Abstract
Cell cycle transitions are subject to regulation by both external signals and internal checkpoints that monitor satisfactory progression of key cell cycle events. In budding yeast, the morphogenesis checkpoint arrests the cell cycle in response to perturbations that affect the actin cytoskeleton and bud formation. Herein, we identify a step in this checkpoint pathway that seems to be directly responsive to bud emergence. Activation of the kinase Hsl1p is dependent upon its recruitment to a cortical domain organized by the septins, a family of conserved filament-forming proteins. Under conditions that delayed or blocked bud emergence, Hsl1p recruitment to the septin cortex still took place, but hyperphosphorylation of Hsl1p and recruitment of the Hsl1p-binding protein Hsl7p to the septin cortex only occurred after bud emergence. At this time, the septin cortex spread to form a collar between mother and bud, and Hsl1p and Hsl7p were restricted to the bud side of the septin collar. We discuss models for translating cellular geometry (in this case, the emergence of a bud) into biochemical signals regulating cell proliferation.
INTRODUCTION
The effects of cell shape and cytoskeletal tension on cell proliferation have been well documented (Sumpio et al., 1987; Chen et al., 1997; Huang and Ingber, 1999), but the molecular mechanisms responsible for sensing these inputs remain unknown. The morphogenesis checkpoint in Saccharomyces cerevisiae provides a tractable system in which to ask how such inputs are translated into cell cycle decisions. This checkpoint arrests the cell cycle when bud formation is impaired by environmental or experimental disturbances, thereby ensuring that unbudded cells do not proceed through mitosis until they have formed a bud (Lew, 2000). Cell cycle arrest is mediated by Swe1p, homologous to Wee1 in Schizosaccharomyces pombe, which phosphorylates and inhibits the cyclin-dependent kinase Cdc28p (Booher et al., 1993; Sia et al., 1998).
The action of Swe1p is counteracted by the phosphatase Mih1p, homologous to Cdc25 in S. pombe (Russell et al., 1989). The checkpoint involves at least two pathways: one leads to Swe1p stabilization (Sia et al., 1998) and the other is thought to inhibit Mih1p through the mitogen-activated protein kinase (MAPK) Slt2p (Harrison et al., 2001). Both pathways are required to promote G2 arrest; herein, we focus on the mechanism of Swe1p stabilization.
Swe1p accumulates during late G1 and S phase and is then degraded so that most of the protein is gone by the time of nuclear division (Sia et al., 1998). However, if cells are exposed to insults or mutations that depolarize actin, Swe1p is stabilized and accumulates (Sia et al., 1998). Swe1p degradation requires the Nim1-family protein kinase Hsl1p and the protein methyltransferase Hsl7p (Ma et al., 1996; Edgington et al., 1999; McMillan et al., 1999a; Lee et al., 2000). Hsl7p interacts directly with both Hsl1p and Swe1p, and mutations that impair either of these interactions cause Swe1p stabilization (Cid et al., 2001; McMillan et al., 2002).
Hsl1p and Hsl7p are localized to a ring on the bud side of the mother-bud neck, and targeting of Hsl7p to that location requires Hsl1p (Barral et al., 1999; Shulewitz et al., 1999; Longtine et al., 2000). When Swe1p first accumulates in late G1, it is localized to the nucleus, but after bud emergence a subpopulation of Swe1p is recruited to the bud side of the neck by Hsl1p and Hsl7p (Longtine et al., 2000). The striking observation that the same proteins (Hsl1p and Hsl7p) are needed for both neck targeting and degradation of Swe1p suggests that Swe1p neck targeting is a requisite step in the Swe1p degradation pathway.
Localization of Hsl1p, Hsl7p, and Swe1p to the neck relies upon the septins (Barral et al., 1999; Shulewitz et al., 1999; Longtine et al., 2000). Septins are filament-forming proteins that organize specialized cortical domains, to which they recruit proteins involved in various processes (Longtine et al., 1996; Gladfelter et al., 2001). In S. cerevisiae, the septins first assemble into a ring at the prebud site, which then expands to form an hourglass-shaped collar on the cytoplasmic face of the plasma membrane at the bud neck. In mutant strains that misorganize septins, localization of Hsl1p and Hsl7p is disrupted. These strains exhibit Swe1p-dependent G2 delays in the cell cycle, suggesting that localization of Hsl1p and Hsl7p to the neck is important for down-regulation of Swe1p (Barral et al., 1999; Longtine et al., 2000). Targeting of Swe1p to the neck presumably serves to mark Swe1p for degradation, although the mechanistic basis for such “marking” remains unknown. In particular, the possible roles of Hsl1p kinase activity and Hsl7p methyltransferase activity in promoting Swe1p degradation are unclear.
In an effort to understand how yeast cells can “sense” perturbations of bud formation, we examined how the Swe1p degradation pathway is affected by insults that delay or block bud emergence. We show that activation of the Hsl1p kinase at the septin cortex is tightly correlated with bud emergence and seems to depend upon bud emergence. We also show that Hsl1p kinase activity is important for the recruitment of Swe1p to the neck, presumably initiating Swe1p degradation. Moreover, deformation of cells allows Hsl7p recruitment even in the absence of bud formation. We suggest that the change in cell shape that accompanies bud emergence triggers a reorganization of the septin cortex, which in turn leads to Hsl1p activation and Swe1p degradation.
MATERIALS AND METHODS
Strains, Plasmids, and Polymerase Chain Reaction (PCR) Manipulations
Standard media and methods were used for plasmid and yeast manipulations (Guthrie and Fink, 1991; Ausubel et al., 1995). S. cerevisiae strains are listed in Table 1.
Table 1.
Yeast strains used in this study
| Straina | Relevant genotype |
|---|---|
| DLY1 | abar1 |
| DLY4339 | aHSL7-3HA:kan cdc12-6::LEU2 |
| DLY4640 | abar1 hsl1K110Rmyc:URA3 |
| DLY4994 | abar1 GAL:MIH1:TRP1 hsl1K110R |
| DLY5000 | abar1 HSL1myc:URA3 HSL7-3HA:kan |
| DLY5002 | abar1 HSL7-3HA:kan hsl1K110R |
| DLY5019 | α bar1 SWE1myc:HIS2 SWE1myc:TRP1 hsl1K110R |
| DLY5334 | abar1 HSL1myc:URA3 HSL7-3HA:kan bed1::URA3 |
| DLY5390 | aHSL7-3HA:kan hsl1K110Rmyc:URA3 |
| DLY5777 | abar1 HSL1myc:URA3 HSL7-3HA:kan GAL1p-SIC1::LEU2 |
| DLY5794 | aHSL7-3HA:kan |
| JMY1289 | abar1 GAL:MIH1:TRP1 hsl1Δ1:URA3 |
| JMY1290 | abar1 GAL:MIH1:TRP1 hsl7Δ1:URA3 |
| JMY1340 | abar1 GAL:MIH1:TRP1 |
| JMY1435 | α bar1 SWE1myc:HIS2 SWE1myc:TRP1 |
| JMY1500 | abar1 HSL1myc:URA3 |
All strains are in the BF264-15Du background (ade1 his2 leu2-3,112 trp1-1a ura3Δns), except DLY4339 which is in the YEF473 background (his3 leu2 trp1 ura3 lys2).
The hsl1Δ1:URA3 and hsl7Δ1:URA3 (Ma et al., 1996), GAL:MIH1: TRP1, mih1ΔTRP1, HSL1myc:URA3, SWE1myc:HIS2, SWE1-myc: TRP1, and HSL7–3HA:kan (McMillan et al., 1999a), and bed1::URA3 (Mondesert and Reed, 1996) alleles were described previously.
