Abstract
Kinesin-I is essential for the transport of membrane-bound organelles in neural and nonneural cells. However, the means by which kinesin interacts with its intracellular cargoes, and the means by which kinesin–cargo interactions are regulated in response to cellular transport requirements are not fully understood. The C terminus of the Drosophila kinesin heavy chain (KHC) was used in a two-hybrid screen of a Drosophila cDNA library to identify proteins that bind specifically to the kinesin tail domain. UNC-76 is an evolutionarily conserved cytosolic protein that binds to the tail domain of KHC in two-hybrid and copurification assays, indicating that kinesin and UNC-76 form a stable complex in vivo. Loss of Drosophila Unc-76 function results in locomotion and axonal transport defects reminiscent of the phenotypes observed in kinesin mutants, suggesting that UNC-76 is required for kinesin-dependent axonal transport. Unc-76 exhibits dosage-sensitive genetic relationships with Khc and Kinesin light chain mutations, further supporting the hypothesis that UNC-76 and kinesin-I work in a common transport pathway. Given the interaction of FEZ1, the mammalian homolog of UNC-76, with protein kinase Cζ, and the role of FEZ1 in axon outgrowth, we propose that UNC-76 helps integrate kinesin activity in response to transport requirements in axons.
INTRODUCTION
Kinesin-I is a plus-end–directed microtubule motor that facilitates the movement of vesicles, messenger ribonucleo-proteins (mRNPs), and organelles. It was first identified as a protein in squid axoplasm that facilitates ATP-dependent vesicle movement along microtubules (Brady, 1985; Vale et al., 1985). Subsequent molecular, genetic, and biochemical studies have shown that kinesin-I is required for intracellular transport in eukaryotes in many cellular contexts (reviewed in Martin et al., 1999b; Goldstein, 2001; Stebbings, 2001). The requirement for kinesin-based transport is especially acute in elongated cell types, such as neurons, in which long-distance transport from the cell body to the synapse must occur. Loss of function of either kinesin subunit results in transport defects in the nervous system, including organelle aggregation, defects in synapse formation, and electrophysiological defects (Saxton et al., 1991; Gho et al., 1992; Hurd and Saxton, 1996; Gindhart et al., 1998).
Kinesin-I is the founding member of a superfamily of motor proteins that share a conserved mechanochemical domain with microtubule-stimulated ATPase activity, but have different cargo binding domains that are specialized for specific cellular functions (Vale and Fletterick, 1997; Hirokawa, 1998; Goldstein and Philip, 1999). For example, the kinesin-I tail domain is composed of two light chains and the C-terminal region of two heavy chains. Biochemical studies and sequence motif predictions helped assemble a model of kinesin–cargo interactions in which protein–protein interaction motifs in the kinesin tail domain bind to proteins on the surface of vesicles, mitochondria, and mRNPs (reviewed in Kamal and Goldstein, 2002; Karcher et al., 2002). In addition to its role in cargo attachment, the kinesin tail domain is also required for negative regulation of the basal ATPase activity of the motor domain (Coy et al., 1999; Stock et al., 1999; Hackney and Stock, 2000). By identifying proteins that bind to the kinesin tail domain, it is possible to understand the nature of kinesin–cargo interactions and how kinesin activity is regulated in response to signal transduction pathways, thus modifying kinesin activity to meet cellular transport requirements.
A number of genetic and biochemical studies have begun to unravel the protein–protein interactions required for kinesin-dependent transport events. These proteins fall into three classes: transmembrane proteins on the vesicle surface that are cargo-bound receptors, scaffold proteins that indirectly link kinesin to cargos, and regulatory proteins that phosphorylate the kinesin tail domain or, like hsp70, remove kinesin from the cargo surface (reviewed in Verhey et al., 2001; Kamal and Goldstein, 2002; Karcher et al., 2002). However, genetic studies have been completed for only two kinesin-associated proteins (KAPs). Sunday driver (SYD) and UNC-16 are the Drosophila and Caenorhabditis elegans homologs of mammalian JIP3/JSAP1, a scaffold protein that binds vesicles, KLC, and components of the jun N-terminal protein kinase (JNK) signaling cascade (Bowman et al., 2000; Byrd et al., 2001); therefore, SYD and its homologs may coordinate regulation of vesicle transport by kinesin with JNK signaling. Amyloid precursor protein-like (APPL) is the Drosophila homolog of amyloid precursor protein (APP), a vesicle protein that, when mutated, causes familial Alzheimer's disease (Luo et al., 1990; Luo et al., 1992). APPL and APP are kinesin cargo receptors that bind to KLC to facilitate vesicle transport (Torroja et al., 1999; Kamal et al., 2000; Gunawardena and Goldstein, 2001; Kamal et al., 2001).
We have completed yeast two-hybrid screens for Drosophila proteins that bind to the kinesin tail domain. One of these proteins, UNC-76, binds to the KHC tail domain, and mutational analysis shows that UNC-76 is essential for axonal transport in the Drosophila nervous system. These results support a model in which the interaction of UNC-76 and kinesin is essential for intracellular transport. UNC-76 homologs are essential for nervous system function in C. elegans, and the mammalian UNC-76 homolog FEZ1 is a direct target of protein kinase C (PKC)ζ-dependent signaling and axon outgrowth (Bloom and Horvitz, 1997; Kuroda et al., 1999). We propose that UNC-76 may directly or indirectly facilitate the regulation of kinesin activity in vivo.
MATERIALS AND METHODS
Yeast Two-Hybrid Analysis
Yeast growth conditions and manipulations were done using established protocols (Ausubel et al., 1997; Parchaliuk et al., 1999). The LexA bait vector pEG202, prey vector pJG4-5, and lacZ reporter plasmid pSH18-34 (Zervos et al., 1993) were used in yeast strain EGY191 (MATα trp1, his3, ura3, and lexAops-LEU2) to express fragments of kinesin heavy chain (KHC) and kinesin light chain (KLC). The bait fusion pEG-KST, containing aa 675–975 of Drosophila KHC, was used to screen the RFLY1 (Finley et al., 1996) Drosophila embryonic cDNA library. Putative KHC interactors were characterized by polymerase chain reaction (PCR) amplification by using primers BCO1 and BCO2 (Ausubel et al., 1997) and restriction fragment length polymorphism mapping of cDNA inserts by using HinfI and HaeIII, and controls were performed to demonstrate that observed interactions were bait and prey specific. The pEG-stalk and pEG-tail constructs encode aa 675–850 and 850–975 of Drosophila KHC, respectively. They were PCR amplified from pJG-KST (Bowman et al., 2000) by using the primers KHC stalk (5′ CCGGCTCGAGCTGGCGGAGTGATCCACCGTCCTC 3′) and BCO1 to amplify the pEG-stalk insert, and KHC tail (5′ GGCCGAATTCATCCGAAAGAATGTCGTAAACGAG 3′) and BCO2 to amplify the pEG-tail insert; PCR products were subcloned into the EcoRI and XhoI sites of pEG202.
Sequence Analysis
Clones representing each restriction fragment length polymorphism class were sequenced by the University of Massachusetts Boston Sequencing Facility. BLASTN comparison of interactor KAPH4 to the Drosophila genome database showed that KAPH4 encodes aa 277–474 of the annotated Drosophila gene CG3981. The cDNA for CG3981 corresponds to expressed sequence tag project clone LD08195 (GenBank accession no. AY069376), which is 2963 base pairs in length and encodes a 474 amino acid polypeptide. Alignment of Drosophila UNC-76, C. elegans UNC-76, and FEZ1 was performed using ClustalW 1.8 (Jeanmougin et al., 1998) and Boxshade 3.21. Secondary structure and sequence motif prediction were performed using COILS (Lupas et al., 1991), Garnier secondary structure prediction (Garnier et al., 1978), PSORT (Nakai and Horton, 1999), InterPro (Apweiler et al., 2001), and PhosphoBase (Kreegipuu et al., 1999).
