Abstract
ER-associated degradation (ERAD) removes defective and mis-folded proteins from the eukaryotic secretory pathway, but mutations in the ER lumenal Hsp70, BiP/Kar2p, compromise ERAD efficiency in yeast. Because attenuation of ERAD activates the UPR, we screened for kar2 mutants in which the unfolded protein response (UPR) was induced in order to better define how BiP facilitates ERAD. Among the kar2 mutants isolated we identified the ERAD-specific kar2-1 allele (Brodsky et al. J. Biol. Chem. 274, 3453–3460). The kar2-1 mutation resides in the peptide-binding domain of BiP and decreases BiP's affinity for a peptide substrate. Peptide-stimulated ATPase activity was also reduced, suggesting that the interdomain coupling in Kar2-1p is partially compromised. In contrast, Hsp40 cochaperone-activation of Kar2-1p's ATPase activity was unaffected. Consistent with UPR induction in kar2-1 yeast, an ERAD substrate aggregated in microsomes prepared from this strain but not from wild-type yeast. Overexpression of wild-type BiP increased substrate solubility in microsomes obtained from the mutant, but the ERAD defect was exacerbated, suggesting that simply retaining ERAD substrates in a soluble, retro-translocation-competent conformation is insufficient to support polypeptide transit to the cytoplasm.
INTRODUCTION
Before being delivered to their ultimate locations, secreted proteins are monitored by a quality control “machine” associated with the endoplasmic reticulum (ER; reviewed by Ellgaard et al., 1999); aberrant polypeptides may be retro-translocated from the ER to the cytoplasm and destroyed by the proteasome in a process termed ER-associated degradation (ERAD; McCracken and Brodsky, 1996). The importance of defining the molecular mechanism of ERAD is underscored by the fact that several human diseases arise from the accumulation or accelerated degradation of ERAD substrates and because some bacterial toxins and viruses coopt the ERAD pathway to exert their effects (reviewed in Thomas et al., 1995; Brodsky and McCracken, 1999; Aridor and Hannan, 2000; Fewell et al., 2001).
ERAD may result from inefficient protein folding, so it is not surprising that molecular chaperones are required for this process. Chaperones prevent the formation of off-pathway intermediates or directly catalyze folding and have been proposed to “judge” whether a nascent protein will ultimately fold or whether it should be targeted for degradation (reviewed by Hayes and Dice, 1996; Hartl, 1996; Horwich et al., 1999; Plemper and Wolf, 1999; Römisch, 1999; Wickner et al., 1999; Fewell et al., 2001; Höhfeld et al., 2001). Hsp70 (heat shock proteins with a molecular mass of ∼70 kDa) molecular chaperones hydrolyze ATP concomitant with the binding of peptides with overall hydrophobic character (Flynn et al., 1991; Blond-Elguindi et al., 1993; Rüdiger et al., 1997); therefore, Hsp70s might retain the solubility of unfolded, retro-translocating polypeptides during their voyage from the ER to the cytoplasm via the Sec61p translocation channel or might “gate” this channel (Wiertz et al., 1996; Pilon et al., 1997; Plemper et al., 1997; Johnson and Haigh, 2000; Tsai et al., 2002).
In accordance with these hypotheses, several lines of evidence support a role for the lumenal Hsp70, BiP, in ERAD in both mammals and yeast. First, the proteolysis of ERAD substrates coincides with the rate at which they are released from BiP in the mammalian ER (Knittler et al., 1995; Beggah et al., 1996; Skowronek et al., 1998; Chillaron and Haas, 2000), and BiP associates preferentially with exposed regions of an unfolded ERAD substrate (Schmitz et al., 1995). Second, the degradation of an ERAD substrate is slowed when yeast contain a mutant allele in the gene encoding BiP, KAR2 (kar2-113; Plemper et al., 1997), and two ERAD substrates in yeast, CPY* and pαF, aggregate in microsomes prepared from another kar2 mutant shifted to the nonpermissive temperature (kar2–203; Nishikawa et al., 2001). The delivery of an ERAD substrate to the proteasome is also reduced in microsomes prepared from kar2-1 and kar2-133 mutants; however, in contrast to the kar2 mutants used in these other studies, the kar2-1 and kar2-133 alleles compromise ERAD efficiency but do not affect protein translocation (import) into the ER, suggesting that the roles for BiP during protein translocation and retro-translocation are distinct (Brodsky et al., 1999) and that a biochemical analysis of the corresponding Kar2-1 and Kar2-133 mutant proteins might elucidate which biochemical activities of BiP are required for ERAD.
To better define the action of BiP during ERAD we sought kar2 mutants in which the unfolded protein response (UPR) was induced and identified the kar2-1 allele. We also present a biochemical analysis of the Kar2-1 and Kar2-133 mutant proteins and the physiological consequences of Kar2-1/133p expression in yeast.