To construct hsl1K110R, we first introduced the mutation into the HSL1-containing plasmid pNE32 (Edgington et al., 1999) by replacing the N-terminal NcoI fragment spanning residues 27–133 of Hsl1p with a corresponding overlap-PCR–generated fragment encoding the K110R change (AAA lys110 codon altered to CGA arg codon, introducing a BstB1 site). The construct was sequenced to verify that the desired mutation (and no others) had been introduced. An EcoRI fragment containing HSL1 promoter and sequences encoding the N-terminal 600 residues of hsl1K110R was then transformed (together with a LEU2-marked carrier plasmid) into a strain containing hsl1-Δ2::URA3 (Ma et al., 1996), in which sequences encoding the N-terminal 495 residues of Hsl1p are replaced with URA3. Colonies in which the hsl1-Δ2::URA3 allele had been replaced with hsl1K110R sequences by homologous recombination were identified by their resistance to 5-fluoroorotic acid and confirmed by PCR analysis of genomic DNA. The C-terminal tagging of this allele with the myc epitope was performed as described for HSL1myc: URA3 (McMillan et al., 1999a).
To construct HSL7G386A, we first introduced the mutation into pDLB1545 (containing a HindIII-SacI HSL7 fragment from 600 base pairs upstream of the ATG to 400 base pairs downstream of the stop codon in YCplac111; Gietz and Sugino, 1988). We replaced an internal HpaI-SacII fragment spanning residues 174–573 of Hsl7p with a corresponding overlap-PCR–generated fragment encoding the G386A change (GGA gly386 codon altered to GCC ala codon, and the neighboring AGA arg387 codon altered to CGG arg codon to introduce a SmaI site). The construct (pDLB1608) was sequenced to verify that the desired mutation (and no others) had been introduced.
To express GST-Hsl7p and GST-Hsl7pG386A in Escherichia coli, we first cloned sequences encoding the entire HSL7 open reading frame (wild type or mutant) plus 400 base pairs downstream as a NdeI-SacI fragment into the corresponding sites in pUNI-10 (Liu et al., 1998). The genes were then excised as EcoRI-SacI fragments and cloned into the corresponding sites of pGEX-KG (Pharmacia, Peapack, NJ), yielding pDLB2211 (GST-Hsl7p) and pDLB2212 (GST-Hsl7pG386A).
Cell Cycle Synchrony, Latrunculin-A (Lat-A) Treatment, and Shmoo Formation
Cells were synchronized in G1 by pheromone arrest (incubation of bar1 cells growing exponentially with 40 ng/ml α-factor for 3 h at 30°C) and release, or by centrifugal elutriation performed as described previously (Lew and Reed, 1993) except that cells were grown in 4% sucrose to achieve a higher cell density. Lat-A (Molecular Probes, Eugene, OR) was added directly to the medium from a 20 mM stock in dimethyl sulfoxide to a final concentration of 100 μM.
For the shmoo formation experiment (Figure 7), some technical aspects of the experiment deserve mention, especially because of an apparent conflict with a similar experiment published previously (Cid et al., 2001). We found that cells that were transferred from pheromone-containing medium (causing G1 arrest and shmoo formation) into Lat-A–containing medium (causing actin depolymerization) frequently failed to overcome the G1 arrest. This was particularly problematic when the cells were supersensitive to pheromone (as with bar1 mutant MATa cells). To circumvent this problem, we used BAR1 cells and allowed a 20-min “recovery” period between washing out the pheromone and adding in Lat-A. In the work of Cid et al. (2001), Hsl7p was localized to the spindle pole body when it was not at the neck (i.e., during telophase and G1). We have never detected Hsl7p at that location but that may simply be due to insufficient sensitivity, because Cid et al. (2001) examined cells that overexpressed Hsl7p (either massively or mildly), whereas we used a single integrated copy of HSL7-HA. Using a protocol similar to the one that we used involving pheromone arrest, shmoo formation, and release into medium-supplemented Lat-A, Cid et al. (2001) reported that Hsl7p remained associated with the spindle pole body, and contrary to our findings, was not detected within the shmoo after release. No controls were shown to indicate whether the Lat-A–treated cells had exited G1, but given our experience, we suspect that those cells, which were bar1 mutant MATa cells and were not given a recovery period, did not escape from the G1 arrest and therefore failed to assemble septin rings and kept Hsl7p at the spindle pole body. Consistent with this interpretation, the Hsl7p remained as a single spot, suggesting that the spindle pole body had not duplicated and separated as would be expected if the cells had successfully entered the cell cycle.
Figure 7.
Septin spreading and Hsl7p recruitment in the absence of ongoing polarized growth or bud emergence. (A and B) Cells of strain DLY5794 (MATa BAR1 HSL7-HA) were arrested in G1 by incubation with 5 μg/ml α-factor at 24°C for 4 h and then washed and incubated in fresh medium. Cells were fixed at the indicated times and stained to visualize the septin Cdc11p. Cells were scored according to whether they displayed septin staining at the base of the shmoo projection (A, cell 1; B, open circles), septin rings (A, cells 2 and 3; B, filled circles), or both (B, squares). A, cell 1 is from the arrested culture; cell 2 illustrates a ring within the shmoo projection; and cell 3 illustrates a ring away from the shmoo projection, which may also have some remnant staining at the base of the projection. For B, 200 shmoos were scored at each time point. Cells that were still round (i.e., not shmoos) and had not made clear projections were not scored. (C–E) Cells of strain DLY5794 arrested as described above were released into fresh medium for 20 min before addition of 100 μM Lat-A to depolymerize actin (preventing bud emergence). One hour later, the cells were fixed and double stained to visualize septins and Hsl7p-HA. (C) Cells in which the septin ring assembled within the shmoo projection: in these cells, the septins expanded to form a broader collar rather than a narrow ring, and Hsl7p was frequently recruited to the distal rim of the septin collar (arrows). Arrowheads indicate shmoo tip. The middle cell had made two projections (pointing up and to the right, respectively). (D) Cells in which the ring assembled in a locally flat part of the cell cortex, away from the shmoo: these cells did not recruit Hsl7p to the septin ring (arrows). Arrowheads indicate shmoo tip. (E) Shmoos were scored according to whether the septin ring assembled within (left) or away from (right) the shmoo projection, and whether Hsl7p was (black) or was not (white) recruited to the septin ring. 200 shmoos were scored.
Immunofluorescence and Microscopy
Immunofluorescence detection was as described previously (Longtine et al., 2000), except that fixation was for 75 min at 23°C. For double-staining experiments, the primary (mouse anti-myc 9E10 and rabbit anti-Cdc11; Santa Cruz Biotechnology, Santa Cruz, CA; and mouse anti-HA 12CA5; Roche Diagnostics, Indianapolis, IN) and secondary (goat anti-mouse Cy2 or Cy3, and goat anti-rabbit Cy2 or Cy3; Jackson Immunoresearch Laboratories, West Grove, PA) antibodies for the two epitopes were applied simultaneously.
Microscopy was performed with an Axioskop (Carl Zeiss, Thorn-wood, NY) with standard fluorescence and differential interference contrast optics. Images were captured with a cooled charge-coupled device camera (Princeton Instruments, Princeton, NJ) interfaced with MetaMorph software (Universal Imaging, Silver Springs, MD).
