Abstract
Although there has been a recent explosion in the identification of budding yeast kinetochore components, the physical interactions that underlie kinetochore function remain obscure. To better understand how kinetochores attach to microtubules and how this attachment is regulated, we sought to characterize the interactions among kinetochore proteins, especially with respect to the microtubule-binding Dam1 complex. The Dam1 complex plays a crucial role in the chromosome-spindle attachment and is a key target for phospho-regulation of this attachment by the Aurora kinase Ipl1p. To identify protein–protein interactions involving the Dam1 complex, and the effects of Dam1p phosphorylation state on these physical interactions, we conducted both a genome-wide two-hybrid screen and a series of biochemical binding assays for Dam1p. A two-hybrid screen of a library of 6000 yeast open reading frames identified nine kinetochore proteins as Dam1p-interacting partners. From 113 in vitro binding reactions involving all nine subunits of the Dam1 complex and 32 kinetochore proteins, we found at least nine interactions within the Dam1 complex and 19 potential partners for the Dam1 complex. Strikingly, we found that the Dam1p–Ndc80p and Dam1p–Spc34p interactions were weakened by mutations mimicking phosphorylation at Ipl1p sites, allowing us to formulate a model for the effects of phosphoregulation on kinetochore function.
INTRODUCTION
To be faithfully segregated during mitosis, duplicated eukaryotic chromosomes must first form bipolar attachments to the mitotic spindle. The DNA-microtubule attachment occurs at a specialized multiprotein structure called the kinetochore. To date, >40 kinetochore proteins have been identified in budding yeast (Cheeseman et al., 2002b). Given the molecular complexity of the kinetochore and its essential role in chromosome segregation, it is important to understand both how the kinetochore is organized and how it is regulated. Recently, phosphorylation by the Ipl1 protein kinase was shown to play an important role in regulating the structure and function of the yeast kinetochore (Cheeseman et al., 2002a; Tanaka et al., 2002). Ip11p is the founding member of the Aurora kinase family (Chan and Botstein, 1993), members of which function in chromosome segregation from yeast to metazoans (Shannon and Salmon, 2002). The nine-subunit Dam1 complex is required for both chromosome segregation and spindle integrity. Mutational analysis of phosphorylation sites in Dam1p clearly demonstrated its role as a key Ipl1 target in vivo (Cheeseman et al., 2002a). However, it was unclear what the molecular consequences of this phosphorylation were. One possible effect of Ipl1p phosphorylation could be to change the physical interactions of Dam1p via phosphorylation. Because the phosphorylation state of the Dam1 complex does not affect its microtubule-binding activity or subunit composition (Cheeseman et al., 2002a), we sought to identify other physical interactions of the Dam1 complex at the kinetochore and to determine whether they are affected by Ipl1 phosphorylation.
Despite the discovery of centromeric DNA-binding components and microtubule-binding components within the kinetochore, how the >40 kinetochore proteins establish the complete connectivity from the DNA to the microtubule is not known. The current understanding of yeast kinetochore organization is primarily derived from two-hybrid interactions, coimmunoprecipitation (coIP), and copurification experiments. The coIP and copurification experiments have provided a powerful means to group kinetochore proteins into discrete subcomplexes, such as the nine-subunit Dam1 complex (Cheeseman et al., 2001a, 2002a; Janke et al., 2002; Li et al., 2002), the four-subunit Ndc80 complex (Janke et al., 2001; Wigge and Kilmartin, 2001), the four-subunit CBF3 complex (Lechner and Carbon, 1991), and the 12-subunit Ctf19 complex (Cheeseman et al., 2002a). Two-hybrid studies have provided additional information about how kinetochore proteins are organized within the subcomplexes and how different subcomplexes are connected to each other (Ortiz et al., 1999; Ito et al., 2000, 2001; Ortiz and Lechner, 2000; Uetz et al., 2000; Janke et al., 2001). However, two-hybrid information is lacking for many proteins, and two-hybrid interactions can be mediated by intermediary proteins.
To examine both the organization of proteins within the kinetochore and the role that phosphorylation plays in the regulation of these protein–protein interactions, we took a dual approach. We conducted a genome-wide two-hybrid screen by using Dam1p as a bait, and we examined Dam1p's direct physical interactions in vitro. To test the effect of phosphorylation on these physical interactions, we also carried out a genome-wide two-hybrid screen with the Dam1p mutants that reflect the dephosphorylated or constitutively phosphorylated state and confirmed the results by in vitro binding assays. Herein, we present a protein interaction map of the yeast kinetochore focusing on those interactions surrounding the Dam1 complex, and we identify a potential mechanism for its phosphoregulation by Ipl1 kinase.
MATERIALS AND METHODS
Plasmids, Yeast Strains, and Growth Conditions
The plasmids and the yeast strains used in this study are listed in Tables 1 and 2, respectively. Yeast media were prepared as described previously (Burke et al., 2000). Synthetic medium (SM) with appropriate nutrients, and yeast extract/peptone medium (YP) were supplemented with 2% glucose or with 2% raffinose or 2% galactose, as indicated. G418 (Mediatech, Herndon, VA) was used at 0.4 mg/ml.
Table 1.
Plasmids used in this study
| Name | Relevant features | Source |
|---|---|---|
| pOAD | Activation domain vector | 1 |
| pOBD2 | DNA-binding domain vector | 1 |
| pBAT4 | E. coli expression vector under T7 promoter (for in vitro translation) | 2 |
| pDD1263 | DUO1 in pBAT4 | 3 |
| pDD1264 | DAM1 in pBAT4 | 3 |
| pDD1265 | SPC34 in pBAT4 | 3 |
| pDD1266 | SPC19 in pBAT4 | 3 |
| pDD1267 | ASK1 in pBAT4 | 3 |
| pDD1268 | DAD1 in pBAT4 | 3 |
| pDD1269 | DAD2 in pBAT4 | 3 |
| pDD1270 | DAD3 in pBAT4 | 3 |
| pDD1271 | DAD4 in pBAT4 | 3 |
| pDD1272 | NDC80 in pBAT4 | 3 |
| pDD1273 | NUF2 in pBAT4 | 3 |
| pDD1274 | SPC24 in pBAT4 | 3 |
| pDD1275 | SPC25 in pBAT4 | 3 |
| pDD1276 | CTF19 in pBAT4 | 3 |
| pDD1277 | OKP1 in pBAT4 | 3 |
| pDD1278 | MCM21 in pBAT4 | 3 |
| pDD1279 | MCM16 in pBAT4 | 3 |
| pDD1280 | MCM22 in pBAT4 | 3 |
| pDD1281 | MCM17 in pBAT4 | 3 |
| pDD1282 | MCM19 in pBAT4 | 3 |
| pDD1283 | MTW1 in pBAT4 | 3 |
| pDD1284 | NDC10 in pBAT4 | 3 |
| pDD1285 | CTF13 in pBAT4 | 3 |
| pDD1286 | CEP3 in pBAT4 | 3 |
| pDD1287 | SKP1 in pBAT4 | 3 |
| pDD1288 | SLK19 in pBAT4 | 3 |
| pDD1289 | CSE4 in pBAT4 | 3 |
| pDD1290 | CBF1 in pBAT4 | 3 |
| pDD1291 | BIR1 in pBAT4 | 3 |
| pDD1292 | SLI15 in pBAT4 | 3 |
| pDD1293 | MPS1 in pBAT4 | 3 |
| pDD1294 | MAD1 in pBAT4 | 3 |
| pDD1295 | MAD2 in pBAT4 | 3 |
| pDD1296 | MAD3 in pBAT4 | 3 |
| pDD1297 | BUB1 in pBAT4 | 3 |
| pDD1298 | BUB2 in pBAT4 | 3 |
| pDD1299 | BUB3 in pBAT4 | 3 |
| pDD1300 | STU2 in pBAT4 | 3 |
| pDD1301 | BIK1 in pBAT4 | 3 |
| pDD1302 | BIM1 in pBAT4 | 3 |
| pDD1303 | dam1(S20A, S257A, S265A, S292A) in pBAT4 | 3 |
| pDD1304 | dam1 (S20D, S257D, S265D, S292D) in pBAT4 | 3 |
| pGAT2 | E. coli expression vector for GST fusion proteins | 2 |
| pDD1305 | SPC19 in pGAT2 | 3 |
| pEG(KT) | 2μ, URA3, leu2-d, GST under GAL1/10 promoter | 4 |
| pDD1306 | DUO1 in pEG(KT) | 3 |
| pDD1017 | DAM1 in pEG(KT) | 5 |
| pDD1307 | SPC34 in pEG(KT) | 3 |
| pDD1310 | ASK1 in pEG(KT) | 3 |
| pDD1016 | TAP tag in pRS426 under GAL1/10 promoter | 6 |
| pDD1312 | DAD1 in pDD1016 | 3 |
| pDD1313 | DAD2 in pDD1016 | 3 |
| pDD1314 | DAD3 in pDD1016 | 3 |
| pDD1315 | DAD4 in pDD1016 | 3 |
| pDD1316 | dam1 (S20A, S257A, S265A, S292A) in pEG(KT) | 3 |
| pDD1317 | dam1 (S20D, S257D, S265D, S292D) in pEG(KT) | 3 |
Table 2.
