Abstract
The function of the Aurora B kinase at centromeres and the central spindle is crucial for chromosome segregation and cytokinesis, respectively. Herein, we have investigated the regulation of human Aurora B by its complex partners inner centromere protein (INCENP) and survivin. We found that overexpression of a catalytically inactive, dominant-negative mutant of Aurora B impaired the localization of the entire Aurora B/INCENP/survivin complex to centromeres and the central spindle and severely disturbed mitotic progression. Similar results were also observed after depletion, by RNA interference, of either Aurora B, INCENP, or survivin. These data suggest that Aurora B kinase activity and the formation of the Aurora B/INCENP/survivin complex both contribute to its proper localization. Using recombinant proteins, we found that Aurora B kinase activity was stimulated by INCENP and that the C-terminal region of INCENP was sufficient for activation. Under identical assay conditions, survivin did not detectably influence kinase activity. Human INCENP was a substrate of Aurora B and mass spectrometry identified three consecutive residues (threonine 893, serine 894, and serine 895) containing at least two phosphorylation sites. A nonphosphorylatable mutant (TSS893–895AAA) was a poor activator of Aurora B, demonstrating that INCENP phosphorylation is important for kinase activation.
INTRODUCTION
Aurora kinases are evolutionarily conserved enzymes that regulate different aspects of cell division. Only a single Aurora kinase is present in Saccharomyces cerevisiae (Francisco et al., 1994; Francisco and Chan, 1994) and Schizosaccharomyces pombe (Petersen et al., 2001; Leverson et al., 2002). In contrast, three distinct Aurora family members, termed Aurora A, B, and C, have been described in metazoan organisms (Adams et al., 2001a; Nigg, 2001; Shannon and Salmon, 2002). A-type Aurora kinases localize to both centrosomes and spindle microtubules (Kimura et al., 1997; Bischoff et al., 1998) and have been implicated in spindle assembly (Dutertre et al., 2002; Kufer et al., 2002; Tsai et al., 2003). The B-type Aurora kinases are present at centromeres in prophase and metaphase, before they relocalize to the central spindle and the midbody in anaphase and telophase (Bischoff et al., 1998; Terada et al., 1998). The C-type Aurora kinases have so far been identified only in mammals (Tseng et al., 1998; Kimura et al., 1999). They are expressed primarily in testis and some tumor cell lines, where they have been localized to spindle poles.
Aurora B exists in a complex with at least two other proteins, inner centromere protein (INCENP) and survivin (Adams et al., 2000; Kaitna et al., 2000; Wheatley et al., 2001a). Aurora B, INCENP, and survivin are so-called chromosomal passenger proteins: they associate with inner centromere regions during prophase, but subsequently relocalize to the midzone of the central spindle and concentrate at the midbody (Adams et al., 2001a). INCENP has a modular organization, and domains required for chromosomal and cytoskeletal interactions have been delineated (Ainsztein et al., 1998; Mackay et al., 1998; Wheatley et al., 2001b). Of particular interest, all metazoan INCENPs, as well as the INCENP homolog of S. cerevisiae (Sli15p), share a highly conserved motif near the C terminus, the so-called IN-box (INCENP conserved box) (Adams et al., 2000, 2001b). Mice lacking INCENP showed very early (32- to 64-cell stage) embryonic lethality (Cutts et al., 1999), and RNA interference (RNAi)-mediated down-regulation of INCENP in Caenorhabditis elegans and Drosophila produced severe mitotic defects (Kaitna et al., 2000; Adams et al., 2001c). Similar defects were also seen upon overexpression of a dominant-negative INCENP mutant in vertebrate cells (Mackay et al., 1998). Thus, INCENP is clearly essential for cell division.
Survivin has originally been proposed to exert an antiapoptotic function (hence its name; Li et al., 1998), but this issue remains controversial (Reed, 2001; Silke and Vaux, 2001). Instead, several lines of evidence point to an important role of survivin in the regulation of late mitotic events (Fraser et al., 1999; Uren et al., 1999, 2000; Speliotes et al., 2000; Kallio et al., 2001). Depletion of C. elegans Aurora B (AIR-2) and survivin (BIR-1) produced very similar phenotypes, indicating that the two proteins interact genetically (Speliotes et al., 2000). Survivin has also been shown to interact with INCENP in yeast two-hybrid experiments and in vitro binding assays, and a dominant-negative mutant of INCENP disrupted the targeting of survivin to centromeres (Wheatley et al., 2001a). Most recently, additional evidence for a physical interaction between survivin and Aurora B/INCENP has also been reported in S. cerevisiae (Cheeseman et al., 2002) and Xenopus egg extracts (Bolton et al., 2002).
Herein, we provide evidence that INCENP and survivin regulate human Aurora B at multiple levels. First, our data suggest that Aurora B kinase activity as well as INCENP and/or survivin contribute to the proper localization of the complex. Second, we show that recombinant human Aurora B is strongly activated upon binding of INCENP, whether or not survivin is part of the complex. Remarkably, this activation results in the phosphorylation of an evolutionarily conserved motif within the C-terminal IN-box of INCENP, and this phosphorylation is critical for full activation of the Aurora B/INCENP complex.
MATERIALS AND METHODS
Cell Culture
HeLa S3 cells were grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (Invitrogen) and 50 μg/ml each of streptomycin and penicillin (Invitrogen). T-Rex-U2OS cells (Invitrogen), and their derivatives were maintained in the same medium with the further addition of 50 μg/ml hygromycin B (Merck, Whitehouse Station, NJ). Sf-9 cells were grown in TC-100 medium (Invitrogen) supplemented with 10% fetal calf serum (Invitrogen) and 50 μg/ml streptomycin and penicillin (Invitrogen).
Cloning and Mutagenesis
Wild-type and kinase-deficient (K106R) human Aurora B cDNAs have been described previously (Meraldi et al., 2002). To produce inducible cell lines expressing Aurora B-myc-His6, wild-type and K106R cDNAs were subcloned into the pcDNA4/TO/myc-His vector (Invitrogen). A human INCENP cDNA was obtained by reverse transcription-polymerase chain reaction (PCR) by using total RNA from HeLa S3 cells. The PCR product was introduced into the pBluescript SK(–) vector and completely sequenced. For baculoviral expression, Aurora B and full-length INCENP were inserted into the transfer vector pUNI10 and then recombined with pHI100-GST or pHI100-His according to published procedures (Liu et al., 1998). Ala mutants of INCENP, 893A894A895A (3A) and 893A894A895A900A (4A), were obtained by site-directed mutagenesis using the QuikChange kit (Stratagene, La Jolla, CA). INCENP mutants 822–919 (C3), 855–919 (C4), 878–919 (C5), 822–897 (C6), 822–877 (C7), and 822–892 (C8) were amplified by PCR and cloned for baculoviral expression into a modified pVL1393 vector (BD Biosciences PharMingen, San Diego, CA), which provides an N-terminal GST-tag followed by a PreScission protease cleavage site.
Antibody Production
A rabbit antibody (C; rabbit 4777) was raised against a peptide spanning the 15 C-terminal residues (NSRRVLPPSALQSVA) of Aurora B, coupled to keyhole limpet hemocyanin. Two rabbit anti-INCENP antibodies were generated against His6-INCENP (231–397) expressed in Escherichia coli (rabbits 55 and 56).
Cell Synchronization
Exponentially growing HeLa S3 cells were blocked for 14 h with thymidine (final concentration 2 mM), washed three times with phosphate-buffered saline (PBS), and incubated with fresh medium. After 13 h, aphidicolin was added (final concentration 1.6 μg/ml) and the plates were incubated for an additional 13 h. Plates were then washed three times with PBS and fresh medium was added. (This moment was defined as time 0). Cells were collected every 2 h thereafter. For arresting cells in prometaphase, nocodazole (final concentration 500 ng/ml) was added 6 h after release of cells from the thymidine/aphidicolin double block, and mitotic cells were collected 6 h later.
Western Blotting and Immunoprecipitation
For Western blotting, cellular proteins were solubilized in 2 × gel sample buffer, boiled for 5 min, and resolved by SDS-PAGE. Proteins were then transferred to nitrocellulose membranes, and these were incubated with antibodies against the N (N: monoclonal antibody [mAb]; BD Biosciences PharMingen) or C terminus of Aurora B (C), INCENP (56), survivin (Novus-Biological, Littleton, CO), cyclin B (USB, Cleveland, OH) and α-tubulin (Sigma-Aldrich, St. Louis, MO), followed by horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse antibody. Immunoreactivity was visualized by enhanced chemiluminescence. For immunoprecipitation, cells were lysed on ice for 30 min in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin, 30 μg/ml DNase, 30 μg/ml RNase, 20 mM β-glycerophosphate, 0.3 mM sodium vanadate) and sonicated for 20 s. The lysates were incubated at 4°C with anti-Aurora B antibody and then with protein A-Sepharose (Bio-Rad, Hercules, CA) for 1 h each. For immunoprecipitation of ectopically expressed Aurora B-myc, lysates were incubated with anti-myc mAb (9E10) bound to protein G-Sepharose for 1 h at 4°C. Samples were spun down and washed twice with radioimmunoprecipitation assay buffer, once with 50 mM Tris-HCl, pH 8.0, 0.4 M NaCl, 1% NP-40, 0.5% deoxycholate, 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin and twice with 10 mM Tris-buffer (10 mM Tris-HCl, pH7.5, 150 mM NaCl, 0.1 mM PMSF).
