Abstract
Genetic evidence suggests that DNA polymerase epsilon (Pol ε) has a noncatalytic essential role during the early stages of DNA replication initiation. Herein, we report the cloning and characterization of the second largest subunit of Pol ε in fission yeast, called Dpb2. We demonstrate that Dpb2 is essential for cell viability and that a temperature-sensitive mutant of dpb2 arrests with a 1C DNA content, suggesting that Dpb2 is required for initiation of DNA replication. Using a chromatin immunoprecipitation assay, we show that Dpb2, binds preferentially to origin DNA at the beginning of S phase. We also show that the C terminus of Pol ε associates with origin DNA at the same time as Dpb2. We conclude that Dpb2 is an essential protein required for an early step in DNA replication. We propose that the primary function of Dpb2 is to facilitate assembly of the replicative complex at the start of S phase. These conclusions are based on the novel cell cycle arrest phenotype of the dpb2 mutant, on the previously uncharacterized binding of Dpb2 to replication origins, and on the observation that the essential function of Pol ε is not dependent on its DNA synthesis activity.
INTRODUCTION
Chromosomal DNA replication in eukaryotic cells requires the activity of at least three DNA polymerases: α, δ, and ε (Waga and Stillman, 1998; Hubscher et al., 2000; Bell and Dutta, 2002). Pol α is tightly associated with a primase activity capable of de novo DNA synthesis, suggesting that this enzyme is responsible for initiation of both leading and lagging strands (Waga and Stillman, 1998). Pol α/primase can only synthesize short primers that must then be extended by the activity of a processive DNA polymerase(s). Biochemical analysis of simian virus 40 (SV40) DNA replication, which has been extensively used as a model system for eukaryotic DNA replication, has shown that primers synthesized by Pol α can be extended by Pol δ and that these two polymerases are sufficient for SV40 replication in vitro (Weinberg et al., 1990; Waga et al., 1994).
A second processive DNA polymerase, Pol ε, is required for cell viability and chromosomal DNA replication in both fission (D'Urso and Nurse, 1997) and budding yeast (Morrison et al., 1990; Araki et al., 1992; Budd and Campbell, 1993). Pol ε has also been implicated in both DNA repair (Jessberger et al., 1993; Wang et al., 1993; Shivji et al., 1995; Holmes, 1999) and cell cycle checkpoint control in eukaryotic cells (Navas et al., 1995). Recently, we have shown that the DNA polymerase and exonuclease domains of Pol ε are dispensable for cell viability in fission yeast (Feng and D'Urso, 2001). Similar observations have been made for the evolutionarily distant yeast, Saccharomyces cerevisiae (Kesti et al., 1999; Dua et al., 2000). These findings have raised important questions regarding the essential role of this replicative enzyme in DNA synthesis. Although the N-terminal catalytic domains are dispensable, the C-terminal half of the enzyme is essential for cell viability and chromosomal replication in fission yeast (D'Urso and Nurse, 1997). In particular, we have shown that temperature-sensitive mutants of Pol ε in fission yeast arrest with a 1C DNA content upon shift to the restrictive temperature, suggesting that Pol ε (presumably the noncatalytic C-terminal half of the protein) is required during the early stages of DNA replication initiation (D'Urso and Nurse, 1997). Nevertheless, its precise molecular function in eukaryotic chromosomal DNA replication remains obscure.
Pol ε has been purified from both budding yeast and human cell extracts as a complex of at least four distinct polypeptides that are conserved from yeast to human (Hamatake et al., 1990; Chui and Linn, 1995). Only the largest 256-kDa subunit contains the domains required for DNA synthesis activity, whereas the function of the smaller subunits, called Dpb2–4, is currently unknown. Deletion of each of the three smaller subunits from the S. cerevisiae genome results in different phenotypes. Only Dpb2 is essential for viability and normal S phase progression (Araki et al., 1991a). Although both Dpb3 and Dpb4 are not required for cell viability, deletion of the genes encoding these proteins does result in partial defects in DNA replication (Araki et al., 1991b; Ohya et al., 2000). Interestingly, both Dpb3 and Dpb4 contain histone-fold motifs, suggesting these proteins might be involved in chromatin remodeling (Araki et al., 1991b; Li et al., 2000; Ohya et al., 2000). Based on the existence of these motifs, it has been proposed these subunits might facilitate replication through heterochromatic regions of DNA (Fuss and Linn, 2002).
To further explore the function of the Pol ε in fission yeast, we have set out to clone and characterize the three additional putative subunits of the Pol ε complex. In this manuscript, we report the cloning and characterization of the second largest subunit encoded by dpb2+. We show that deletion of dpb2 from the yeast genome is lethal and that cells depleted for Dpb2 protein display an S phase delay consistent with an initiation defect. Moreover, we have constructed a temperature-sensitive dpb2 mutant and found that these cells arrest in late G1/early S phase upon shift to the restrictive temperature. This phenotype has never been observed for temperature-sensitive mutants of either Pol α or Pol δ, suggesting that Pol ε and Dpb2 provide a novel function(s) during DNA replication initiation (Hughes et al., 1992; D'Urso et al., 1995; MacNeill et al., 1996; Iino and Yamamoto, 1997; Reynolds et al., 1998; Reynolds and MacNeill, 1999). Consistent with this notion, using a chromatin immunoprecipitation (ChIP) assay, we find that both Dpb2 and Pol ε bind replication origins early in S phase, very near to the time of DNA replication initiation.
EXPERIMENTAL PROCEDURES
Schizosacchromyces pombe Strains and Materials
All S. pombe strains were derived from 972(h–), 975(h+), and 968(h90) by using standard genetic procedures (Moreno et al., 1991; Table 1). S. pombe cosmid p8B7, containing the complete genomic sequence of dpb2+, was obtained from The Sanger Center (Hinxton, United Kingdom). S. pombe strain OGC9 and anti-Mcm6 polyclonal antibodies were kindly provided by H. Masukata (Osaka University, Osaka, Japan).