In Vitro Association of Unc-76 and Kinesin
Epitope-tagged versions of UNC-76 were generated by subcloning the fragment of UNC-76 encoded by the prey plasmid pJG-H4 (aa 277–474; pET-H4) and full-length UNC-76 (pET-UNC-76) into the bacterial expression vector pET30a (Novagen, Madison, WI). The encoded fusion protein contains a 6xHIS-tag for protein purification and an S-tag for fusion protein detection on Western blots. Both fusion proteins were soluble and stable under standard bacterial expression and purification conditions (QIAGEN, Valencia, CA).
His-tag copurification assays were done according to Micheva et al. (1997) with some modifications. Approximately 300 wild-type (ORE-R) or MYC-KLC (Gindhart et al., 1998) flies were homogenized in NP-40 buffer (150 mM NaCl, 1% NP-40, 50 mM Tris-Cl, pH 8.0, 100 μg/ml phenylmethylsulfonyl fluoride) on ice. The homogenate was centrifuged 2 × 10 min at 10,000 × g in an Eppendorf microcentrifuge at 4°C to pellet insoluble debris. The crude supernatant was incubated for 30 min with 25 μl of Ni2+-NTA agarose beads (QIAGEN) at 4°C with rocking to preclear the supernatant. The beads were pelleted by centrifuging for 2 s at 10,000 × g, washed three times with NP-40 buffer, and resuspended in 1× SDS-PAGE sample buffer. His-tagged pET-H4 or pET-UNC-76 were coupled to Ni2+-NTA agarose beads by incubating 50 μl of NP-40 washed beads with 10 μg/ml His-tagged fusion protein for 20 min at room temperature. The beads were pelleted, washed twice in NP-40 buffer, and resuspended in 40 μl of NP-40 buffer. Coated beads (25 μl) were added to the precleared lysate and incubated for 30 min at 4°C with rocking. The beads were pelleted by centrifuging 2 s at 10,000 × g, washed three times with NP-40 buffer, and resuspended in 1× SDS-PAGE sample buffer. Gel samples of the crude supernatant and copurification supernatant fractions were diluted with an equal volume of 2× SDS-PAGE sample buffer.
Antisera Production and Immunochemistry
UNC-76 aa 277–474 (pET-H4) was expressed in Escherichia coli, purified, and used to generate polyclonal antisera 1347 and 1348 in guinea pigs (Pocono Rabbit Farm and Laboratory, Canadensis, PA). Antiserum 1348 was affinity purified using bacterially expressed pGEX-H4, which encodes aa 277–474 of UNC-76 fused to glutathione S-transferase in plasmid pGEX-2T (Guan and Dixon, 1991). For affinity purification of UNC-76 antisera, purified pGEX-H4 protein was cross-linked to glutathione-Sepharose 4B (Amersham Biosciences, Piscataway, NJ) according to established protocols (Bar-Peled and Raikhel, 1996). Affinity purified UNC-76 antisera were used at a 1:100 dilution for immunostaining and a 1:1000 dilution for Western blots. Immunostaining of Drosophila embryos and larvae was performed using standard protocols (Hurd and Saxton, 1996; Gindhart et al., 1998; Rothwell and Sullivan, 2000). KLC antisera (Gindhart et al., 1998) were used at a 1:100 dilution, and synaptotagmin (SYT) antisera (Littleton et al., 1993) were used at a 1:500 dilution. Secondary antibodies conjugated to alkaline phosphatase horseradish peroxidase, fluorescein isothiocyanate (FITC), and Texas Red were obtained from Sigma-Aldrich (St. Louis, MO) and Jackson Immunoresearch Laboratories (West Grove, PA), and used according to manufacturers' instructions. To control for staining variability, fixation and antibody staining of mutant and wild-type larvae (e.g., Unc-76– and Unc-76/+) were performed simultaneously in the same sylgard-coated Petri dish. Each genotype was analyzed on multiple occasions, with two or three larvae of a particular genotype dissected per experiment. Segmental nerves showing representative staining patterns were selected for imaging by confocal microscopy. Antibody detection on Western blots was performed on a Storm 840 PhosphorImager (Amersham Biosciences), by using the ECF chemifluorescence or ECL Plus kit (Amersham Biosciences). Quantitation of bands on Western blots was performed using ImageQuant software (Amersham Biosciences). Duplicate gels were stained with Coomassie Blue and analyzed using NIH Image.
Drosophila Genetics
The Khc and Klc alleles used in this work were described previously (Gindhart et al., 1998; Martin et al., 1999a; Brendza et al., 2000). Khc16 is a null allele, and Df(3L)34ex5 is a small deletion that removes the Klc transcription unit. The Unc-76 P[lacW] alleles l(1)G0360, l(1)G0158, and l(1)G0310, and the Unc-76 duplication chromosomes Dp(1;2;Y)w+ (duplication of 2C9-3D1 on Y chromosome) and Tp(1; 3)wvco (transposition of 2C1-3C5 onto chromosome 3) were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN). The P element insertion site of each Unc-76 allele was identified by plasmid rescue and DNA sequencing of genomic DNA flanking each insertion site (Huang et al., 2000). To generate Unc-76 revertants and deletion mutants, P[lacW]l(1)G0310, w/FM7c, B females were crossed to w/Dp(1;2;Y)w+; Dr/TMS, Sb Δ2-3 males. Dysgenic F1 male progeny were crossed to Sxl/Binsinscy, w B females, and white-eyed female F2 progeny were crossed to Binsinscy males to establish individual lines and screen for X-linked lethality. Putative excision mutants were tested for complementation of l(1)G0310; 33% of the excision events (42/125) failed to complement l(1)G0310. Small deletions within Unc-76 were identified by PCR amplification of Unc-76excision/+ genomic DNA by using primers flanking the P[lacW] l(1)G0310 insertion site. Using this approach, a small deletion (Df(1)ex107) that removes Unc-76 sequences was identified. To precisely map the deletion breakpoints, PCR products were sub-cloned into pBS SK+ (Stratagene, La Jolla, CA) and sequenced. The lethal phase of Unc-76 alleles was determined according to Gindhart et al. (1998).
Brightfield and Fluorescence Microscopy
Embryos were examined using an Olympus BX60 microscope with differential interference contrast optics and a 20× objective. Bright-field images were obtained using an Olympus C-3040 digital camera. Epifluorescence images were captured using an Olympus BX60 microscope, 60× oil immersion objective, FITC filter set, CCD-300T camera (Dage-MTI, Michigan City, IN), Scion AG-5 frame grabber, and Scion Image software (Scion, Frederick, MD). Confocal images were obtained using a Leica TCS-NT confocal imaging system and a Leica DM IRBE inverted microscope with a 40× oil immersion objective. Colocalization of UNC-76 and SYT in segmental nerves was performed using secondary antisera coupled to FITC and Texas Red. Optical sections of 2 μm were collected, and stacks of optical sections were obtained using Leica LCS confocal microscope software. Images were prepared using Adobe Photoshop (Adobe Systems, San Jose, CA).