Purification of Recombinant Proteins
Recombinant baculoviruses coding for glutathione S-transferase (GST)- or His-tagged Aurora B, INCENP, and survivin were generated using the BaculoGold kit (BD Biosciences PharMingen) and used to infect Sf-9 insect cells, according to the manufacturer's protocol. To all complex formation between Aurora B, INCENP, and survivin in vivo, insect cells were coinfected with the appropriate viruses. After 48 h, infected cells were pelleted and lysed in lysis buffer (50 mM Tris-HCl, pH 8.0, 0.4 M NaCl, 1% NP-40, 0.5% deoxycholate, 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin, 20 mM β-glycerophosphate, 0.3 mM sodium vanadate), followed by centrifugation. The supernatants were incubated with glutathione-Sepharose beads for 1 h at 4°C. Beads were washed twice with lysis buffer, once with 50 mM Tris-HCl, pH 8.0, 0.4 M NaCl, 0.1% NP-40, 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin, and once with 10 mM Tris-buffer. Then, samples were analyzed by Western blotting and used for kinase assays.
For in vitro complex formation (experiment shown in Figure 11, C and D), recombinant GST-Aurora B was eluted with 20 mM glutathione solution in 50 mM Tris-HCl, pH 8.0, and dialyzed for 2 × 1 h against 10 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.5 mM EDTA, 1 mM dithiothreitol (DTT), 0.1 mM PMSF. Recombinant GST-INCENP fragments were prepared as follows: proteins bound to glutathione-Sepharose beads were washed with and resuspended in cleavage buffer (50 mM Tris-HCl, pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.01% NP-40). Then, PreScission protease (Amersham Biosciences, Piscataway, NJ) was added and the suspension incubated overnight at 4°C. After centrifugation, a first supernatant was collected. The beads were washed once again with cleavage buffer and the second supernatant combined with the first. Aliquots of purified Aurora B and INCENP proteins were frozen in liquid nitrogen and stored at –80°C.
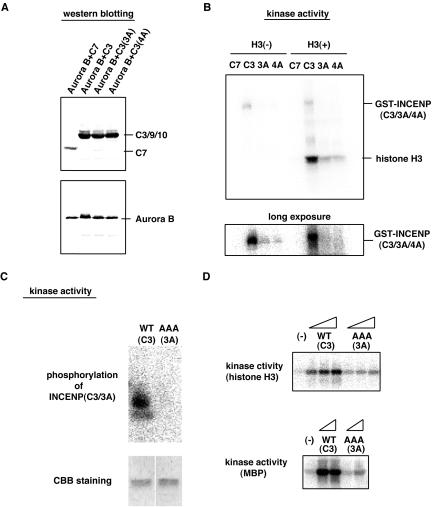
Figure 11.
Ala mutants of INCENP C3 fragment are less effective in Aurora B activation. (A) The GST-INCENP mutants C7, C3, C3(3A), or C3(4A) and His-Aurora B were coexpressed in Sf-9 cells and the complexes purified as described in the legend to Figure 9. INCENP and Aurora B protein levels were determined by Western blotting. (B) Kinase activities associated with the samples described in A were measured with or without exogenous histone H3; the bottom panel shows a prolonged exposure to illustrate phosphorylation of GST-INCENP fragments. (C) GST-Aurora B and INCENP C3 (WT or 3A) were purified separately, and after mixing of the proteins, phosphorylation of INCENP was assayed (top); the presence of equal amounts of INCENP substrate was demonstrated by Coomassie Blue staining (CBB; bottom). (D) The effect of adding increasing amounts of INCENP C3 (WT or 3A) to Aurora B was measured by in vitro kinase assays, with histone H3 (top) or MBP (bottom) as substrates.
In Vitro Kinase Assays
Immunoprecipitated or recombinant kinases were included into a 20-μl reaction containing 0.5 mg/ml histone H3 (or 1 mg/ml myelin basic protein [MBP]), 50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 1 mM EGTA, 1 mM DTT, 5 mM NaF, 5 mM β-glycerophosphate, 0.05 mM sodium vanadate, 0.1 mM ATP, and 1 μCi of γ-[32P]ATP. After 20 min at 30°C, reactions were stopped by the addition of 5 μl of 5× gel sample buffer. Protein samples were separated by SDS-PAGE and phosphate incorporation determined by PhosphorImager.
Short Interfering RNA (siRNA) Treatment
Transfection of T-Rex-U2OS or HeLa S3 cells with siRNA duplex oligonucleotides were performed using oligofectAMINE (Invitrogen), as described previously (Elbashir et al., 2001). The siRNA sequences used for targeting human Aurora B were (AA)CGAGACCTATCGCCGCATC(GT), for human INCENP (AA)GAAGCAGATTGAGCAGAAG(TT), and for human survivin (AA)GAATTAACCCTTGGTGAAT(TT). In all experiments, a duplex targeting the luciferase gene was used as a control (Elbashir et al., 2001).
Immunofluorescence Microscopy
T-Rex-U2OS or HeLa S3 cells grown on coverslips were fixed in 20 mM PIPES, pH 6.8, 0.2% Triton X-100, 1 mM MgCl2, 10 mM EGTA, 4% formaldehyde for 10 min at room temperature, and washed three times with PBS. Cells were then incubated in 3% bovine serum albumin/PBS with primary antibodies. These were used at the following dilutions: anti-Aurora B (N; 1:1000); anti-INCENP (55; 1:1000); anti-survivin (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA); anti-CENP-B (1:1000; gift from W. Earnshaw, University of Edinburgh, Edinburgh, Scotland); anti-α-tubulin (1:1000; Sigma-Aldrich); anti-myc mAb (not diluted). Primary antibodies were visualized using Texas-Red or Alexa green-conjugated secondary antibodies (Molecular Probes, Eugene, OR), and DNA was counterstained with 4,6-diamidino-2-phenylindole (DAPI).
Mass Spectrometry
Coomassie-stained protein bands were in-gel digested by trypsin (sequencing grade; Promega) (Shevchenko et al., 1996) and desalted using homemade miniaturized reversed-phase columns (Gobom et al., 1999). Matrix-assisted laser desorption ionization/time of flight mass spectra were acquired on a Reflex III instrument (Bruker Daltonik, Bremen, Germany) in negative ion reflector mode. As a matrix, 2,5 dihydroxybenzoic acid (Bruker Daltonik) was used. Mass spectra were searched for signals showing a mass difference of 80 mass units (phosphate group) by a self-developed software script. Such phosphopeptide-candidates were submitted to post-source decay fragment ion analysis (Hoffmann et al., 1999). Peptides showing the typical losses of 98 mass units (phosphoric acid) and 80 mass units (phosphate group) were accepted as phosphopeptides. For peptide sequencing by electrospray tandem mass spectrometry, samples were filled into nano electrospray needles (Protana, Odense, Denmark) and analyzed on a QTOF Ultima (Micromass, Manchester, United Kingdom) quadrupole time-of-flight instrument.
RESULTS
Analysis of Aurora B Function by a Dominant-Negative Approach
To study the function and regulation of human Aurora B kinase, we produced stably transformed T-Rex-U2OS cell lines that allow the inducible expression of wild-type or catalytically inactive mutant Aurora B (K106R) under tetracycline control. After induction of the transgenes for 24 h, both wild-type and K106R mutant Aurora B were expressed to similar levels (Figure 1A). Using an anti-myc antibody directed against the C-terminal myc epitope tag, both proteins were then immunoprecipitated and assayed for kinase activity. When isolated from nocodazole-arrested cells, wild-type Aurora B readily phosphorylated histone H3 (Figure 1A) and MBP (our unpublished data), but only little activity was associated with the kinase isolated from an asynchronously growing cell population. As expected, the K106R mutant displayed no kinase activity even when immunoprecipitated from nocodazole-treated cells (Figure 1A).
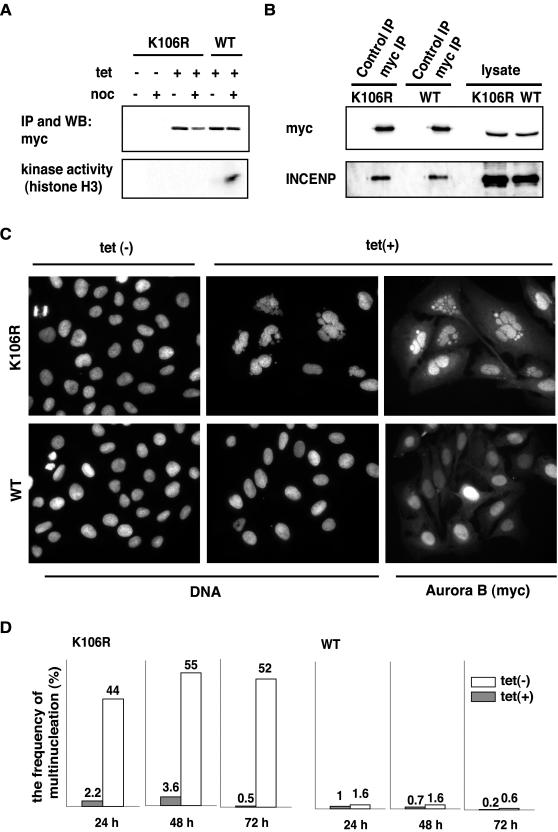
Figure 1.