Table 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| 972 | h- | Moreno et al., 1991 |
| 567 | h- ade6-704 leu1-32 ura4-D18 | Moreno et al., 1991 |
| GD28 | h-/h+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 | This study |
| GD131 | h-/h+ dpb2+/dpb2::ura4+ ade6-M210/ade6-M216 leu1-32/leu1-32 ura4-D18/ura4-D18 | This study |
| GD149 | h+ dpb2::ura4+ leu1-32 ura4-D18 int pJK148(nmt81)-dpb2+ | This study |
| GD242 | h- dpb2::4xflag-dpb2+(sup3-5) ade6-704 leu1-32 ura4-D18 | This study |
| GD254 | h- cdc25-22 dpb2::4xflag-dpb2+(sup3-5) ade6-704 leu1-32 ura4-D18 | This study |
| GD280 | h- dpb2::ura4+ leu1-32 ura4-D18 int pJK148(nmt81)-dpb2∋413-417 | This study |
| GD261 | h- cdc10-129 dpb2::4xflag-dpb2+(sup3-5) ade6-704 leu1-32 ura4-D18 | This study |
| GD237 | h- cdc25-22 cdc20::ura4+ ade6-M216 leu1-32 ura4-D18 int pJK148 (nmt1) 4xflag-cdc20C1 | This study |
| GD205 | h- cdc25-22 ade6-704 leu1-32 ura4-D18 | This study |
S. pombe Methods
All media and growth conditions, unless otherwise noted, were as described previously (Moreno et al., 1991). DNA content was determined by fluorescence-activated cell sorting (FACS) as described previously (Sazer and Sherwood, 1990). To block cells at the G2/M transition, cells containing the cdc25-22 allele were incubated in minimal media at the restrictive temperature of 36°C for 4 h. Reentry into the cell cycle was induced by rapidly chilling cells to the permissive temperature of 25°C.
Molecular Cloning and the Generation of the Heterozygous Diploid Strain GD131 (dpb2+/dpb2::ura4+) and the Haploid Strain GD149 [dpb2::ura4+ int pJK148-(nmt81)dpb2+]
One to two kilobases of flanking genomic sequence, upstream and downstream of the dpb2+ open reading frame, respectively, were amplified by polymerase chain reaction (PCR). The amplified products were cloned into pKS(+)-bluescript on either side of a 1.8-kb HindIII fragment containing the ura4+ gene, generating plasmid pKS(+)Δdpb2::ura4+. Digestion of this plasmid with EcoRI/XhoI yielded a 4.3-kb fragment containing ura4+ and dpb2+ flanking sequences that was then transformed into the diploid strain GD28. Stable ura4+ integrants were selected and deletion of dpb2+ was confirmed by Southern blot analysis.
To generate GD149, dpb2+ was amplified from the cosmid p8B7 by using forward primer dpb2-g(F) (5′-cggcgcatatgaacaattccattacgg-3′) and the reverse primer dpb2-b(R) (5′-gtccggcccgggttctattattcggggctcag-3′), tagged with NdeI and XmaI sites, respectively. The product was then cloned into pRep81 at the NdeI and XmaI sites. The resulting plasmid was then digested with PstI and SacI, yielding a 4.0-kb fragment containing dpb2+ flanked by the nmt81 promoter and terminator sequences. This PstI/SacI fragment was then cloned into the integrative vector pJK148, linearized within the leu1+ gene and transformed into the diploid strain GD131. Stable leu+ transformants were selected, induced to sporulate, and haploid progeny prototrophic for both uracil and leucine were selected (strain GD149).
Epitope-tagging of Chromosomal dpb2+
Four tandem copies of the FLAG epitope were first introduced in frame at the amino terminus of dpb2+ in pRep81-dpb2+ as follows. First, two annealed oligonucleotides of the following sequences, 5′-tatggactacaaggacgacgatgacaaggattacaaagatgacgacgataagct-3′ and 5′-taagcttatcgtcgtcatctttgtaatccttgtcatcgtcgtccttgtagtcca-3′, were cloned into the NdeI site of the plasmid pRep1-dpb2+. The transformants were screened for the correct orientation of the insertion, resulting in the fusion of two copies of FLAG in frame with dpb2+. Consequently, the insertion also led to the elimination of the previous NdeI site at the beginning of the dpb2+ open reading frame and the simultaneous introduction of a new NdeI site at the beginning of the two copies of FLAG. The same procedure was repeated to introduce another two copies of FLAG in frame with the 2xflag-dpb2+, resulting in the plasmid pRep1-4xflag-dpb2+. Then, ∼1 kb of genomic DNA immediately upstream of the ATG (including the dpb2+ promoter sequences) was amplified from cosmid p8B7 by using primers tagged with XhoI and NdeI restriction sites. The amplified product was then combined with the NdeI/XmaI fragment of pRep1-4xflag-dpb2+, and in a triple-way ligation, cloned into pKS-bluescript at the XhoI and XmaI sites. The resulting plasmid, called pKS(+)-4xflag-dpb2+ was then digested BamHI/SmaI to delete the C-terminal half of dpb2+, which was then replaced with a BamHI/EcoRV fragment containing the sup3-5 gene. This plasmid, called pKS(+)-4xflag-dpb2N(sup3-5) was linearized at the BglII site within the dpb2+ gene and transformed into strain 567. Replacement of the wild-type dpb2+ gene with 4xflag-dpb2+ was confirmed by both PCR and Western blot analysis of 4x-FLAG Dpb2 by using anti-FLAG monoclonal antibodies (our unpublished data).
Epitope Tagging of cdc20C1
To tag cdc20C1 gene with 4xflag, a fragment containing the nmt1 promoter and 4xflag epitope was amplified from pRep1-4xflag-dpb2+ by using the primers nmtP-1F(PstI) (5′ agcttgcatgccctgcaggtcg 3′) and 4xFLAG-R(XhoI) (5′ ggtaactcgagcaatggaattgttcataag 3′). This fragment was then cloned at the PstI and XhoI sites in pRep3X. The resulting plasmid was named pRep3X-4xflag. A 2.9-kb XhoI/XmaI fragment encoding the C-terminal half of Pol ε from amino acid position 1319–2199 (Cdc20C1) was cloned into the plasmid pRep3X-4xflag. Finally, a 5.1-kb PstI/SacI fragment from plasmid pRep3X-4xflag-cdc20C1 was cloned into pJK148, generating pJK148-nmt1–4xflag-cdc20C1, which was then integrated at the leu1 locus as described previously (Keeney and Boeke, 1994).
Site-directed Mutagenesis of dpb2+
To delete amino acid residues 413–417 in Dpb2, the N- and C-terminal halves of dpb2+ were PCR amplified from plasmid pRep81-dpb2+, by using primers dpb2-g(F) with dpb2Δ413-417-R (5′-cccatttgtcgtccaagggacaaatatgaattttgtcttttcgca-3′) and dpb2-b(R) with dpb2Δ413-417-F (5′-tgcgaaaagacaaaattcatatttgtcccttggacgacaaatggg-3′), respectively. The two amplified products were combined and used as a template for PCR amplification of dpb2Δ413-417 using primers dpb2-g(F) and dpb2-b(R). The gene was then cloned into pRep81 at the NdeI and XmaI sites, generating pRep81-dpb2Δ413-417. The PstI/SacI fragment from pRep81-dpb2Δ413-417 was then cloned into pJK148 and integrated into strain GD131 as described for the generation of strain GD149 (see above).