RESULTS
UNC-76 Binds to the Tail Domain of Drosophila KHC
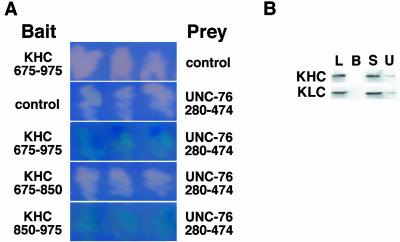
We performed a yeast two-hybrid screen, by using the carboxy-terminal 300 aa of Drosophila KHC to screen a Drosophila embryonic cDNA library for genes that encode KAPs. Screening 1.8 × 107 transformants (Ura+, His+, Trp+) resulted in the isolation of 170 putative KHC interactors (Leu+, LacZ+). Six clones identified in the screen encode aa 277–474 of annotated gene CG3981 (Adams et al., 2000). This clone binds to the Drosophila KHC tail domain (aa 850–975) but not to the stalk domain, the KLC repeats, or to control bait proteins (Figure 1A; our unpublished data). CG3981 is similar to the C. elegans protein UNC-76 and rat FEZ1 (Bloom and Horvitz, 1997; Kuroda et al., 1999); therefore, we will refer to gene CG3981 as Unc-76. ClustalW alignment reveals three blocks of sequence conservation among the homologs at aa 140–202, 345–413, and 421–474 of Drosophila UNC-76 (Figure 2A). The KHC-interacting region of UNC-76 includes the second and third conserved domains. Structure and domain predictions show that UNC-76 is acidic, primarily α-helical, and may contain a small α-helical coiled-coil between aa 350–388, but did not reveal any other notable structural motifs. The second conserved domain (aa 347–407) is similar to the schwannomin (NF2) binding domain in SCHIP-1 (60.1% conserved residues; Goutebroze et al., 2000). NF2 is an evolutionarily conserved tumor suppressor protein involved in tight junction formation (Trofatter et al., 1993).
Figure 1.
UNC-76 interacts with the KHC tail domain in the yeast two-hybrid assay and in copurification assays. (A) Yeast cells containing a lacZ reporter gene and various combinations of LexA DNA binding domain (baits, left column) and B42 activation domain (preys, right column) fusion proteins were grown on CM Gal/Raff Xgal plates. Colonies in which reporter gene activation is enhanced by specific bait-prey interactions are blue, whereas colonies in which a bait-prey interaction do not occur are white. Colonies that contain the KHC stalk and tail domains (aa 675-976) or the KHC tail domain (aa 850-975) bait fusions and the UNC-76 prey fusion enhance reporter gene transcription, but colonies containing the KHC stalk domain (aa 675-850) and UNC-76 do not. Control, LexA DNA binding domain bait or B42 activation domain prey. (B) Western analysis of protein fractions from UNC-76 copurification assay. 6xHIS-tagged full-length UNC-76 was bound to Ni2+-NTA agarose beads and incubated with detergent-soluble extracts of adult flies containing a transgenic copy of myc epitope-tagged KLC. KHC and KLC copurify with UNC-76 beads (lane U), but not with Ni2+-NTA agarose beads alone (lane B). KHC, blot probed with anti-KHC antibody; KLC, blot probed with anti-myc antibody to detect transgenic KLC; L, detergent-soluble lysate; B, proteins that copurify with Ni2+-NTA beads; S, supernatant from UNC-76 copurification assay; U, proteins that copurify with 6xHIS-UNC-76 beads.
Figure 2.
Alignment of Drosophila UNC-76 with its homologs and genetic map of Unc-76 locus. (A) CLUSTALW 1.8 alignment of Drosophila UNC-76, C. elegans UNC-76, and rat FEZ1 is shown. Identical aa in two of three homologs are shaded black, conserved amino acid substitutions are shaded gray, and gaps are represented by dashed lines. Three regions of high sequence similarity are boxed, and the region of similarity with NF1-binding protein SCHIP-1 is underlined. The sequence of Drosophila UNC-76 is available from GenBank under accession no. NP_569981. (B) Map of the Unc-76 genetic interval and location of Unc-76 mutations used in this analysis. The Unc-76 gene is transcribed from left to right in this diagram; the gene is oriented so Unc-76 is transcribed toward the centromere of the X chromosome. Exons are represented as boxes; coding regions are filled, whereas the 5′ and 3′-untranslated region are not. The location of P element insertions l(1)G0360, l(1)G0158, and l(1)G0310 are shown; they are inserted at +9, +28, and +172 of the Unc-76 transcript, respectively. The portion of the Unc-76 transcript deleted by Df(1)ex107 (+3 to +581) is shown by a solid line under Unc-76 exon 1. Df(1)ex107 deletes the Unc-76 translation start site. Bar, 1 kb.
To test whether Drosophila UNC-76 can bind to native kinesin, bacterially expressed 6xHIS-tagged UNC-76 was incubated with Ni2+-NTA agarose beads, and then the 6xHIS-UNC-76–coated beads were used in a copurification assay with detergent-soluble extracts from MYC-KLC Drosophila adults (Gindhart et al., 1998). Western analysis of supernatant and bead fractions with antibodies to KHC and KLC confirm that endogenous fly kinesin binds to 6xHIS-UNC-76 beads, but not control beads lacking protein (Figure 1B) or containing protein encoded by the bacterial expression vector alone. Similar results were observed using the portion of UNC-76 encoded by the two-hybrid clone, thus confirming that the kinesin binding site is located between UNC-76 aa 277–474. The copurification of KLC with 6xHIS-UNC-76 indicates that UNC-76 binds to native kinesin containing both light and heavy chains, because the UNC-76 binding site is in the tail domain of the heavy chain (Figure 1A). These results support two-hybrid data demonstrating the direct interaction of KHC and UNC-76.
Unc-76 Is Essential for Drosophila Development
To determine the in vivo function of UNC-76, mutations in the Unc-76 gene were identified, and the resulting phenotypes were studied at the organismal and cellular level. The Unc-76 gene is located at cytological interval 2C9 on the X chromosome (Adams et al., 2000), distal to the embryonic polarity gene corkscrew (csw; Figure 2B). A collection of lethal P[lacW] insertion mutations, mapped to this interval by chromosomal in situ hybridization, was mapped precisely by cloning and sequencing genomic DNA flanking the insertion site. Three P[lacW] insertions were identified in the Unc-76 5′-untranslated region. All three insertions are recessive lethal, fail to complement each other and deletion mutations that remove Unc-76, but are complemented by genetic duplications containing Unc-76. The P[lacW] insertion mutation l(1)G0310 was used to generate Unc-76 deletions and revertants. Flies heterozygous for l(1)G0310 were crossed to flies containing a stable source of P transposase to induce excision of the P[lacW] transposon. Precise excision events restore wild-type gene structure, but imprecise excision events often result in small deletions flanking the original insertion site. More than 100 l(1)G0310 excisions were tested by complementation and PCR analysis. Precise excisions of l(1)G0310 are viable and complement lethal Unc-76 alleles, whereas imprecise excisions, such as the small deletion Df(1)ex107 (Figure 2B), are recessive lethal and do not complement other Unc-76 alleles. Collectively, these results indicate that the lethality associated with the P element insertions in Unc-76 is caused by loss of Unc-76 function. More importantly, these results demonstrate that Unc-76 is essential for Drosophila development.