MATERIALS AND METHODS
Strains and Plasmids
Yeast strains used for these studies were as follows: MS10 (a ura3-52 leu2-3112 ade2-101); MS1111 (a ura3-52 leu2-3112 ade2-101 kar2-1); and MS193 (a ura3-52 leu2-3112 ade2-101 kar2-133). The kar2-1 mutant was first described by Polaina and Conde (1982) and the kar2-133 mutant was obtained by plasmid shuffle, as outlined in Vogel et al. (1990). DNA sequence analysis identified the mutations in the kar2-1 and kar2-133 open reading frames. To obtain kar2 mutants in which the UPR was activated, the following strains were constructed. First, a KAR2 disruption cassette was obtained by amplifying the HIS3 gene from pRS303 with primers BiPDel1 (5′-G G A C A C G A A A A G G G T T C T C T G G A A G A T A T A A A T A T G G C T A T G C T C T T G G C C T C C T C T A G-3′) and BiPDel2 (5′ C A C T G T T A C T G A G T A C C T T A A C C C C A G T C T C TATACTCTTCTCGTTCAGAATGACACG-3′) and that contain a region of homology to the KAR2 gene on each end. This disruption cassette was used to transform the yeast wild-type diploid strain YPH501 (a/α, his3-Δ200/his3-Δ200, leu2-Δ1/leu2-Δ1, ura3-52/ura3-52, trp1-Δ63/trp1-Δ63) and His+ transformants in which one KAR2 allele was disrupted were confirmed by PCR using primers HISend (5′-GTGATTAACGTCCACACAGG-3′) and BiPend (5′-GGGATGAGATGAGATGAGATG-3′). The resulting strain was transformed with the single-copy, KAR2-containing plasmid, pMR397, which is marked with URA3. The transformed diploid strain was starved for nitrogen for 5 d to induce sporulation and tetrads were dissected and grown on rich medium. After replica plating the resulting colonies on various selective media to screen for auxotrophies, the MMY8–2 strain (α, kar2::HIS3, his3-Δ200, leu2-Δ1, ura3-52, trp1-Δ63, pMR397) was isolated. MMY8–2 was then transformed with pMZ11 (CEN4, TRP1, UPRE::LacZ) to monitor UPR activation through β-galactosidase activity (Zhou and Schekman, 1999). The resulting strain is called MMY9 and was used for the screen described below.
BiP was overproduced in wild-type and kar2-1 yeast from the 2μ URA-marked plasmid, pMR109, which contained the ClaI-SalI (KAR2) fragment from pMR48 (Rose et al., 1989). Cells with pMR109 were selected on synthetic complete medium lacking uracil (SC-ura) and supplemented with glucose to a final concentration of 2%. All manipulations involving yeast were performed using standard protocols (Rose et al., 1990).
Isolation of kar2 Mutants Exhibiting an Enhanced UPR
Plasmid pMR713 (CEN4, LEU2, KAR2) was mutagenized with hydroxylamine (Rose et al., 1990) and transformed into MMY9 (see above). The resulting transformants, which grew on medium lacking tryptophan, uracil, and leucine, were replica-plated on 5-FOA at 30°C to select for yeast lacking pMR397. Approximately 35,000 transformants were screened for constitutive activation of the UPR by monitoring β-galactosidase activity at 30°C with an agarose overlay containing X-Gal (100 μg/ml) and 0.2% Sarkosyl as a permeating agent (Kabani et al., 2000a). Candidate colonies were restruck to reconfirm UPR activation, and the corresponding pMR713 mutagenized plasmids from the rescreened colonies (∼120) were isolated and individually transformed back into MMY9. After replica-plating on 5-FOA and testing for β-galactosidase activity to confirm that UPR activation was plasmid-based, 20 mutants were retained. These resulting pMR713 plasmids were also introduced into MMY8–2 and the transformants were used for further analysis (see below). As a control, MMY8–2 was transformed with untreated pMR713 and replica-plated on 5-FOA to cure yeast of the pMR397 plasmid and is referred to as wild type where indicated.
ERAD, Translocation, and UPR Assays
The degradation of unglycosylated proalpha factor (pαF) in yeast ER-derived microsomes was measured as described (McCracken and Brodsky, 1996). In brief, 35S-labeled prepro-alpha factor (ppαF) lacking the core consensus glycosylation sites was translocated into microsomes, after which the microsomes were harvested, washed twice, and resuspended in a chase reaction either containing or lacking an ATP-regenerating mix and yeast cytosol at a final concentration of 5 mg/ml. Reactions were incubated at 30°C and were quenched after 20 min by the addition of trichloroacetic acid (TCA) to a final concentration of 20%. The percentage of pαF remaining was determined by phosphorimage analysis after SDS-PAGE of electrophoresed reaction products.
In vitro translocation assays were prepared as above except that after a 40-min translocation assay the reactions were divided and incubated in the presence or absence of trypsin in order to determine the amount of protected, and thus translocated, pαF. Samples were processed and translocation efficiency was calculated as described (Brodsky and Schekman, 1993). Translocation of preBiP and ppαF was assayed in vivo after incorporation of 35S-methionine into total cellular protein and immunoprecipitation of BiP and ppαF using specific antiserum and Protein A-sepharose, as previously published (Morrow and Brodsky, 2001).