ChIP Analysis
The ChIP assay was performed as described previously (Ogawa et al., 1999) with minor modifications. Fifty milliliters of fission yeast cells (107/ml) were cross-linked in 1% formaldehyde at room temperature for 30 min. Cells were washed once with 40 ml of double distilled H2O followed by resuspension in 100 μl of lysis buffer (50 mM HEPES, pH 7.9; 140 mM NaCl; 1 mM EDTA; 1% Triton X-100; 0.1% sodium deoxycholate; 1 mM phenylmethylsulfonyl fluoride; 10 μg/ml aprotinin; 1 μg/ml leupeptin; 1 μg/ml pepstatin). Glass beads (0.5 g) were then added. Cells were then broken with three 20-s pulses in a Savant FP120 bead beater with intermittent chilling on ice. After addition of 900 μl of lysis buffer, the cell lysate was transferred to a new tube and sonicated until the cellular DNA was sheared to an average length of 0.5–1 kb. The lysate was then clarified by centrifugation (12,000g, 15 min) at 4°C, and 90% of the supernatant was incubated with anti-FLAG or anti-Mcm6p antibodies bound to magnetic beads (Dynal Biotech, Oslo, Norway). Purification of DNA from the immunoprecipitates and the remaining 10% of supernatant (Input control) was performed as described previously (Strahl-Bolsinger et al., 1997). Purified DNA was dissolved in 30 μl of double distilled H2O and 1 μl of each DNA sample (IP or Input DNA) was used for each PCR reaction. The nucleotide sequences of the PCR primers used were as follows: ars2004-F, 5′-atggtagatggagaaacggg-3′; ars2004-R, 5′-cacggcatctttcttcacga-3′; ars3002-F, 5′-ttggcgctaaacaatctctg-3′; ars3002-R, 5′-tccttgtcgaactcaattgc-3′; nonARS-F, 5′-tcgaagatcctaccgctttc-3′; nonARS-R, 5′-gattcacataacccgctagc-3′. The ars2004, ars3002, and nonARS primers were used at 0.3, 0.3, and 0.5 μM, respectively. Reaction conditions for the PCR were as described by Ogawa et al., (1999): 95°C, 9 min; 30 cycles of (95°C, 1 min; 53°C, 1 min; 72°C, 2 min); 72°C, 7 min; 4°C, ∞. Amplified products were separated on a 2.5% agarose gel and stained with 0.5 μg/ml ethidium bromide. Gel images and quantification of signals were analyzed using an AlphaImager 2000 (Alpha Innotech, San Leandro, CA).
RESULTS
S. pombe dpb2+ Is Essential for Cell Viability
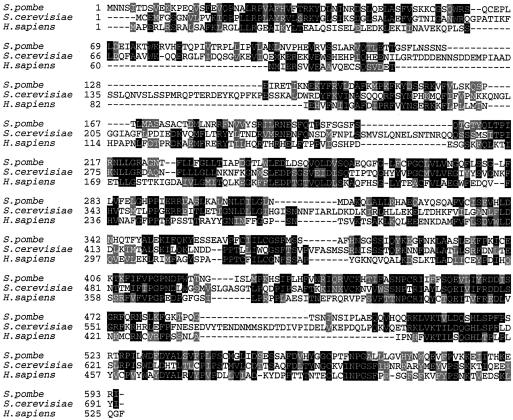
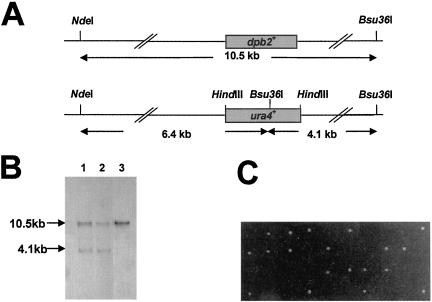
We have identified a gene in the S. pombe genome database that is predicted to encode a protein with 31% identity to S. cerevisiae DPB2 and human DPE2 (Figure 1). To test whether this gene, which we have named dpb2+, is essential for cell viability, we replaced the complete open reading frame with a single copy of the ura4+ gene, generating the diploid strain GD131. Replacement of dpb2+ with ura4+ was confirmed by Southern blot analysis (Figure 2, A and B). The diploid strain GD131 was induced to sporulate, and the germinating spores were subjected to tetrad analysis. Spores deleted for dpb2+ fail to form colonies, demonstrating that dpb2+ is essential for cell viability (Figure 2C). Microscopic examination of germinating spores reveals that cells deleted for dpb2+ undergo one to two rounds of cell division before arresting with an elongated “cdc” phenotype (our unpublished data), indicating that dpb2+ is essential for normal cell cycle progression.
Figure 1.
Protein sequence alignment of S. pombe dpb2+, S. cerevisiae DPB2, and human DPE2. Fission yeast dpb2+ shares 31% identity with both the human and S. cerevisiae genes. Identical residues are indicated in black, whereas homologous residues are indicated in gray. Sequence alignment was performed using ClustalW 1.8.
Figure 2.
dpb2+ is essential for cell viability. (A) Strategy for deleting dpb2+ by gene replacement. The 1.76-kb open reading frame of dpb2+ was replaced with the 1.8-kb ura4+ gene. A HindIII fragment containing the ura4+ gene was cloned into pBluescript. A 1.2- and 2-kb fragment corresponding to sequences upstream and downstream of the dpb2+ open reading frame were cloned into the EcoRI/HindIII and HindIII/XhoI sites of pBluescript-ura4+, respectively. The resulting plasmid was digested with EcoRI and XhoI and the excised fragment was transformed into the diploid strain GD28. Positive transformants were selected on minimal medium minus uracil. (B) Southern blot analysis of heterozygous diploid +/Δ (lanes 1 and 2) and homozygous diploid +/+. DNA was digested by NdeI and Bsu36I and probed with the EcoRI and HindIII fragment. (C) Tetrad analysis of the heterozygous diploid strain (+/Δ) revealed a two viable:two nonviable segregation as expected for an essential gene disruption. All viable spores were confirmed to be ura–.