Unc-76 Mutations Exhibit Phenotypes Similar to Khc and Klc Mutations
The loss of Unc-76 function results in lethality at the transition between second and third instar at approximately 5 d of development. The P[lacW] alleles alone or in combination with Df(1)ex107 die at the L2-L3 transition, suggesting that these mutations are null alleles of Unc-76. Individuals lacking Unc-76 function exhibit a progressive paralysis phenotype reminiscent of the paralysis observed in null alleles of Khc and Klc (Saxton et al., 1991; Hurd and Saxton, 1996; Gindhart et al., 1998). First instars look normal and do not exhibit impaired locomotion or feeding behavior. However, progressive loss of vigor is observed during the second instar, resulting in complete paralysis and death at the onset of L3. The longest-living Unc-76 mutants have mouthparts characteristic of third instars, but lack the ability to shed their second instar cuticle. This “L3 trapped in an L2 body” phenotype is also observed in Klc null mutants. The “tail flipping” locomotion defect that is the hallmark of weak loss-of-function Khc and Klc alleles (Hurd and Saxton, 1996; Gindhart et al., 1998; Martin et al., 1999a) is not observed in Unc-76 mutants, although uncoordinated crawling behavior is seen before onset of paralysis. However, the locomotion defects characteristic of moderate Khc and Klc alleles are readily observed only in L3 larvae. Because the available Unc-76 mutations are null alleles that die before L3, the ability of Unc-76 mutations to evoke tail flipping cannot be tested directly. The similarity between the organismal phenotypes of Unc-76 and Khc is also observed in C. elegans, in which unc-76 and the KHC homolog unc-116 both exhibit an uncoordinated phenotype (Patel et al., 1993; Bloom and Horvitz, 1997; Byrd et al., 2001). Unlike Drosophila, however, nematode unc-76 and unc-116 are not required for viability.
Loss of function mutations in Drosophila fast axonal transport motor complexes kinesin-I, kinesin-II, and dynein result in defects in the trafficking of axonal cargos (Hurd and Saxton, 1996; Gindhart et al., 1998; Bowman et al., 1999; Martin et al., 1999a; Ray et al., 1999). These trafficking defects include axon clogs, which are aggregates of membrane-bounded anterograde cargos such as synaptic vesicle precursors, mitochondria, and prelysosomal vesicles (Hurd and Saxton, 1996). Because Unc-76 mutations cause neuromuscular phenotypes similar to those observed when axonal microtubule motor function is disrupted, we examined neuromuscular preparations of Unc-76 mutant and control larvae for disruption of axonal transport. Immunostaining with antisera to the synaptic vesicle precursor marker SYT shows that the segmental nerves of Unc-76 mutant larvae contain aggregates of SYT immunoreactivity, whereas Unc-76/+ control larvae have a diffuse, punctate staining pattern (Figure 3). The aggregates of SYT staining observed in Unc-76 mutant larvae are similar to those observed in Khc and Klc mutant larvae and seem to be axon clogs correlated with the disruption of transport in the Drosophila nervous system. Like kinesin mutants, the segmental nerves of Unc-76 mutants contain aggregates of KLC immunoreactivity, but tubulin, which is a component of slow axonal transport (Terada et al., 2000), is not found in SYT-containing aggregates (our unpublished data). The similarity of the Unc-76, Khc, and Klc neuromuscular and organelle jam phenotypes, in conjunction with two-hybrid and copurification experiments demonstrating the physical association of UNC-76 and KHC, strongly suggests that UNC-76 is an important component of the kinesin transport pathway in the Drosophila nervous system.
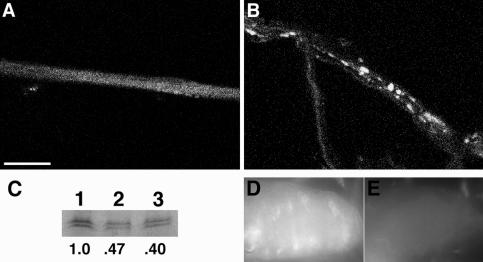
Figure 3.
Unc-76 mutations cause axonal transport defects in the Drosophila nervous system. Larvae heterozygous (A) or homozygous (B) for Unc-76 null allele l(1)G0310 were incubated with antisera specific for the synaptic vesicle precursor marker SYT to compare the accumulation pattern of fast anterograde transports in wild-type and Unc-76 mutant backgrounds. Note that large aggregates of SYT immunoreactivity are found in the segmental nerves of Unc-76 mutants (B), but SYT accumulation is diffuse in heterozygotes (A). Bar, 25 μm. (C) UNC-76 protein concentration is reduced in Unc-76 heterozygous adults. Whole cell extracts of wild-type (ORE-R, lanes 1 and 2) and Unc-76 heterozygote (l[1]G0310/+, lane 3) were electrophoresed, blotted, and incubated with UNC-76 antisera. The equivalent of one adult female is present in lanes 1 and 3, whereas 0.5 fly equivalent is present in lane 2 as a control. The numbers below each lane represent the ratio of the pixel volume of each lane divided by the pixel volume of lane 1 (wild-type). (D and E) UNC-76 antisera staining of wild-type (D) and Unc-76– (E) second instar CNS. Note that UNC-76 staining is more intense in ventral nerve cord of wild-type larva, whereas staining is at background levels in Unc-76– ventral nerve cord. Epifluorescence images were captured at 60× magnification. Anterior is to the left.
UNC-76 Is Not Associated with Axon Clogs in the Drosophila Nervous System
An important question is whether UNC-76 functions as a kinesin-cargo adaptor. Proteins that help kinesin attach to cargos should be transported down axons by kinesin-dependent fast axonal transport. To test this hypothesis, we generated specific antisera to UNC-76, and studied its accumulation in embryos and larvae. The UNC-76 antisera recognize a doublet at ∼70 kDa, the intensity of which is reduced by half in adults heterozygous for the Unc-76 mutation l(1)G0310 (Figure 3C) and the deletion mutant Df(1)ex107 (our unpublished data). UNC-76 antisera specificity for immunostaining experiments was demonstrated by incubating wild-type and l(1)G0310 second instar neuromuscular preparations with affinity-purified UNC-76 antisera; wild-type larvae display UNC-76 staining in the larval central nervous system (CNS) (Figure 3D), but only background staining is observed in l(1)G0310 individuals (Figure 3E). Specific accumulation of UNC-76 is first observed in the developing nervous system at stage 14 of embryogenesis, persisting through the end of embryogenesis (Figure 4). This result is consistent with RNA in situ hybridization experiments that showed Unc-76 transcript localization in the embryonic CNS (our unpublished data). UNC-76 accumulation is observed within neurons but is not found in glial cells. Accumulation is observed in both the longitudinal (parallel to embryo midline) and commissural (cross midline) axons (Figure 4, A and B); later in development, however, UNC-76 is observed only in longitudinal axons and the brain lobes (Figure 4, C and D). The localization of UNC-76 to axon tracks of the CNS during embryonic development suggests that the interaction between UNC-76 and kinesin is restricted to the nervous system and that kinesin-UNC-76 interactions can occur in axons.
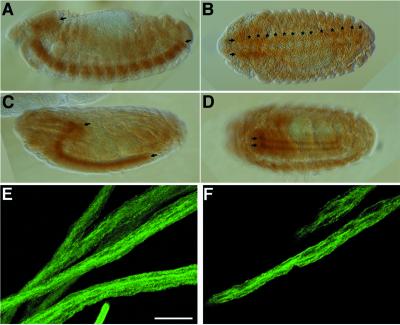
Figure 4.
Immunolocalization of UNC-76 in the embryonic and larval nervous system. (A–D) UNC-76 is found in axons of the embryonic CNS. Anterior is to the left. (A and B) Lateral (A) and horizontal (B) views of a stage 14 embryo. (A) Shows that UNC-76 protein is restricted to the CNS (arrows) at this stage of development, whereas (B) shows that UNC-76 accumulation is detectable in both longitudinal (arrows) and commissural (asterisks) axons. (C and D) Lateral and horizontal views of UNC-76 staining at stage 17. UNC-76 is found within longitudinal axons of CNS (arrows) late in embryogenesis, but commissural axon staining is greatly reduced (D) compared with earlier in development (B). (E and F) UNC-76 staining in segmental nerves of wild-type third instars. (E) A projection of 29 optical sections, each 2 μm in thickness, whereas F is one optical section from the projection. Note that UNC-76 accumulation is diffuse within segmental nerves. Bar (E and F), 25 μm.