The UPR was assayed quantitatively by measuring β-galactosidase activity in extracts (Rose and Botstein, 1983) prepared from yeast transformed with pJC104 (kindly provided by Dr. Peter Walter, University of California, San Francisco). Plasmid pJC104 encodes the lacZ gene behind four repeats of the unfolded protein response element (UPRE; Mori et al., 1992; Cox et al., 1993).
Protein Purification and Other Biochemical Assays
Hexahistidine-tagged derivatives of wild-type, Kar2-1p, and Kar2-133p were generated and purified by nickel affinity and conventional chromatography as described for other mutant forms of Kar2p (McClellan et al., 1998). The proteins were estimated to be >95% pure as determined by SDS-PAGE and Coomassie Brilliant Blue staining. Single-turnover ATPase assays (Sullivan et al., 2000) of [α-32P]ATP-BiP complexes indicated that the rate of ATP hydrolysis by hexahistidine-tagged yeast BiP (0.12–0.21 min–1) was similar to that reported for hexahistidine-tagged mammalian BiP (0.30 min–1; Chevalier et al., 1998). Steady state ATPase assays were performed as previously published (McClellan et al., 1998).
The glutathione-S-transferase (GST)-Sec63p-J fusion protein was purified as described (Corsi and Schekman, 1997), and a GST-Jem1p-J was purified from the Escherichia coli TG1 strain transformed with pSNJ20 (S. Nishikawa S. and T. Endo, unpublished data). The pSNJ20 plasmid allows the expression of the C-terminal 115 residues of Jem1p fused at the C terminus of GST. The fusion protein also contains a hexahistidine-tag at the C terminus. Bacteria were grown in Luria broth (LB) containing 50 μg/ml ampicillin at 26°C to midlog phase and isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and the cells were incubated further for 1 h. The bacteria were harvested, washed with water, and resuspended in Buffer A (50 mM HEPES-KOH, pH 7.4, 5 mM β-mercaptoethanol, 200 mM NaCl, 10 mM imidazole). Lysozyme was added to a final concentration of 0.1 mg/ml and the cells were incubated on ice for 30 min. Protease inhibitors (phenylmethylsulfonylfluoride, 1 mM; leupeptin, 1 μg/ml; pepsatin A, 1 μg/ml) were added and the cells were disrupted six times by sonication for 30 s with a 1-min incubation on ice between each sonication. Triton X-100 was added to a final concentration of 0.1%, the broken cells were centrifuged for 10 min at 13,000 rpm in a Sorvall SA600 rotor, and the resulting supernatant was centrifuged at 20,000 rpm in a Sorvall SA600 rotor for 20 min. The cleared lysate was loaded onto a 5 ml Ni-NTA agarose column (Qiagen, Valencia, CA) that had been equilibrated in buffer A, and the column was washed with 30 ml of the following: (1) Buffer A; (2) Buffer A containing 1% Triton X-100 and 5% glycerol; (3) Buffer A containing 1 M NaCl and 5% glycerol; (4) Buffer A containing 5 mM MgCl2, 5 mM ATP, and 300 mM NaCl; (5) Buffer A containing 0.5 M Tris-HCl, pH 7.4, and 300 mM NaCl; (6) Buffer A containing 300 mM NaCl, 5% glycerol, and 25 mM imidazole; and (7) Buffer A containing 300 mM NaCl, 5% glycerol, and 50 mM imidazole. The GST-Jem1p protein was eluted with a 15 × 15-ml linear gradient of Buffer A containing 300 mM NaCl, 5% glycerol, and imidazole from 50–250 mM; 1-ml fractions were collected and analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. Peak fractions were pooled and dialyzed against 20 mM HEPES-KOH, pH 7.4, 50 mM KCl, and 5% glycerol, and then snap-frozen in liquid nitrogen and stored at –80°C. The GST-Jem1p was >95% pure as assessed by Coomassie Brilliant Blue staining.
GST-fusion protein pull-down assays were performed as published (Kabani et al., 2000b, 2002). Cell extracts for Western blots were prepared as reported (Morrow and Brodsky, 2001), and the proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes (0.22-μM pore diameter; Schleicher and Schuell, Keene, NH). Quantitative immunoblots were performed using anti-BiP rabbit antiserum (Brodsky and Schekman, 1993) and anti-Sec61p rabbit antiserum (Stirling et al., 1992), used as a loading control, both of which were decorated with 125I-Protein A (Amersham Biosciences, Piscataway, NJ). Images were obtained and quantified using a Fuji phosphorimager and MacBas software (v. 2.4; Fuji Medical Systems, Stamford, CT). Carbonate extraction on ∼1 equivalent of ER-derived microsomes was accomplished as described in Fujiki et al. (1982), and the pellet and supernatant fractions were analyzed by SDS-PAGE and immunoblot analysis using Enhanced Chemiluminescence (Pierce, Rockford, IL). The Sec63p-BiP complex was purified from octylglucoside-solubilized yeast microsomes by diethylaminoethane, Superose-6, and hydroxylapatite column chromatography as previously published (Brodsky and Schekman, 1993).