Dpb2 Is Required for Normal S Phase Progression
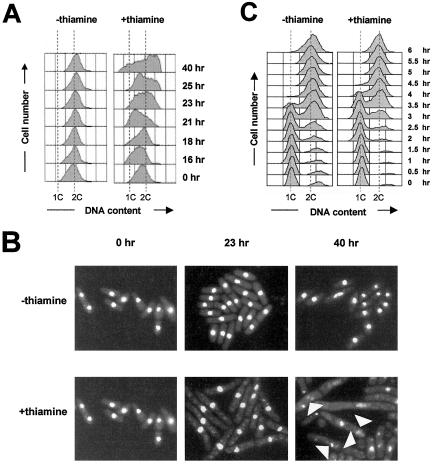
Based on the observations that budding yeast DPB2 is required for chromosomal DNA replication (Araki et al., 1991a), we anticipated that S. pombe cells lacking dpb2+ would also be defective for DNA synthesis. To test this possibility, we first analyzed DNA content in Δdpb2 germinating spores by FACS. Although spores lacking dpb2+ proceed through S phase more slowly (our unpublished data), DNA synthesis seemed to be completed before cell cycle arrest, suggesting that sufficient amounts of Dpb2 protein were present (presumably contributed by the mother cell during sporulation) to support limited amounts of DNA synthesis. Therefore, we sought an alternative method to analyze DNA replication in the absence of Dpb2 protein. To this end, we constructed a Δdpb2 strain containing an integrated copy of dpb2+ under the control of the thiamine repressible nmt81 promoter (strain GD149). Cells were grown in the absence of thiamine for several generations and then split into two cultures, one of which was treated with thiamine to repress dpb2+ expression. FACS analysis demonstrates that cells grown in the absence of thiamine display a 2C DNA content peak typical of exponentially growing S. pombe cells (Figure 3A, left). In contrast, the culture treated with thiamine gradually accumulated cells with less than 2C DNA content, suggesting DNA replication was impaired (Figure 3A, right). Eventually these cells ceased to divide and elongated with a cdc phenotype (Figure 3B). Moreover, 4,6-diamidino-2-phenylindole staining of nuclei revealed that chromosomes became fragmented after incubation in the presence of thiamine, suggesting that Dpb2 might be important for maintaining chromosomal integrity after DNA synthesis arrest (Figure 3B, 40 h).
Figure 3.
Cells depleted for Dpb2 are defective in S phase progression. (A) A culture of haploid GD149 cells was grown to log phase, the culture was then diluted and one-half was treated with 10 μg/μl thiamine to repress dpb2+ gene expression. Samples were collected at the indicated times and DNA content analyzed by flow cytometry. (B) Cells stained with the DNA binding dye 4,6-diamidino-2-phenylindole after incubation with thiamine at 0, 23, and 40 h. Arrows indicate abnormal nuclear morphology, including missegregated chromosomes and anucleate cells. (C) DNA replication initiation is delayed in cell cycle synchronized cultures depleted for Dpb2. Wild-type (972) or GD149 cells were grown in the absence of nitrogen to arrest cells in G1. During nitrogen starvation, thiamine was added (10 μg/μl) to the media to repress transcription from the nmt81 promoter. After 20 h or when >90% of the cells showed a G1 arrest, cells were induced to reenter the cell cycle by addition of fresh media containing nitrogen, as well as thiamine to maintain transcriptional repression of dpb2+. Cells were collected every 30 min for 6 h and the DNA content analyzed by flow cytometry. The positions of 1C and 2C DNA content are indicated.
To further examine the role of Dpb2 during chromosomal DNA replication, Dpb2 protein was depleted in cell cycle-synchronized cultures. Exponentially growing GD149 cells grown in the absence of thiamine were synchronized in G0/G1 by nitrogen starvation. During starvation, half of the culture was treated with thiamine to inhibit dpb2+ expression. After arrest in G0/G1, cells were stimulated to reenter the cell cycle by addition of fresh media containing nitrogen with or without thiamine (+ or – thiamine). Cells were collected every 30 min and DNA content analyzed by FACS. As shown in Figure 3C, compared with the control (–thiamine), cells treated with thiamine (+thiamine) proceed through S phase more slowly. However, these cells still seem to complete a substantial amount of DNA replication. The persistence of the 1C DNA content peak in cells depleted for Dpb2 (compare +thiamine and –thiamine at 3.5 h) suggest that initiation of DNA replication is defective in these cells. These data are consistent with Dpb2 being essential for initiation and possibly elongation of DNA synthesis.
A Temperature-sensitive dpb2 Mutant Arrests in Early S Phase after Shift to the Restrictive Temperature
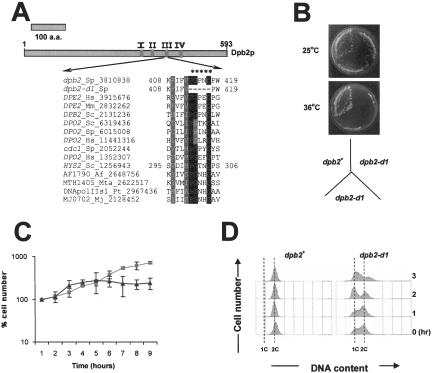
The experiments described above were designed to test whether Dpb2 is required for DNA replication. Although the data support this hypothesis, the inability to deplete all the cellular Dpb2 made it difficult to address its precise role in S phase. Therefore, we decided to generate a dpb2 temperature-sensitive mutant that would allow more rapid and complete inactivation of the protein. To select a target area for mutagenesis, we compared all known Dpb2 proteins to identify conserved amino acid residues. As previously reported, the regulatory B-type subunits of eukaryotic DNA polymerases, which includes Dpb2, belong to a protein superfamily (Makiniemi et al., 1999) that contains a calcineurin-like phosphatase domain similar to those found in archaeal X-type DNA polymerases (Aravind and Koonin, 1999). Although some of the key residues required for phosphatase activity are no longer present in the eukaryotic proteins (Aravind and Koonin, 1999), some of the remaining conserved residues might be important to stabilize the protein. In fact, several temperature-sensitive mutations found in eukaryotic DNA polymerase B-type subunits have been localized to this region of the protein (Makiniemi et al., 1999). For example, in the HYS2/POL31 gene of S. cerevisiae, a mutation converting an Asp to Asn at amino acid position 304 renders the protein temperature sensitive (Hashimoto et al., 1998).
Therefore, we set out to mutate key residues in the corresponding domain (motif III) of S. pombe dpb2+ (Figure 4A). Based on the analysis of hys2 D304N mutation, we first mutated the corresponding Asp417 in dpb2+ to either Asn or Ala by site-directed mutagenesis. Neither of these mutations had any effect on yeast cell cycle progression or cell viability (our unpublished data). Likewise, mutation of the highly conserved Pro413 to Ala had no effect on Dpb2 function (our unpublished data). Finally, deletion of all five residues from amino acid position 413–417 rendered the protein temperature sensitive. As shown in Figure 4B, mutant dpb2-d1 (Δ413–417) failed to form colonies at the restrictive temperature of 36°C. More importantly, after shift to the restrictive temperature, dpb2-d1 divides once before arresting with a near 1C DNA content (Figure 4, C and D), suggesting that Dpb2 is required during initiation, or at least during an early step in replication elongation (see DISCUSSION).
Figure 4.