Yeast strains used in this study
| Name | Genotype | Source |
|---|---|---|
| PJ69-4a | MATa, leu2-3,112, ura3-52, trp1-901, his3-200, gal4Δ, gal80Δ, LϒS2::GAL1-HIS3, GAL2-ADE2, met2::GAL7-lacZ. | 1 |
| PJ69-4α | MATα, leu2-3,112, ura3-52, trp1-901, his3-200, gal4Δ, gal80Δ, LϒS2::GAL1-HIS3, GAL2-ADE2, met2::GAL7-lacZ. | 1 |
| DDY1810 | MATa, leu2, ura3-52, trp1, prb1-1122, pep4-3, pre1-451 | 2 |
| DDY2369 | MATa, leu2, ura3-52, trp1, prb1-1122, pep4-3, pre1-451, DAD1-STag-TEV-ZZ::KanMX | 2 |
| DDY2469 | MATa, leu2, ura3-52, trp1, prb1-1122, pep4-3, pre1-451, SPC24-STag-TEV-ZZ::KanMX | 2 |
| DDY2481 | MATa, his3Δ200, leu2-3,112, ade2-1, spc34Δ::HIS3, ura3-52::spc34(T199D)::URA3 | 2 |
| DDY2496 | MATa, his3Δ200, leu2-3,112, ura3-52, lys2-801, dam1(S257D, S265D, S292D)::KanMX | 2 |
1, Yeast Resource Center; 2, Drubin/Barnes Laboratory. All Drubin/Barnes laboratory strains are derived from strain S288C.
Two-Hybrid Assays
For the genome-wide two-hybrid screens, wild-type DAM1, dam1 mutated to remove all of the phosphorylation sites (dam1 S to A), and dam1 mutated to mimic the fully Ipl1-phosphorylated state (dam1 S to D) were cloned into the Gal4 DNA binding domain vector pOBD2 (Yeast Resource Center, Seattle, WA) and two-hybrid screens were performed as described previously (Uetz et al., 2000).
Genes to be tested directly in the two-hybrid assays were cloned into the DNA binding domain vector (pOBD2, the baits) or the activation domain vector (pOAD, the preys). The baits were expressed in PJ69-4α strain and the preys were expressed in PJ69-4a strain. The positive interactions were detected by selection on synthetic complete medium lacking leucine, tryptophan, and histidine, and containing 3 mM 3-amino-1,2,4-triazole (Sigma-Aldrich, St. Louis, MO). All of the vectors and yeast strains are available from the Yeast Resource Center (Seattle, WA).
Purification of Glutathione S-Transferase (GST)-Fusion Proteins from Bacteria
GST-fusion proteins were expressed in Escherichia coli BL21(DE3) from the vector pGAT2 (Peranen et al., 1996). The bacteria were grown at 37°C until the OD600 reached 0.5 and were then induced with 0.4 mM isopropyl β-d-thiogalactoside at 28°C for 4 h. The cell pellet was washed with water and resuspended in HEK-T buffer (50 mM HEPES pH7.5, 1 mM EDTA, 100 mM KCl, 1% Triton X-100), with 1 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitors. The cells were lysed with lysozyme, sonicated three times for 30 s, and centrifuged in an SA-600 rotor at 10,000 rpm for 20 min at 4°C. The supernatant was passed through a glutathione-agarose (Sigma-Aldrich) column, and the bound GST-fusion proteins were eluted in elution buffer (20 mM glutathione, 100 mM Tris-HCl pH 8.0, 120 mM NaCl, 1% Triton X-100) and dialyzed into HEK-T.
Purification of GST-Fusion Proteins from Yeast
GST-fusion proteins were expressed from pEG(KT) (Mitchell et al., 1993) under the galactose-inducible promoter and purified from DDY1810 as described previously (Rodal et al., 1999). The yeast cells were induced with galactose at 30°C for 8 h, harvested, frozen in liquid nitrogen, and stored at –80°C. Cell pellets were lysed in liquid nitrogen in a Waring Blender and thawed in HEK-T buffer (50 mM HEPES pH7.5, 1 mM EDTA, 100 mM KCl, 1% Triton X-100), with 1 mM PMSF and protease inhibitors. The lysate was sonicated four times for 30 s each time, and centrifuged in an SA-600 rotor at 10,000 rpm for 20 min. The supernatant was filtered through cheesecloth and passed over SP Sepharose Fast Flow (Amersham Biosciences, Piscataway, NJ) for GST-Duo1p, GST-Dam1p, GST-Spc34p, and GST-Ndc80p, or Q Sepharose Fast Flow (Amersham Biosciences) for GST-Ask1p. Bound GST-fusion proteins were eluted with 40 ml of HEK500-T buffer (50 mM HEPES pH7.5, 1 mM EDTA, 500 mM KCl, 1% Triton X-100), and the eluate was bound to glutathione-agarose (Sigma-Aldrich). Finally, GST-fusion proteins were eluted in elution buffer (20 mM glutathione, 100 mM Tris-HCl pH 8.0, 120 mM NaCl, 1% Triton X-100) and dialyzed into HEK-T. The protein concentration was determined with the BCA protein assay reagent kit (Pierce Chemical, Rockford, IL) by using bovine serum albumin (Sigma-Aldrich) as a standard.
To ensure that each purified GST-fusion protein did not bring along other components in the complex, we performed immunoblots on the six GST-fusion proteins and tested for the presence of copurifying endogenous Duo1p and Dam1p with the respective antibodies. There was no detectable Duo1p or Dam1p present in the purified GST-Spc34p and GST-Ask1p, no Duo1p in the purified GST-Dam1p, and no Dam1p in the purified GST-Duo1p (GST-Spc19p was purified from E. coli) (our unpublished data). Therefore, the GST pull-down results reflected the direct binding between any of the two subunits.
Purification of Calmodulin Binding Peptide (CBP)-Fusion Proteins from Yeast
DAD1, DAD2, DAD3, and DAD4 genes were cloned into pDD1016 (a generous gift of Erin O'Shea, University of California, San Francisco, San Francisco, CA) to overexpress CBP-TEV-ProA fusions under the Gal promoter. The proteins were purified according to the tandem affinity purification method as described previously (Rigaut et al., 1999), with the following modification. The frozen yeast cells were lysed in lysis buffer (50 mM bis-Tris propane pH 7.0, 100 mM KCl, 5 mM EDTA, 5 mM EGTA, 10% glyecerol, 1% Triton X-100, 1 mM PMSF, 1× protease inhibitor mixture). The lysate was sonicated at three times for 50 s and centrifuged at 10,000 rpm for 20 min in an SA-600 rotor. The supernatant was adjusted to 400 mM KCl, and 1 ml of IgG agarose (Sigma-Aldrich) was added. After binding for 3 h at 4°C, the resin was washed with 20 ml of lysis buffer (plus 400 mM KCl), and washed with 20 ml of TEV cleavage buffer (10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, 1 mM dithiothreitol) before TEV treatment. Finally, the calmodulin beads (Stratagene, La Jolla, CA) with bound proteins were washed with calmodulin binding buffer (10 mM β-mercaptoethanol, 10 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM Mg-acetate, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40) and the beads were used in the binding reactions. All steps were carried out at 4°C.
Purification of the Dam1p Complex and Ndc80 Complex
The Dam1 and Ndc80 complexes were purified as described previously (Cheeseman et al., 2001a, 2002a).