The oligomeric state of pαF was assessed as described (Nishikawa et al., 2001). In brief, washed microsomes containing pαF were obtained after an in vitro translocation assay (see above), the membranes were solubilized with Triton X-100, and the clarified extract was loaded onto a 5–40% sucrose gradient in 0.1% Triton X-100 and centrifuged at 145,000 × g for 20 h at 4°C. After the gradient was fractionated pαF was detected and quantified by SDS-PAGE and phosphorimager analysis.
Spectroscopy
The CALLQSRLLLSAPRRAAATARY (APPY) peptide was synthesized, purified, labeled, and analyzed by mass spectrometry as described in Montgomery et al. (1999). The CLLLSAPRR (p5) peptide (Pierpaoli et al., 1998) was synthesized by solid phase methods on PerSeptive Biosystems (Framingham, MA) automated peptide synthesizer and purified by reverse-phase chromatography on a C18 column and analyzed by mass spectrometry. The affinity of yeast BiP for F-APPY (fluorescein-labeled APPY) and the binding of AEDANS-labeled p5 to BiP were measured by fluorescence anisotropy as described by Montgomery et al. (1999) after incubating 0.02–0.05 μM of F-APPY or p5-AEDANS with the indicated concentrations of BiP overnight at 4°C. The BiP used for these experiments was dialyzed for 16 h against 20 mM HEPES, pH 7.4, 100 mM KCl, 5 mM MgCl2, 0.8 mM DTT and concentrated using Centricon microconcentrators (Amicon, Beverly, MA). Circular Dichroism (CD) spectroscopy of yeast BiP (final concentration 1.6 μM) was performed on an Aviv CD Spectrometer (Model 202; Lakewood, NJ) using a 0.1-cm path-length. The temperature of the cuvette was maintained at 26°C.
RESULTS
Isolation of UPR-activating kar2 Mutants
BiP is required for both ERAD and for protein translocation (import) into the ER (reviewed in Fewell et al., 2001). Therefore, we desired allele-specific kar2 mutants to more specifically define the action of BiP during ERAD. Our screen for such alleles is based on that used by Zhou and Schekman (1999) to isolate sec61 mutants that were ERAD-defective but translocation-proficient, and the premise for the screen derives from the fact that defects in ERAD result in UPR induction (Cassagrande et al., 2000; Friedlander et al., 2000; Ng et al., 2000; Travers et al., 2000).
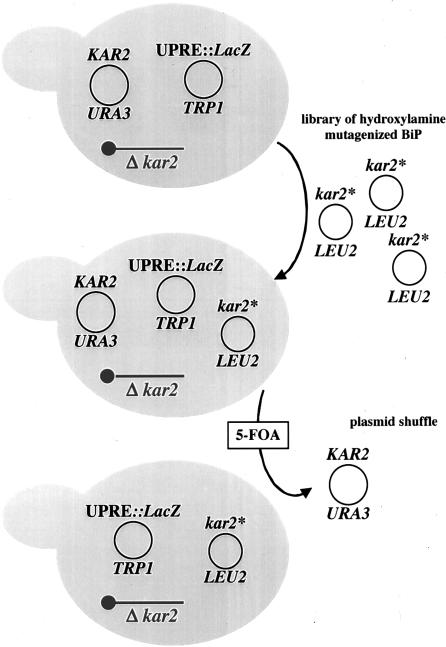
The MMY9 strain bears a chromosomal deletion of the KAR2 gene and is viable because it contains pMR397, a single-copy uracil-selectable plasmid that expresses KAR2 under the control of its own promoter (Figure 1). MMY9 also contains plasmid pMZ11 that can be used to report on the UPR by measurements of β-galactosidase activity (Cox et al., 1993; Zhou and Schekman, 1999). A library of mutagenized kar2 alleles (kar2*) was then transformed into this strain and pMR397 was counterselected on medium containing 5-FOA. After β-galactosidase activity was determined using an agar-overlay (see MATERIALS AND METHODS), kar2*-containing plasmids from candidate colonies were purified and their ability to induce the UPR was reconfirmed. Twenty of the retransformed mutants exhibited UPR activation and were retained for further analysis.
Figure 1.
The screen for UPR-induced kar2 mutant yeast (see text for details).