Temperature-sensitive mutant dpb2-d1 arrests in late G1-early S phase after shift to the restrictive temperature. (A) Amino acid sequence alignment of the most highly conserved region (motif III) of the putative phosphatase domain with a protein super family consisting of the B subunits of eukaryotic DNA polymerases and selected members of the regulatory subunits of the archael X-family DNA polymerases. The location of the conserved domains (motif I-IV) is shown for S. pombe Dpb2. Asterisks indicate the amino acids 413–417, deleted in the mutant dpb2-d1. Identical residues are shown in black. Conservative changes are indicated in gray. Species abbreviations are as follows: Af, Archaeoglobus fulgidus; Hs, Homo sapiens; Mj, Methanococcus jannaschii; Mm, Mus musculus; Mta, Methanothermobacter thermoautotrophicum; Pt, Pyrococcus furiosus; Sc, S. cerevisiae; and Sp, S. pombe. (B) Cells carrying wild-type dpb2+ or dpb2-d1 (Δ413–417) were streaked on minimal medium and incubated at 25°C or 36°C. Plates were photographed after 4 d. (C) Cell number analysis in dpb2-d1 cells after shift to the restrictive temperature. Wild-type 972 cells or dpb2-d1 (Δ413–417) were grown at 25°C in minimal medium to mid-log phase and shifted to 36°C for 8 h. Please note that after shift to the restrictive temperature, dpb2-d1 is limited to one population doubling (typical of an early S phase arrest), whereas the wild-type culture continues to grow exponentially. (D) dpb2-d1 cells arrest with a 1C DNA content after shift to restrictive temperature. dpb2+ (top) and dpb2-d1 (bottom) were grown at 25°C overnight. The following day, cells were shifted to the restrictive temperature of 36°C, collected at the indicated times, and measured for DNA content by flow cytometry. The position of 1C and 2C DNA content are indicated.
Dpb2 Protein Binds to DNA Replication Origins Early in S Phase
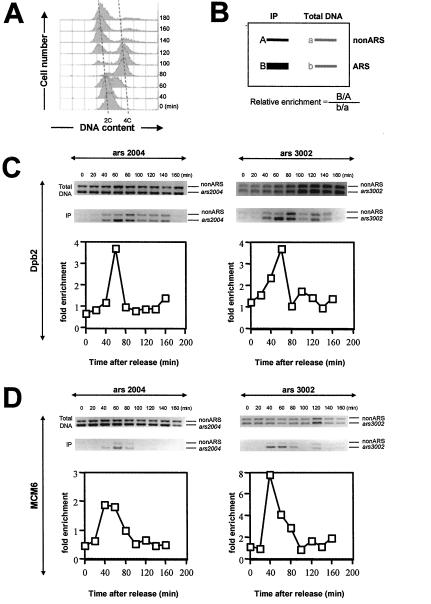
If Dpb2 is required for initiation of DNA replication, then this protein should interact with replication origins at the beginning of S phase. To test this possibility, we examined whether S. pombe Dpb2 associates with origins of replication during the cell cycle by using a ChIP assay coupled with PCR. For these experiments, we N-terminally tagged the endogenous copy of dpb2+ with four tandem copies of the FLAG epitope. This strain also includes the temperature-sensitive cdc25 allele to allow synchronization in G2 (GD254). Consistent with data from other laboratories (Ogawa et al., 1999; Wuarin et al., 2002), DNA replication begins ∼60–80 min after release from the cdc25 block (Figure 5A). It should be pointed out that DNA replication begins before cell separation after release from the cdc25 block point, and therefore initiation of DNA replication correlates with a shift from 2C to 4C DNA content. As cells separate, most cells have completed DNA replication, resulting in the accumulation of cells with a 2C DNA content peak at later times in the experiment. We used primers previously designed by Ogawa et al., 1999 to amplify sequences specific to origin DNA (ars2004 or ars3002) and to nonARS DNA, located ∼25 kb centromere-proximal to ars2004 on chromosome II (Ogawa et al., 1999). Similar primers have been successfully used for the analysis of Orc1 and Mcm binding to origin DNA (Ogawa et al., 1999; Takahashi and Masukata, 2001; Takahashi et al., 2003). These primer sets were used in quantitative PCR on DNA prepared either from immunoprecipitated chromatin fractions or from total DNA isolated from whole cell soluble extracts. To control for loading differences or variance in amplification from the different primer sets, we normalized the relative enrichment at ARS (autonomous replicating sequence) sequences to the nonARS PCR product (Figure 5B).
Figure 5.
4xFLAG-Dpb2 binds ARS elements early in S phase. (A) Analysis of DNA content by flow cytometry indicates that DNA synthesis begins at 60–80 min after release from the G2 block. (B) Relative enrichment ratio of immunoprecipitated ARS DNA to nonARS DNA was calculated and plotted from the raw data in C. (C and D) ChIP analyses with antibody to the FLAG-epitope for Dpb2 (C) or antibody to Mcm6 (D) were used to measure the levels of Dpb2 and Mcm6 at ARS loci. DNA isolated from the immunoprecipitated chromatin (IP) or from whole cell soluble extracts (Total DNA) was subjected to PCR to amplify DNA fragments from either ars2004, ars3002 or from nonARS DNA. Please note that the peak of Dpb2 binding to ARS elements occurs 60 min after release from the G2 block (C) in contrast to Mcm6 binding which peaks at 40 min postrelease (D). No significant chromatin binding was observed when cross-linking agent was omitted, if cell lysates were prepared in the absence of FLAG-tagged protein, or if antibodies were excluded from the immunoprecipitation reaction (our unpublished data).
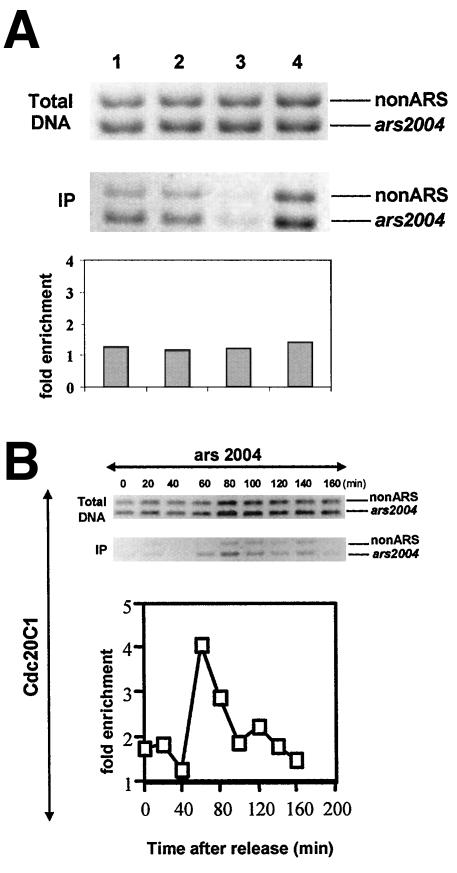
Our data demonstrate that Dpb2 binds preferentially to origins of replication ∼60 min after release from the cdc25 block (Figure 5C), very near to the time of DNA replication initiation (Figure 5A). Although the level of enrichment (three- to fivefold) of Dpb2 binding to ARS elements is similar to that observed for Mcm6 (Figure 5D), a component of the prereplicative complex (preRC), the peak of Dpb2 association with origin DNA seems to occur slightly later than that of Mcm6. Mcm6 is thought to bind to origin DNA during the later stages of mitosis or in early G1 (Ogawa et al., 1999). Therefore, our data are consistent with Dpb2 binding to replication origins in late G1 or early S phase after assembly of the preRC. To test whether Dpb2 can interact with origin DNA in the absence of preRCs, we examined Dpb2 binding to origin DNA in cdc10-129 cells at 36°C. Cdc10 is required for expression of the MCM loading factors Cdc18 and Cdt1, and in cdc10-V50 cells at 36°C, Mcm4 fails to associate with origin-associated DNA sequences (Wuarin et al., 2002). No specific association of Dpb2 to ARS-associated chromatin was observed in cdc10-129 cells at 36°C, suggesting that the binding of Dpb2 to origin DNA is dependent on prior assembly of the preRC (Figure 6A). It should be noted that ChIP analysis is not sensitive enough to detect enrichment of Dpb2 or Mcm6 binding to origin DNA in exponentially growing populations where very few cells are in S phase. Therefore, considering that the amount of Dpb2 association to chromatin detected in cdc10-arrested cells is similar to the amount detected in either G2-arrested (cdc25) or exponentially growing 972 cells, we conclude that very little, if any, Dpb2 protein is bound to chromatin in the absence of Cdc10.