General Immunoblot Procedures
Immunoblot analysis was performed using standard SDS-PAGE, and proteins were transferred to nitrocellulose membranes (Protran BA83; Schleicher & Schuell, Keene, NH). Membranes were blocked for 1 h with Tris-buffered saline/0.05% Tween 20 containing 5% nonfat milk, followed by overnight incubation with affinity-purified antibodies. Rabbit anti-GST antibody was used at a dilution of 1:2000, anti-Duo1p antibody (Hofmann et al., 1998) was used at a dilution of 1:2000, anti-Dam1p antibody (Cheeseman et al., 2001a) was used at a dilution of 1:1000, and affinity-purified anti-Ndc80p antibody was used at a dilution of 1:10,000 (a generous gift from Arshad Desai, Ludwig Institute of Cancer Research, San Diego, CA). Rabbit IgG was used at a dilution of 1:10,000 from a 2% stock solution (ICN Biomedicals, Costa Mesa, CA). Anti-rabbit horseradish peroxidase-conjugated secondary antibodies (Amersham Biosicences) were used at a dilution of 1:5000.
In Vitro-coupled Transcription/Translation
Genes to be in vitro transcribed/translated were cloned into an E. coli expression vector, pBAT4 (Peranen et al., 1996). In vitro-coupled transcription and translation of proteins was performed using the Promega TnT quick coupled transcription/translation system according to the manufacturer's guidelines (Promega, Madison, WI)
Binding Assay
Purified GST-fusion proteins (or CBP-fusion proteins) were bound to the glutathione-agarose beads (or calmodulin beads) at a final concentration of 0.1 μg/μl. For each binding reaction, 5 μl of in vitro-translated product was diluted into 20 μl of HEK-T buffer (or calmodulin binding buffer) containing 1 mg/ml bovine serum albumin (Sigma-Aldrich), and then prespun at 14,000 × g for 10 min in a tabletop centrifuge. The supernatant was mixed with 25 μl of GST-fusion proteins on the beads, and the mixture was incubated at 25°C for 30 min with occasional mixing. After the incubation, the reaction was spun at 14,000 × g for 1 min in a tabletop centrifuge and the supernatant and the beads were separated. The beads were washed three times with 300 μl of HEK-T buffer. The supernatant and bead samples were separated on 16% tricine gels, which were processed for autoradiography after electrophoresis. The gels were scanned using a PhosphorImager SI450 (Amersham Biosciences), and the images were analyzed using ImageQuant version 1.2 software (Amersham Biosciences).
To determine binding affinities for interactions involving wild-type or mutant Dam1p, the in vitro-coupled transcription/translation (IVT) prespun supernatant was diluted 1:1 with HEK-T buffer. Diluted IVT (10 μl) was mixed with 10 μl of GST-Dam1p (range 0.35–9.5 μM) or 10 μl of 10 μM GST, and incubated for 20 min at room temperature. Glutathione-agarose beads (30 μl, 50% slurry in HEK-T) were added to the reaction and incubated for 20 min with occasional mixing at room temperature. After the incubation, the supernatant and beads were separated by centrifugation and subsequently handled as describe above.
Chromatin Immunoprecipitation
Immunoprecipitation of formaldehyde cross-linked chromatin was performed essentially as described previously (Enquist-Newman et al., 2001).
Affinity-purified rabbit anti-Duo1p antibody (1.2 mg/ml) was used at a 1:200 dilution. Affinity-purified guinea pig anti-Dam1p antibody (1 mg/ml) was used at a dilution of 1:150. Affinity-purified rabbit anti-Ndc80p (8.2 mg/ml) antibody was used at a dilution of 1:1500. Immune complexes were isolated on protein A-Sepharose CL-4B beads (Amersham Biosciences). Polymerase chain reaction (PCR) reactions were amplified using BioExact DNA polymerase (Bioline, Randolph, MA) for 24 cycles. PCR products were resolved on 2.5% agarose gels and visualized with ethidium bromide. Stained PCR products were imaged with a Gel Doc 1000 system (Bio-Rad, Hercules, CA) and quantified using ImageQuant image analysis software (Amersham Biosciences).
RESULTS
A Genome-Wide Two-Hybrid Screen for Dam1p-interacting Proteins
To provide a context for understanding the function and regulation of the Dam1 spindle/kinetochore complex, we first focused on its microtubule-binding subunit Dam1p, which is a critical in vivo target of the Ipl1 kinase (Cheeseman et al., 2002a). We began by conducting a yeast two-hybrid screen against a genome-wide array of ∼6000 yeast open reading frames (Uetz et al., 2000) by using Dam1p as a bait. For these studies, we cloned wild-type DAM1 into the Gal4 DNA binding domain vector pOBD2. Positives identified in the genome-wide screen were combined into a single microarray for further studies (see below). Screens using both the genome-wide array and the microarray were performed at least twice. The results from these screens are summarized in Table 3 (second column) and in Supplementary Table 1. Nine out of 26 interacting proteins identified were components of the spindle or kinetochore. The other proteins are either relevant ones that had not previously been shown to have related functions, or could be false-positives (Supplementary Table 1).
Table 3.
Genome-wide two-hybrid screens using wild-type and mutant DAM1 as baits
|
Dam1 Baits
|
|||
|---|---|---|---|
| Categories of the identified proteins | Wild type DAM1 | dam1 (S to A) | dam1 (S to D) |
| Dam1 complex | Dam1 | Dam1 | Dam1 |
| Duo1a,b,c | Duo1 | Duo1 | |
| Spc34b | Spc34 | ||
| Spc19 | Spc19 | Spc19 | |
| Dad1b | Dad1 | Dad1 | |
| Dad2 | Dad2 | weak | |
| Ndc80 complex | Ndc80b | Ndc80 | |
| Spindle/kinetochore | Bim1 | Bim1 | |
| Ipl1 complex | Sli15d | ||
| Ctf19 complex | (Mcm16b) | Mcm16 | |
This table only lists the proteins that are known to be involved in mitotic kinetochore or spindle. The rest of the dataset is included in the Supplemental Table 1.
Ndc80p as an activation domain hybrid appears as a mid-frequency false positive in the genome-wide two-hybrid assay and could not be reliably used to assess interactions with the Dam1 baits. Therefore, the results shown here reflect the combination of using Ndc80p as a DNA binding domain fusion and Dam1 proteins as activation domain hybrids.
Dad3 and Dad4, the two recently identified subunits of the Dam1 complex, were not included in the genome-wide array. Therefore, their two-hybrid interactions with Dam1p were not tested.
Two-hybrid interactions that had been reported in previous studies are referenced with superscript letters. The references are listed below.
Table notes: blank cells represent no interactions. “weak” indicates the protein shows weak interaction with wild-type or mutant Dam1p.
Sources of the identified interactions are as follows: a (Uetz et al., 2000); b (Ito et al., 2001); c (Hofmann et al., 1998); d (Kang et al., 2001). (MCM16b): This interaction was not detected in our two-hybrid screen.
Of the nine interacting spindle or kinetochore proteins identified, six are subunits of the Dam1 complex. The identification of two-hybrid interactions with multiple subunits of the Dam1 complex is consistent with the tight association of these proteins observed during Dam1 complex purification (Cheeseman et al., 2001a; Janke et al., 2002; Li et al., 2002), and with the interactions identified during previous genome-wide two-hybrid analyses (Ito et al., 2000; Uetz et al., 2000; Ito et al., 2001). It is important to note that not all of these interactions are necessarily direct; they may be bridged by other subunits of the Dam1 complex. Interactions were also identified with a number of other components of the mitotic spindle and the kinetochore. These proteins include Ndc80p, Sli15p, and Bim1p. Although the former two kinetochore proteins were reported to interact with Dam1p (Ito et al., 2001; Kang et al., 2001), the microtubule-associated Bim1p was reported to interact with Duo1p (Uetz et al., 2000; Ito et al., 2001), which in turn interacts with Dam1p. In summary, the present and previous two-hybrid studies (Table 3) indicate that the Dam1 complex makes numerous physical interactions at the spindle and kinetochore.
