from focal adhesions. This result can be explained by a regulatory function of the N-terminal region. The N-terminal region might prevent F-actin binding by the C-terminal 60 residues in dependence of ligand binding by the PH-domain. The PH-domain ligand, proposed to be PI(3,4)P2, would control the restricted actin array association. According to this scenario, the PH-domain and the C-terminally truncated fragment might be expected to associate with the plasma membrane of extending lamellipodia. However, no clear indication for such a subcellular localization was noticed for these two fragments, indicating that there is also a region C-terminal of the PH-domain needed for full specificity of SWAP-70 localization. This is in accordance with the nonfilamentous plasma membrane localization of SWAP(1-525) at leading edges and areas of loose actin arrays. The C-terminal 60 residues that are missing in SWAP(1-525) are responsible for F-actin association. This association seems to be indirect or dependent on a cofactor, because no direct binding to F-actin could be demonstrated.
The overexpression of SWAP-70 induced alterations in actin organization and cell morphology, indicating that SWAP-70 does play a role in the control of actin organization. SWAP-70 may keep the loose actin filament arrays in a transitional state. Targeting of an excess of SWAP-70 to the loose actin filament arrays may cause their depolymerization, because constructs that were not properly targeted did not alter the actin organization. Aluminum fluoride may counteract the observed changes of actin organization by stabilization of the actin filaments. It has been reported that aluminum fluoride inhibits turnover and depolymerization of actin filaments in vitro (Combeau and Carlier, 1988). Alternatively, aluminum fluoride might reverse the effects of an excess of SWAP-70 by activation of a signaling pathway opposing the SWAP-70 activity.
We propose that SWAP-70 regulates the actin cytoskeleton and cell motility as an effector or adaptor in response to agonist stimulated PI 3-kinase activity, possibly by acting as a protein that prevents premature bundling of actin filaments in protruding zones.
Supplementary Material
Acknowledgments
We thank Angelika Freitag for valuable technical assistance, Christine Drexhage and Nicole Gründker for collection of the SWAP(1-525) data, and Dr. Heiko Düβmann for help with adaptation of the videos. We acknowledge the gift of the GFP-antibody from Prof. Dr. Markus Maniak (University of Kassel, Kassel, Germany). This work was supported by the Deutsche Forschungsgemeinschaft (Ba 1354/5-1) and the Austrian Science Research Council (project P14660-PAT).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-01-0043. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-01-0043.
Abbreviations used: 3′-phosphoinositide, refers to PI(3)P, PI(3,4)P2, PI(3,5)P2, and PI(3,4,5)P3; GFP, green fluorescent protein; GST, glutathione S-transferase; HA, hemagglutinin; PH, pleckstrin homology; PI 3-kinase, phosphoinositide 3-kinase; PI(3,4)P2, phosphatidylinositol 3,4-bisphosphate; PI(3,4,5)P3, phosphatidylinositol 3,4,5-trisphosphate.
The online version of this article contains video material for some figures. Online version is available at www.molbiolcell.org.