Biochemical Procedures
Procedures for harvesting and lysis of yeast cells, SDS-PAGE, immunoprecipitation, and immunoblotting, were as described previously (McMillan et al., 1999a), except that 6% low-bis (Barral et al., 1999) gels were used to better resolve phosphorylated Hsl1p and Hsl7p species. Hsl1p kinase assays were performed as described for Cdc28p (McMillan et al., 1999b) except that no histone H1 substrate was added.
Procedures for harvesting and lysis of bacterial cells, purification of glutathione S-transferase (GST)-tagged proteins and elution with glutathione were as described previously (McMillan et al., 1999b; Bose et al., 2001). For methyltransferase assays, ∼1 μg of GST-Hsl7p or GST-Hsl7pG386A was incubated with 10 μg of histone H2A (Roche Diagnostics) and 15 μl of [3H]S-adenosylmethionine (PerkinElmer Life Sciences, Boston, MA) in methylation buffer (150 mM NaCl, 50 mM Tris-HCl pH 8.0, 1% NP-40) at 30°C for 60 min. The reaction was terminated by boiling in sample buffer; samples were resolved on a 15% polyacrylamide gel; and the gel was fixed, treated with EN3HANCE (PerkinElmer Life Sciences), dried, and exposed to BioMax MS film with Transcreen LE intensifying screen (Eastman Kodak, Rochester, NY) for 3 d at –80°C.
RESULTS
Role of Hsl1p and Hsl7p Catalytic Activities in Swe1p Regulation
To address how Swe1p degradation is regulated by the morphogenesis checkpoint, we first need to understand how Swe1p is targeted for degradation in unperturbed cells. Having established a requirement for Hsl1p and Hsl7p in this process (McMillan et al., 1999a), we wished to examine the role of their respective catalytic activities. In the absence of Hsl1p or Hsl7p, Swe1p is stabilized and accumulates to higher than normal levels, but Swe1p activity is counteracted by the phosphatase Mih1p so the overall effect of the Swe1p increase on the cell cycle can be quite minor. Cells lacking Mih1p are sensitized to the level of Swe1p, and if Swe1p is stabilized in these cells (as occurs upon deletion of HSL1 or HSL7) they undergo a lethal G2 arrest associated with the development of very elongated buds (McMillan et al., 1999a). We used the viability of mih1Δ cells as a sensitive bioassay for the function of catalytically inactive versions of Hsl1p and Hsl7p.
Mutation of lys 110 to arg in kinase subdomain II (Hanks and Hunter, 1995) of Hsl1p severely reduced Hsl1p autophosphorylation in vitro (Figure 1A). Hsl1pK110R was expressed at comparable levels to wild-type Hsl1p (Figure 2A), but failed to rescue the viability of strains lacking Mih1p (Barral et al., 1999; Figure 1B). Like hsl1Δ mih1Δ strains, hsl1K110R mih1Δ cells arrested with very elongated buds (Figure 1B), suggesting that Hsl1p kinase activity is important for Swe1p regulation.
Figure 1.
Role of Hsl1p and Hsl7p catalytic activities in Swe1p regulation. (A) Myc-tagged wild-type or K110R mutant Hsl1p proteins immunoprecipitated from yeast lysates (strains JMY1500 and DLY4640) were incubated together with [γ-32P]ATP to assess Hsl1p autophosphorylation (top) or immunoblotted to assess protein abundance (bottom). (B) The GAL1p-MIH1 strains JMY1340 (HSL1), JMY1289 (hsl1Δ), and DLY4994 (hsl1K110R) were grown on galactose-containing medium and cells were streaked out onto dextrose-containing medium to repress Mih1p expression. Microcolonies were photographed after 2 d at 30°C. (C) GST-tagged wild-type or G386A mutant Hsl7p proteins purified using glutathione-Sepharose were incubated together with [3H]S-adenosylmethionine and histone H2A to assess histone methylation (top) or stained with Coomassie Blue to assess protein abundance (middle, histone H2A; bottom, GST-Hsl7p). (D) The GAL1p-MIH1 strain JMY1290 (hsl7Δ) was transformed with plasmids containing HSL7 (pDLB1545), empty vector (YCplac111), or HSL7G386A (pDLB1608) and microcolonies were analyzed as described above.
Figure 2.
Role of Hsl1p kinase activity in phosphorylation and localization of Hsl1p, Hsl7p, and Swe1p. (A) Cells of strains DLY5000 (HSL1-myc HSL7-HA) and DLY5390 (hsl1K110R-myc HSL7-HA) were grown to exponential phase, harvested, and lysed. Top, Western blot of myc-tagged Hsl1p or Hsl1pK110R. Bottom, Western blot of hemagglutinin (HA)-tagged Hsl7p from HSL1 or hsl1K110R strains. (B) Localization of Hsl1p, Hsl7p, and Swe1p in strains containing wild-type (top) or catalytically inactive (bottom) Hsl1p. Left, localization of Hsl1p-myc or Hsl1pK110R-myc in strains DLY5000 and DLY5390. Middle, localization of Swe1p-myc in strains JMY1435 and DLY5019. Right, localization of Hsl7p-HA in strains DLY5794 and DLY5002.
The physiological target(s) of Hsl7p methyltransferase activity are unknown, but Hsl7p can methylate histone H2A in vitro (Lee et al., 2000). Mutation of gly 386 to ala in methyltransferase motif I (Ma, 2000) of Hsl7p abolished detectable H2A methylation (Figure 1C), but HSL7G386A fully rescued the viability of strains lacking Mih1p (Figure 1D). Thus, Hsl7p methyltransferase activity seems to be dispensable for Swe1p regulation.
What are the targets of Hsl1p kinase activity? In S. pombe, the related kinase Nim1 phosphorylates Wee1 (Coleman et al., 1993; Parker et al., 1993; Wu and Russell, 1993), but attempts to detect phosphorylation of Swe1p by Hsl1p in vitro have thus far been unsuccessful, even though the same preparations of Hsl1p readily phosphorylate both Hsl1p itself and Hsl7p (our unpublished observations; Cid et al., 2001). In vivo, Hsl1p but not Hsl1pK110R undergoes extensive hyperphosphorylation (Figure 2A; Barral et al., 1999), and Hsl7p undergoes Hsl1p-dependent phosphorylation (Figure 2A; McMillan et al. 1999a), so both Hsl1p and Hsl7p seem to be bona fide targets of Hsl1p kinase activity.
To assess the role of Hsl1p kinase activity in more detail, we examined the localization of Hsl1p, Hsl7p, and Swe1p in cells containing Hsl1pK110R as the only source of Hsl1p. Hsl1p kinase activity was dispensable for the efficient targeting of Hsl1p to the neck (Figure 2B). However, Swe1p remained in the nucleus and was almost never detected at the neck in hsl1K110R cells (Figure 2B). In the case of Hsl7p, an intermediate result was obtained: only 42% of budded hsl1K110R cells had detectable levels of Hsl7p at the neck (compared with >95% of budded HSL1 cells), and many of those cells had severely reduced Hsl7p staining at the neck compared with HSL1 cells (Figure 2B, arrow). This was not due to a reduction in the total level of Hsl7p, which was similar in HSL1 and hsl1K110R cells (Figure 2A). These results suggest that the failure of hsl1K110R cells to target Swe1p to the neck (Figure 2B) underlies their inability to down-regulate Swe1p (Figure 1B).