Direct Binding Assays for Dam1 Complex Protein–Protein Interactions
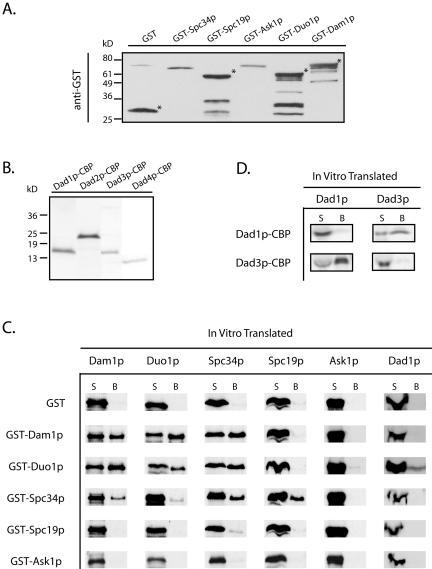
The genome-wide two-hybrid study with Dam1p as bait identified six subunits of the Dam1 complex. To determine which of these interactions reflects direct protein–protein interactions and to learn more about the subunit organization of the Dam1 complex, we tested for direct physical interactions among the nine subunits of this complex by using an in vitro binding assay (see MATERIALS AND METHODS). For this binding assay, one subunit in the form of a GST or CBP fusion protein (Figure 1, A and B) was bound to the glutathione-agarose (or calmodulin-Sepharose) beads and tested for its ability to pull down a second subunit that was translated in vitro. Purified GST protein was used as a control to ensure that the binding was not mediated through the GST portion of the fusion protein. As seen in Figure 1C, GST did not interact with any of the subunits of the Dam1 complex, although association with trace amounts of IVT-Spc34p and IVT-Spc19p was observed. The results for 81 in vitro binding reactions involving the nine subunits of the Dam1 complex, and the corresponding two-hybrid information, are shown in Figure 1, C and D, and summarized in Figure 2A. Five pairs of two-hybrid interactions were confirmed in reciprocal binding assays as were the self-interactions of Dam1p and Duo1p (Figure 1C). Other novel interactions identified included the self-interaction of Spc34p (Figure 1C), and the interaction between Dad1p and Dad3p, which was also confirmed reciprocally (Figure 1D). However, using this assay, we did not identify significant interactions for Ask1p, Dad2p, or Dad4p. The absence of interactions with Ask1p, Dad2p, and Dad4p might suggest that these proteins require multiple subunits to become active or to form a binding interface. This hypothesis is supported by the observation that Ask1p, Dad2p, and Dad4p seem to form a subcomplex when multiple subunits of the Dam1 complex are coexpressed (Miranda and Harrison, unpublished data).
Figure 1.
Purified GST and CBP fusion proteins used in this study and results of in vitro binding assays involving subunits of the Dam1 complex. (A) Western blots of the purified GST-fusion proteins. Top, purified GST-fusion proteins that are detected by anti-GST antibody and marked with asterisks. The lower bands in GST-Spc19p, GST-Duo1p and GST-Dam1p are degradation products. (B) Commassie staining of the purified Dad1p-CBP (15 kDa), Dad2p-CBP (19.5 kDa), Dad3p-CBP (14.8 kDa), and Dad4p-CBP (12.2 kDa) after separation on a 16% tricine gel. (C) Autoradiography of the pairwise binding reactions among the subunits of the Dam1 complex. The IVT proteins were labeled with [35S]methionine. The 35S signals in B fractions represent positive binding interactions. (D) Dad1p–Dad3p interactions confirmed in reciprocal binding assays. S, supernatant; B, glutathione-agarose beads.
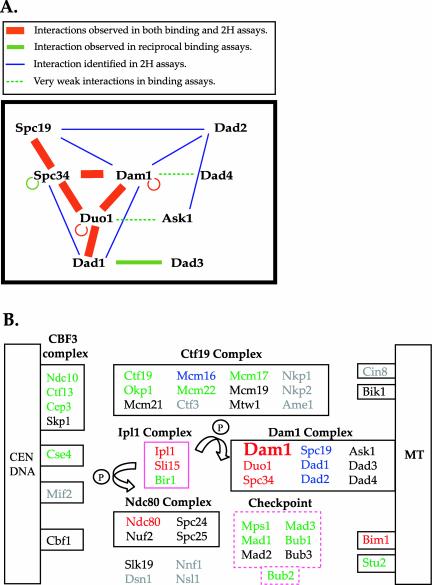
Figure 2.
Schematic representation of protein interactions involving the Dam1 complex and the kinetochore proteins. (A) Model of Dam1 complex organization reflecting interactions identified in this study and in previously published two-hybrid assays (Ito et al., 2000, 2001; Uetz et al., 2000). (B) Interactions between Dam1p of the Dam1 complex and other kinetochore and spindle proteins. Kinetochore components that belong to discrete subcomplexes are grouped in solid boxes and the regulatory proteins are boxed in pink. The checkpoint proteins are grouped in the pink dashed box to show their related functions, not to imply physical association. Proteins interacting with Dam1p are color coded in this figure: red, interacting proteins that are identified in both two-hybrid and binding assays; green, interacting proteins that are identified only in binding assays; blue, interacting proteins that are identified only in two-hybrid screens; black, proteins that did NOT show any interactions with Dam1p; gray, proteins that were not tested in binding assays. MT, microtubules.
Together with the two-hybrid data, these binding assays have identified multiple physical interactions within the Dam1 complex. From the two complementary analyses, we propose the following model for interactions within the Dam1p complex (Figure 2A). Strikingly, several proteins seem to make multiple connections with other subunits in the complex. This extensive network of interactions might generate the extremely tight association observed for subunits of this complex (Cheeseman et al., 2001a).
Components of Dam1 Complex Interact with Many Kinetochore, Spindle, and Regulatory Proteins
Previous studies indicated that the Dam1 complex functions in spindle integrity and is critical for the attachment of kinetochores to microtubules (Hofmann et al., 1998; Jones et al., 1999; Cheeseman et al., 2001a,b; Janke et al., 2002; Li et al., 2002). Both the spindle and the kinetochore are complex proteinaceous structures, and the functions of the Dam1 complex may require interactions with other proteins on the spindle and at the kinetochore. The two-hybrid data presented above, together with data derived from previous studies, suggest that the Dam1 complex may interact with the Ndc80 and the Ctf19 complexes at the kinetochore and interact with Bim1p along the spindle or at the kinetochore (Supplementary Table 2). As mentioned above, the two-hybrid information is lacking or incomplete for many kinetochore proteins (Supplementary Table 3, two-hybrid assays), and the two-hybrid analyses may not represent direct association of prey and bait proteins, especially because all of the kinetochore and spindle proteins are present in the nucleus. Therefore, we took a systematic approach to test for direct physical interactions between the Dam1 complex and 32 other kinetochore, spindle, and mitotic checkpoint-related proteins. Each protein was expressed by in vitro coupled transcription/translation and then tested for interactions in the binding assay. The binding assays were carried out as described above by using purified GST-Dam1p. The results for 32 protein combinations are shown in Figure 2B and in Supplementary Table 2 (the binding assays).
It has been demonstrated that Ndc80p is required for the association of the Dam1 complex with the kinetochore (Janke et al., 2002). In our binding assay, Ndc80p was the only protein in the four-subunit Ndc80 complex that showed binding with Dam1p (Figure 2B, Ndc80 complex). In contrast, multiple interactions between the Dam1 and Ctf19 complexes were observed using this assay. Of the eight proteins of the 12-subunit Ctf19 complex tested, four were able to bind to Dam1p (Figure 2B, Ctf19 complex). These data differ from those obtained in the two-hybrid studies in which only two interactions (Spc34p-Mcm22p and Dam1p-Mcm16p, see Supplementary Table 2) were identified between these two complexes. Nevertheless, our results are consistent with the fact that numerous genetic interactions between dam1-1 and mutants of the Ctf19 complex have been detected (Cheeseman et al., 2001a).
Surprisingly, despite the lack of previous reports of a physical interaction between the Dam1 complex and the DNA-binding CBF3 complex, we found that Ndc10p, Ctf13p, and Cep3p of the CBF3 complex were pulled down by GST-Dam1p (Figure 2B). Although a role for this interaction remains to be determined, a genetic interaction has been reported between ask1-3 and ndc10-1 (Li et al., 2002).
The in vitro binding assays also identified interactions between the Dam1 complex and other kinetochore or spindle proteins that did not belong to a discrete subcomplex (Figure 2B). The potential partners include the histone H3-like Cse4p, the microtubule-associated Bim1p, and Stu2p. Among these, Bim1p has been shown to interact with Dam1p in our two-hybrid screen.