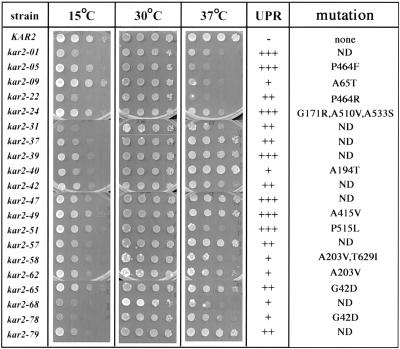
We next assayed the growth properties of the kar2 mutants at various temperatures. Although each mutant exhibited wild-type growth at 30°C (Figure 2) and 35°C (our unpublished data), several strains were thermosensitive at 37°C (e.g., kar2-51), cryosensitive at 15°C (e.g., kar2-40), or both (e.g., kar2-68). Also shown in Figure 2 is the relative strength of the UPR activation in each strain, although no obvious correlation between growth phenotype and UPR was observed. For example, both kar2-49 and kar2-51 exhibit high UPR activation, whereas only kar2-51 is thermosensitive (Figure 2).
Figure 2.
Temperature-sensitive phenotypic analysis of the UPR-inducing kar2 mutants. Serial dilutions from log-phase cultures of the wild-type and the indicated mutant strains were inoculated on rich medium and incubated at the indicated temperatures for 3–6 d. The relative strength of UPR activation in the various mutants at 30°C is also indicated; –, no activation; + to +++, varying degrees of UPR activation as determined by the length of time required for the mutant colonies to turn blue in agar-overlay plate assays for β-galactosidase activity (see MATERIALS AND METHODS for details). The nomenclature of the mutants in this figure does not correspond to previously isolated kar2 mutant alleles, but the amino acid position(s) and predicted substitution(s) in the mutant alleles are indicated. Nucleotide substitutions that do not change the encoded amino acid sequence from the wild-type allele (i.e., silent mutations) are not shown. ND, DNA sequence analysis of the mutant allele was not completed.
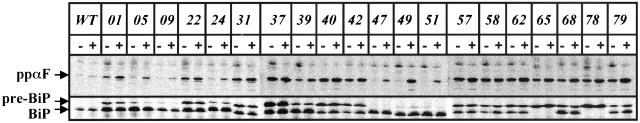
To assay protein translocation, we examined whether cytoplasmic precursors of two secreted proteins, prepro-α-Factor (ppαF) and preBiP, accumulated in each strain at 26°C or after a 5-min shift at 37°C. ppαF is translocated posttranslationally into the ER, whereas BiP uses both the co- and posttranslational pathways (Ng et al., 1996). As shown in Figure 3 the kar2 mutants displayed varying translocation defects, ranging from no defect at 26°C and a very mild defect after temperature shift (e.g., kar2–51) to a strong block at 26 and 37°C (e.g., kar2-40). Interestingly, a band corresponding only to preBiP was observed in the kar2-65 and kar2-78 mutants; a similar result was obtained when the molecular mass of BiP was examined in ER-derived microsomes prepared from these strains by immunoblot analysis (our unpublished data). This result suggested that the Kar2-78 and Kar2-65 mutant proteins might possess an uncleaved signal sequence. Indeed, DNA sequence analysis revealed that both kar2-65 and kar2-78 bear a mutation of glycine 42 to an aspartic acid at the predicted signal sequence cleavage site (Rose et al., 1989; Figure 2).
Figure 3.
Analysis of ppαF and preBiP translocation in the kar2 mutant strains. Each strain was grown at 26°C to midlog phase and total protein was labeled with 35S-methionine/cysteine for 5 min at either 26° (–) or 37°C (+). Cell extracts were prepared and ppαF and BiP were immunoprecipitated, resolved by SDS-PAGE, and visualized by phosphorimager analysis.
To determine whether retention of a signal sequence affected BiP solubility, we performed sodium carbonate extraction on microsomes obtained from the wild-type and kar2-01 strains (in which signal sequence cleavage was apparent; Figure 3) and the kar2-65 and kar2-78 strains. Wild-type BiP and Kar2-01p were exclusively or predominantly found in the soluble fraction, as expected; in contrast, Kar2-65p and Kar2-78p were primarily membrane-associated. Moreover, we noted that ∼50% of Kar2-65p and Kar2-78p were sensitive to exogenous protease when microsomes were prepared from strains expressing these mutant alleles (our unpublished data). Because a strong ppαF translocation defect was noted in kar2-65 and kar2-78 yeast (Figure 3), we suggest that a proportion of the signal sequence-containing BiP mutant proteins are incompletely translocated or fail to translocate and/or that BiP activity is reduced when its signal sequence is retained. In either case, both translocation and retro-translocation (ERAD) would be compromised and the UPR would be activated.
The kar2-51 Mutant Bears the Same Amino Acid Substitution as the ERAD-specific kar2-1 Mutant
On the basis of the results presented in Figures 2 and 3, we identified several mutants that were translocation-defective and might have exhibited an induced UPR because of further effects on ERAD and/or protein folding. For example, defects in ERAD, protein folding, and translocation were previously noted for kar2 mutant strains containing amino acid substitutions in the ATPase domain (e.g., kar2-113 and kar2-159; Brodsky et al., 1995; Simons et al., 1995; Plemper et al., 1997; see below). Consistent with this result, DNA sequence analysis uncovered an A203V mutation in kar2-62 and an A194T mutation in kar2-40 (Figure 2), both of which affect amino acids in BiP's ATPase domain.