Timing of Hsl1p and Hsl7p Localization and Hyperphosphorylation in Unperturbed Cells
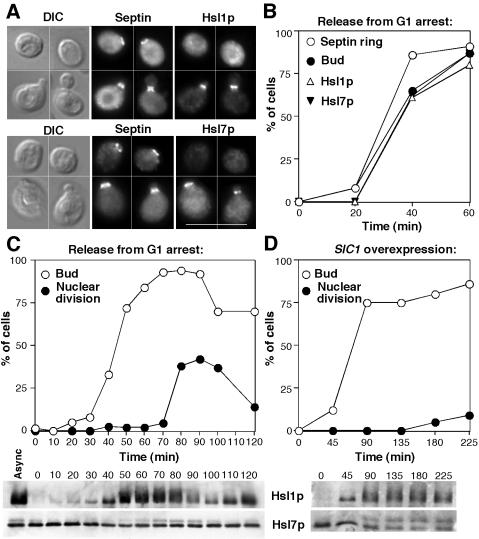
Hsl1p abundance varies during the cell cycle (Tanaka and Nojima, 1996; McMillan et al., 1999a; Burton and Solomon, 2000), whereas Hsl7p abundance is constant (McMillan et al., 1999a). We found that Hsl1p and Hsl7p recruitment to the septins was tightly correlated with bud emergence (Figure 3, A and B). Even cells with very small buds generally displayed robust Hsl1p and Hsl7p staining at the neck, whereas neither protein was detected at septin rings in unbudded cells (Figure 3A). These findings are in agreement with a recent study on Hsl7p localization, which additionally detected localization of Hsl7p to the SPB during G1, before its recruitment to the septin cortex (Cid et al., 2001).
Figure 3.
Timing of Hsl1p and Hsl7p localization and phosphorylation. (A) Asynchronous cells of strain DLY5000 (HSL1-myc HSL7-HA) were double stained to visualize the septin Cdc11p and either Hsl1p-myc (top) or Hsl7p-HA (bottom). Bar, 10 μm. (B and C) Cells of strain DLY5000 were synchronized by pheromone-induced G1 arrest/release in two separate experiments. (B) Aliquots of one culture were fixed and stained to examine the septin ring formation, Hsl1p recruitment, Hsl7p recruitment, and bud formation, as indicated. More than 200 cells were scored for each sample. (C) Aliquots of the other culture were lysed, separated by SDS-PAGE, and immunoblotted to detect Hsl1p-myc (top) or Hsl7p-HA (bottom). Parallel aliquots were fixed, stained with 4,6-diamidino-2-phenylindole to visualize DNA, and scored for bud emergence and nuclear division (n = 200). (D) Cells of strain DLY5777 (HSL1-myc HSL7-HA GAL1p-SIC1) were grown to exponential phase in sucrose-containing medium and arrested in G1 with pheromone. Galactose was added to 2% final concentration to induce Sic1p expression (this strain contains multiple integrants of the GAL1p-SIC1 construct, which produce enough Sic1p to block cell cycle progression), and 1 h later the cells were washed and resuspended in galactose-containing medium lacking pheromone. All operations were performed at 30°C. Bud emergence, nuclear division, and Hsl1p and Hsl7p phosphorylation were then assessed after release from arrest.
Phosphorylation of Hsl1p (Barral et al., 1999) and Hsl7p (McMillan et al., 1999a) cause mobility shifts of the proteins upon analysis by SDS-PAGE, and we used these shifts to assess the timing of phosphorylation during the cell cycle. In synchronized cells, Hsl1p accumulation began just before bud emergence, and Hsl1p hyperphosphorylation occurred shortly thereafter, remaining steady until nuclear division, after which Hsl1p was degraded (Figure 3C). The correlation between bud emergence and Hsl1p phosphorylation might reflect a causal relationship, whereby bud emergence triggers Hsl1p activation. However, bud emergence is approximately coincident with the initial activation of Cdc28p by B-type cyclins, which triggers other cell cycle events, including DNA replication and SPB separation (Lew et al., 1997). To assess whether Clb/Cdc28p activity, rather than bud emergence, might be the relevant trigger for Hsl1p phosphorylation, we repeated the synchrony experiment under conditions in which Clb/Cdc28p activation was blocked by the inhibitor Sic1p. As described previously, cells overexpressing Sic1p formed elongated buds and did not undergo nuclear division (Figure 3D), consistent with arrest at the G1/S boundary. However, a clear Hsl1p mobility shift occurred after bud emergence (Figure 3D), indicating that significant Hsl1p phosphorylation can occur independent of Clb/Cdc28p.
Hsl7p phosphorylation was also detected just after bud emergence in synchronized cells (Figure 3C), independent of Clb/Cdc28p (Figure 3D). These data are consistent with previous findings that cells arrested due to Sic1p are still able to localize Hsl7p (Cid et al., 2001) and Swe1p (McMillan et al., 2002) to the neck.
In summary, recruitment of Hsl1p and Hsl7p to the septin cortex occurred just after bud emergence and coincided with (or was shortly followed by) phosphorylation of Hsl1p and Hsl7p, which was largely independent of Clb/Cdc28p. Together with previous studies, these findings suggest a pathway for degradation of Swe1p in unperturbed cells in which the septin cortex recruits and activates Hsl1p kinase, promoting efficient recruitment of Hsl7p and Swe1p to the bud side of the neck, where Swe1p becomes marked for destruction in a still mysterious manner.
Effect of Actin Depolymerization on Hsl1p and Hsl7p
Which step in the Swe1p degradation pathway is blocked by the morphogenesis checkpoint? To address this question, we treated cells with the actin-depolymerizing drug Lat-A and examined the behavior of Hsl1p and Hsl7p. Before bud emergence, the septins assemble into a ring at the prebud site, and this process is unaffected by Lat-A treatment (Ayscough et al., 1997). In unbudded Lat-A–treated cells, Hsl1p was recruited to the septin ring even though the actin depolymerization blocked bud emergence (Figure 4A). In striking contrast, Hsl7p was never recruited to the septin ring in unbudded Lat-A–treated cells (Figure 4B). Thus, it seems that actin depolymerization in unbudded cells disrupts an early step in the Swe1p degradation pathway, involving the recruitment of Hsl7p to the septins by Hsl1p.
Figure 4.
Effect of actin depolymerization on Hsl1p and Hsl7p. (A) Cells of strain DLY5000 (HSL1-myc HSL7-HA) were grown to exponential phase and treated with 100 μM Lat-A for 1 h at 30°C and then fixed and double stained to visualize the septin Cdc11p and either Hsl1p-myc (top) or Hsl7p-HA (bottom). Bar, 10 μm. (B) G1 cells of strain DLY5000 were isolated by centrifugal elutriation and inoculated into fresh medium at 30°C. Lat-A or dimethyl sulfoxide (control) was added at 45 min (before bud emergence) and the incubation was continued for a further 2 h, and cells were processed to detect Hsl1p-myc (top) or Hsl7p-HA (bottom) by Western blot. (C) Cells of strain DLY5000 were grown to exponential phase at 30°C, treated with 100 μM Lat-B, and processed to detect Hsl1p-myc (top) or Hsl7p-HA (bottom) by Western blot. At later times, the population was skewed toward unbudded cells because some division took place but bud emergence was blocked. (D) Cells of strain DLY4339 (cdc12-6 HSL7-HA) were grown to exponential phase at 23°C, shifted to 37°C for the indicated times, and processed to detect Hsl7p-HA by Western blot.