A catalytically inactive Aurora B acts as a dominant-negative mutant. (A) Inducible T-Rex U2OS cell lines (Invitrogen) harboring myc-tagged K106R mutant or wild-type (wt) human Aurora B transgenes were treated with or without tetracycline (1 μg/ml final concentration). After 8 h, nocodazole (or dimethyl sulfoxide) was added and cells were incubated for an additional 16 h. Myc-tagged Aurora B was immunoprecipitated with anti-myc antibody, and protein amounts and kinase activities in immunoprecipitates were determined by Western blotting (top) and in vitro kinase assays (bottom). (B) Myc-tagged K106R or wild-type Aurora B were immunoprecipitated with anti-myc antibody from lysates used in A. Immunoprecipitates were subjected to Western blotting with antibodies against myc-Aurora B and INCENP; whole-cell lysates were analyzed in parallel. (C) K106R and wild-type Aurora B expressing cells were treated for 24 h with or without tetracycline and then analyzed by immunofluorescence microscopy with anti-myc antibodies. DNA was counterstained with DAPI. (D) Histogram shows a quantitative analysis of the extent of multinucleation at different times of wild-type or K106R mutant Aurora B expression.
At the cellular level, the expression of the K106R mutant produced a striking multinucleation phenotype (Figure 1C). The frequency of multinucleation depended on the length of expression of the transgene, and by 48 h this phenotype was seen in 50–60% of all cells (Figure 1D). The remainder of the cells divided normally, presumably reflecting differences in expression levels. Almost no multinucleation was seen when cells were not treated with tetracycline or induced to express the wild-type Aurora B (Figure 1, C and D). Both wild-type and K106R mutant Aurora B interacted with endogenous INCENP, as demonstrated by coimmunoprecipitation experiments (Figure 1B). These data confirm that the catalytically inactive Aurora B acts as a dominant-negative mutant whose overexpression severely impairs mitotic progression (Terada et al., 1998). In the cell line analyzed herein, most mitotic cells harboring the dominant-negative Aurora B mutant were found to be in prophase and prometaphase-like states, whereas anaphase or telophase cells were difficult to find.
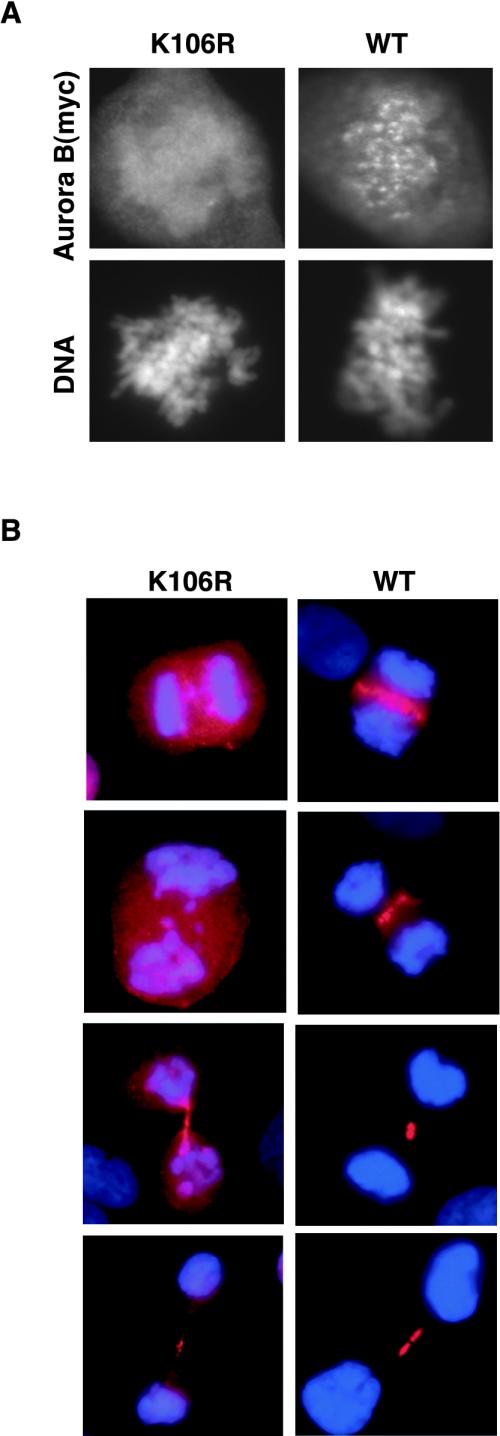
In parallel to the multinucleation phenotype, we also observed an impaired localization of the catalytically inactive Aurora B. Whereas the ectopically expressed wild-type Aurora B showed the expected relocalization from centromeres (Figure 2A) to the central spindle (Figure 2B), the K106R mutant Aurora B frequently failed to localize properly to either centromeres or the spindle midzone (Figure 2). Few, if any, of the prometaphase cells expressing mutant Aurora B showed centromere staining (Figure 2A). Cells at later stages of mitosis (anaphase/telophase) were difficult to find, because the expression of mutant Aurora B severely impaired mitotic progression, but in these cases, mutant Aurora B showed no, or at most faint, association with spindle structures (Figure 2B). As suggested in recent studies (Murata-Hori and Wang, 2002; Murata-Hori et al., 2002), inactive Aurora B may localize transiently to chromosomes and spindle structures. However, in the mitotic cells analyzed herein, the bulk of the mutant protein seemed to be distributed diffusely, indicating that proper localization of Aurora B to centromeres, the central spindle and the midbody requires kinase activity. Finally, in some cells expressing only low amounts of mutant Aurora B, the protein displayed (near-) normal localization (our unpublished data). These latter cells showed no evidence of mitotic aberration.
Figure 2.
Mislocalization of the K106R Aurora B mutant in mitosis. T-Rex U2OS cell lines expressing myc-tagged Aurora B wild-type and K106R mutant were fixed 24 h after induction and subjected to immunofluorescence microscopy, by using the anti-myc antibody. DNA was counterstained with DAPI. (A) prometaphase (B) anaphase and telophase stages. Red, myc; blue, DAPI.
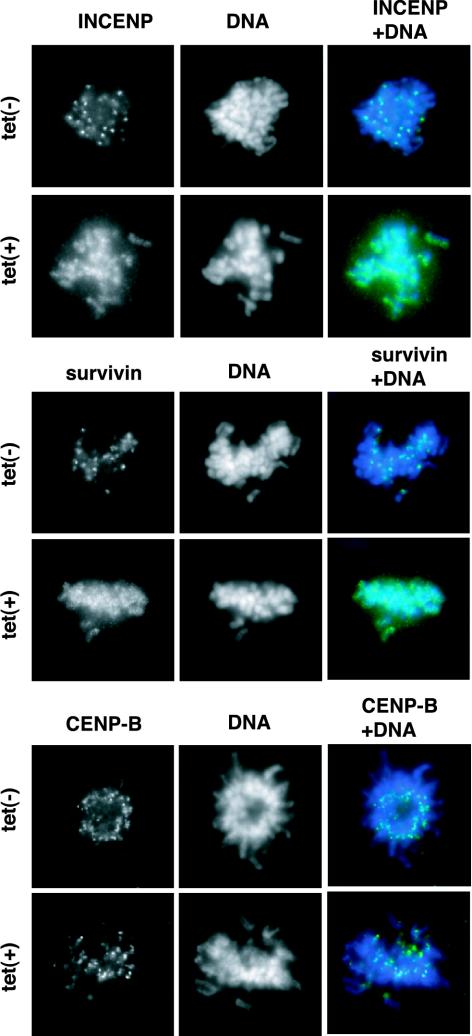
To determine whether INCENP and survivin also depended on Aurora B activity for proper localization, the cells expressing the K106R mutant were stained with antibodies directed against INCENP or survivin. Although the kinetochore marker CENP-B localized properly, neither INCENP nor survivin could be detected at centromeres (Figure 3). These data suggest that catalytically inactive Aurora B sequesters both INCENP and survivin and prevents their association with centromeres. The expression of wild-type Aurora B did not interfere with INCENP or survivin localization (our unpublished data).
Figure 3.
Localization of INCENP and survivin in cells expressing the K106R mutant of Aurora B. T-Rex U2OS cell lines expressing myc-tagged K106R mutant Aurora B were incubated for 10 h in the presence or absence of tetracycline, fixed and then stained with antibodies against INCENP, survivin, or CENP-B (green). DNA was counterstained with DAPI (blue). Prometaphase stage cells are shown.