References
- Ballestrem, C., Wehrle-Haller, B., and Imhof, B.A. (1998). Actin dynamics in living mammalian cells. J. Cell Sci. 111, 1649–1658. [DOI] [PubMed] [Google Scholar]
- Bartles, J.R. (2000). Parallel actin bundles and their multiple actinbundling proteins. Curr. Opin. Cell Biol. 12, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe, T., Wabl, M., Akhmedov, A.T., and Jessberger, R. (1998). A B-cell-specific DNA recombination complex. J. Biol. Chem. 273, 17025–17035. [DOI] [PubMed] [Google Scholar]
- Borggrefe, T., Masat, L., Wabl, M., Riwar, B., Cattoretti, G., and Jessberger, R. (1999). Cellular, intracellular, and developmental expression patterns of murine SWAP-70. Eur. J. Immunol. 29, 1812–1822. [DOI] [PubMed] [Google Scholar]
- Burridge, K., and Chrzanowska-Wodnicka, M. (1996). Focal adhesions, contractility, and signaling. Annu. Rev. Cell. Dev. Biol. 12, 463–518. [DOI] [PubMed] [Google Scholar]
- Chieregatti, E., Gärtner, A., Stöffler, H.E., and Bähler, M. (1998). Myr 7 is a novel myosin IX-RhoGAP expressed in rat brain. J. Cell Sci. 111, 3597–3608. [DOI] [PubMed] [Google Scholar]
- Chung, C.Y., Funamoto, S., and Firtel, R.A. (2001). Signaling pathways controlling cell polarity and chemotaxis. Trends Biochem. Sci. 26, 557–566. [DOI] [PubMed] [Google Scholar]
- Combeau, C., and Carlier, M.F. (1988). Probing the mechanism of ATP hydrolysis on F-actin using vanadate and the structural analogs of phosphate BeF3– and AlF4–. J. Biol. Chem. 263, 17429–17436. [PubMed] [Google Scholar]
- de Weers, M., Mensink, R.G., Kraakman, M.E., Schuurman, R.K., and Hendriks, R.W. (1994). Mutation analysis of the Bruton's tyrosine kinase gene in X-linked agammaglobulinemia: identification of a mutation which affects the same codon as is altered in immunodeficient xid mice. Hum. Mol. Genet. 3, 161–166. [DOI] [PubMed] [Google Scholar]
- Devreotes, P.N., and Zigmond, S.H. (1988). Chemotaxis in eucaryotic cells: a focus on leucocytes and Dictyostelium. Annu. Rev. Cell Biol. 4, 649–686. [DOI] [PubMed] [Google Scholar]
- Ferguson, K.M., Kavran, J.M., Sankaran, V.G., Fournier, E., Isakoff, S.J., Skolnik, E.Y., and Lemmon, M.A. (2000). Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell 6, 373–384. [DOI] [PubMed] [Google Scholar]
- Franke, T.F., Kaplan, D.R., Cantley, L.C., and Toker, A. (1997). Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science 275, 665–668. [DOI] [PubMed] [Google Scholar]
- Gibson, T.J., Hyvönen, M., Musacchio, A., Saraste, M., and Birney, E. (1994). PH domain: the first anniversary. Trends Biochem. Sci. 19, 349–353. [DOI] [PubMed] [Google Scholar]
- Golemis, E.A., and Khazak, V. (1997). Alternative yeast two-hybrid systems. The interaction trap and interaction mating. Methods Mol. Biol. 63, 197–218. [DOI] [PubMed] [Google Scholar]
- Gossen, M., and Bujard, H. (1992). Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne, P., Sechi, A., Benesch, S., and Small, J.V. (2001). Scar/WAVE is localised at the tips of protruding lamellipodia in living cells. FEBS Lett. 492, 215–220. [DOI] [PubMed] [Google Scholar]
- Haslam, R.J., Koide, H.B., and Hemmings, B.A. (1993). Pleckstrin domain homology. Nature 363, 309–310. [DOI] [PubMed] [Google Scholar]
- Heath, J.P., and Holifield, B.F. (1993). On the mechanisms of cortical actin flow and its role in cytoskeletal organisation of fibroblasts. Symp. Soc. Exp. Biol. 47, 35–56. [PubMed] [Google Scholar]
- Isakoff, S.J., Cardozo, T., Andreev, J., Li, Z., Ferguson, K.M., Abagyan, R., Lemmon, M.A., Aronheim, A., and Skolnik, E.Y. (1998). Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J. 17, 5374–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, K., Nagase, T., Suyama, M., Miyajima, N., Tanaka, A., Kotani, H., Nomura, N., and Ohara, O. (1998). Prediction of the coding sequences of unidentified human genes. X. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 5, 169–176. [DOI] [PubMed] [Google Scholar]
- Kavran, J.M., Klein, D.E., Lee, A., Falasca, M., Isakoff, S.J., Skolnik, E.Y., and Lemmon, M.A. (1998). Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 273, 30497–30508. [DOI] [PubMed] [Google Scholar]