To examine Hsl1p and Hsl7p phosphorylation in unbudded Lat-A–treated cells, we isolated early G1 cells using centrifugal elutriation, and exposed them to Lat-A (or dimethyl sulfoxide as a control). Neither Hsl1p nor Hsl7p phosphorylation was detected in the Lat-A–treated sample (Figure 4B). Staining with fluorescent phalloidin confirmed that actin was depolymerized in the Lat-A–treated samples, and staining for Hsl1p confirmed that >50% of the unbudded Lat-A–treated cells had recruited Hsl1p to the septin ring in this experiment. Thus, although Hsl1p was recruited to the septin ring, it did not promote Hsl1p or Hsl7p phosphorylation in unbudded Lat-A–treated cells.
In budded cells where Hsl1p and Hsl7p recruitment had already taken place, both Hsl1p and Hsl7p remained at the neck after Lat-A treatment (Longtine et al., 2000). As shown in Figure 4C, actin depolymerization did not greatly affect Hsl1p or Hsl7p phosphorylation in asynchronous, mostly budded cells. It is not clear from this whether Hsl1p activity was inhibited, because the proteins were phosphorylated before actin depolymerization, and we do not know their rate of dephosphorylation in vivo. In a separate experiment, we found that Hsl7p was dephosphorylated within 30 min upon shift of a Ts septin mutant (cdc12-6) to the restrictive temperature (Figure 4D), indicating that Hsl7p dephosphorylation is rapid under these conditions. Therefore, we suspect that the observed maintenance of Hsl7p phosphorylation in cells with depolymerized actin is due to persisting activity of Hsl1p. A caveat to this conclusion is that dephosphorylation may not be as fast when Hsl7p is at the neck (as in the actin depolymerization experiment) as it is when Hsl7p is displaced (as in the septin shift experiment).
In summary, actin depolymerization allows recruitment of Hsl1p to the septin ring, but blocks phosphorylation of Hsl1p and Hsl7p, as well as recruitment of Hsl7p to the septin ring, in unbudded cells. In budded cells, the localization and phosphorylation of Hsl1p and Hsl7p are not dramatically affected by actin depolymerization.
Effect of Delayed Bud Emergence on Hsl1p and Hsl7p
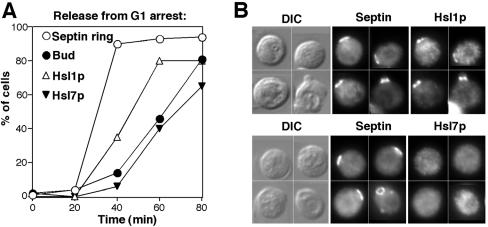
In addition to preventing bud formation, actin depolymerization triggers a stress response involving the MAPK Slt2p (Harrison et al., 2001). The failure to activate Hsl1p in unbudded Lat-A–treated cells might therefore be due either to the failure to form a bud or to a stress response. To ask whether blocking bud emergence was sufficient to block Hsl1p activation and Hsl7p recruitment to the septins, we used the mnn10Δ/bed1Δ (bud emergence delayed) mutant. In this mutant, actin polarization and septin ring assembly occur without obvious defects (Mondesert and Reed, 1996), and Slt2p is not activated above wild-type levels (our unpublished observations), but subsequent bud emergence is significantly delayed due to a primary defect in protein glycosylation. In bed1Δ cells, Hsl1p was recruited to the septin ring before bud emergence, but Hsl7p was not recruited to the septin cortex until just after the eventual emergence of the bud (Figure 5). Thus, blocking bud emergence is sufficient to prevent Hsl7p recruitment even in the absence of overt actin perturbation.
Figure 5.
Timing of Hsl1p and Hsl7p recruitment to the septin cortex in bed1Δ mutants. (A) Cells of strain DLY5334 (bed1Δ HSL1-myc HSL7-HA) were synchronized in G1 by pheromone and released into fresh medium at t = 0, and fixed at intervals to examine septin ring formation, Hsl1p recruitment, Hsl7p recruitment, and bud formation. More than 200 cells were scored for each sample. (B) Asynchronous cells of strain DLY5334 were double stained to visualize the septin Cdc11p and either Hsl1p-myc (top) or Hsl7p-HA (bottom).
Hsl1p hyperphosphorylation was also delayed in bed1Δ cells, even though Hsl1p was localized to the septin ring in the unbudded cells (Figure 6). This result indicates that recruitment of Hsl1p to the septins is not sufficient for its activation, which seems to require bud formation.
Figure 6.
Timing of Hsl1p phosphorylation in bed1Δ mutants. Early G1 cells were isolated by centrifugal elutriation, inoculated into fresh medium, and fixed to examine Hsl1p recruitment to the septin cortex and bud formation. More than 200 cells were scored for each sample. The phosphorylation state of Hsl1p in the same samples was examined by Western blotting (bottom panels). (A) Strain DLY5000 (BED1 HSL1-myc). (B) Strain DLY5334 (bed1Δ HSL1-myc).
What Aspect of Bud Emergence Controls Hsl1p Activation and Hsl7p Recruitment?
Bud emergence involves polarized growth in the vicinity of the bud site. As a consequence of polarized growth, there is a change in the local cell shape to generate the bud. Distinguishing whether the polarized growth itself or the resulting cell shape is responsible for Hsl1p activation and Hsl7p recruitment is nontrivial, because polarized growth necessarily alters local cell shape, and because the only known way to alter yeast cell shape is through polarized growth. However, we devised an experiment to ask whether ongoing polarized growth was required for Hsl7p recruitment, or whether an altered cell shape due to polarized growth in the past would suffice to allow these events to take place.
To alter cell shape in the absence of Hsl1p activation, we treated cells with mating pheromone, which arrests the cell cycle in G1 and triggers polarized growth, resulting in the formation of “shmoo” projections, which fuse during conjugation with a mating partner. We then released the cells from the pheromone arrest and added Lat-A to block polarized growth (see MATERIALS AND METHODS for details). G1-arrested cells do not express Hsl1p, but upon release into the cell cycle, these cells accumulated Hsl1p and assembled septin rings at the cortex (see above). However, they could not embark on further polarized growth or bud formation because of the absence of F-actin.
As described previously (Longtine et al., 1998), shmoos contained cortical septin assemblies at the base of the shmoo projection that frequently looked like parallel septin “bars” (Figure 7A, cell 1) and contributed to shaping the shmoo (Giot and Konopka, 1997). These septins disappeared after release from the arrest, and morphologically distinct septin rings formed at the cortex (Figure 7B). Occasionally, cells were observed that stained for septins both at the base of the shmoo and at a distinct ring (Figure 7B), suggesting that the disappearance of one structure is not linked to the appearance of the other. The new septin rings assembled either within the locally tubular shmoo projection (55% of shmoos, n = 200) or at locally flat sites away from the shmoo projection (45% of shmoos, n = 200). Shmoos that assembled septins in a locally flat geometry away from the shmoo projection displayed narrow septin rings similar to those in unbudded proliferating cells (Figure 7A, cell 3). In contrast, cells that assembled septins within the locally tubular geometry of the shmoo projection allowed the septin cortex to spread into a broader collar more similar to that observed at the bud neck (Figure 7A, cell 2).
To assess whether the new septin rings formed at different locations could recruit Hsl7p in the absence of further polarized growth, we added Lat-A to the cells after release from the G1 arrest, and examined Hsl7p localization 1 h later. Strikingly, the ability of these cells to recruit Hsl7p to the septin cortex was linked to the local geometry within which the new septin ring was formed. Cells that formed narrow septin rings away from the shmoo projection did not recruit Hsl7p (Figure 7, D and E), whereas cells that formed septin collars within the shmoo projection were able to recruit Hsl7p to the distal rim of the septin collar (Figure 7, C and E). (Results from a similar previously published experiment [Cid et al., 2001] are in apparent conflict with this result, but see MATERIALS AND METHODS for a likely resolution). Thus, Hsl7p recruitment (and presumably Hsl1p activation) can occur in cells that assemble the septins within a locally tubular (presumably more “neck-like”) context even in the absence of further polarized growth or bud emergence.