Additional proteins that are localized at the kinetochore and that serve a regulatory role include the Ipl1 complex and the mitotic checkpoint proteins. At the restrictive temperature, duo1 and dam1 mutants showed a mitotic checkpoint-dependent G2/M arrest and there were genetic interactions between checkpoint mutants and both duo1 and dam1 mutants (Cheeseman et al., 2001b). Using the in vitro binding assays, we found that Dam1p interacted with four kinetochore-associated checkpoint proteins and with Bub2p (Figure 2B). The physical and genetic interactions between the Dam1 and Ipl1 kinase complexes have been well documented (Kang et al., 2001; Cheeseman et al., 2002a). In addition to confirming the binding between Dam1p and Ipl1p/Sli15p (Kang et al., 2001), we found that Dam1p bound directly to Bir1p, recently shown to be a component of the Ipl1 complex (Cheeseman et al., 2002a). Because there is a genetic interaction between bir1Δ and dam1-1 mutants, Bir1p may be involved in mediating the effects of Ipl1 kinase on the Dam1 complex.
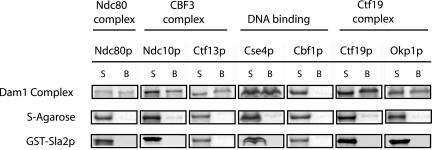
Intact Dam1 Complex Interacts with Components of the Ndc80, CBF3, and Ctf19 Complexes and with Cse4p
Although many physical interactions were identified as described above, one possibility is that the interactions between Dam1p and the Ndc80, CBF3, and Ctf19 complexes occurred because purified Dam1p has exposed domains that are not normally accessible to solvent when Dam1p is present in the Dam1 complex. Therefore, we purified the intact Dam1 complex from yeast extracts and tested it for interactions with components of the Ndc80, CBF3, and Ctf19 complexes. As shown in Figure 3, the Dam1 complex was able to pull down in vitro translated Ndc80p of the Ndc80 complex, Ndc10p and Ctf13p of the CBF3 complex, Okp1p and Ctf19p of the Ctf19 complex, and the histone H3-like Cse4p. The DNA-binding protein Cbf1p was included as a negative control; it did not bind to the Dam1 complex, similar to the results obtained using purified GST-Dam1p. We further demonstrated that the interactions were not due to nonspecific coiled-coil interactions because none of these proteins showed interactions with a cell cortex protein, Sla2p, which possesses a large coiled-coil region (Figure 3).
Figure 3.
Intact Dam1 complex binds to components of the Ndc80, CBF3, and Ctf19 complexes. Six proteins translated in vitro were pulled down by intact Dam1 complex immobilized on the S-agarose beads (top). None of these proteins associated with the S-agarose beads alone (middle) or with the coiled-coil rich GST-Sla2p (bottom). Cbf1p was included as a negative control because it did not bind to either GST-Duo1p or GST-Dam1p. Each binding reaction contained ∼2.6 nmol of Dam1 complex on the beads and was carried out in duplicate. S, supernatant; B, S-agarose beads.
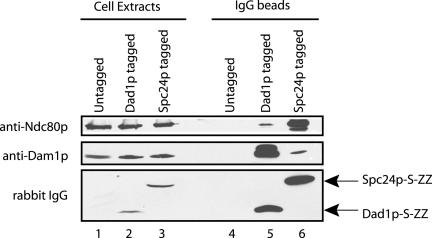
Interactions of Dam1 and Ndc80 Complexes in the Yeast Extracts
To further verify the physical interactions described above, we attempted to coimmunoprecipitate the Dam1 complex and interacting proteins from yeast extracts. However, Ndc80p was not found in the immunoprecipitates of Duo1p, Dam1p, Dad1p-13myc, Dad1p-GFP, and vice versa. These results were similar to the observation of Janke et al. (2002) who demonstrated that the Dam1 and Ndc80 complexes are discrete. Nonetheless, we found evidence for association of the Dam1 and Ndc80 complexes during early steps of their purification by affinity chromatography under low salt conditions (100 mM KCl). Cell extracts of a Dad1p-tagged strain (Dad1p-S tag-ZZ) were incubated with IgG-Sepharose to enrich for the Dam1 complex on beads. Although the majority of Ndc80p was in the flow through, we detected Ndc80p on the IgG beads in a Dam1 complex-dependent manner (Figure 4, lanes 4 and 5). Similar results were obtained in a reciprocal experiment by using cell extracts of an Spc24p-tagged strain. Although the Ndc80 complex was enriched on IgG beads, a small amount of Dam1p was also found associated with the beads. The association of Dam1p depended on the presence of the Ndc80 complex (Figure 4, lanes 4 and 6) and was disrupted under high salt conditions (300 mM KCl) (our unpublished data).
Figure 4.
Coprecipitation of the Dam1 complex and the Ndc80 complex from cell extracts. Ndc80p coprecipitated with the Dam1 complex on IgG-Sepharose beads in the cell extracts of a Dad1p-tagged strain (Dad1p-S tag-ZZ), but not in the cell extracts of an untagged strain. Similarly, Dam1p coprecipitated with the Ndc80 complex on IgG beads in the cell extracts of an Spc24p-tagged strain (Spc24p-S tag-ZZ), but not in the cell extracts of an untagged strain. Cell extracts from all three strains were prepared under identical conditions as described in the purification of Dam1 and Ndc80 complexes (see MATERIALS AND METHODS), except that the cell extracts were maintained at 100 mM KCl while incubating with IgG-Sepharose beads. The beads were then washed extensively with lysis buffer containing 100 mM KCl. The cell extracts and IgG precipitates were separated on 10% SDS-PAGE gels for immunoblotting with anti-Dam1p antibodies or rabbit IgG, and on an 8.5% SDS-PAGE gel for immunoblotting with anti-Ndc80p antibodies. Rabbit IgG was used to detect the ZZ domain of protein A present in the Dad1p and Spc24p tandem affinity purification tags.
A Genome-Wide Two-Hybrid Screen with dam1(SA) and dam1(SD) Mutants as Baits
With at least 40 proteins at the kinetochore and a web of potential interactions that exist among them, it is important to not only identify interactions among the proteins but also to determine how the interactions are regulated. To determine whether Dam1p phosphorylation affects any of the physical interactions of the Dam1 complex described above, we conducted a second genome-wide two-hybrid screen by using dam1 phosphorylation site mutants. For these studies, we used a dam1 mutant, which has all of the known Ipl1p phosphorylation sites removed (S20A, S257A, S265A, S292A; designated as S to A), and a dam1 mutant, which mimics the constitutively phosphorylated state (S20D, S257, S265D, S292D; designated as S to D) (Cheeseman et al., 2002a), as the DNA-binding hybrid, and screened against a library of ∼6000 yeast open reading frames, each expressed as an activation domain fusion protein (Table 3). Previous studies on the Dam1 complex purified from yeast extracts suggested that its subunit composition was unaffected by the phosphorylation state (Cheeseman et al., 2001). Consistent with this conclusion, dam1p (S to A) showed the same physical interactions with subunits of the Dam1 complex as the wild-type Dam1p. dam1p (S to D) also showed a similar range of interactions; however, the strength of the interactions with Dad2p and Spc34p seemed reduced compared with wild type. These results suggest that the physical interactions within the Dam1 complex remain largely unchanged in vivo regardless of phosphorylation state, although interactions with Spc34p and Dad2p may be altered.
In vivo, Ipl1p associates in a trimeric complex with the inner centromere protein (INCENP)-related protein Sli15p and the survivin-like protein Bir1p (Cheeseman et al., 2002). Interestingly, although we observed direct binding interactions between Dam1 and all three components of the Ipl1 complex, we only found two-hybrid interactions between wild-type Dam1p and Sli15p. In addition, although wild-type Dam1p interacted strongly with Sli15p, neither dam1p (S to A) nor dam1p (S to D) showed an interaction with Sli15p.
Finally, we also identified a variety of physical interactions between Dam1p and components of the kinetochore and the mitotic spindle including Ndc80p, Bim1p, and Mcm16p. Interestingly, all of these interactions were affected by phosphorylation state. Bim1p failed to interact with the S to D form of Dam1p, whereas Mcm16p interacted specifically with dam1p (S to D), but not with wild-type Dam1p or dam1p (S to A). Ndc80p as an activation domain hybrid occurs as a mid-frequency false positive in the genome-wide two-hybrid assay and could not be reliably used to assess interactions with the Dam1p DNA-binding domain mutants. Therefore, we generated a fusion of the DNA-binding domain and Ndc80p. Under this condition, Ndc80p showed positive interactions with both the wild-type and S to A form of Dam1p, but not with the S to D form of Dam1p. In summary, these results suggest that the phosphorylation state of the Dam1 complex may function in part to control physical interactions with other proteins.