The accumulation of UNC-76 in axons is consistent with the model that UNC-76 is a kinesin-cargo adapter. A prediction of this model is that UNC-76 should be found associated with membrane-bound cargos in axons. The segmental nerves of the Drosophila larva provide an excellent model system for studying axonal transport, because the nonmyelinated segmental nerves consist of 30–40 sensory and motor axons that enervate each hemisegment. Axon clogs observed when vesicle transport is disrupted contain both anterograde and retrograde membrane cargos, as well as cargo-bound motor proteins. Presumably, if UNC-76 is a kinesin-cargo adapter in axons, then UNC-76 should colocalize with axon clogs. To test this hypothesis, we compared the localization pattern of UNC-76 and SYT in segmental nerves. Previous findings demonstrated that the presynaptic vesicle markers SYT and cysteine string protein colocalize with kinesin, kinesin-II, and dynein in axon clogs (Gindhart et al., 1998). Intriguingly, UNC-76 staining in segmental nerves is diffuse (Figure 4, E and F) and is not enriched in SYT-containing axon clogs of Khc/+; Klc/+ individuals (Figure 5), suggesting that UNC-76 is not associated with membrane cargos transported by fast axonal transport. Although it is difficult to determine the specific location of UNC-76 within axons by using immunofluorescence, it seems to be distributed uniformly, suggesting it is in the axoplasm.
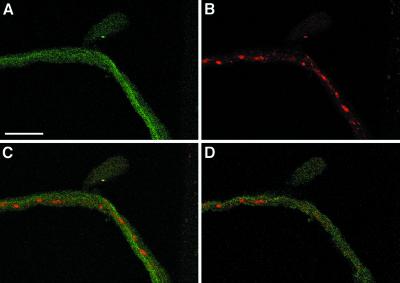
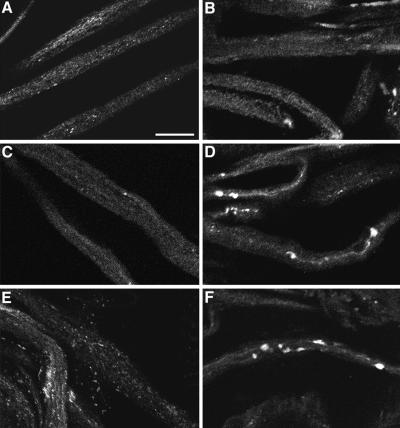
Figure 5.
UNC-76 and SYT do not colocalize in segmental nerves. (A) Shows the accumulation of UNC-76 in a nine-section optical projection (each section is 2 μm) of a segmental nerve from a Khc16/+; Df(3L)34ex5/+ individual that exhibits locomotion defects and SYT axon clogs (B). Note that UNC-76 accumulation is fairly uniform, whereas SYT is found in axon clogs (B). (C) Overlay of images in A and B shows that UNC-76 and SYT are largely nonoverlapping; upon viewing a single 2-μm optical section (D), it is apparent that UNC-76 is not detected in axon clogs. Bar, 25 μm.
Dosage-dependent Interactions between Unc-76, Khc, and Klc
To more fully understand the functional relationship between UNC-76 and kinesin in axons, we studied the effect of altering Unc-76 dosage in Khc and Klc heterozygotes. The reduction of KHC or KLC function by 50% does not have a noticeable effect on axonal transport or locomotion of Drosophila larvae. However, larvae with half the normal Khc or Klc dosage provide a sensitized genotype for the identification of mutations that disrupt axonal transport (Martin et al., 1999). To test whether altering Unc-76 dosage enhances Khc or Klc, males of the genotype Unc76–/Dp(Unc-76+) were crossed to Khc–/+ or Klc–/+ females, progeny larvae were scored for locomotion defects, and the presence of axonal clogs in larvae of each genotypic class was assessed by SYT immunolocalization of neuromuscular preparations (Figure 6). Two genetic duplications were used: Dp(1;2;Y)w+ is a duplication of segment 2C-3D onto the Y chromosome, and Tp(1;3)wvco is a transposition of 2C-3C onto chromosome 3. This approach ensured that Unc-76 would be expressed in the appropriate spatiotemporal domain, but at a higher level than wild type, thus minimizing the potential for pleiotropic effects caused by ectopic Unc-76 expression. No enhancement of Khc or Klc was observed when Unc-76 dosage was reduced by half (Figure 6C; our unpublished data). In contrast, increasing the dosage of Unc-76, in combination with reducing Khc or Klc dosage, results in strong axon clog and locomotion phenotypes (Figure 6, D and F). This interaction is observed using both Unc-76 duplications. The Unc-76 duplication on the Y chromosome has a stronger effect, because it elicits both locomotion and axon clog phenotypes in Khc heterozygotes, whereas the chromosome 3 duplication causes only the axon clog phenotype. The enhancement of Khc by increased Unc-76 dosage was more pronounced than its enhancement of Klc; this is similar to observations for Appl (Gunawardena and Goldstein, 2001). The genetic interaction between Dp(Unc-76) and kinesin mutations was more pronounced when the Khc or Klc mutant allele was inherited maternally. This result is not unexpected, given the large maternal contribution of kinesin and the perdurance of maternally supplied kinesin for several days.
Figure 6.
Increasing the genetic dosage of Unc-76 enhances Klc and Khc mutant phenotypes. SYT accumulation in segmental nerves of larvae of the following genotypes: (A) Dp(1;2;Y)w+/+; (Unc-76 duplication); (B) Df(34ex5)/+ (Klc deletion); (C) l(1)G0310/+ (Unc-76 null allele); Df(34ex5)/+; (D) Dp(1;2;Y)w+/+; Df(34ex5)/+; (E) Khc16/+ (Khc null allele); and (F) Df(1;2;Y)w+/+; Khc16/+. Bar, 25 μm.
DISCUSSION
A number of studies have demonstrated unequivocally that the microtubule motor kinesin-I binds to its intracellular cargos by protein–protein interactions and that kinesin transport is regulated by phosphorylation-dependent signaling pathways. We have used the yeast two-hybrid system to identify KAP-encoding genes that potentially mediate or regulate kinesin–cargo interactions and then used reverse genetic and cell biology tools available in Drosophila to reveal the in vivo relationship between KAPs and kinesin. We have identified an evolutionarily conserved protein, UNC-76, that binds specifically to the KHC tail domain in the yeast two-hybrid system and copurification assays. Immunostaining and genetic analyses demonstrate that UNC-76 function is required for axonal transport in the Drosophila nervous system. The dosage-dependent enhancement of kinesin mutant phenotypes by Unc-76 suggests that the binding of KHC by UNC-76 is relevant in vivo. Collectively, these results support a model in which UNC-76 has an essential function in kinesin transport pathways.
UNC-76 Function in the Nervous System
The analysis of UNC-76 function in other model systems is consistent with our observation that it has an essential role in the Drosophila nervous system. The C. elegans gene unc-76 was identified by the uncoordinated crawling behavior of loss-of-function unc-76 mutants (Hedgecock et al., 1985; Bloom and Horvitz, 1997). The UNC-76 protein accumulates in all C. elegans neurons at all stages of development (Bloom and Horvitz, 1997). Mutants have subtle defects in axon outgrowth and bundling in axon fascicles, but the growth and differentiation of individual axons along the body wall are unaffected. We have not observed a requirement for Unc-76 in Drosophila nervous system development, because locomotion defects in larvae lacking zygotic Unc-76 are not observed until mid-second instar, ∼3 d posthatching. Similar locomotion defects in Khc and Klc mutants do not become apparent until the second instar, presumably due to the perdurance of maternally supplied kinesin; it is possible that the maternal contribution of UNC-76 mRNA or protein is sufficient for embryogenesis and early larval development. UNC-76 protein accumulates in a dynamic pattern in axons during embryonic CNS development but is not detected in motor or sensory axons of the embryo. However, by the second instar, UNC-76 is detected in segmental nerves, which contain both motor and sensory axons. We cannot eliminate the possibility that UNC-76 accumulates in axons of the embryonic peripheral nervous system, because other proteins, such as PTP69D, clearly have a role in embryonic motor neuron development but cannot be detected in motor neurons of wild-type embryos (Desai et al., 1996).