Figure 6.
Dpb2 fails to associate with origin DNA in cdc10-129 cells at 36°C. (A) Exponentially growing 972 cells (lane 1), G2 arrested GD254 cells (lane 2), or G1 arrested GD261 cells (lanes 3 and 4) were subjected to ChIP analysis for the presence of chromatin bound Dpb2. Either magnetic beads conjugated to anti-FLAG antibodies (lanes 1, 2 and 4) or magnetic beads alone (lane 3) were used for immunoprecipitation. The relative ratio of immunoprecipitated DNA to total DNA was plotted. No enrichment of Dpb2 binding to ars2004 is observed in cdc10-129 cells arrested in G1 (lane 4). The levels of binding to either non-ARS or ARS-associated chromatin in cdc10 cells is equivalent to the level detected in either G2 arrested (lane 2) or exponentially growing cells (lane 1). Very little chromatin is precipitated from cdc10 cells in the absence of antibodies (lane 3). (B) ChIP analysis using anti-FLAG antibodies for detection of Pol ε-Cterm at ars2004.
In a previous study, we demonstrated that fission yeast cells are viable in the absence of the N-terminal, but not C-terminal half of the enzyme (Feng and D'Urso, 2001). This result was surprising because the N terminus of Pol ε contains all the domains necessary for polymerase activity. Thus, we concluded that the noncatalytic C-terminal half of Pol ε is likely to be required during assembly of the IC. To provide additional evidence in support of this hypothesis, we set out to investigate whether the C termini of Pol ε can bind specifically to origin DNA in cells lacking the N-terminal sequences. For these experiments, we used the GD273 strain that contains four tandem copies of the FLAG epitope N-terminally tagged to Pol ε-Cterm under the control of the thiamine repressible nmt1 promoter (Feng et al., 1989). After release from the cdc25 block, we observed that the C-terminal half of Pol ε binds preferentially to ARS associated sequences at 60 min postrelease, similar to the results obtained for Dpb2 (Figure 6B).
DISCUSSION
Previously, we have demonstrated that the large catalytic subunit of DNA polymerase epsilon, encoded by cdc20, is essential for chromosomal DNA replication in S. pombe. Based on the observation that temperature-sensitive mutants of cdc20 arrest in late G1 or early S phase, we suggested that Pol ε is required for an early step in DNA replication (D'Urso and Nurse, 1997). Interestingly, the essential features of the Pol ε catalytic subunit do not reside in the N-terminal half of the enzyme that contains the highly conserved DNA polymerase domains, but rather in the C-terminal half of the enzyme where there are no known catalytic functions (Fuss and Linn, 2002). The observation that mutants in Pol ε arrest in late G1 or early S phase combined with the fact that the enzyme's polymerase activity is dispensable, led us to propose that Pol ε might be important for assembly of the initiation complex (Feng and D'Urso, 2001).
Consistent with this hypothesis, we demonstrate that both the C-terminal half of Pol ε, and its smaller putative subunit, Dpb2, bind preferentially to replication origins early in S phase. Furthermore, we demonstrate that like cdc20 mutants (Feng and D'Urso, 2001), a temperature-sensitive mutant of dpb2 arrests with a 1C DNA content after shift to the restrictive temperature, suggesting that Dpb2 is required for the onset of S phase. Although it is currently impossible to rule out that DNA replication initiation occurs in cdc20 or dpb2 mutants, it is interesting that early S phase arrest phenotype has never been reported for mutants defective in other DNA polymerases or their associated subunits, all of which arrest with a near 2C DNA content (Hughes et al., 1992; Waseem et al., 1992; MacNeill et al., 1996; MacNeill and Fantes, 1997).
Regardless of whether Pol ε is directly involved in DNA replication initiation, it is clear from our studies that Cdc20 and Dpb2 provide an essential function that does not rely on their ability to synthesize DNA. One possibility is that Pol ε binds replication origins early in S phase and is important for either recruiting or stabilizing the interaction of other replication proteins to DNA. Along these lines, it is interesting that in budding yeast, Pol ε can interact with replication origins in the absence of Pol α (Masumoto et al., 2000). Moreover, association of Pol α with ARS-associated chromatin requires Dpb11, a protein that both genetically and physically interacts with Pol ε (Masumoto et al., 2000). Similarly, in Xenopus cell-free extracts, association of Pol ε with chromatin during DNA replication can occur in the absence of Pol α or replication protein A (RPA) (Mimura et al., 2000). Based on these findings, Mimura et al. 2000 proposed that Pol ε might associate with chromatin before synthesis of RNA primers by Pol α.Weare currently testing whether the binding of Pol α or RPA to replication origins occurs before, or is dependent on, the prior loading of Dpb2 or Pol ε.
Recent studies using a Xenopus cell-free replication system have suggested that neither Pol ε or Pol δ is required for initiation of DNA replication, but both are necessary for efficient chain elongation (Waga et al., 2001). However, it is unclear whether the limited amounts of DNA synthesis detected in the absence of Pol δ or Pol ε in this system represent true initiation events, or simply reflect inappropriate binding and synthesis by Pol α in the absence of other polymerases.