Disruption of the Aurora B/INCENP/Survivin Complex by RNAi
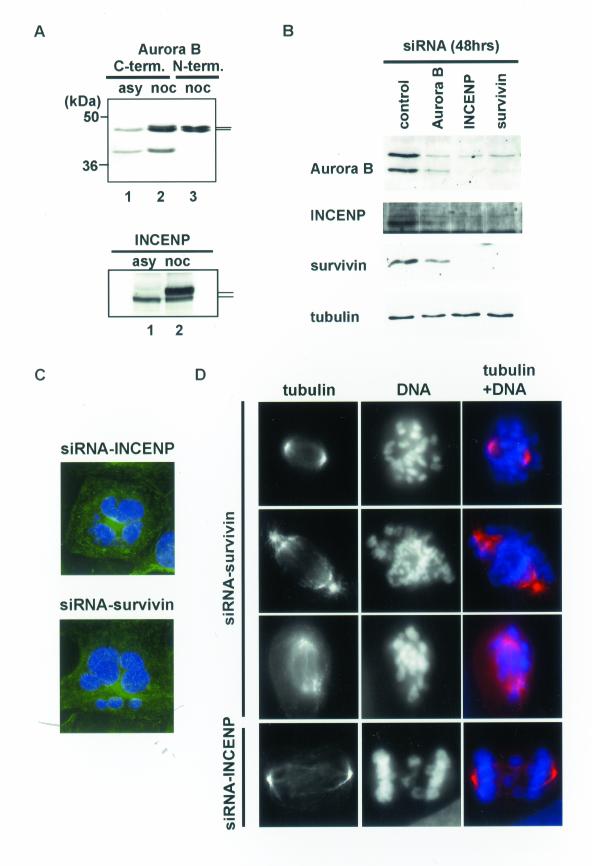
To better understand the mutual influence of Aurora B, INCENP, and survivin on the assembly and localization of the kinase complex, we targeted all three proteins by siRNA (Elbashir et al., 2001). Experiments were performed on U2OS (Figures 4 and 5) and HeLa S3 cells (our unpublished data), with similar results. Two distinct antibodies were used to monitor the levels of Aurora B (Figure 4A). One antibody, directed against the N terminus of Aurora B, produced a closely spaced doublet band of the expected size (Figure 4A, lane 3), but the second antibody, raised against the C terminus, also recognized an additional protein of lower molecular weight (Figure 4A, lanes 1 and 2). Western blotting after siRNA treatment of cells with a duplex targeting Aurora B revealed that the levels of both proteins decreased in parallel (Figure 4B), suggesting that the smaller immunoreactive protein represents either a cleavage product or a splice variant of Aurora B. The physiological role, if any, of this protein is not presently clear.
Figure 4.
Depletion of INCENP or survivin by siRNA results in mitotic defects and multinucleation. (A) Aurora B and INCENP protein levels in asynchronously growing (lane 1) or nocodazole treated (lanes 2 and 3) HeLa S3 cell extracts. To detect the Aurora B protein by Western blotting, two distinct antibodies were used, one directed against the C terminus (lanes 1 and 2), the other against the N terminus (lane 3). (B) Aurora B, INCENP, surviving, and α-tubulin levels after 48 h treatment with siRNA directed to Aurora B, INCENP, or survivin, as indicated. The GL-2 duplex was used for control. (C) Depletion of INCENP or survivin caused multinucleation. U2OS cells were fixed 48 h after siRNA treatment and stained with anti-α-tubulin antibodies (green). DNA was counterstained with DAPI (blue). (D) Mitotic defects in INCENP- or survivin-depleted cells, 48 h after treatment with siRNA. α-Tubulin staining: red; DNA staining: blue.
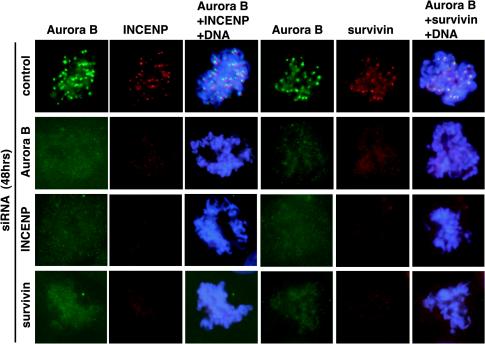
Figure 5.
Lack of centromere localization of Aurora B/INCENP/survivin after depletion of individual proteins by siRNA. After treatment of T-Rex-U2OS cells for 48 h by siRNA duplexes directed against Aurora B, INCENP, or survivin, all three proteins were localized by immunofluorescence microscopy. DNA was counterstained with DAPI.
Interestingly, both Aurora B and INCENP showed an electrophoretic retardation in M phase samples (Figure 4A). Many kinases are activated by phosphorylation of a threonine residue within the so-called T-loop (or activation loop), and this seems to be true also for Aurora family members (Walter et al., 2000; Murnion et al., 2001; Chen and Tang, 2002; Littlepage et al., 2002). The relevant residue in Aurora B, T232, is located within a motif that fits the emerging consensus for Aurora B phosphorylation sites (see below), suggesting that the observed upshift could result from autophosphorylation of the T-loop. The shift in INCENP is also likely to reflect phosphorylation, as inferred from previous studies on the Xenopus homolog (Stukenberg et al., 1997; Bolton et al., 2002). In the case of survivin, no evidence for cell cycle dependent alterations in gel electrophoretic mobility could be observed (our unpublished data; Figure 6).
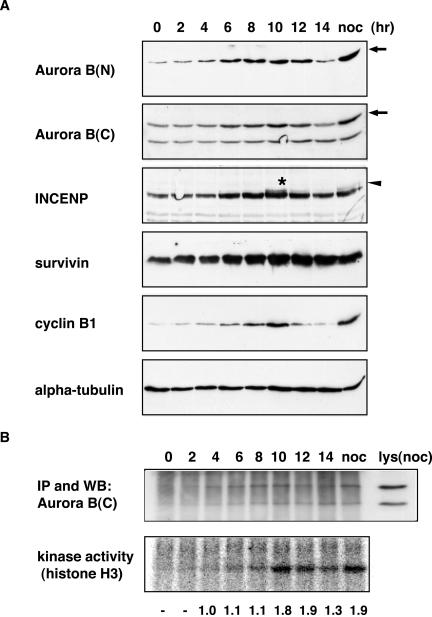
Figure 6.
Cell cycle analysis of Aurora B, INCENP, and survivin. (A) Aurora B, INCENP, survivin, cyclin B, and α-tubulin protein levels through the cell cycle were determined by Western blotting. HeLa S3 cells were synchronized by an arrest-release protocol from an aphidicolin/thymidine block at the G1/S boundary (time 0). For comparison, cells were also analyzed after a nocodazole induced prometaphase arrest (noc). Arrows point to the full-length, wild-type Aurora B, whereas the arrowhead and asterisk denote the slower migrating form of INCENP. (B) Aurora B kinase activity at the corresponding cell cycle stages was measured in immunoprecipitates, by using histone H3 as a substrate, and analyzed using a PhosphorImager (bottom). Amounts of immunoprecipitated Aurora B protein were determined by Western blotting (top). A sample of the nocodazole-treated cell lysate before immunoprecipitation [lys(noc)] was analyzed in parallel. The numbers indicate the relative specific activity of Aurora B, with the value at the 4-h time point arbitrarily set to 1.0.
Depletion of any one subunit of the Aurora B/INCENP/survivin complex by siRNA caused a substantial reduction not only in the corresponding target but also in the two other subunits, albeit to a variable extent (Figure 4B). The abundance of α-tubulin was not reduced, attesting to the specificity of these effects (Figure 4B). At the cellular level, a phenotype was observed that was very similar to that seen after overexpression of the K106R Aurora B mutant in T-Rex-U2OS cells. Beginning at 24 h and prominent by 48 h after transfection with siRNA duplexes most interphase cells displayed multiple nuclei and/or micronuclei (Figure 4C). Mitotic cells showed mainly prometaphase-like phenotypes, with apparent defects in both chromosome congression and spindle morphology (Figure 4D). Anaphase and telophase stages were also abnormal, frequently showing lagging chromosomes (Figure 4D). In addition, a significant number of apoptotic cells were seen (our unpublished data). Results were qualitatively similar regardless of which subunit was targeted by siRNA, but the phenotypes seen after depletion of Aurora B were less pronounced than those seen after depletion of INCENP and survivin. This may reflect a lower efficiency of the Aurora B siRNA treatment.
Immunofluorescent staining of mitotic cells revealed that the depletion of any one of the components of the Aurora B/INCENP/survivin complex by siRNA abolished the centromere association of all three proteins (Figure 5). Taken at face value, this suggests that the formation of the entire Aurora B/INCENP/survivin complex is required for efficient localization, confirming and extending previous studies from C. elegans (Speliotes et al., 2000) and Drosophila (Adams et al., 2001c). However, this interpretation is complicated by the observed reduction in protein levels of the entire complex (Figure 4B; see DISCUSSION).
Cell Cycle Regulation of the Aurora B Kinase Complex
To examine the regulation of Aurora B activity during the cell cycle, HeLa S3 cells were synchronized and Aurora B, INCENP and survivin protein levels determined by Western blotting; in parallel, Aurora B kinase activity was measured in immunoprecipitates (Figure 6). Aurora B, INCENP, and survivin protein levels showed very similar changes during cell cycle progression, whereas α-tubulin levels, analyzed as a loading control, were constant. All three subunits of the Aurora B complex were low in G1/S, increased during G2 and M, and then declined. In comparison, cyclin B1 accumulated with similar kinetics but then disappeared earlier and more abruptly (Figure 6A), suggesting that it is degraded before Aurora B.
Maximal Aurora B kinase activity was observed in immunoprecipitates prepared from M phase samples, at 10–12 h after release from the G1/S block (Figure 6B). At this time point, the specific activity of Aurora B was nearly twice as high as that measured at the 4- to 8-h time points, indicating that the cell cycle-dependent increase in protein abundance was not sufficient to explain the rather abrupt activation of Aurora B. Instead, we emphasize that at the 10-h time point a population of INCENP with a reduced electrophoretic mobility became apparent (Figure 6; see also Figure 4A). This mobility shift of INCENP occurred at about the same time as Aurora B activation, raising the possibility that INCENP-modification could contribute to Aurora B activation.