The mammalian homolog of UNC-76, FEZ1, was identified in a yeast two-hybrid screen for proteins that bind the regulatory domain of rat PKCζ (Kuroda et al., 1999). FEZ1 is expressed at high levels in the nervous system of embryos and adults. In COS-7 cultured cells, FEZ1 is found associated with both cytosolic and membrane fractions, but its phosphorylation by PKCζ causes the redistribution of membrane-bound FEZ1 to the cytosol. In addition, the phosphorylation of FEZ1 by PKCζ stimulates neurite outgrowth in PC12 cultured cells, suggesting that PKCζ-dependent neuron differentiation may require FEZ1. Interestingly, antisense inhibition experiments in cultured rat hippocampal neurons demonstrate that kinesin is required for full extension of neurites (Ferreira et al., 1992), thus suggesting a possible relationship between PKCζ signaling, FEZ1 activity, and kinesin-dependent neurite outgrowth. Growth of axons occurs rapidly during Drosophila larval development, especially between L2 and L3, when the larva increases in length severalfold (Jan and Jan, 1976). Perhaps Unc-76 mutant lethality at the L2-L3 transition indicates that its function is required for axon outgrowth in the developing larval nervous system.
Is UNC-76 a Cargo Adapter or Kinesin Regulator?
Atypical PKCs (ι/λ and ζ) are essential for nerve growth factor-dependent neurite outgrowth and mitogen-activated protein kinase activation in PC12 cells (Reinhold and Neet, 1989; Lloyd and Wooten, 1992; Wooten et al., 1994). Perhaps UNC-76 acts as a control point for the regulation of kinesin activity by PKCζ and its effectors. Posttranslational modification of UNC-76 by PKCζ could regulate UNC-76 binding to KHC, or UNC-76 could recruit regulatory proteins or cargos to the kinesin tail domain, as has been suggested for the KLC-binding scaffold proteins JIP1, JIP2, and JIP3/JSAP1 (SYD), which bind kinesin and members of the JNK signaling cascade (Ito et al., 1999; Yasuda et al., 1999; Bowman et al., 2000; Kelkar et al., 2000; Verhey et al., 2001). Another plausible model is that UNC-76 is a cargo adapter for cytosolic nonmembrane cargos that escape entrapment in axon clogs. Analysis of kinesin-dependent axon clogs has shown that they are composed of membrane-bound organelles (Hurd and Saxton, 1996; Martin et al., 1999); the presence of soluble nonmembrane cargos, such as mRNPs or protein complexes, was not tested directly. Recent experiments show that KHC homolog KIF5B is present in soluble complexes with NF1 (neurofibromin) and NF2 (merlin) in HeLa cells (Hakimi et al., 2002), supporting a model in which kinesin is required for NF1 and NF2 transport to the cell periphery. UNC-76 contains a domain similar to the NF2 binding domain of SCHIP-1 (Goutebroze et al., 2000); an intriguing possibility is that UNC-76 is a bipartite cargo adaptor that mediates kinesin–NF2 interactions. Overexpression of UNC-76 may enhance kinesin mutant phenotypes by reducing the kinesin pool available for vesicle transport. UNC-76 may target kinesin for inactivation by signal transduction pathways (regulatory model), or mask the kinesin tail domain's ability to bind membrane cargo receptors (titration model).
The in vivo relationship between UNC-76 and PKCζ in Drosophila is unclear. The closest Drosophila homolog to PKCζ is encoded by the DaPKC gene (Wodarz et al., 2000). During embryogenesis, DaPKC is expressed in epithelial tissues and developing neuroblasts. DaPKC protein accumulates at the apical cortex of polarized cells, and is part of a protein complex with bazooka (BAZ) essential for apical-basal epithelial polarity; however, there is no known role for DaPKC in axonal transport. We are currently investigating the nature of UNC-76 interactions with DaPKC and other regulatory proteins, and possible functional relationships between those interactions and kinesin activity. Drosophila UNC-76 contains a 120 aa N-terminal region that is not evolutionarily conserved (Figure 2); perhaps this region can serve as an interface for Drosophila-specific regulatory interactions.
Parallels and Orthagonals between UNC-76, APPL, and SYD
The Unc-76 loss-of-function phenotype is similar to those observed for Appl and syd, which encode proteins that bind to the kinesin tail domain (Torroja et al., 1999; Bowman et al., 2000; Gunawardena and Goldstein, 2001). Mutations in all three genes result in the disruption of axonal transport in larval segmental nerves, resulting in aggregates of membrane-bound cargos. APPL is the homolog of APP, a transmembrane protein that is a kinesin cargo receptor through its interaction with the tetratricopeptide (TPR) repeats of KLC (Kamal et al., 2000, 2001). SYD is the Drosophila homolog of JIP3/JSAP1 and has the dual role of kinesin cargo receptor through KLC binding and scaffold for components of the JNK signaling cascade. UNC-76 differs from APPL and SYD in that it binds to the tail domain of KHC, not the TPR repeats of KLC. Furthermore, UNC-76 does not seem to be a membrane cargo receptor, because it is not a transmembrane protein and is not enriched in axon clogs in the segmental nerves of kinesin mutant larvae. However, the possibility that UNC-76 is a receptor, adapter, or facilitator of kinesin binding for cargos that do not accumulate in axon clogs cannot be eliminated. Interestingly, it has been shown that the first 119 aa of C. elegans UNC-76 are sufficient for targeting to axons, and fusions of the first 197 aa of UNC-76 to β-gal or green fluorescent protein are directed to axons in C. elegans (Bloom and Horvitz, 1997). The part of Drosophila UNC-76 that binds to KHC does not include this region. Perhaps there are multiple binding sites for kinesin in UNC-76, or UNC-76 can be transported into neurons by another motor protein or through indirect association with kinesin. The axon targeting domain of UNC-76 is intriguing in light of the recent identification of mouse GRIP1, which binds to KHC homologs KIF5a/b/c and targets kinesin to dendrites (Setou et al., 2002). The amino terminus of UNC-76 may have an analogous role in axon targeting of kinesin or other motor proteins.
The similar phenotypes and dosage-dependent genetic interactions of Unc-76, Khc, and Klc support a model in which UNC-76 is an important component of the kinesin transport pathway. Individuals lacking UNC-76 display locomotion phenotypes and axon clogs identical to the phenotypes displayed by strong alleles of either kinesin subunit. Furthermore, increasing the genetic dosage of Unc-76 enhances the phenotype of Khc and Klc mutants. This may be caused by the titration of free kinesin away from other binding partners, such as APPL and SYD. There are several similarities between the genetic interactions of Unc-76 and Appl with Khc and Klc. Overexpression of APPL and UNC-76 enhances Khc more than Klc, enhancement of the Khc and Klc phenotypes is more pronounced when the kinesin mutant allele is inherited maternally, and reduction of Appl or Unc-76 dosage does not enhance Khc or Klc (Torroja et al., 1999; Gunawardena and Goldstein, 2001). The phenotype of Khc mutants may be more severe than Klc because all kinesin cargos require the activity of the KHC-encoded kinesin motor domain, whereas some cargos may bind directly to the KHC tail, thus not requiring KLC for transport. Similarly, loss of Appl or Unc-76 function may disrupt the transport of a subset of kinesin cargos. The increase of Unc-76 and Appl dosage could reduce the concentration of soluble kinesin available for cargo binding to reduce kinesin-dependent transport to subthreshold levels. Although it is possible that another gene in the Unc-76 duplication enhances kinesin mutant phenotypes, this seems unlikely given the small number of genes identified in previous genetic screens for mutations causing larval locomotion defects (Martin et al., 1999a; Gunawardena and Goldstein, 2001). However, it is important to define more precisely the functional domains of UNC-76 that interact with kinesin and other proteins, such as PKCζ.