Phosphorylation Affects Dam1p Interactions In Vitro
The two-hybrid screen with dam1 phosphorylation site mutants suggested that a subset of physical interactions made by Dam1p are affected by its phosphorylation state. We sought to verify these effects by using in vitro binding assays. We first tested the phosphorylation dependency of the Dam1p–Sli15p and Dam1p–Mcm16p interactions as suggested by the two-hybrid results. In contrast to our two-hybrid results (Table 3), we found that IVT-Sli15p bound to both the wild-type GST-Dam1p and the S to D mutant with the same affinity (Supplementary Figure 1B) and that IVT-Mcm16p bound to neither the wild-type nor the S to D mutant of Dam1p (Supplementary Figure 1C).
Next, we tested the interaction between purified intact Ndc80 complex and IVT wild-type Dam1p, dam1p (S to A), and dam1p (S to D) mutants. Strikingly, with 3 nmol of Ndc80 complex bound to the S-agarose beads, wild-type Dam1p showed at least 5.5 times greater binding to Ndc80 complex than did the dam1p (S to D) mutant (Figure 5). With half of the amount of Ndc80 complex on the beads, the dam1p (S to D) mutant showed a twofold reduction in binding compared with wild-type Dam1p (our unpublished data). On the other hand, the dam1p (S to A) mutant consistently showed virtually identical binding as the wild type Dam1p (Figure 5). These binding data support the conclusion that constitutive Dam1p phosphorylation weakens the interaction between Dam1p and Ndc80p.
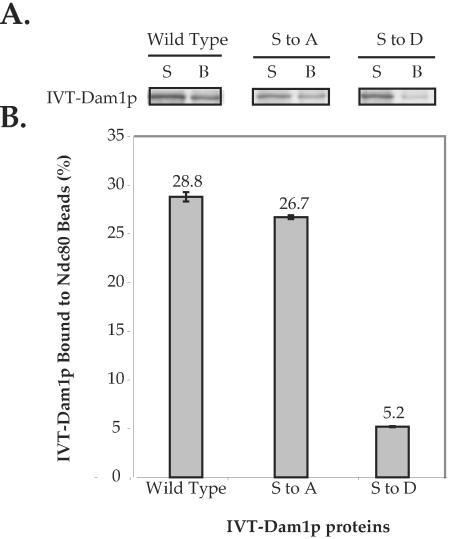
Figure 5.
Intact Ndc80 complex shows reduced binding to dam1p (S to D). Intact Ndc80 complex immobilized on S-agarose beads bound similar amounts of in vitro translated wild-type Dam1p and dam1p (S to A), but showed much weaker binding to dam1p (S to D). Each binding reaction contained ∼3 nmol of Ndc80 complex on the beads purified from 16 g of yeast cells and was carried out in duplicate samples. The IVT Dam1 proteins did not bind to S-agarose beads alone. (A) Autoradiography shows the IVT Dam1 proteins in supernatant and bead fractions in each binding reaction. The quantification of 35S signal in the bead fraction is shown in the corresponding bar graph in B. S, supernatant; B, S-agarose beads. (B) Bar graph shows the percentage of IVT Dam1p bound to Ndc80 complex-associated beads.
Furthermore, we compared the binding affinity between IVT-Ndc80p and wild-type GST-Dam1p and the S to D mutant. Using 0.14 μM of the GST fusion proteins, wild-type Dam1p bound twice as much IVT Ndc80p as the S to D mutant (Supplementary Figure 1A). We also tested the phosphorylation-dependent interaction between Dam1p and Spc34p, another protein that showed decreased interaction with dam1p (S to D) mutant in our two-hybrid screen. Similar to Ndc80p, more IVT Spc34p was pulled down by wild-type Dam1p than by the S to D mutant when the concentration of GST fusion proteins was below 1 μM (Supplementary Figure 1A). To ensure that the differences in the binding affinities with Ndc80p and Spc34p were due to the phosphorylation site mutations and were not due to conformational changes induced by multiple mutations in Dam1p, we tested the interaction between Dam1p and Duo1p, which was not affected by those mutations in the two-hybrid screen. We found that IVT-Duo1p bound to both the wild-type GST-Dam1p and the S to D mutant to the same extent at all concentrations tested (Supplementary Figure 1B). Therefore, we conclude that the binding affinity between Dam1p and Ndc80p, and between Dam1p and Spc34p, is specifically reduced by Dam1p phosphorylation. These data corroborate our two-hybrid findings. The observation that the dam1 (S to D) mutant exhibited a much decreased, but not completely abolished, interaction with Ndc80p is consistent with the observation that dam1 (S to D) mutant prey sometimes showed a detectable interaction with the NDC80 bait in the absence of 3 mM 3-amino-1,2,4-triazole (our unpublished data). This result also corresponds well with the fact the dam1 (S to D) mutant showed poor growth, rather than a lethal mutant phenotype (Cheeseman et al., 2002a), suggesting that the kinetochore-microtubule connection is not completely disrupted.
The constitutive phosphorylation of Dam1p results in weaker Dam1p–Ndc80p and Dam1p–Spc34p interactions and slower growth. We predict that this effect would be exacerbated by Ipl1p phosphorylation of Ndc80p and Spc34p, which have also been demonstrated to be Ipl1 targets in vivo (Cheeseman et al., 2002a). Indeed, a severe synthetic growth defect is observed when dam1 (S257D, S265D, S290D) is combined with an ndc80 constitutive phosphorylation mutant (Kang and Chan, unpublished data). Moreover, we also found synthetic lethality between spc34 (T199D) and dam1 (S257D, S265D, S292D) mutants. Together, these data suggest that the physical interactions between Dam1 and Ndc80 complexes, and the interactions between subunits of the Dam1 complex, are crucial for cell viability and that these interactions are regulated by the dynamic cycles of phosphorylation and dephosphorylation.
Phosphorylation Affects Dam1p Interaction with the Kinetochore In Vivo
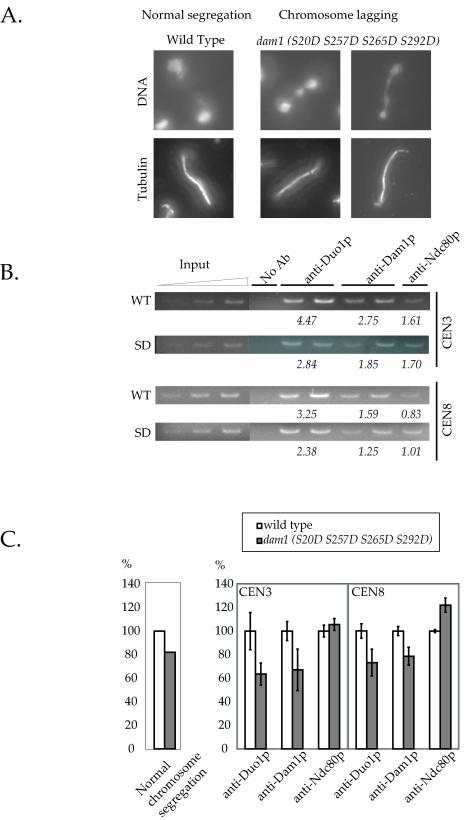
The in vivo consequence of Dam1p phosphorylation was previously demonstrated by a chromosome lagging phenotype in the dam1 (S20D S257D S265D) mutant, which suggested a deficient chromosome-microtubule attachment (Cheeseman et al., 2002a). We found that this chromosome lagging phenotype was even more dramatic in the dam1 (S20D S257D S265D S292D) mutant (Figure 6A). Approximately 20% of the cells in an asynchronous culture showed this phenotype (Figure 6C, left). To test whether the defects in the attachment are due to the weakened interaction between the Dam1 complex with the dam1p (S to D) mutant and the Ndc80 complex, we conducted chromatin immunoprecipitation analysis, comparing the amount of Dam1 complex associated with the centromeres in the wild-type and the dam1 (S20D S257D S265D S292D) mutant (Figure 6B). Strikingly, the amount of Dam1 complex associated with the centromeres in the dam1 (S20D S257D S265D S292D) mutant was reduced by 25–35% compared with that in the wild type (Figure 6, B and C, anti-Duo1p and anti-Dam1p). In contrast, the amount of Ndc80p associated with the centromeres was unchanged in the wild type and the mutant (Figure 6, B and C, anti-Ndc80p). Together, these data strongly suggest that phosphorylation of the Dam1 complex weakens its interaction with the kinetochore, most likely through decreased binding between Dam1p and Ndc80p.