Role of INCENP and Survivin in the Activation of Aurora B
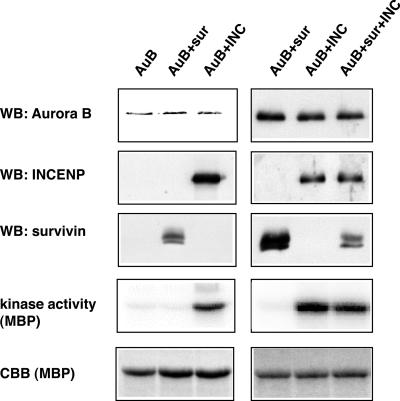
To examine the role of INCENP and survivin in Aurora B activation, Aurora B kinase complexes were reconstituted, using the baculovirus expression system. The three proteins were coexpressed in various combinations in Sf-9 insect cells, and complexes were then purified through an N-terminal GST-tag on Aurora B. As shown in Figure 7, Aurora B formed binary complexes with both INCENP and survivin. When all three subunits were coexpressed, both INCENP and survivin copurified with GST-Aurora B (Figure 7, right). We presume that this reflects formation of a ternary complex but cannot rigorously exclude a mixture of two binary complexes. Determination of kinase activities associated with these complexes, by using MBP as an exogenous substrate, revealed that INCENP caused a 7- to 10-fold activation of Aurora B, whereas survivin produced no detectable effect (Figure 7). We have not been able to accurately determine the stoichiometry of these complexes but emphasize that no influence of survivin on Aurora B activity could be detected over a wide range of survivin/Aurora B ratios (our unpublished data).
Figure 7.
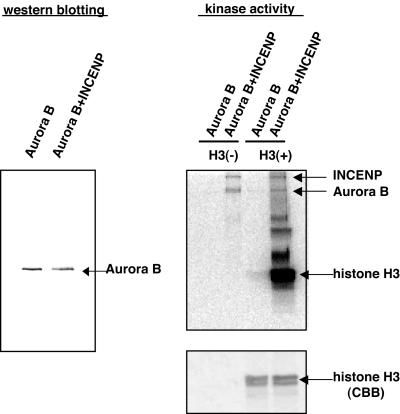
Effects of INCENP and survivin on Aurora B kinase activity. GST-Aurora B, GST-Aurora B/His-survivin, GST-Aurora B/His-INCENP, and GST-Aurora B/His-survivin/His-INCENP complexes were coexpressed in Sf-9 cells and purified using glutathione-Sepharose. Aurora B protein levels were determined by Western blotting, and kinase activity was measured in in vitro kinase assays, with or without MBP as an exogenous substrate. Phosphate incorporation was analyzed using a PhosphorImager. INCENP and survivin copurifying with Aurora B were also detected by Western blotting. MBP was visualized by Coomassie Blue staining (CBB).
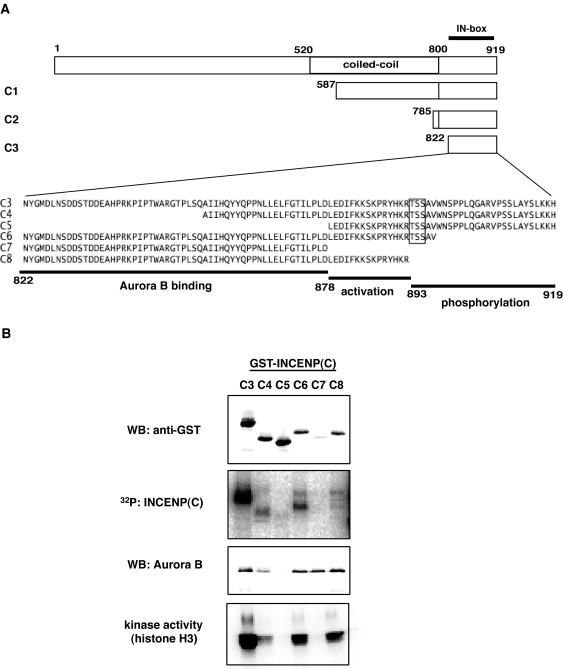
We have also examined the ability of INCENP to activate Aurora B with regard to histone H3, a purported physiological substrate of this kinase. Again, the Aurora B/INCENP complex was much more active than Aurora B alone (Figure 8). Furthermore, both INCENP and Aurora B became phosphorylated in these reactions. This indicates that INCENP is both an activator and a substrate of Aurora B. To determine which domains within INCENP were important for Aurora B activation, a series of INCENP deletion mutants was produced (Figure 9A). These mutants were then assayed for their ability to bind to Aurora B, to be phosphorylated by Aurora B, and to activate Aurora B (Figure 9B). Because Aurora B had previously been reported to bind to the C-terminal region of INCENP in other species (Kaitna et al., 2000), our analysis was focused on the C-terminal domain of INCENP. Initial experiments revealed that both the C1 (587–919) and the C2 (785–919) fragments were able to bind and activate Aurora B as well as to serve as substrates (our unpublished data). Thus, smaller fragments, C3-C8, were analyzed (Figure 9, A and B). C3 (822–919), which lacks the predicted coiled-coil region but contains the IN-box, bound to and activated Aurora B, and this fragment was also phosphorylated efficiently. In contrast, fragments C4 (855–919) and C5 (878–919) produced only weak and marginal activation, presumably because both mutants showed only weak binding to Aurora B. Fragments C6 (822–897), C7 (822–877), and C8 (822–892) all interacted with Aurora B to a similar extent, but C7 was unable to confer kinase activation. These data allow the following conclusions to be drawn. First, because the C3 mutant activated Aurora B as efficiently as full-length INCENP, the coiled-coil region (residues 520–800) does not seem to be necessary for activation. Second, because C7 was unable to activate Aurora B, although it bound as efficiently as C6 and C8, a domain of ∼15 residues (878–892) is required for Aurora B activation.
Figure 8.
Phosphorylation of INCENP by Aurora B. GST-Aurora B and GST-Aurora B/His-INCENP complexes were purified using glutathione-Sepharose, and Aurora B protein levels were determined by Western blotting (left). The complexes were then used for in vitro kinase reactions, with or without histone H3 as an exogenous substrate. Phosphate incorporation was detected using a PhosphorImager (right, top) and histone H3 visualized by CBB (bottom). Note the incorporation of phosphate into both INCENP and Aurora B (arrows).
Figure 9.
Mapping of INCENP domains required for activation of Aurora B. (A) Schematic representation of INCENP deletion mutants (C1–C8). The amino acid sequences of deletion mutants C3–C8 are indicated using the single-letter code. The regions of INCENP involved in Aurora B binding (822–877) and activation (878–892), as well as the domain serving as a substrate for phosphorylation (893–919), were deduced from the data shown in B. (B) The GST-INCENP deletion mutants (C3–C8) were coexpressed with His-Aurora B in Sf-9 cells and isolated on glutathione-Sepharose beads. The GST-INCENP mutants and coprecipitating His-Aurora B were then detected by Western blotting with anti-GST and anti-Aurora B antibodies, respectively. Phosphorylation of the INCENP mutants and histone H3 kinase activity of all samples were tested in parallel.
Phosphorylation of INCENP Enhances Aurora B Activity
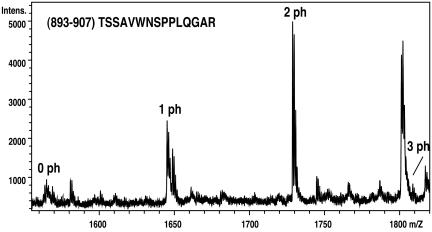
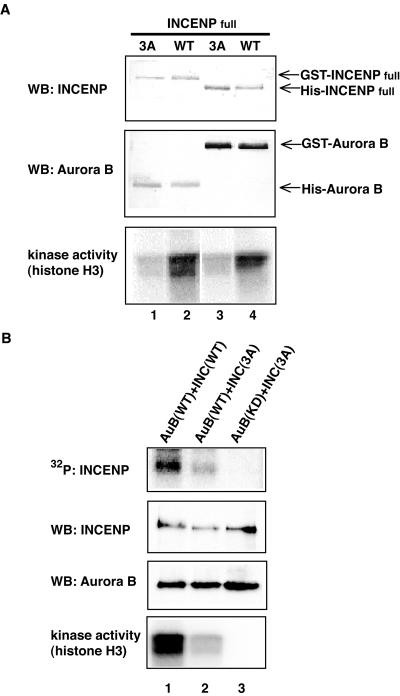
In a next series of experiments, we sought to identify the Aurora B phosphorylation site(s) in the C terminus of INCENP. To this end, phosphorylated C2 (785–919) and C3 (822–919) were analyzed by mass spectrometry (Figure 10). Both fragments contained a single phosphopeptide, TSSAVWNSPPLQGAR (893–907). The doubly phosphorylated species of this peptide was the most abundant one, although a minor component containing three phosphates could also be detected (Figure 10). Fragmentation analysis led us to conclude that at least two residues within the TSS motif were phosphorylated, but we could not determine the exact sites. In line with these results, the INCENP fragments containing the TSS motif (C3 and C6) were phosphorylated by Aurora B, whereas those lacking the motif (C7 and C8) were not (Figure 9B). To further confirm the above-mentioned mass spectrometry data, point mutations were introduced at T893, S894, S895, and S900. Mutated C3 fragments with three (3A; T893A, S894A, and S895A) or four (4A; T893A, S894A, S895A, and S900A) residues changed to alanine still bound to Aurora B (Figure 11A), but they were not phosphorylated to any significant extent (Figure 11B). No differences could be observed between the 3A and 4A mutants, confirming that Aurora B phosphorylates the C-terminal region of INCENP primarily on the three residues making up the TSS motif.