Acknowledgments
We thank Andrea Pereira (University of Massachusetts Medical Center) for providing access to the confocal microscope; J. Troy Littleton for providing anti-SYT antisera; the Indiana Stock Center for providing fly stocks; Russ Finley for providing yeast strains, plasmids, and the RFLY1 library; Erica Golemis for providing yeast strains; David Schultz for providing control bait constructs; Alex Rivest for performing RNA in situ hybridization experiments; Courtney Tanzi, Richard White, and Adan Colon-Carmona for reading of the manuscript; and Shermali Gunawardena, Richard White, and Brian White for useful discussions. J.G. is supported by National Science Foundation awards 9974835 and 0097685, and University of Massachusetts Boston. R.G. is supported by the American Heart Association Award 0050589N (to A.P.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-12-0800. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-12-0800.
References
- Adams, M.D., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- Apweiler, R., et al. (2001). The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1997). Current Protocols in Molecular Biology, New York: John Wiley & Sons.
- Bar-Peled, M., and Raikhel, N.V. (1996). A method for isolation and purification of specific antibodies to a protein fused to the GST. Anal. Biochem. 241, 140–142. [DOI] [PubMed] [Google Scholar]
- Bloom, L., and Horvitz, H.R. (1997). The Caenorhabditis elegans gene unc-76 and its human homologs define a new gene family involved in axonal outgrowth and fasciculation. Proc. Natl. Acad. Sci. USA 94, 3414–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, A.B., Kamal, A., Ritchings, B.W., Philip, A.V., McGrail, M., Gindhart, J.G., and Goldstein, L.S. (2000). Kinesin-dependent axonal transport is mediated by the sunday driver (SYD) protein. Cell 103, 583–594. [DOI] [PubMed] [Google Scholar]
- Bowman, A.B., Patel-King, R.S., Benashski, S.E., McCaffery, J.M., Goldstein, L.S., and King, S.M. (1999). Drosophila roadblock and Chlamydomonas LC 7, a conserved family of dynein-associated proteins involved in axonal transport, flagellar motility, and mitosis. J. Cell Biol. 146, 165–180. [PMC free article] [PubMed] [Google Scholar]
- Brady, S.T. (1985). A novel brain ATPase with properties expected for the fast axonal transport motor. Nature 317, 73–75. [DOI] [PubMed] [Google Scholar]
- Brendza, R.P., Sheehan, K.B., Turner, F.R., and Saxton, W.M. (2000). Clonal tests of conventional kinesin function during cell proliferation and differentiation. Mol. Biol. Cell 11, 1329–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd, D.T., Kawasaki, M., Walcoff, M., Hisamoto, N., Matsumoto, K., and Jin, Y. (2001). UNC-16, a JNK-signaling scaffold protein, regulates vesicle transport in C. elegans. Neuron 32, 787–800. [DOI] [PubMed] [Google Scholar]
- Coy, D.L., Hancock, W.O., Wagenbach, M., and Howard, J. (1999). Kinesin's tail domain is an inhibitory regulator of the motor domain. Nat. Cell Biol. 1, 288–292. [DOI] [PubMed] [Google Scholar]
- Desai, C.J., Gindhart, J.G.J., Goldstein, L.S.B., and Zinn, K. (1996). Receptor tyrosine phosphatases are required for motor axon guidance in the Drosophila embryo. Cell 84, 599–609. [DOI] [PubMed] [Google Scholar]
- Ferreira, A., Niclas, J., Vale, R.D., Banker, G., and Kosik, K.S. (1992). Suppression of kinesin expression in cultured hippocampal neurons using antisense oligonucleotides. J. Cell Biol. 117, 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley, R.L., Jr., Thomas, B.J., Zipursky, S.L., and Brent, R. (1996). Isolation of Drosophila cyclin D, a protein expressed in the morpho-genetic furrow before entry into S phase. Proc. Natl. Acad. Sci. USA 93, 3011–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier, J., Osguthorpe, D.J., and Robson, B. (1978). Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J. Mol. Biol. 120, 97–120. [DOI] [PubMed] [Google Scholar]
- Gho, M., McDonald, K., Ganetzky, B., and Saxton, W.M. (1992). Effects of kinesin mutations on neuronal functions. Science 258, 313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindhart, J.G., Jr., Desai, C.J., Beushausen, S., Zinn, K., and Goldstein, L.S. (1998). Kinesin light chains are essential for axonal transport in Drosophila. J. Cell Biol. 141, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, L.S. (2001). Kinesin molecular motors: transport pathways, receptors, and human disease. Proc. Natl. Acad. Sci. USA 98, 6999–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, L.S., and Philip, A.V. (1999). The road less traveled: emerging principles of kinesin motor utilization. Annu. Rev. Cell Dev. Biol. 15, 141–183. [DOI] [PubMed] [Google Scholar]
- Goutebroze, L., Brault, E., Muchardt, C., Camonis, J., and Thomas, G. (2000). Cloning and characterization of SCHIP-1, a novel protein interacting specifically with spliced isoforms and naturally occurring mutant NF2 proteins. Mol. Cell. Biol. 20, 1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, K.L., and Dixon, J.E. (1991). Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192, 262–267. [DOI] [PubMed] [Google Scholar]
- Gunawardena, S., and Goldstein, L.S. (2001). Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron 32, 389–401. [DOI] [PubMed] [Google Scholar]
- Hackney, D.D., and Stock, M.F. (2000). Kinesin's IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat. Cell Biol. 2, 257–260. [DOI] [PubMed] [Google Scholar]
- Hakimi, M.-A., Speicher, D.W., and Shiekhattar, R. (2002). The motor protein kinesin-1 links neurofibromin and merlin in a common cellular pathway for neurofibromatosis. J. Biol. Chem. 277, 36909–36912. [DOI] [PubMed] [Google Scholar]
- Hedgecock, E.M., Culotti, J.G., Thomson, J.N., and Perkins, L.A. (1985). Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 111, 158–170. [DOI] [PubMed] [Google Scholar]
- Hirokawa, N. (1998). Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279, 519–526. [DOI] [PubMed] [Google Scholar]
- Huang, A.M., Rehm, E.J., and Rubin, G.M. (2000). Recovery of DNA sequences flanking P-element insertions: inverse PCR and plasmid rescue. In: Drosophila Protocols, ed. W. Sullivan, M. Ashburner, and R.S. Hawley, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 429–437. [DOI] [PubMed]
- Hurd, D.D., and Saxton, W.M. (1996). Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics 144, 1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, M., Yoshioka, K., Akechi, M., Yamashita, S., Takamatsu, N., Sugiyama, K., Hibi, M., Nakabeppu, Y., Shiba, T., and Yamamoto, K.I. (1999). JSAP1, a novel jun N-terminal protein kinase (J.N.K)-binding protein that functions as a scaffold factor in the JNK signaling pathway. Mol. Cell. Biol. 19, 7539–7548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan, L.Y., and Jan, Y.N. (1976). Properties of the larval neuromuscular junction in Drosophila melanogaster. J. Physiol. 262, 189–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin, F., Thompson, J.D., Gouy, M., Higgins, D.G., and Gibson, T.J. (1998). Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23, 403–405. [DOI] [PubMed] [Google Scholar]