DISCUSSION
Hsl7p Methyltransferase Activity Is Dispensable for Its Role in Swe1p Down-Regulation
We found that a mutation that abolished the ability of Hsl7p to methylate histone H2A in vitro did not impair Hsl7p function in Swe1p down-regulation in vivo. The catalytic domain of S. cerevisiae Hsl7p has diverged significantly from that of other protein methyltransferases (including the Hsl7p homologs Skb1p in S. pombe and JBP1 in humans) (Ma, 2000), perhaps suggesting that it no longer acts enzymatically in S. cerevisiae, and retains only a residual catalytic activity. Alternatively, it may be that Hsl7p methyltransferase activity is important for some other Hsl7p function that was not assayed in our work. Recent studies have suggested that Skb1p participates in the response to osmotic stress in S. pombe (Bao et al., 2001) and that JBP1 is part of the “methylosome” complex involved in RNA splicing in mammals (Friesen et al., 2001). It is unclear whether Hsl7p plays similar roles in S. cerevisiae, but there are observations (such as the original isolation of hsl7 mutants in a synthetic lethal screen with histone H3 tail mutants [Ma et al., 1996] and the localization of Hsl7p to the SPB during G1 [Cid et al., 2001)] that are not readily explained by the known role of Hsl7p in Swe1p down-regulation, and it remains quite plausible that other roles requiring the methyltransferase activity remain to be discovered.
Hsl1p Kinase Activity Is Important for Targeting Hsl7p and Swe1p to the Neck
Unlike Hsl7p methyltransferase activity, Hsl1p kinase activity was essential for Swe1p down-regulation. Catalytically inactive Hsl1pK110R was localized to the neck just like wild-type Hsl1p and was able to recruit some Hsl7p to the neck. In contrast, a previous study found that Hsl1pK110A failed to localize to the neck (Mizunuma et al., 2001). At present, we cannot account for the difference between that study and our own, but Shulewitz and Thorner (personal communication) found that GFP-Hsl7p could still be targeted to the neck in hsl1K110R cells, consistent with our findings.
Although Hsl1pK110R was localized effectively to the neck, the localization of Hsl7p to the neck was somewhat reduced in hsl1K110R strains. Because Hsl7p interacts directly with the nonkinase domain of Hsl1p (Cid et al., 2001), it was not anticipated that Hsl7p recruitment by Hsl1p would be affected by Hsl1p kinase activity. This result, along with others discussed below, suggests that Hsl7p recruitment to the septin cortex is under more stringent regulation than was previously appreciated. Neck targeting of Swe1p was more severely affected than was neck targeting of Hsl7p in hsl1K110R cells. In principle, the reduced localization of Hsl7p could have a disproportionate effect on Swe1p localization (e.g., if it takes two or more molecules of Hsl7p to tether one molecule of Swe1p at the neck). Alternatively, Hsl1p kinase activity might play an additional role in Swe1p neck targeting (e.g., hyperphosphorylated Hsl1p might interact with Swe1p at the neck). The finding that Hsl1p catalytic activity is required for targeting Swe1p to the neck as well as for down-regulating Swe1p strengthens the hypothesis that Swe1p neck targeting is critical for Swe1p degradation.
Hsl1p Activation and Hsl7p Recruitment to the Septin Cortex Depend on Bud Emergence
Previous work indicated that incubation of septin-mutant cells at restrictive temperature eliminated Hsl1p activity (Barral et al., 1999). Furthermore, even relatively mild perturbations of the septin cortex caused delocalization of Hsl1p from the neck, as well as a Swe1p-dependent cell cycle delay (Longtine et al., 2000). From these observations, it is generally accepted that Hsl1p recruitment to the septin cortex is crucial for its activation and function in Swe1p down-regulation. Our new results indicate that although recruitment to the septins may be necessary for Hsl1p activation, recruitment alone is not sufficient. In particular, conditions that delayed (bed1Δ mutants) or blocked (Lat-A treatment) bud emergence prevented Hsl1p kinase activation and Hsl7p recruitment, even though Hsl1p was itself recruited to the septin ring.
What is the additional requirement for Hsl1p activation? Whereas Lat-A treatment causes complete actin depolymerization (Ayscough et al., 1997), the actin cytoskeleton in bed1Δ mutants is well polarized and seems intact (Mondesert and Reed, 1996). Thus, it seems unlikely that the failure to activate Hsl1p in unbudded cells is simply due to actin perturbation. Moreover, Hsl1p hyperphosphorylation and Hsl7p recruitment in bed1Δ mutants occurred efficiently just after the eventual emergence of a bud. These observations strongly suggest that Hsl1p is activated in response to bud emergence. Similar arguments lead to the conclusion that Hsl7p is recruited to the septin cortex in response to bud emergence.
How might bud emergence promote Hsl1p activation? There are some interesting parallels between the behavior of Hsl1p described here and that of the related Gin4p kinase in S. cerevisiae. Gin4p is localized to the septin ring before bud emergence (Longtine et al., 1998), but Gin4p hyperphosphorylation only occurs later in the cell cycle (Altman and Kellogg, 1997). Gin4p hyperphosphorylation involves the self-association of two Gin4p molecules, in a manner that depends on the minor septin Shs1p (Mortensen et al., 2002). It is attractive to speculate that Hsl1p might be activated in a similar manner, with dimerization triggered somehow by bud emergence.
In a previous study, we suggested that the morphogenesis checkpoint monitored actin perturbation per se, rather than bud emergence (McMillan et al., 1998). This was based on the finding that some cells could still respond to Lat-A treatment by arresting the cell cycle in a Swe1p-dependent manner even after they had formed a bud. This response may involve Swe1p stabilization in budded cells treated with Lat-A, but such stabilization must occur by a distinct mechanism from the one described here for unbudded cells, because Hsl7p localization and phosphorylation were unaffected by Lat-A in budded cells. It seems plausible that Swe1p is subject to more than one form of regulation in response to perturbations of morphogenesis, such that bud emergence controls Hsl7p recruitment by Hsl1p, and actin perturbation in budded cells controls some other step in Swe1p regulation.
Whereas kinase-dead Hsl1pK110R retained a diminished ability to recruit Hsl7p to the septin cortex in budded cells, no detectable Hsl7p recruitment was seen in unbudded Lat-A–treated or bed1Δ cells. These observations suggest that the inactivity of Hsl1p in unbudded cells cannot fully account for the complete lack of detectable Hsl7p at the septin rings in those cells. Conceivably, Hsl1pK110R might have retained some residual kinase activity in vivo, sufficient to promote a reduced level of Hsl7p recruitment. Alternatively (and, we believe, more likely), the Hsl7p interaction site on Hsl1p in unbudded cells may be masked, either by an intramolecular inhibitory interaction or by some accessory factor. Activation of Hsl1p upon bud emergence would be associated with relief of that inhibitory interaction, allowing Hsl7p recruitment.
How Do Yeast Cells “Know” Whether They Have a Bud?