Figure 10.
Identification of major phosphorylation sites within INCENP by mass spectrometry. Matrix-assisted laser desorption ionization/time of flight mass spectrum of a tryptic digest of the INCENP C2 fragment (785–919) phosphorylated by Aurora B in vitro. A selected mass range containing the unphosphorylated (0 ph), singly (1 ph), doubly (2 ph), and triply phosphorylated (3 ph) peptide (893–907) TSSAVWNSPPLQGAR is shown. The identity and phosphorylation states of these peptides were confirmed using postsource decay analysis and electrospray tandem mass spectrometry (our unpublished data) as described in MATERIALS AND METHODS. The spectrum was recorded in negative ion reflector mode using 2,5 dihydroxybenzoic acid as matrix.
To determine whether phosphorylation by Aurora B influences INCENP function, we compared the ability of wild-type and triple Ala mutant versions (3A) of both C3 (Figure 11) and full-length INCENP (Figure 12) to activate Aurora B. Kinase assays were performed on Aurora B/INCENP complexes that had either been formed in Sf-9 cells in vivo (Figures 11, A and B, and 12), or reconstituted from recombinant proteins in vitro (Figure 11, C and D). In all cases, the 3A mutants were much less effective in Aurora B activation than wild-type versions of INCENP. These data indicate that the phosphorylation of TSS by Aurora B enhances the ability of INCENP to activate Aurora B. However, as shown in Figure 9B, the mutant C8 (which lacks the TSS motif) was nearly as efficient in activating Aurora B as C6 (which contains the motif and hence can be phosphorylated). This indicates that deletion of the INCENP C terminus (898–919) produces a similar effect as TSS phosphorylation. We conclude that maximal activation of Aurora B by wild-type INCENP requires the phosphorylation of the TSS motif, but that this requirement is alleviated upon removal of the extreme C terminus of INCENP.
Figure 12.
INCENP phosphorylation and Aurora B activation. (A) Full-length INCENP with mutated TSS motif barely activates Aurora B. Complexes of Aurora B and either wild-type or 3A mutant full-length INCENP were formed in Sf-9 cells and purified using glutathione-Sepharose beads. Recovery of proteins was determined by Western blotting (top and middle) and kinase activity of the complexes measured using histone H3 as a substrate (bottom). Lane 1, GST-INCENP (3A)/His-Aurora B; lane 2, GST-INCENP (WT)/His-Aurora B; lane 3, His-INCENP (3A)/GST-Aurora B; and lane 4, His-INCENP (WT)/GST-Aurora B. (B) Residual phosphorylation of INCENP with mutated TSS motif. Phosphate incorporation into INCENP was tested by subjecting Aurora B/INCENP complexes to in vitro kinase reactions. Protein levels of Aurora B and INCENP were determined by Western blotting, and the kinase activity of each complex was measured with histone H3 as substrates. Lane 1, GST-Aurora B(WT)/His-INCENP(WT); lane 2, GST-Aurora B(WT)/His-INCENP(3A); and lane 3, GST-Aurora B(K106R)/His-INCENP(3A).
When the TSS motif was altered to AAA, phosphorylation of the C-terminal fragment C3 by Aurora B was virtually abolished (Figure 11). In contrast, a full-length INCENP bearing these mutations was still phosphorylated by Aurora B, albeit to a lesser extent, whereas no phosphorylation could be seen upon coexpression of INCENP with the catalytically inactive Aurora B mutant, ruling out contamination by insect cell kinases (Figure 12B). This indicates that Aurora B phosphorylates INCENP also outside the C-terminal domain.
In an attempt to test the role of INCENP phosphorylation on the TSS motif in human cells, we have transiently transfected full-length wild-type and 3A-mutant INCENP into HeLa S3 and U2OS cells (our unpublished data). We found that both proteins localized primarily to the nucleus, although, at high expression levels, they also produced microtubule bundling in the cytoplasm. In mitotic cells, both proteins still localized to prometaphase centromeres and the midbody. No adverse effects could be observed on either the localization of endogenous Aurora B or mitotic progression. Although we cannot exclude subtle phenotypes (e.g., an increased rate of chromosome missegregation), these data indicate that the 3A mutant INCENP did not exert a dominant negative effect. In control experiments, overexpression of an INCENP mutant comprising the N-terminal domain (residues 1–290) severely impaired cell division and caused frequent DNA bridges, consistent with previous results (Mackay et al., 1998).
INCENP Phosphorylation and Aurora B Consensus Sites
The region flanking the TSS motif is highly conserved among INCENP proteins from various species, with the notable exception of Sli15p from budding yeast. As shown in Figure 13, the TSS motif is conserved among vertebrates, whereas invertebrates (D. melanogaster and C. elegans) feature a SS motif. Furthermore, in all these species the INCENP phosphoacceptor sites are preceded by two basic amino acids (KR), suggesting that basic residues contribute to a consensus motif defining Aurora B substrates. Indeed, several presumed substrates of Aurora B, including histone H3 (Hsu et al., 2000; Adams et al., 2001c; Giet and Glover, 2001; Goto et al., 2002; Sugiyama et al., 2002) and CENP-A (Zeitlin et al., 2001), contain similar phosphoacceptor sites, and putative autophosphorylation sites within the activation loop of Aurora B also conform to this consensus.
Figure 13.
Aurora B phosphorylation sites in INCENP. The C termini of INCENP proteins of all listed species, except for budding yeast, show a highly conserved phosphorylation motif. This motif conforms to an emerging consensus motif for Aurora B phosphorylation sites (see text).
DISCUSSION
The B-type Aurora kinases are critical regulators of chromosome segregation and cytokinesis in organisms ranging from yeast to human. They are implicated in regulating chromosome structure and cohesion, kinetochore–microtubule interactions, microtubule dynamics, the spindle checkpoint, and mitotic exit (Shannon and Salmon, 2002). Yet, our understanding of the regulation of these kinases remains incomplete and several critical substrates probably await identification. Herein, we have investigated the consequences of overexpressing wild-type and catalytically inactive Aurora B in human cells and examined the effects of depleting Aurora B and its complex partners INCENP and survivin by siRNA. Furthermore, we have studied the expression and activity of Aurora B kinase during the human cell cycle and obtained evidence for an involvement of posttranslational mechanisms in kinase activation. Aurora B complexes with INCENP and survivin were then reconstituted from recombinant baculoviruses and subjected to biochemical analyses. These studies, and the use of mass spectrometry, have revealed an evolutionarily conserved mechanism for Aurora B activation by INCENP binding and phosphorylation.
Probing Aurora B Function by siRNA and Dominant-Negative Approaches
Our functional studies confirm the notion that Aurora B is critical for multiple aspects of mitotic progression. Expression of the kinase-deficient mutant exerted a dominant-negative effect and resulted in a terminal phenotype characterized by multinucleation, in agreement with previous findings (Terada et al., 1998; Tatsuka et al., 1998). The molecular mechanisms leading to this phenotype remain to be completely understood, but we emphasize that most mitotic cells showed either a prometaphase-like state with poorly aligned chromosomes or an aberrant anaphase state characterized by lagging chromosomes (Figure 2B). This points to both a chromosome congression defect and the lack of a functional spindle assembly checkpoint, in agreement with recent studies performed in other species (Biggins and Murray, 2001; Adams et al., 2001c; Kallio et al., 2002; Murata-Hori and Wang, 2002; Tanaka et al., 2002).
Our results suggest that both Aurora B kinase activity and the formation of the Aurora B/INCENP/survivin complex contribute to its proper localization. Recently, Murata-Hori and Wang reported that a green fluorescent protein-tagged version of a catalytically inactive Aurora B showed at least partial localization to centromeres and central spindles (Murata-Hori and Wang, 2002; Murata-Hori et al., 2002). In our stable cell lines, we also observed (near-) correct localization of mutant Aurora B in those cells that expressed low amounts of this protein. In the majority of cells, however, the catalytically inactive Aurora B did not efficiently localize to either the centromere or the midzone, presumably due to higher expression levels (Figure 2B). Thus, it is plausible that mutant Aurora B may localize correctly at low expression levels, but mislocalize at higher levels. Endogenous INCENP and survivin also failed to localize properly in the presence of mutant Aurora B (Figure 3). Thus, phosphorylation of an as yet unidentified Aurora B substrate seems to be essential for efficient association of the Aurora B/INCENP/survivin complex with the centromere.