Figure 6.
The dam1p (S to D) mutant shows reduced interaction with centromeres. (A) The chromosome lagging phenotype of dam1 (S20D S257D S262D S292D) mutant was quantified. Wild-type and mutant cells were grown to log phase at 25°C and processed for tubulin immunofluorescence microscopy and DNA staining. Cells (200) were counted for each strain. The result is shown in (C, left bar graph). (B) Association of the Dam1 complex with centromeres in wild-type and dam1 (S20D S257D S262D S292D) mutant cells was examined by chromatin immunoprecipitation analysis. Centromeres from chromosome 3 and chromosome 8 were amplified by PCR from total input chromatin (input), mock-treated no antibody controls (no antibody), and duplicated Duo1p, Dam1p, and Ndc80p immunoprecipitates (only one Ndc80p sample is shown herein). The PCR products were quantified. The values underneath the gel lanes correspond to the percentage of total centromeric DNA that is immunoprecipitated with the indicated antibody. The values are the average from duplicate samples. (C, right bar graph) Comparison of centromeric DNA immunoprecipitated in wild-type and dam1 (S20D S257D S265D S292D) mutant. The amount of centromeric DNA immunoprecipitated from the mutant is expressed as the percentage of that from the wild-type.
DISCUSSION
Mechanism of Phosphoregulation of the Yeast Kinetochore
Previous studies have demonstrated that the Ipl1 kinase is a key regulator of the budding yeast kinetochore. Specifically, it facilitates the establishment of kinetochore biorientation by promoting turnover of kinetochore-microtubule connections. It has been hypothesized that Ipl1p causes frequent detachment of kinetochores from microtubules in the absence of tension (Tanaka et al., 2002). Because the Dam1 and Ndc80 complexes have been identified as in vivo targets of Ipl1p (Cheeseman et al., 2002a) and because the phosphorylation state of the Dam1 complex does not affect its microtubule binding activity, one possible mechanism of kinetochore detachment would be to disrupt a subset of protein–protein interactions within the kinetochore, thereby leading to the detachment of kinetochores from spindle microtubules (Cheeseman et al., 2002a). Four lines of evidence support the conclusion that the physical interactions between Dam1p and Ndc80p are weakened by Ipl1 kinase-mediated phosphorylation. First, we found the two-hybrid interaction between Dam1p and Ndc80p was greatly reduced or abolished when the dam1 (S to D) mutant (which mimics the constitutively Ipl1p phosphorylated state) was used as a prey in combination with the NDC80 bait. Second, we demonstrated that purified intact Ndc80 complex bound more tightly to wild-type Dam1p than to the dam1p (S to D) mutant. Third, the dam1 (S to D) mutant shows a severe synthetic growth defect in combination with an ndc80 (S to D) mutant (Kang and Chan, unpublished data). And finally, mutant dam1p (S to D) showed reduced association with the centromeres that corresponds well with the chromosome lagging phenotype, whereas the Ndc80p-centromere association remained unchanged.
Although the possibility exists that the differential interaction was due to conformational changes induced in Dam1p by multiple phosphorylation site mutations, three factors lead us to conclude that this is not the case. First, we found that the amount of IVT Ndc80p that bound to GST-Dam1p was also reduced by the addition of Ipl1p (our unpublished data). Second, the two-hybrid and the in vitro binding assays demonstrated that the differential physical interactions of the dam1p mutant were observed only with specific partners (i.e., Ndc80p and Spc34p, but not Duo1p). Third, the phosphorylation site mutants cause very specific chromosome segregation defects, in contrast to other loss of function dam1 alleles, which show both aberrant spindle structures and chromosome missegregation phenotypes (Cheeseman et al., 2001b, 2002a). This suggests that the dam1 phosphorylation sites mutants are functional for a subset of Dam1p activities. Therefore, the data presented above strongly support our hypothesis that phosphorylation of essential kinetochore components, such as the Dam1 and Ndc80 complexes, weakens their physical interactions and therefore results in the detachment of the kinetochore from the microtubule (Figure 7).
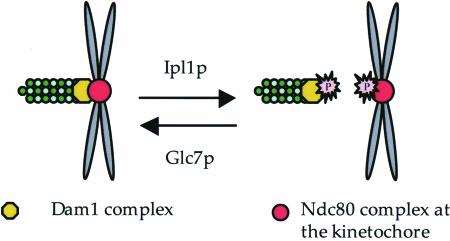
Figure 7.
A model for phosphoregulation at the yeast kinetochore. Interactions between the Dam1 complex and the Ndc80 complex are proposed to be weakened when Dam1p and Ndc80p are phosphorylated by Ipl1p, facilitating formation of bipolar connections between chromosomes and the spindle.
Our previous published work showed that dam1 S to D mutants partially suppress the ipl1-2 temperature sensitivity and kinetochore structure defects (Cheeseman et al., 2002a). These results supported our conclusion that Dam1p is a bona fide downstream target of the Ipl1 kinase. Our current model cannot fully account for how dam1 constitutive phosphorylation mutations suppress the ipl1-2 kinetochore assembly defect, because we postulate herein that phosphorylation weakens kinetochore subunit interactions. Here is a possible scenario. The loading of kinetochore proteins at centromeres is partially dependent on microtubule interactions (Enquist-Newman et al., 2001; Li et al., 2002). Furthermore, establishing new spindle interactions requires breaking old interactions, and this is mediated by Ipl1 kinase phosphorylation of the Dam1 complex. Therefore, Ipl1p phosphorylation may in some cases promote kinetochore assembly by facilitating formation of new kinetochore attachments.
An additional mechanism by which the Ipl1p kinase may affect the yeast kinetochore might be through regulation of the assembly of the Dam1 complex. The Dam1p–Spc34p interaction was weakened by Ipl1p-specific phosphorylation in both the two-hybrid screen and the in vitro binding assay. In addition, the growth defect caused by constitutive Dam1p phosphorylation is aggravated by constitutive Spc34p phosphorylation, as seen by the synthetic lethality between dam1 (S257D, S265D, S292D) and spc34 (T199D) mutants. Because the subunit composition of purified Dam1 complex is unaffected by the phosphorylation state (Cheeseman et al., 2001a), it is possible that the assembly of the entire Dam1 complex is compromised. Alternatively, the weakened Dam1p–Spc34p interaction might result in conformational change in the Dam1 complex that in turn affects the interaction between the Dam1 complex and other kinetochore components.
Dam1p Interactions Identified in Both In Vitro Binding and Two-Hybrid Assays
To elucidate the protein–protein interactions that underlie the mechanism of chromosome-microtubule attachment at the yeast kinetochore, we constructed a protein interaction map focusing on the microtubule-binding Dam1 kinetochore complex. By two complementary approaches, two-hybrid analysis and in vitro binding assays, we identified interactions among the subunits of the Dam1 complex, and interactions between the Dam1 complex and other kinetochore or spindle components. We also demonstrated the association of the Dam1 and Ndc80 complexes in yeast extracts.
Six interacting partners for Dam1p were common to both assays: Duo1p, Dam1p, Spc34p, Ndc80p, Sli15p, and Bim1p (Figure 2B, highlighted in red). As discussed above, the Ndc80p–Dam1p and the Spc34p–Dam1p interactions seem to be regulated by phosphorylation. Duo1p represents an internal positive control for all of the assay conditions because its interaction with Dam1p has been well-documented using both in vitro and two-hybrid assays (Hofmann et al., 1998; Ito et al., 2000, 2001; Uetz et al., 2000). Because the Dam1 complex was shown to contain a single copy of each subunit (Cheeseman et al., 2001a), the self-interaction of Dam1p raises the possibility that this complex dimerizes. Whether dimerization might be important for the role of the Dam1 complex at the spindle, the kinetochore, or both, remains to be determined. Other Dam1p binding partners include the Sli15p subunit of the Ipl1 complex and Bim1p, a microtubule-associated protein. The Dam1p–Sli15p interaction may play an important role in targeting the Ipl1 kinase to the Dam1 complex. Because Bim1p has been implicated in kinetochore function based on its genetic interactions (Tong et al., 2001), it will be important to determine whether its interaction with Dam1p occurs at the kinetochore, along the spindle, or at both locations.