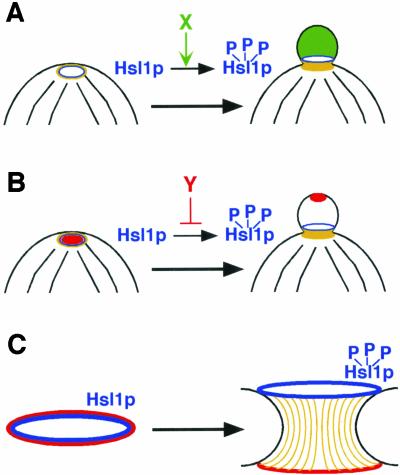
How might bud emergence influence Hsl1p activation (in a broad sense, encompassing both enzymatic activity and ability to recruit Hsl7p)? Below, we consider three models that could explain this behavior.
First, it is possible that a hypothetical “factor X” is delivered to the cortex during polarized growth and that factor X activates Hsl1p (Figure 8A). To account for the observed Hsl7p recruitment to septin rings in shmoos in the absence of ongoing polarized growth, this model requires the additional assumption that factor X persists for an extended period at sites of previous polarized growth (i.e., the shmoo projections). To account for the finding that unbudded bed1Δ cells did not recruit Hsl7p to the septin rings until bud emergence, this model postulates that factor X is not delivered to the prebud site before bud emergence. The appeal of this model is lessened by the observation that, so far as we are aware, all of the many proteins that have been shown to localize to sites of polarized growth in small buds and shmoos also localize to the prebud site.
Figure 8.
Models for Hsl1p activation in response to bud emergence. (A and B) Part of a mother cell is illustrated on the left, just before bud emergence, with the presumptive bud site at the top. The same cell after bud emergence is indicated on the right. Black lines represent polarized actin cables, and the yellow ring (left) or collar (right) represents the septins. Hsl1p is depicted in blue. (A) Hsl1p phosphorylation (indicating activation) is triggered by a factor X (green) that is recruited during polarized growth (but not to the presumptive bud site before bud emergence). (B) Hsl1p phosphorylation is inhibited by a factor Y (red) that is recruited to sites of polarized growth (including the presumptive bud site). Inhibition is short-range, so that after bud emergence the distance between factor Y at the bud tip and Hsl1p at the neck is sufficient to permit Hsl1p activation. (C) Hsl1p activation is correlated with the spreading of the septin ring seen in unbudded cells (left) to a collar after bud emergence (right). On spreading of the septins (yellow) into a collar, Hsl1p (blue) localizes to the bud side of the neck whereas some other proteins (red) localize to the mother side of the neck. The spreading may involve a septin reorganization (yellow bars) that directly activates Hsl1p. Alternatively, activation may be achieved by separating Hsl1p from an inhibitor (red) that stays on the mother side of the neck.
Second, it is possible that a hypothetical “factor Y” localizes together with Cdc42p at sites of polarized growth (including prebud sites) and acts as a short-range inhibitor of Hsl1p (Figure 8B). In unbudded bed1Δ or Lat-A–treated cells, the proximity of this factor to Hsl1p on the septin ring allows factor Y to inhibit Hsl1p and prevent Hsl7p recruitment. However, in budded cells, the increased distance between factor Y at the bud tip and Hsl1p at the neck allows Hsl1p activation. This model can account for the observed Hsl7p recruitment to septin rings in shmoos so long as the distance between the septin ring and the shmoo tip is sufficient to mitigate factor Y's inhibitory effect, which seems plausible. This model is appealing because there are many proteins known to display this pattern of localization and because it effectively uses the geometric change that accompanies bud emergence to dictate whether Hsl1p is activated.
Neither of the models discussed above takes note of the apparent link between Hsl1p activation and septin behavior. As mentioned above, assembled septins are critical for Hsl1p activity. Moreover, we have observed a correlation between septin spreading (from a narrow ring to a three-dimensional collar) and Hsl1p activation (as inferred from Hsl7p recruitment), both during bud formation and in shmoos that assemble septin rings in Lat-A. Finally, it is striking that upon spreading of the septins to form a collar, Hsl1p and Hsl7p become restricted to the bud side of the collar. Thus, it is attractive to speculate that septin spreading is the immediate cause of Hsl1p activation and Hsl7p recruitment (Figure 8C).
Notably, recent work has demonstrated that there is a dramatic change in septin dynamics at around the time of septin spreading during bud emergence (Dobbelaere et al., 2003) (Kozubowski and Tatchell, personal communication; Bi and Pringle, personal communication). In particular, septins in the narrow ring are exchanging with those in the soluble pool, whereas septins in the collar are stable and do not exchange. It is easy to envisage that the change in septin organization responsible for the altered dynamics could also activate Hsl1p. In addition, it is worth noting that proteins recruited to the initial septin ring subsequently become separated from each other along the mother-bud axis within the septin collar (Gladfelter et al., 2001). Thus, spreading of a septin ring to form a collar physically separates Hsl1p from other, potentially inhibitory, proteins (Figure 8C).
The proposal that septin spreading from a ring to a collar promotes Hsl1p activation raises an obvious follow-up question: how does bud emergence promote septin spreading? As suggested for Hsl1p above, one could imagine that a hypothetical factor X delivered to the cortex during bud emergence might induce septin spreading or that a hypothetical factor Y colocalized with Cdc42p might block septin spreading until bud emergence separates it from the septin ring. Alternatively, septin spreading could simply result from the change in local cell shape, from flat to tubular, that accompanies bud emergence. Indeed, it is difficult to visualize how a comparable “spreading” could occur without a locally tubular cortex to accommodate the septin collar. For this reason, we favor the hypothesis that septin spreading is an automatic consequence of generating a locally tubular cell cortex. Hsl1p may have evolved from an ancestral septin-dependent kinase to use this shape-dependent change in septin organization to act as a checkpoint sensor for bud emergence.
Acknowledgments
We thank Mark Shulewitz and Jeremy Thorner, Erfei Bi and John Pringle, and Lukasz Kozubowski and Kelly Tatchell for communication of unpublished results. Thanks to Sally Kornbluth, Tony Means, Joe Heitman, Bruce Nicklas, and Robin Wharton for critical comments on versions of this manuscript and to members of the Lew laboratory for stimulating input. This work was funded by grants from the National Institutes of Health (GM-53050) and the Leukemia and Lymphoma Society (to D.J.L.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-03-0154. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-03-0154.
References
- Altman, R., and Kellogg, D. (1997). Control of mitotic events by Nap1 and the Gin4 kinase. J. Cell Biol. 138, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A. and Struhl, K. (eds.). (1995) Current Protocols in Molecular Biology. New York: John Wiley & Sons.