We also observed that depletion of any one subunit of the Aurora B/INCENP/survivin complex by siRNA abolished centromere-staining by antibodies against all three components (Figure 5). The most straightforward interpretation of this result is that all three subunits are required for correct localization of the complex, in line with results from previous RNAi studies in C. elegans (Speliotes et al., 2000) and Drosophila (Adams et al., 2001c) and a recent antisense experiments in human cells (Chen et al., 2002). However, because depletion of Aurora B, INCENP or survivin reduced the levels of all three components of the complex (Figure 4), one could also argue that all three subunits are required for stability, rather than localization, of the complex. This is difficult to exclude, but we favor an explanation for the data in Figure 4B that takes into account the terminal phenotype produced by these siRNA experiments. Depletion of any one subunit in fact produces multinucleated cells in G1 phase (Figure 4C) in which the three subunits of the Aurora B complex are known to be low (Figure 6).
Cell Cycle Regulation of the Aurora B Complex
Our analysis of the expression of Aurora B, INCENP, and survivin during the human cell cycle revealed that all three subunits are coordinately expressed, with lowest levels in G1, followed by steady increases toward the onset of mitosis. Thus, the abundance of INCENP and survivin is unlikely to limit the activity of Aurora B in human cells. At the G2/M transition, Aurora B kinase activity increased sharply, and concomitantly, INCENP became modified. This suggested that modification of INCENP could conceivably contribute to the activation of the complex. Previous studies have identified Xenopus INCENP as a candidate substrate of Cdk1 (Stukenberg et al., 1997), but the functional consequences of INCENP phosphorylation by Cdk1 are not known. In this study, we demonstrate that phosphorylation of human INCENP by Aurora B contributes to the activation of the complex (see below).
Reconstitution of Active Aurora B Complexes in Sf-9 Insect Cells
To further study the roles of INCENP and survivin in the activation of Aurora B, recombinant proteins were expressed from baculoviruses in insect cells. Both INCENP and survivin were able to bind Aurora B. However, survivin was unable to confer kinase activation, regardless of whether or not INCENP was present in the complex. This result contrasts with a recent report that survivin activates Aurora B through binding to its catalytic domain (Chen et al., 2002). We cannot presently explain this discrepancy, but note that another study implicates the noncatalytic, regulatory domain of Aurora B in survivin binding (Bolton et al., 2002). Our in vitro data suggest that survivin regulates other aspects of Aurora B function, although they do not exclude a contribution to the regulation of catalytic activity in vivo. In complexes isolated from Xenopus egg extracts, Aurora B activity could be modulated by removal and back-addition of survivin (Bolton et al., 2002). However, simple binding of survivin is unlikely to account for cell cycle-dependent differences in Aurora B activity, because interphasic and mitotic Aurora B complexes contained similar amounts of survivin (Bolton et al., 2002).
Most interestingly, binding of INCENP strongly stimulated Aurora B activity, regardless of whether complexes were allowed to form within Sf-9 cells in vivo or reconstituted from purified recombinant proteins in vitro. Thus, a binary complex of human Aurora B and INCENP displays activity, confirming and extending results obtained for yeast and invertebrate complexes (Kang et al., 2001; Bishop and Schumacher, 2002). By analyzing a series of INCENP deletion mutants, we identified a C-terminal domain (residues 822–892) as the minimal region required for Aurora B activation. This region overlaps with the IN-box proposed previously to comprise a binding domain for Aurora B kinases (Adams et al., 2001a; Leverson et al., 2002). However, our present mapping data refine our understanding of this interaction domain. In particular, whereas residues 822–877 were required for complex formation with Aurora B, residues 878–892 were critical for activation but, interestingly, not required for binding.
Concomitant with activation of Aurora B, strong phosphorylation of INCENP was observed. By mass spectrometry, a tripeptide (TSS: T893, S894, and S895) located within the C-terminal IN-box was identified as the major site of phosphorylation. These results contribute to define a consensus motif for Aurora B phosphorylation sites (Figure 13). Mutation of the three TSS residues to alanines (3A) substantially reduced the ability of both full-length INCENP and a C-terminal INCENP fragment (C3) to activate Aurora B. To test the role of TSS phosphorylation in vivo, we transiently expressed both wild-type and 3A-INCENP in human cells. These assays failed to reveal a striking dominant-negative effect associated with the 3A-INCENP mutant, but it would be premature to exclude subtle phenotypes. It is possible that the 3A-INCENP mutant could not be expressed to sufficient levels to interfere with endogenous wild-type INCENP. Alternatively, the 3A-INCENP mutant may still confer sufficient activation to Aurora B to permit functionality (Figure 12). Although the regulatory influence of INCENP phosphorylation may be difficult to demonstrate by overexpression studies, this modification is likely to be critical for Aurora B function at physiological protein levels.
While this manuscript was in preparation, phosphorylation of the C terminus of INCENP (ICP-1) was reported to stimulate Aurora B (AIR-2) activity in C. elegans (Bishop and Schumacher, 2002). Our present results are in excellent agreement with this report, and the phosphorylation sites identified by mass spectrometry in human INCENP match the sites inferred from site-directed mutagenesis in C. elegans ICP-1. However, it is noteworthy that vertebrates contain three phosphorylatable residues, whereas Drosophila and C. elegans only contain two (see alignment in Figure 13). Together, the studies performed on human INCENP and C. elegans ICP-1 concur to demonstrate that phosphorylation of a conserved motif within the IN-box represents an evolutionarily conserved mechanism for activation of the Aurora B kinase.
Our data suggest a two-step model for the activation of Aurora B by INCENP. They indicate that INCENP binding first stimulates the activity of Aurora B, but that phosphorylation of INCENP is then required for full activation of the complex. In support of this view, a C-terminal INCENP fragment lacking the TSS motif was a fairly powerful activator of Aurora B (Figure 9), suggesting that phosphorylation of the TSS motif is primarily required to change the position or conformation of the IN-box in such a way as to confer maximal activity to the complex. It will be interesting to see whether this model is borne out by future structural analyses of Aurora B/INCENP complexes.
Acknowledgments
We thank Dr. William Earnshaw (University of Edinburgh, Edinburgh, Scotland) for a kind gift of anti-CENP-B antibodies, Monika Matzner and Albert Ries for technical help, and all members of the laboratory for valuable discussion. This work was supported by the Fonds der Chemischen Industrie (to E.A.N), the Max-Planck-Society and a European Molecular Biology Oraganization long-term fellowship (to R.H).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-11-0769. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-11-0769.
References
- Adams, R.R., Carmena, M., and Earnshaw, W.C. (2001a). Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11, 49–54. [DOI] [PubMed] [Google Scholar]
- Adams, R.R., Eckley, D.M., Vagnarelli, P., Wheatley, S.P., Gerloff, D.L., Mackay, A.M., Svingen, P.A., Kaufmann, S.H., and Earnshaw, W.C. (2001b). Human INCENP colocalizes with the Aurora-B/AIRK2 kinase on chromosomes and is overexpressed in tumour cells. Chromosoma 110, 65–74. [DOI] [PubMed] [Google Scholar]
- Adams, R.R., Maiato, H., Earnshaw, W.C., and Carmena, M. (2001c). Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153, 865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, R.R., Wheatley, S.P., Gouldsworthy, A.M., Kandels-Lewis, S.E., Carmena, M., Smythe, C., Gerloff, D.L., and Earnshaw, W.C. (2000). INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 10, 1075–1078. [DOI] [PubMed] [Google Scholar]
- Ainsztein, A.M., Kandels-Lewis, S.E., Mackay, A.M., and Earnshaw, W.C. (1998). INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J. Cell Biol. 143, 1763–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., and Murray, A.W. (2001). The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15, 3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, J.R., et al. (1998). A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17, 3052–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, J.D., and Schumacher, J.M. (2002). Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B kinase stimulates Aurora B kinase activity. J. Biol. Chem. 277, 27577–27580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton, M.A., Lan, W., Powers, S.E., McCleland, M.L., Kuang, J., and Stukenberg, P.T. (2002). Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol. Biol. Cell 13, 3064–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., Anderson, S., Jwa, M., Green, E.M., Kang, J., Yates, J.R., Chan, C.S., Drubin, D.G., and Barnes, G. (2002). Phosphoregulation of kinetochore-microtubule attachments by the aurora kinase ipl1p. Cell 111, 163–172. [DOI] [PubMed] [Google Scholar]