- Kamal, A., Almenar-Queralt, A., LeBlanc, J.F., Roberts, E.A., and Goldstein, L.S. (2001). Kinesin-mediated axonal transport of a membrane compartment containing beta-secretase and presenilin-1 requires APP. Nature 414, 643–648. [DOI] [PubMed] [Google Scholar]
- Kamal, A., and Goldstein, L.S. (2002). Principles of cargo attachment to cytoplasmic motor proteins. Curr. Opin. Cell Biol. 14, 63–68. [DOI] [PubMed] [Google Scholar]
- Kamal, A., Stokin, G.B., Yang, Z., Xia, C.H., and Goldstein, L.S. (2000). Axonal transport of amyloid precursor protein is mediated by direct binding to the kinesin light chain subunit of kinesin-I. Neuron 28, 449–459. [DOI] [PubMed] [Google Scholar]
- Karcher, R.L., Deacon, S.W., and Gelfand, V.I. (2002). Motor-cargo interactions: the key to transport specificity. Trends Cell Biol. 12, 21–27. [DOI] [PubMed] [Google Scholar]
- Kelkar, N., Gupta, S., Dickens, M., and Davis, R.J. (2000). Interaction of a mitogen-activated protein kinase signaling module with the neuronal protein JIP3. Mol. Cell. Biol. 20, 1030–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreegipuu, A., Blom, N., and Brunak, S. (1999). PhosphoBase, a database of phosphorylation sites: release 2.0. Nucleic Acids Res. 27, 237–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda, S., Nakagawa, N., Tokunaga, C., Tatematsu, K., and Tanizawa, K. (1999). Mammalian homologue of the Caenorhabditis elegans UNC-76 protein involved in axonal outgrowth is a protein kinase C zeta-interacting protein. J. Cell Biol. 144, 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littleton, J.T., Bellen, H.J., and Perin, M.S. (1993). Expression of synaptotagmin in Drosophila reveals transport and localization of synaptic vesicles to the synapse. Development 118, 1077–1088. [DOI] [PubMed] [Google Scholar]
- Lloyd, E.D., and Wooten, M.W. (1992). pp42/44MAP kinase is a component of the neurogenic pathway utilized by nerve growth factor in PC12 cells. J. Neurochem. 59, 1099–1109. [DOI] [PubMed] [Google Scholar]
- Luo, L., Tully, T., and White, K. (1992). Human amyloid precursor protein ameliorates behavioral deficit of flies deleted for Appl gene. Neuron 9, 595–605. [DOI] [PubMed] [Google Scholar]
- Luo, L.Q., Martin-Morris, L.E., and White, K. (1990). Identification, secretion, and neural expression of APPL, a Drosophila protein similar to human amyloid protein precursor. J. Neurosci. 10, 3849–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas, A., Van Dyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Martin, M., Iyadurai, S.J., Gassman, A., Gindhart, J.G., Jr., Hays, T.S., and Saxton, W.M. (1999a). Cytoplasmic dynein, the dynactin complex, and kinesin are interdependent and essential for fast axonal transport. Mol. Biol. Cell 10, 3717–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M.A., Hurd, D.D., and Saxton, W.M. (1999b). Kinesins in the nervous system. Cell Mol. Life Sci. 56, 200–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva, K.D., Kay, B.K., and McPherson, P.S. (1997). Synaptojanin forms two separate complexes in the nerve terminal. Interactions with endophilin and amphiphysin. J. Biol. Chem. 272, 27239–27245. [DOI] [PubMed] [Google Scholar]
- Nakai, K., and Horton, P. (1999). PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–36. [DOI] [PubMed] [Google Scholar]
- Parchaliuk, D.L., Kirkpatrick, R.D., Agatep, R., Simon, S.L., and Gietz, R.D. (1999). Yeast two-hybrid system: part C-characterizing positives. Tech. Tips Online, P01714.
- Patel, N., Thierry-Mieg, D., and Mancillas, J.R. (1993). Cloning by insertional mutagenesis of a cDNA encoding Caenorhabditis elegans kinesin heavy chain. Proc. Natl. Acad. Sci. USA 90, 9181–9185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, K., Perez, S.E., Yang, Z., Xu, J., Ritchings, B.W., Steller, H., and Goldstein, L.S. (1999). Kinesin-II is required for axonal transport of choline acetyltransferase in Drosophila. J. Cell Biol. 147, 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold, D.S., and Neet, K.E. (1989). The lack of a role for protein kinase C in neurite extension and in the induction of ornithine decarboxylase by nerve growth factor in PC12 cells. J. Biol. Chem. 264, 3538–3544. [PubMed] [Google Scholar]
- Rothwell, W.F., and Sullivan, W. (2000). Fluorescent analysis of Drosophila embryos. In: Drosophila Protocols, ed. W. Sullivan, M. Ashburner, and R.S. Hawley, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 141–157.
- Saxton, W.M., Hicks, J., Goldstein, L.S., and Raff, E.C. (1991). Kinesin heavy chain is essential for viability and neuromuscular functions in Drosophila, but mutants show no defects in mitosis. Cell 64, 1093–1102. [DOI] [PubMed] [Google Scholar]
- Setou, M., Seog, D.H., Tanaka, Y., Kanai, Y., Takei, Y., Kawagishi, M., and Hirokawa, N. (2002). Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature 417, 83–87. [DOI] [PubMed] [Google Scholar]
- Stebbings, H. (2001). Cytoskeleton-dependent transport and localization of mRNA. Int. Rev. Cytol. 211, 1–31. [DOI] [PubMed] [Google Scholar]
- Stock, M.F., Guerrero, J., Cobb, B., Eggers, C.T., Huang, T.G., Li, X., and Hackney, D.D. (1999). Formation of the compact confomer of kinesin requires a COOH-terminal heavy chain domain and inhibits microtubule-stimulated ATPase activity. J. Biol. Chem. 274, 14617–14623. [DOI] [PubMed] [Google Scholar]
- Terada, S., Kinjo, M., and Hirokawa, N. (2000). Oligomeric tubulin in large transporting complex is transported via kinesin in squid giant axons. Cell 103, 141–155. [DOI] [PubMed] [Google Scholar]
- Torroja, L., Chu, H., Kotovsky, I., and White, K. (1999). Neuronal overexpression of APPL, the Drosophila homologue of the amyloid precursor protein (APP), disrupts axonal transport. Curr. Biol. 9, 489–492. [DOI] [PubMed] [Google Scholar]
- Trofatter, J.A., et al. (1993). A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell 72, 791–800. [DOI] [PubMed] [Google Scholar]
- Vale, R.D., and Fletterick, R.J. (1997). The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 13, 745–777. [DOI] [PubMed] [Google Scholar]
- Vale, R.D., Reese, T.S., and Sheetz, M.P. (1985). Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey, K.J., Meyer, D., Deehan, R., Blenis, J., Schnapp, B.J., Rapoport, T.A., and Margolis, B. (2001). Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J. Cell Biol. 152, 959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz, A., Ramrath, A., Grimm, A., and Knust, E. (2000). Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol. 150, 1361–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten, M.W., Zhou, G., Seibenhener, M.L., and Coleman, E.S. (1994). A role for zeta protein kinase C in nerve growth factor-induced differentiation of PC12 cells. Cell Growth Differ. 5, 395–403. [PubMed] [Google Scholar]
- Yasuda, J., Whitmarsh, A.J., Cavanagh, J., Sharma, M., and Davis, R.J. (1999). The JIP group of mitogen-activated protein kinase scaffold proteins. Mol. Cell. Biol. 19, 7245–7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervos, A.S., Gyuris, J., and Brent, R. (1993). Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell 72, 223–232. [DOI] [PubMed] [Google Scholar]