- Ayscough, K.R., Stryker, J., Pokala, N., Sanders, M., Crews, P., and Drubin, D.G. (1997). High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137, 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, S., Qyang, Y., Yang, P., Kim, H., Du, H., Bartholomeusz, G., Henkel, J., Pimental, R., Verde, F., and Marcus, S. (2001). The highly conserved protein methyltransferase, Skb1, is a mediator of hyperosmotic stress response in the fission yeast Schizosaccharomyces pombe. J. Biol. Chem. 276, 14549–14552. [DOI] [PubMed] [Google Scholar]
- Barral, Y., Parra, M., Bidlingmaier, S., and Snyder, M. (1999). Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher, R.N., Deshaies, R.J., and Kirschner, M.W. (1993). Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 12, 3417–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose, I., Irazoqui, J.E., Moskow, J.J., Bardes, E.S., Zyla, T.R., and Lew, D.J. (2001). Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J. Biol. Chem. 276, 7176–7186. [DOI] [PubMed] [Google Scholar]
- Burton, J.L., and Solomon, M.J. (2000). Hsl1p, a Swe1p inhibitor, is degraded via the anaphase-promoting complex. Mol. Cell. Biol. 20, 4614–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.S., Mrksich, M., Huang, S., Whitesides, G.M., and Ingber, D.E. (1997). Geometric control of cell life and death. Science 276, 1425–1428. [DOI] [PubMed] [Google Scholar]
- Cid, V.J., Shulewitz, M.J., McDonald, K.L., and Thorner, J. (2001). Dynamic localization of the Swe1 regulator Hsl7 during the Saccharomyces cerevisiae cell cycle. Mol. Biol. Cell 12, 1645–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, T.R., Tang, Z., and Dunphy, W.G. (1993). Negative regulation of the Wee1 protein kinase by direct action of the nim1/cdr1 mitotic inducer. Cell 72, 919–929. [DOI] [PubMed] [Google Scholar]
- Dobbelaere, J., Gentry, M.S., Hallberg, R.L., and Barral, Y. (2003). Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell 4, 345–357. [DOI] [PubMed] [Google Scholar]
- Edgington, N.P., Blacketer, M.J., Bierwagen, T.A., and Myers, A.M. (1999). Control of Saccharomyces cerevisiae filamentous growth by cyclin-dependent kinase Cdc28. Mol. Cell. Biol. 19, 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen, W.J., Paushkin, S., Wyce, A., Massenet, S., Pesiridis, G.S., Van Duyne, G., Rappsilber, J., Mann, M., and Dreyfuss, G. (2001). The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 21, 8289–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and Sugino, A. (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six base pair restriction sites. Gene 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Giot, L., and Konopka, J.B. (1997). Functional analysis of the interaction between Afr1p and the Cdc12p septin, two proteins involved in pheromone-induced morphogenesis. Mol. Biol. Cell 8, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter, A.S., Pringle, J.R., and Lew, D.J. (2001). The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4, 681–689. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., Fink, G.R., Simon, M.I., and Abelson, J.N. (eds.). (1991) Guide to Yeast Genetics and Molecular Biology. Methods in Enzymology, Vol 194. San Diego, CA; Academic Press. [PubMed]
- Hanks, S.K., and Hunter, T. (1995). Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. [Review]. FASEB J. 9, 576–596. [PubMed] [Google Scholar]
- Harrison, J.C., Bardes, E.S., Ohya, Y., and Lew, D.J. (2001). A role for the Pkc1p/Mpk1p kinase cascade in the morphogenesis checkpoint. Nat. Cell Biol. 3, 417–420. [DOI] [PubMed] [Google Scholar]
- Huang, S., and Ingber, D.E. (1999). The structural and mechanical complexity of cell growth control. Nat. Cell Biol. 1, E131–E138. [DOI] [PubMed] [Google Scholar]
- Lee, J.H., Cook, J.R., Pollack, B.P., Kinzy, T.G., Norris, D., and Pestka, S. (2000). Hsl7p, the yeast homologue of human JBP1, is a protein methyltransferase. Biochem. Biophys. Res. Commun. 274, 105–111. [DOI] [PubMed] [Google Scholar]
- Lew, D.J. (2000). Cell-cycle checkpoints that ensure coordination between nuclear and cytoplasmic events in Saccharomyces cerevisiae. Curr. Opin. Genet. Dev. 10, 47–53. [DOI] [PubMed] [Google Scholar]
- Lew, D.J., and Reed, S.I. (1993). Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120, 1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew, D.J., Weinert, T., and Pringle, J.R. (1997). Cell cycle control in Saccharomyces cerevisiae. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Cell Cycle and Cell Biology, ed. J.R. Pringle, J.R. Broach, and E.W. Jones, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 607–695.
- Liu, Q., Li, M.Z., Leibham, D., Cortez, D., and Elledge, S.J. (1998). The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 8, 1300–1309. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., DeMarini, D.J., Valencik, M.L., Al-Awar, O.S., Fares, H., De Virgilio, C., and Pringle, J.R. (1996). The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 8, 106–119. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., Fares, H., and Pringle, J.R. (1998). Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 143, 719–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., Theesfeld, C.L., McMillan, J.N., Weaver, E., Pringle, J.R., and Lew, D.J. (2000). Septin-dependent assembly of a cell-cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 4049–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X.J. (2000). Cell-cycle regulatory proteins Hsl7p/Skb1p belong to the protein methyltransferase superfamily. Trends Biochem. Sci. 25, 11–12. [DOI] [PubMed] [Google Scholar]
- Ma, X.-J., Lu, Q., and Grunstein, M. (1996). A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 10, 1327–1340. [DOI] [PubMed] [Google Scholar]
- McMillan, J.N., Longtine, M.S., Sia, R.A.L., Theesfeld, C.L., Bardes, E.S.G., Pringle, J.R., and Lew, D.J. (1999a). The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol. Cell. Biol. 19, 6929–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan, J.N., Sia, R.A.L., Bardes, E.S.G., and Lew, D.J. (1999b). Phosphorylation-independent inhibition of Cdc28p by the tyrosine kinase Swe1p in the morphogenesis checkpoint. Mol. Cell. Biol., 19, 5981–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan, J.N., Sia, R.A.L., and Lew, D.J. (1998). A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J. Cell Biol., 142, 1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan, J.N., Theesfeld, C.L., Harrison, J.C., Bardes, E.S., and Lew, D.J. (2002). Determinants of Swe1p degradation in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 3560–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizunuma, M., Hirata, D., Miyaoka, R., and Miyakawa, T. (2001). GSK-3 kinase Mck1 and calcineurin coordinately mediate Hsl1 down-regulation by Ca2+ in budding yeast. EMBO J. 20, 1074–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondesert, G., and Reed, S.I. (1996). BED1, a gene encoding a galactosyltransferase homologue, is required for polarized growth and efficient bud emergence in Saccharomyces cerevisiae. J. Cell Biol. 132, 137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen, E.M., McDonald, H., Yates 3rd., J., and Kellogg, D.R. (2002) Cell Cycle-dependent assembly of a Gin4-Septin Complex. Mol. Biol. Cell 13, 2091–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, L.L., Walter, S.A., Young, P.G., and Piwnica-Worms, H. (1993). Phosphorylation and inactivation of the mitotic inhibitor Wee1 by the nim1/cdr1 kinase. Nature 363, 736–738. [DOI] [PubMed] [Google Scholar]
- Russell, P., Moreno, S., and Reed, S.I. (1989). Conservation of mitotic controls in fission and budding yeast. Cell 57, 295–303. [DOI] [PubMed] [Google Scholar]
- Shulewitz, M.J., Inouye, C.J., and Thorner, J. (1999). Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 7123–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia, R.A.L., Bardes, E.S.G., and Lew, D.J. (1998). Control of Swe1p degradation by the morphogenesis checkpoint. EMBO J. 17, 6678–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpio, B.E., Banes, A.J., Levin, L.G., and Johnson, G. (1987). Mechanical stress stimulates aortic endothelial cells to proliferate. J. Vasc. Surg. 6, 252–256. [PubMed] [Google Scholar]
- Tanaka, S., and Nojima, H. (1996). Nik1: a Nim1-like protein kinase of S. cerevisiae interacts with the Cdc28 complex and regulates cell cycle progression. Genes Cells 1, 905–921. [DOI] [PubMed] [Google Scholar]
- Wu, L., and Russell, P. (1993). Nim1 kinase promotes mitosis by inactivating Wee1 tyrosine kinase. Nature 363, 738–741. [DOI] [PubMed] [Google Scholar]