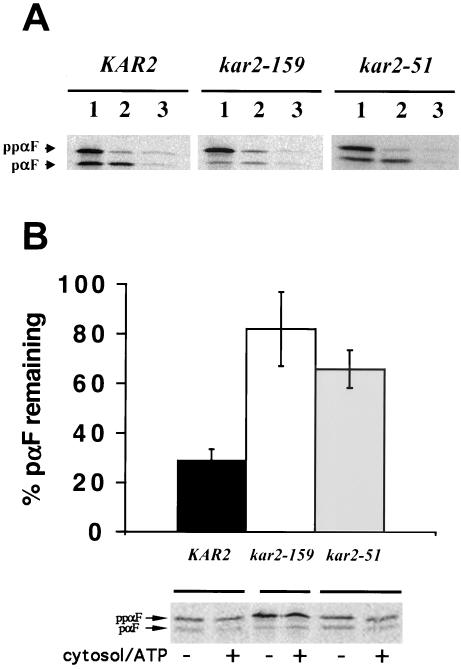
We also identified a few strains that were translocation-proficient at permissive temperatures but exhibited an increased UPR, two criteria expected for ERAD-specific kar2 mutants. Several of these candidates possess >1 amino acid changes as determined by DNA sequence analysis of the inserts in the corresponding kar2-containing plasmids (our unpublished data). However, we identified a point mutation of proline 515 to a leucine in kar2-51, which is the same mutation found on the chromosome in kar2-1 yeast, a previously identified ERAD-specific mutant (Brodsky et al., 1999). To confirm that yeast containing the plasmid-borne kar2-51 mutation were also ERAD-defective but proficient for translocation, we performed in vitro translocation and ERAD assays with microsomes from the wild-type and kar2-51 strains. As a control, we assayed these processes in microsomes prepared from the kar2-159 strain, which contains a mutation in the ATPase domain, severely affecting the ability of Kar2-159p to bind ATP, and which consequently is defective for both ERAD and translocation (Brodsky et al., 1995, and our unpublished data; see MATERIALS AND METHODS). We found that the ppαF translocation efficiency in microsomes derived from wild-type and kar2-51 yeast were identical (∼30%; Figure 4A), whereas the translocation efficiency in kar2-159–derived microsomes was ∼9%. Microsomes obtained from either kar2-51 or kar2-159 yeast exhibited reduced retro-translocation and degradation of unglycosylated pαF, an in vitro ERAD-substrate (Figure 4B; McCracken and Brodsky, 1996). We conclude that kar2-51 is ERAD-defective but translocation-proficient, thus supporting the premise of our screen.
Figure 4.
The kar2-51 mutant (kar2-1) is translocation-proficient but ERAD-defective in vitro. (A) Microsomes derived from the indicated strains were incubated with in vitro–translated 35S-labeled ΔGppαF (McCracken and Brodsky, 1996) for 40 min at 30°C. Equal amounts of each reaction were either (1) untreated or treated with (2) trypsin or (3) trypsin and Triton X-100, and incubated on ice for 30 min before proteins were TCA-precipitated and resolved by SDS-PAGE and visualized by phosphorimager analysis. The amount of trypsin-protected (translocated) signal sequence-cleaved pαF was 30, 9 and 28% in wild-type, kar2-159, and kar2-51 microsomes, respectively (as averaged from three independent experiments). A representative experiment is shown. (B) 35S-labeled ΔGppαF was translocated into microsomes derived from the indicated strains for 1 h at 20°C, the microsomes were washed, and cytosol and ATP were added to one half of the reactions, whereas the other half was incubated with buffer. After 20 min at 30°C the reactions were quenched with TCA, and precipitated proteins were resolved by SDS-PAGE and phosphorimager analysis to determine the percentage of pαF degraded. The buffer control was set to 100% in each experiment. Data represent the means of three independent experiments (±SD). A representative gel from one experiment is also shown.
For the remainder of this study, we chose to focus on the kar2-1/kar2-51 mutant and on the kar2-133 mutant, which is also ERAD-defective but translocation-proficient (Brodsky et al., 1999). A further analysis of the other mutants will be reported elsewhere.