Recently, it was reported that in human cells, Pol ε fails to colocalize with PCNA or active replication foci early in S phase. Although Pol ε foci are observed as early as late G1, in early S phase these foci do not colocalize with sites of 5-bromo-deoxyuridine incorporation or proliferating cell nuclear antigen (PCNA). Consistent with our yeast studies, these observations suggest that Pol ε may have a unique function in late G1 or early S phase that is independent of DNA synthesis. The authors speculated that the presence of Pol ε foci in late G1 or early S phase might represent DNA repair foci or inactive replication foci containing Pol ε (Fuss and Linn, 2002). However, we would like to offer an alternative explanation that these foci might represent sites where replication complexes are being assembled. The adjacent PCNA foci might represent active replication forks that have migrated away from the assembly sites. Interestingly, although Pol ε foci did not colocalize with PCNA during early S phase, they did seem to colocalize with active replication foci during late S phase. Considering that most late-replicating DNA regions are believed to be associated with heterochromatin, it was suggested that Pol ε might have a unique role in replicating these regions through facilitating chromatin remodeling (Fuss and Linn, 2002). This hypothesis is supported by two observations. First, the mouse homolog of Dpb2, called Dpe2, interacts with SAP18, a polypeptide that associates with the transcriptional corepressor Sin3. These two proteins have been shown to induce repression of transcription in reporter assays that might involve recruitment of remodeling factors such as histone deacetylase to chromatin (Wada et al., 2002). Second, human Pol ε p17, a homolog of the Pol ε subunit Dpb4, has been found as a component of human chromatin accessibility complex (CHRAC), a chromatin-remodeling factor (Poot et al., 2000).
Finally, our analysis of Pol ε function in fission yeast may provide an explanation for why Pol ε is dispensable for SV40 viral DNA replication. In the case of SV40, the virus encodes its own initiator protein, called T antigen. T antigen has multiple roles in promoting DNA replication, including site directed binding to the origin, ATP-dependent unwinding of DNA strands, and recruitment of DNA polymerase α/primase and possibly topoisomerase I to the site of initiation (Fanning, 1992; Fanning and Knippers, 1992; Simmons et al., 1996). Perhaps similar to the function of the origin recognition complex (ORC) and mini-chromosome maintenance complexes (MCMs), the essential nonreplicative function of Pol ε is provided by T antigen, and therefore is not required for viral DNA replication. We speculate that this function might be related to T antigen's ability to recruit Pol α/primase and possibly other factors to DNA. Therefore, the association of Pol ε and Dpb2 to ARS-associated chromatin might represent a key step in the assembly of the replication initiation complex.
Acknowledgments
We thank B. Burhaus, K. Downey, A. So, and M.G. Spiga for critically reviewing the manuscript; J. Diffley and J. Huberman for helpful discussions; and H. Masukata for providing strains and antibodies for ChIP analysis. W.F. was supported by a predoctoral research fellowship from the American Heart Association. L.R.-M. is supported by a postdoctoral fellowship from the American Heart Association. G.D. is supported by American Chemical Society research project grant RPG-00-262-01-GMC and by National Institutes of Health, National Cancer Institute grant IR0I-CA-099034.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-02-0088. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-02-0088.
Abbreviations used: ChIP, chromatin immunoprecipitation; Pol, DNA polymerase; PCR, polymerase chain reaction.
References
- Araki, H., Hamatake, R.K., Johnston, L.H., and Sugino, A. (1991a). DPB2, the gene encoding DNA polymerase II subunit B, is required for chromosome replication in Saccharomyces cerevisiae. Proc. Nat. Acad. Sci. USA 88, 4601–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki, H., Hamatake, R.K., Morrison, A., Johnson, A.L., Johnston, L.H., and Sugino, A. (1991b). Cloning DPB3, the gene encoding the third subunit of DNA polymerase II of Saccharomyces cerevisiae. Nucleic Acids Res. 19, 4867–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki, H., Ropp, P.A., Johnson, A.L., Johnston, L.H., Morrison, A., and Sugino, A. (1992). DNA polymerase-II, the probable homolog of mammalian DNA polymerase-epsilon, replicates chromosomal DNA in the yeast Saccharomyces-cerevisiae. EMBO J. 11, 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L., and Koonin, E.V. (1999). DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 27, 4658–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S.P., and Dutta, A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374. [DOI] [PubMed] [Google Scholar]
- Budd, M.E., and Campbell, J.L. (1993). DNA polymerases delta and epsilon are required for chromosomal replication in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui, G.S., and Linn, S. (1995). Purification of mammalian polymerases: DNA polymerase epsilon. Methods Enzymol. 262, 93–98. [DOI] [PubMed] [Google Scholar]
- D'Urso, G., Grallert, B., and Nurse, P. (1995). DNA polymerase alpha, a component of the replication initiation complex, is essential for the checkpoint coupling S phase to mitosis in fission yeast. J. Cell Sci. 108, 3109–3118. [DOI] [PubMed] [Google Scholar]
- D'Urso, G., and Nurse, P. (1997). S. pombe cdc20 encodes DNA polymerase ε is required for the chromosomal replication but not for the S phase checkpoint. Proc. Natl. Acad. Sci. USA 94, 12491–12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua, R., Edwards, S., Levy, D.L., and Campbell, J.L. (2000). Subunit interactions within the Saccharomyces cerevisae DNA polymerase epsilon (pol epsilon) complex. Demonstration of a dimeric pol. J. Biol. Chem. 275, 28816–28825. [DOI] [PubMed] [Google Scholar]
- Fanning, E. (1992). Simian virus 40 large T antigen: the puzzle, the pieces, and the emerging picture. J. Virol. 66, 1289–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning, E., and Knippers, R. (1992). Structure and function of simian virus 40 large tumor antigen. Annu. Rev. Biochem. 61, 55–85. [DOI] [PubMed] [Google Scholar]
- Feng, P.S., Xu, G.L., Zhao, Y., and Huang, Y.H. (1989). Continuous fermentation with yeast (Shizosaccharomyces pombe) floccules for ethanol production. Chin. J. Biotechnol. 5, 55–63. [PubMed] [Google Scholar]
- Feng, W., and D'Urso, G. (2001). Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol. Cell. Biol. 21, 4495–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss, J., and Linn, S. (2002). Human DNA polymerase epsilon colocalizes with proliferating cell nuclear antigen and DNA replication late, but not early, in S phase. J. Biol. Chem. 277, 8658–8666. [DOI] [PubMed] [Google Scholar]
- Hamatake, R.K., Hasegawa, H., Clark, A.B., Bebenek, K., Kunkel, T.A., and Sugino, A. (1990). Purification and characterization of DNA polymerase II from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 265, 4072–4083. [PubMed] [Google Scholar]