In Vitro Binding Assays Identified Novel Partners of the Dam1 Complex
Many subunits of the Ctf19 and CBF3 complexes, as well as other kinetochore proteins, did not show positive interactions in any of the high-throughput two-hybrid screens, or in our two-hybrid screens (Supplementary Table 3). This lack of two-hybrid interactions might represent the false negatives in these screens, resulting from the lack of expression of fusion proteins in the array, or a lack of functionality for the fusion proteins (Drees et al., 2001). We therefore tested for direct physical associations involving these proteins and included >90% (41 proteins) of the known kinetochore components in our Dam1p-binding assays. Surprisingly, we found that the inner kinetochore (DNA-binding) proteins Ndc10p, Ctf13p, Cep3, and Cse4p interact with Dam1p in vitro (Figure 2B). The Dam1 complex is thought to function at the outer kinetochore based on its ability to bind to microtubules and based on its lack of two-hybrid interactions with the inner kinetochore proteins. To rule out the possibility that the binding occurred because Dam1p did not exist in its native complex, we demonstrated that intact Dam1 complex also interacted with Ndc10p, Ctf13p, and Cse4p. In light of these binding results, it is possible that the Dam1 complex associates with the CBF3 complex directly in vivo. This association might occur at the kinetochore to stabilize the linkage between the inner and outer kinetochore in parallel with the Ctf19 complex, or along the anaphase spindle, or at the spindle midzone, where Ndc10p and the Dam1 complex have been observed (Goshima and Yanagida, 2000; Tanaka et al., 2002; Buvelot et al., 2003). Undoubtedly, much more work is needed before we fully understand how the kinetochore is assembled and regulated.
The binding assays also identified interactions between the Dam1 complex and several checkpoint proteins, such as Mps1p, Mad1p, Mad3p, Bub1p, and Bub2p. These checkpoint proteins (except Bub2p) are thought to be recruited to the kinetochore upon activation of the spindle assembly checkpoint. However, only one two-hybrid interaction, Mad1p-Spc25p (Newman et al., 2000), has previously been found between checkpoint and kinetochore proteins (see Supplementary Table 3). Herein, we presented evidence for additional potential links for the targeting of checkpoint proteins to the kinetochore. Further investigation is required to determine whether or how these interactions might contribute to the checkpoint signaling.
In conclusion, numerous protein–protein interactions at the kinetochore have been identified in our systematic study. It is now important to test their individual contributions to kinetochore function and to determine whether these interactions are spatially and/or temporally regulated.
Supplementary Material
Acknowledgments
We thank C. Chan, J. Kang, S.C. Harrison, and J.J. Miranda for generously sharing unpublished results; M. Duncan for purified GST protein; Y. Sun for purified GST-Sla2p protein; A. Desai for the anti-Ndc80p antibody; and L. Hsu and M. Schlissel for assistance with the PhosphorImager. We also thank J. Wong, S. Westermann, and Y. Nakajima for discussions and critical reading of the manuscript; and A. Engqvist-Goldstein, C. Zhang, and Y. Sun for discussions and advice. This work was supported by a grant to G. Barnes from the National Institute of General Medical Sciences (GM-47842) and a grant to the Yeast Resource Center from the National Center for Research Resources of the National Institutes of Health (P41 RR11823). S.F. is an investigator of the Howard Hughes Medical Institute.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-11-0765. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-11-0765.
Abbreviations used: GST, glutathione S-transferase; IVT, in vitro-coupled transcription/translation.
The online version of this article contains supplementary tabular material. Online version is available at www.molbiolcell.org.
References
- Burke, D., Dawson, D., and Stearns, T. (2000). Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Buvelot, S., Tatsutani, S.Y., Vermaak, D., and Biggins, S. (2003). The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, C.S.M., and Botstein, D. (1993). Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics 135, 677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., Anderson, S., Jwa, M., Green, E., Kang, J.-S., Yates, J.R., Chan, C.S.M., Drubin, D.G., and Barnes, G. (2002a). Phosphoregulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111, 163–172. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., Brew, C., Wolyniak, M., Desai, A., Anderson, S., Muster, N., Yates, J.R., Huffaker, T.C., Drubin, D.G., and Barnes, G. (2001a). Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 155, 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., Drubin, D.G., and Barnes, G. (2002b). Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157, 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., Enquist-Newman, M., Müller-Reichert, T., Drubin, D.G., and Barnes, G. (2001b). Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 152, 197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees, B.L., et al. (2001). A protein interaction map for cell polarity development. J. Cell Biol. 154, 549–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist-Newman, M., Cheeseman, I.M., Van Goor, D., Drubin, D.G., Meluh, P., and Barnes, G. (2001). Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell 12, 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and Yanagida, M. (2000). Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100, 619–633. [DOI] [PubMed] [Google Scholar]
- Hofmann, C., Cheeseman, I.M., Goode, B.L., McDonald, K.L., Barnes, G., and Drubin, D.G. (1998). Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol. 143, 1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. [Comment in Proc. Natl. Acad. Sci. USA 98, 4277–4278 UI: 21192614]. Proc. Natl. Acad. Sci. USA 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, T., Tashiro, K., Muta, S., Ozawa, R., Chiba, T., Nishizawa, M., Yamamoto, K., Kuhara, S., and Sakaki, Y. (2000). Toward a protein-protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl. Acad. Sci. USA 97, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., Ortiz, J., Lechner, J., Shevchenko, A., Magiera, M.M., Schramm, C., and Schiebel, E. (2001). The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20, 777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., Ortiz, J., Tanaka, T.U., Lechner, J., and Schiebel, E. (2002). Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.H., Bachant, J.B., Castillo, A.R., Giddings, T.H., and Winey, M. (1999). Yeast Dam1p is required to maintain spindle integrity during mitosis and interacts with the Mps1p kinase. Mol. Biol. Cell 10, 2377–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.-s., Cheeseman, I.M., Kallstrom, G., Velmurugan, S., Barnes, G., and Chan, C.S.M. (2001). Functional cooperation of Dam1, Ipl1, and the inner centromere protein (IN.C.E.N.P)-related protein Sli15 during chromosome segregation. J. Cell Biol. 155, 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner, J., and Carbon, J. (1991). A 240-kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64, 717–726. [DOI] [PubMed] [Google Scholar]
- Li, Y., Bachant, J., Alcasabas, A.A., Wang, Y., Qin, J., and Elledge, S.J. (2002). The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16, 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, D.A., Marshall, T.K., and Deschenes, R.J. (1993). Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast 9, 715–722. [DOI] [PubMed] [Google Scholar]
- Newman, J.R.S., Wolf, E., and Kim, P.S. (2000). A computationally directed screen identifying interacting coiled coils from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97, 13203–13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, J., and Lechner, J. (2000). The budding yeast kinetochore: less simple than expected. Protoplasma 211, 12–19. [Google Scholar]
- Ortiz, J., Stemmann, O., Rank, S., and Lechner, J. (1999). A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13, 1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peranen, J., Rikkonen, M., Hyvoenen, M., and Kaariainen, L. (1996). T7 vectors with modified T7/lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 236, 371–373. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., Seraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- Rodal, A.A., Tetreault, J.W., Lappalainen, P., Drubin, D.G., and Amberg, D.C. (1999). Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 145, 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, K.B., and Salmon, E.D. (2002). Chromosome dynamics: new light on Aurora B kinase function. Curr. Biol. 12, R458–R460. [DOI] [PubMed] [Google Scholar]
- Tanaka, T.U., Rachidi, N., Janke, C., Pereira, G., Galova, M., Schiebel, E., Stark, M.J.R., and Nasmyth, K. (2002). Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108, 317–329. [DOI] [PubMed] [Google Scholar]
- Tong, A.H.Y., et al. (2001). Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364–2368. [DOI] [PubMed] [Google Scholar]
- Uetz, P., et al. (2000). A. comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Wigge, P.A., and Kilmartin, J.V. (2001). The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152, 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.