The kar2-1 and kar2-133 Mutations Reside in the Peptide-binding Domain
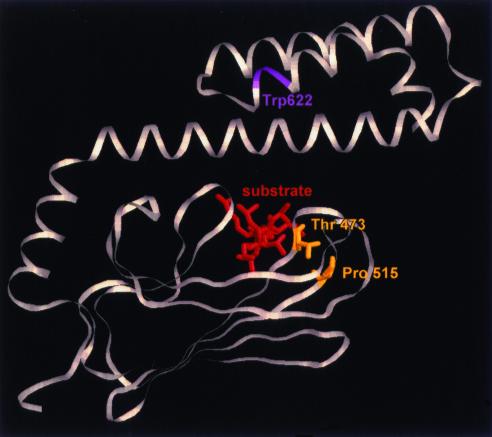
DNA sequence analysis of the kar2-1 and kar2-133 mutations indicates that a proline at position 515 (P-470 in DnaK) is converted to a leucine and a threonine at position 473 (T-428 in DnaK) is replaced by a phenylalanine, respectively. These residues are highly conserved among Hsp70s and lie near the tips of loops that connect β-sheets and that in turn forge the platform of the Hsp70 substrate-binding domain (Figure 5; Zhu et al., 1996; our unpublished data). The mutation in kar2-1 is in loop 6, whereas in kar2-133 the mutation is in loop 3. Notably, the side chain of the threonine mutated in kar2-133 is predicted to form a hydrogen bond to the polypeptide backbone at the tip of loop 5,6. Thus, we propose that the kar2-1 and kar2-133 mutations affect the structure of the peptide-binding domain similarly.
Figure 5.
Mutations in kar2-1 and kar2-133 reside in the peptide-binding domain of BiP. The yeast BiP and E. coli DnaK amino acid sequences were aligned and mutated residues in kar2-1 (P515L) and kar2-133 (T473F) were mapped onto the three-dimensional structure of the DnaK peptide-binding domain (Zhu et al., 1996). The mutated residues are highlighted in yellow, and the approximate location of the lone tryptophan (at position 622) in yeast BiP is indicated in magenta. The bound substrate is displayed in red and is oriented perpendicular to the page.
Yeast Expressing Kar2-1p and Kar2-133p Induced the Unfolded Protein Response
The strain used for the screen in which kar2-1 was reisolated contained kar2 on a single-copy plasmid. To ensure that experiments were free of potential plasmid copy-number artifacts, subsequent in vivo experiments were conducted with haploid yeast harboring a chromosomal copy of kar2-1. The kar2-133 strain and wild-type yeast were examined in parallel. UPR induction in the kar2-1 and kar2-133 mutants was confirmed after introduction of a UPR reporter plasmid (see MATERIALS AND METHODS): The UPR was enhanced 6–7-fold in the mutant strains compared with wild-type yeast when cells were grown at 26°C and was activated further after a shift to 37°C for 1 h (Table 1). In accordance with these data, we observed that the levels of Kar2-1p and Kar2-133p were ≥2-fold higher than wild-type BiP in microsomes prepared from the respective strains and that this difference became more pronounced after a 37°C shift.
Table 1.
Kar2-1p and Kar2-122p expression induces the UPR
| Strain/protein | KAR2/BiP | kar2-1/Kar2-1p | kar2-133/Kar2-133p |
|---|---|---|---|
| UPRa (26°C) | 840 | 6010 | 4910 |
| (37°C) | 720 | 7220 | 6280 |
| Relative amountb (26°C) | 1.0 | 2.0 | 3.0 |
| (37°C) | 1.0 | 2.3 | 4.5 |
The UPR (in calculated units) in the indicated strains was measured from cells that were grown exclusively at 26°C or that had been shifted to 37°C for 1 hr, as described in the “MATERIALS AND METHODS”.
The amount of BiP, Kar2-1p, and Kar2-133p was measured in cell extracts by quantitative immunoblot analysis from strains that were grown exclusively at 26°C or that had been shifted to 37°C for 1 hr.
The Purified Kar2-1 and Kar2-133 Proteins Are Not Grossly Mis-folded
To begin to elucidate the molecular basis of the kar2-1 and kar2-133 ERAD defects we purified wild-type BiP, Kar2-1p, and Kar2-133p by nickel affinity and ion exchange column chromatography (see MATERIALS AND METHODS), and then we performed two assays to assess the global conformation of the mutant proteins. First, CD spectroscopy was performed and the spectral characteristics for the wild-type and mutant proteins were identical (our unpublished data). Second, we examined the digestion patterns of wild-type BiP and Kar2-1p and Kar2-133p after limited proteolysis in the presence of ATP or ADP and observed that the mutant proteins, like wild-type BiP, exhibited an ATP-dependent conformational change (McClellan et al., 1998; J. Endres and J.L.B., unpublished data); these results are consistent with the fact that the single turnover and steady-state ATPase activities of wild-type BiP and Kar2-1p and Kar2-133p are similar (see MATERIALS AND METHODS; Figures 6 and 7; below).
Figure 6.