- Hashimoto, K., Nakashima, N., Ohara, T., Maki, S., and Sugino, A. (1998). The second subunit of DNA polymerase III (delta) is encoded by the HYS2 gene in Saccharomyces cerevisiae. Nucleic Acids Res. 26, 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, A.M., and Haber, J.E. (1999). Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96, 415–424. [DOI] [PubMed] [Google Scholar]
- Hubscher, U., Nasheuer, H.P., and Syvaoja, J.E. (2000). Eukaryotic DNA polymerases, a growing family. Trends Biochem. Sci. 25, 143–147. [DOI] [PubMed] [Google Scholar]
- Hughes, D.A.M., MacNeill, S.A. and Fantes, P.A. (1992). Molecular cloning and sequence analysis od cdc27+ required for G2-M transition in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet 231, 401–410. [DOI] [PubMed] [Google Scholar]
- Iino, Y., and Yamamoto, M. (1997). The Schizosaccharomyces pombe cdc6 gene encodes the catalytic subunit of DNA polymerase delta. Mol. Gen. Genet. 254, 93–97. [DOI] [PubMed] [Google Scholar]
- Jessberger, R., Podust, V., Hubscher, U., and Berg, P. (1993). A mammalian protein complex that repairs double-strand breaks and deletions by recombination. J. Biol. Chem. 268, 15070–15079. [PubMed] [Google Scholar]
- Keeney, J.B., and Boeke, J.D. (1994). Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136, 849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesti, T., Flick, K., Keranen, S., Syvaoja, J.E., and Wittenberg, C. (1999). DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell 3, 679–685. [DOI] [PubMed] [Google Scholar]
- Li, Y., Pursell, Z.F., and Linn, S. (2000). Identification and cloning of two histone fold motif-containing subunits of HeLa DNA polymerase epsilon. J. Biol. Chem. 275, 23247–23252. [DOI] [PubMed] [Google Scholar]
- MacNeill, S.A., and Fantes, P.A. (1997). Genetic and physiological analysis of DNA replication in fission yeast. Methods Enzymol. 283, 440–459. [DOI] [PubMed] [Google Scholar]
- MacNeill, S.A., Moreno, S., Reynolds, N., Nurse, P., and Fantes, P.A. (1996). The fission yeast Cdc1 protein, a homologue of the small subunit of DNA polymerase delta, binds to Pol3 and Cdc27. EMBO J. 15, 4613–4628. [PMC free article] [PubMed] [Google Scholar]
- Makiniemi, M., Pospiech, H., Kilpelainen, S., Jokela, M., Vihinen, M., and Syvaoja, J.E. (1999). A novel family of DNA-polymerase-associated B subunits. Trends Biochem. Sci. 24, 14–16. [DOI] [PubMed] [Google Scholar]
- Masumoto, H., Sugino, A., and Araki, H. (2000). Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol. 20, 2809–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura, S., Masuda, T., Matsui, T., and Takisawa, H. (2000). Central role for cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells 5, 439–452. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–723. [DOI] [PubMed] [Google Scholar]
- Morrison, A.L., Araki, H., Clark, A.B., Hamatake, R.K., and Sugino, A. (1990). A third essential DNA polymerase in S. cerevisiae. Cell 62, 1142–1151. [DOI] [PubMed] [Google Scholar]
- Navas, T.A., Zhou, Z., and Elledge, S.J. (1995). DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell 80, 29–39. [DOI] [PubMed] [Google Scholar]
- Ogawa, Y., Takahashi, T., and Masukata, H. (1999). Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol. Cell. Biol. 19, 7228–7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya, T., Maki, S., Kawasaki, Y., and Sugino, A. (2000). Structure and function of the fourth subunit (Dpb4p) of DNA polymerase epsilon in Saccharomyces cerevisiae. Nucleic Acids Res. 28, 3846–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot, R.A., Dellaire, G., Hulsmann, B.B., Grimaldi, M.A., Corona, D.F., Becker, P.B., Bickmore, W.A., and Varga-Weisz, P.D. (2000). HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 19, 3377–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, N., and MacNeill, S.A. (1999). Characterisation of XlCdc1, a Xenopus homologue of the small (PolD2) subunit of DNA polymerase delta; identification of ten conserved regions I-X based on protein sequence comparisons across ten eukaryotic species. Gene 230, 15–22. [DOI] [PubMed] [Google Scholar]
- Reynolds, N., Watt, A., Fantes, P.A., and MacNeill, S.A. (1998). Cdm1, the smallest subunit of DNA polymerase d in the fission yeast Schizosaccharomyces pombe, is non-essential for growth and division. Curr. Genet. 34, 250–258. [DOI] [PubMed] [Google Scholar]
- Sazer, S., and Sherwood, S.W. (1990). Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci. 97, 509–516. [DOI] [PubMed] [Google Scholar]
- Shivji, M.K., Podust, V.N., Hubscher, U., and Wood, R.D. (1995). Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry 34, 5011–5017. [DOI] [PubMed] [Google Scholar]
- Simmons, D.T., Melendy, T., Usher, D., and Stillman, B. (1996). Simian virus 40 large T antigen binds to topoisomerase I. Virology 222, 365–374. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger, S., Hecht, A., Luo, K., and Grunstein, M. (1997). SIR2 and SIR3 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11, 83–93. [DOI] [PubMed] [Google Scholar]
- Takahashi, T., and Masukata, H. (2001). Interaction of fission yeast ORC with essential adenine/thymine stretches in replication origins. Genes Cells 6, 837–849. [DOI] [PubMed] [Google Scholar]
- Takahashi, T., Ohara, E., Nishitani, H., and Masukata, H. (2003). Multiple ORC-binding sites are required for efficient MCM loading and origin firing in fission yeast. EMBO J. 22, 964–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada, M., Miyazawa, H., Wang, R.S., Mizuno, T., Sato, A., Asashima, M., and Hanaoka, F. (2002). The second largest subunit of mouse DNA polymerase epsilon, DPE2, interacts with SAP18 and recruits the Sin3 corepressor protein to DNA. J. Biochem. 131, 307–311. [DOI] [PubMed] [Google Scholar]
- Waga, S., Bauer, G., and Stillman, B. (1994). Reconstitution of complete SV40 DNA replication with purified replication factors. J. Biol. Chem. 269, 10923–10934. [PubMed] [Google Scholar]
- Waga, S., Masuda, T., Takisawa, H., and Sugino, A. (2001). DNA polymerase varepsilon is required for coordinated and efficient chromosomal DNA replication in Xenopus egg extracts. Proc. Natl. Acad. Sci. USA 98, 4978–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga, S., and Stillman, B. (1998). The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67, 721–751. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Wu, X., and Friedberg, E.C. (1993). DNA repair synthesis during base excision repair in vitro is catalyzed by DNA polymerase epsilon and is influenced by DNA polymerases alpha and delta in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waseem, N.H., Labib, K., Nurse, P., and Lane, D. (1992). Isolation and analysis of the fission yeast gene encoding polymerase δ accessory protein PCNA. EMBO J. 11, 5111–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg, D.H., Collins, K.L., Simancek, P., Russo, A., Wold, M.S., Virshup, D.M., and Kelly, T.J. (1990). Reconstitution of simian virus-40 DNA-replication with purified proteins. Proc. Natl. Acad. Sci. USA 87, 8692–8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin, J., Buck, V., Nurse, P., and Millar, J.B.A. (2002). Stable association of mitotic Cyclin B/Cdc2 to replication origins prevents endoreplication. Cell 11, 419–459. [DOI] [PubMed] [Google Scholar]