- Machesky, L.M., and Hall, A. (1997). Role of actin polymerization and adhesion to extracellular matrix in rac- and rho-induced cytoskeletal reorganization. J. Cell Biol. 138, 913–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masat, L., Caldwell, J., Armstrong, R., Khoshnevisan, H., Jessberger, R., Herndier, B., Wabl, M., and Ferrick, D. (2000). Association of SWAP-70 with the B cell antigen receptor complex. Proc. Natl. Acad. Sci. USA 97, 2180–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meili, R., Ellsworth, C., Lee, S., Reddy, T.B.K., Ma, H., and Firtel, R.A. (1999). Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 18, 2092–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameh, L.E., et al. (1997). A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J. Biol. Chem. 272, 22059–22066. [DOI] [PubMed] [Google Scholar]
- Resnitzky, D., Gossen, M., Bujard, H., and Reed, S.I. (1994). Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol. Cell. Biol. 14, 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert, P., Weiner, O.D., Wang, F., Bourne, H.R., and Servant, G. (2000). Leukocytes navigate by compass: roles of PI3Kγ and its lipid products. Trends Cell Biol. 10, 466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim, K. et al. (1996). Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton′s tyrosine kinase. EMBO J. 15, 6241–6250. [PMC free article] [PubMed] [Google Scholar]
- Schafer, D.A., Welch, M.D., Machesky, L.M., Bridgman, P.C., Meyer, S.M., and Cooper, J.A. (1998). Visualization and molecular analysis of actin assembly in living cells. J. Cell Biol. 143, 1919–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant, G., Weiner, O.D., Herzmark, P., Balla, T., Sedat, J.W., and Bourne, H.R. (2000). Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science 287, 1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, J.V. (1988). The actin cytoskeleton. Electron Microsc. Rev. 1, 155–174. [DOI] [PubMed] [Google Scholar]
- Small, J.V., Rottner, K., Kaverina, I., and Anderson, K.I. (1998). Assembling an actin cytoskeleton for cell attachment and movement. Biochim. Biophys. Acta 1404, 271–281. [DOI] [PubMed] [Google Scholar]
- Small, J.V., Stradal, T., Vignal, E., and Rottner, K. (2002). The lamellipodium: where motility begins. Trends Cell Biol. 12, 112–120. [DOI] [PubMed] [Google Scholar]
- Thomas, C.C., Dowler, S., Deak, M., Alessi, D.R., and van Aalten, D.M.F. (2001). Crystal structure of the phosphatidylinositol 3,4-bisphosphate-binding pleckstrin homology (PH) domain of tandem PH-domain-containing protein 1 (TAPP1): molecular basis of lipid specificity. Biochem. J. 358, 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Kaay, J., Beck, M., Gray, A., and Downes, P. (1999). Distinct phosphatidylinositol 3-kinase lipid products accumulate upon oxidative and osmotic stress and lead to different cellular responses. J. Biol. Chem. 274, 35963–35968. [DOI] [PubMed] [Google Scholar]
- Verkhovsky, A.B., Svitkina, T.M., and Borisy, G.G. (1995). Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J. Cell Biol. 131, 989–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky, A.B., Svitkina, T.M., and Borisy, G.G. (1998). Self-polarization and directional motility of cytoplasm. Curr. Biol. 9, 11–20. [DOI] [PubMed] [Google Scholar]
- Wang, Y.-L. (1985). Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling, J. Cell Biol. 101, 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer, C.M., Desai, A., Bulinski, J.C., and Salmon, E.D. (1998). Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr. Biol. 8, 1227–1230. [DOI] [PubMed] [Google Scholar]
- Weiner, O.D., Servant, G., Welch, M.D., Mitchison, T.J., Sedat, J.W., and Bourne, H.R. (1999). Spatial control of actin polymerization during neutrophil chemotaxis. Nat. Cell Biol. 1, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond, S.H. (1981). Consequences of chemotactic peptide receptor modulation for leukocyte orientation. J. Cell Biol. 88, 644–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.