- Chen, J., Jin, S., Tahir, S.K., Zhang, H., Liu, X., Sarthy, A.V., McGonigal, T.P., Liu, Z., Rosenberg, S.H., and Ng, S.C. (2003). Survivin enhances Aurora-B kinase activity and localizes Aurora-B in human cells. J. Biol. Chem. 278, 486–490. [DOI] [PubMed] [Google Scholar]
- Chen, S.H., and Tang, T.K. (2002). Mutational analysis of the phosphorylation sites of the Aie1 (Aurora-C) kinase in vitro. DNA Cell Biol. 21, 41–46. [DOI] [PubMed] [Google Scholar]
- Cutts, S.M., Fowler, K.J., Kile, B.T., Hii, L.L., O'Dowd, R.A., Hudson, D.F., Saffery, R., Kalitsis, P., Earle, E., and Choo, K.H. (1999). Defective chromosome segregation, microtubule bundling and nuclear bridging in inner centromere protein gene (Incenp)-disrupted mice. Hum. Mol. Genet. 8, 1145–1155. [DOI] [PubMed] [Google Scholar]
- Dutertre, S., Descamps, S., and Prigent, C. (2002). On the role of aurora-A in centrosome function. Oncogene 21, 6175–6183. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Francisco, L., and Chan, C.S. (1994). Regulation of yeast chromosome segregation by Ipl1 protein kinase and type 1 protein phosphatase. Cell Mol. Biol. Res. 40, 207–213. [PubMed] [Google Scholar]
- Francisco, L., Wang, W., and Chan, C.S. (1994). Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Mol. Cell Biol. 14, 4731–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, A.G., James, C., Evan, G.I., and Hengartner, M.O. (1999). Caenorhabditis elegans inhibitor of apoptosis protein (IAP) homologue BIR-1 plays a conserved role in cytokinesis. Curr. Biol. 9, 292–301. [DOI] [PubMed] [Google Scholar]
- Giet, R., and Glover, D.M. (2001). Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152, 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobom, J., Nordhoff, E., Mirgorodskaya, E., Ekman, R., and Roepstorff, P. (1999). Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 34, 105–116. [DOI] [PubMed] [Google Scholar]
- Goto, H., Yasui, Y., Nigg, E.A., and Inagaki, M. (2002). Aurora-B phosphorylates histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells 7, 11–17. [DOI] [PubMed] [Google Scholar]
- Hoffmann, R., Metzger, S., Spengler, B., and Otvos, L., Jr. (1999). Sequencing of peptides phosphorylated on serines and threonines by post-source decay in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Mass Spectrom. 34, 1195–1204. [DOI] [PubMed] [Google Scholar]
- Hsu, J.Y., et al. (2000). Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291. [DOI] [PubMed] [Google Scholar]
- Kaitna, S., Mendoza, M., Jantsch-Plunger, V., and Glotzer, M. (2000). Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol. 10, 1172–1181. [DOI] [PubMed] [Google Scholar]
- Kallio, M.J., McCleland, M.L., Stukenberg, P.T., and Gorbsky, G.J. (2002). Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 12, 900–905. [DOI] [PubMed] [Google Scholar]
- Kallio, M.J., Nieminen, M., and Eriksson, J.E. (2001). Human inhibitor of apoptosis protein (IAP) survivin participates in regulation of chromosome segregation and mitotic exit. FASEB J. 15, 2721–2723. [DOI] [PubMed] [Google Scholar]
- Kang, J., Cheeseman, I.M., Kallstrom, G., Velmurugan, S., Barnes, G., and Chan, C.S. (2001). Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 155, 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., Kotani, S., Hattori, T., Sumi, N., Yoshioka, T., Todokoro, K., and Okano, Y. (1997). Cell cycle-dependent expression and spindle pole localization of a novel human protein kinase, Aik, related to Aurora of Drosophila and yeast Ipl1. J. Biol. Chem. 272, 13766–13771. [DOI] [PubMed] [Google Scholar]
- Kimura, M., Matsuda, Y., Yoshioka, T., and Okano, Y. (1999). Cell cycle-dependent expression and centrosome localization of a third human aurora/Ipl1-related protein kinase, AIK3. J. Biol. Chem. 274, 7334–7340. [DOI] [PubMed] [Google Scholar]
- Kufer, T.A., Sillje, H.H., Korner, R., Gruss, O.J., Meraldi, P., and Nigg, E.A. (2002). Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 158, 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverson, J.D., Huang, H.K., Forsburg, S.L., and Hunter, T. (2002). The Schizosaccharomyces pombe aurora-related kinase Ark1 interacts with the inner centromere protein Pic1 and mediates chromosome segregation and cytokinesis. Mol. Biol. Cell 13, 1132–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., Ambrosini, G., Chu, E.Y., Plescia, J., Tognin, S., Marchisio, P.C., and Altieri, D.C. (1998). Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 396, 580–584. [DOI] [PubMed] [Google Scholar]
- Littlepage, L.E., Wu, H., Andresson, T., Deanehan, J.K., Amundadottir, L.T., and Ruderman, J.V. (2002). Identification of phosphorylated residues that affect the activity of the mitotic kinase Aurora-A. Proc. Natl. Acad. Sci. USA 99, 15440–15445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Li, M.Z., Leibham, D., Cortez, D., and Elledge, S.J. (1998). The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 8, 1300–1309. [DOI] [PubMed] [Google Scholar]
- Mackay, A.M., Ainsztein, A.M., Eckley, D.M., and Earnshaw, W.C. (1998). A dominant mutant of inner centromere protein (INCENP), a chromosomal protein, disrupts prometaphase congression and cytokinesis. J. Cell Biol. 140, 991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi, P., Honda, R., and Nigg, E.A. (2002). Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53–/– cells. EMBO J. 21, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata-Hori, M., Tatsuka, M., and Wang, Y.L. (2002). Probing the dynamics and functions of aurora B kinase in living cells during mitosis and cytokinesis. Mol. Biol. Cell 13, 1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata-Hori, M., and Wang, Y. (2002). The kinase activity of Aurora B is required for kinetochore-microtubule interactions during mitosis. Curr. Biol. 12, 894–899. [DOI] [PubMed] [Google Scholar]
- Murnion, M.E., Adams, R.R., Callister, D.M., Allis, C.D., Earnshaw, W.C., and Swedlow, J.R. (2001). Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J. Biol. Chem. 276, 26656–26665. [DOI] [PubMed] [Google Scholar]
- Nigg, E.A. (2001). Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2, 21–32. [DOI] [PubMed] [Google Scholar]
- Petersen, J., Paris, J., Willer, M., Philippe, M., and Hagan, I.M. (2001). The S. pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J. Cell Sci. 114, 4371–4384. [DOI] [PubMed] [Google Scholar]
- Reed, J.C. (2001). The Survivin saga goes in vivo. J. Clin. Investig. 108, 965–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, K.B., and Salmon, E.D. (2002). Chromosome dynamics: new light on aurora B kinase function. Curr. Biol. 12, R458–R460. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Silke, J., and Vaux, D.L. (2001). Two kinds of BIR-containing protein -inhibitors of apoptosis, or required for mitosis. J. Cell Sci. 114, 1821–1827. [DOI] [PubMed] [Google Scholar]
- Speliotes, E.K., Uren, A., Vaux, D., and Horvitz, H.R. (2000). The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol. Cell 6, 211–223. [DOI] [PubMed] [Google Scholar]
- Stukenberg, P.T., Lustig, K.D., McGarry, T.J., King, R.W., Kuang, J., and Kirschner, M.W. (1997). Systematic identification of mitotic phosphoproteins. Curr. Biol. 7, 338–348. [DOI] [PubMed] [Google Scholar]
- Sugiyama, K., Sugiura, K., Hara, T., Sugimoto, K., Shima, H., Honda, K., Furukawa, K., Yamashita, S., and Urano, T. (2002). Aurora-B associated protein phosphatases as negative regulators of kinase activation. Oncogene 21, 3103–3111. [DOI] [PubMed] [Google Scholar]
- Tanaka, T.U., Rachidi, N., Janke, C., Pereira, G., Galova, M., Schiebel, E., Stark, M.J., and Nasmyth, K. (2002). Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108, 317–329. [DOI] [PubMed] [Google Scholar]
- Tatsuka, M., Katayama, H., Ota, T., Tanaka, T., Odashima, S., Suzuki, F., and Terada, Y. (1998). Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 58, 4811–4816. [PubMed] [Google Scholar]
- Terada, Y., Tatsuka, M., Suzuki, F., Yasuda, Y., Fujita, S., and Otsu, M. (1998). AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J. 17, 667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, M.Y., Wiese, C., Cao, K., Martin, O., Donovan, P., Ruderman, J., Prigent, C., and Zheng, Y. (2003). A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat. Cell Biol. 5, 242–248. [DOI] [PubMed] [Google Scholar]
- Tseng, T.C., Chen, S.H., Hsu, Y.P., and Tang, T.K. (1998). Protein kinase profile of sperm and eggs: cloning and characterization of two novel testis-specific protein kinases (AIE1, AIE2) related to yeast and fly chromosome segregation regulators. DNA Cell Biol. 17, 823–833. [DOI] [PubMed] [Google Scholar]
- Uren, A.G., Beilharz, T., O'Connell, M.J., Bugg, S.J., van Driel, R., Vaux, D.L., and Lithgow, T. (1999). Role for yeast inhibitor of apoptosis (IAP)-like proteins in cell division. Proc. Natl. Acad. Sci. USA 96, 10170–10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren, A.G., Wong, L., Pakusch, M., Fowler, K.J., Burrows, F.J., Vaux, D.L., and Choo, K.H. (2000). Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr. Biol. 10, 1319–1328. [DOI] [PubMed] [Google Scholar]
- Walter, A.O., Seghezzi, W., Korver, W., Sheung, J., and Lees, E. (2000). The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene 19, 4906–4916. [DOI] [PubMed] [Google Scholar]
- Wheatley, S.P., Carvalho, A., Vagnarelli, P., and Earnshaw, W.C. (2001a). INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr. Biol. 11, 886–890. [DOI] [PubMed] [Google Scholar]
- Wheatley, S.P., Kandels-Lewis, S.E., Adams, R.R., Ainsztein, A.M., and Earnshaw, W.C. (2001b). INCENP binds directly to tubulin and requires dynamic microtubules to target to the cleavage furrow. Exp. Cell Res. 262, 122–127. [DOI] [PubMed] [Google Scholar]
- Zeitlin, S.G., Shelby, R.D., and Sullivan, K.F. (2001). CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 155, 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]