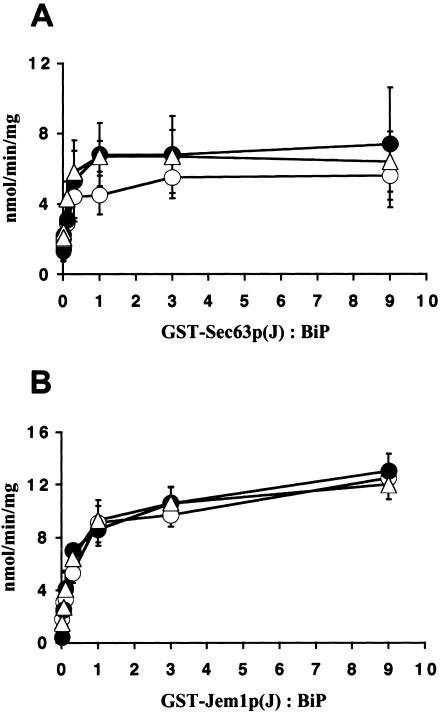
Kar2-1p and Kar2-133p interact functionally with the J domains of Sec63p and Jem1p. Steady state hydrolysis of ATP by wild-type BiP (○), Kar2-1p (•), and Kar2-133p (▵) was measured using 5 μg of BiP and either the (A) GST-Sec63p-J or (B) GST-Jem1p-J fusion proteins. ATPase activity is plotted as nmol of ATP hydrolyzed/min/mg BiP vs. the molar ratio of the J domain–containing fusion proteins to BiP.
Figure 7.
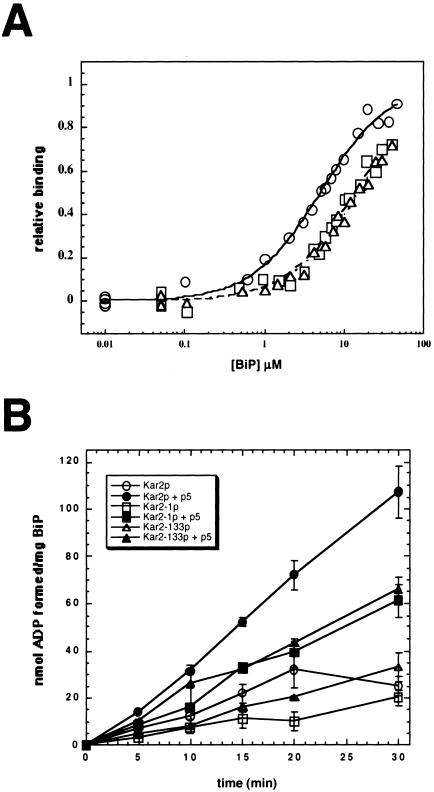
Kar2-1p and Kar2-133p exhibit reduced peptide affinity and interdomain coupling. (A) Fluorescence anisotropy measurements of F-APPY binding to wild-type BiP (○), Kar2-1p (□), and Kar2-133p (▵), and a best-fit analysis of the data using a single binding site was performed as described by Montgomery et al. (1999). (B) Steady state ATPase assays were performed after assembling reactions on ice in either the presence (filled symbols) or absence (open symbols) of a 1000-fold molar excess of p5: Wild-type BiP (circles), Kar2-1p (squares), and Kar2-133p (triangles).
Kar2-1p and Kar2-133p Interact Functionally with the J Domains of Sec63p and Jem1p
The interaction between BiP and the J-domain of Sec63p in the ER is essential for protein translocation (Brodsky and Schekman, 1993; Brodsky et al., 1995; Lyman and Schekman, 1995; Corsi and Schekman, 1997; Matlack et al., 1997; McClellan et al., 1998), and a mutation in SEC63 that abrogates this interaction has been reported to modestly reduce the degradation of a soluble ERAD substrate (Plemper et al., 1997). Therefore, to determine if Kar2-1p and Kar2-133p associate functionally with the J domain of Sec63p, the wild-type and mutant proteins were incubated with increasing concentrations of a GST-Sec63p-J domain fusion protein that was used previously to report on the interaction of BiP with Sec63p (Corsi and Schekman, 1997; McClellan et al., 1998). We found that the steady state ATPase activities of wild-type BiP and Kar2-1p and Kar2-133p were enhanced identically in a concentration-dependent manner by the Sec63p J domain (Figure 6A). We also assessed the ability of wild-type, Kar2-1p and Kar2-133p to associate with Sec63p after purification of the BiP-Sec63p-Sec71p-Sec72p complex from detergent-solubilized microsomes prepared from wild-type, kar2-1, and kar2-133 mutant yeast (Brodsky and Schekman, 1993). After ion exchange (DEAE) and gel filtration (Super-soe-6) column chromatography, the complex eluted from hydroxylapatite on a 0.2–0.5 M phosphate gradient and the ratio of BiP to Sec63p that coeluted was comparable regardless of whether the microsomes derived from the wild-type or kar2 mutant yeast (our unpublished data).
BiP also associates with two other J domain–containing proteins in the ER, Scj1p and Jem1p. Deletion of both proteins, but neither one alone, attenuates ERAD and increases ERAD substrate aggregation, suggesting that Scj1p and Jem1p function redundantly to facilitate polypeptide retrotranslocation (Nishikawa et al., 2001). To measure the interaction of Jem1p with wild-type and the mutant Kar2 proteins, we incubated increasing concentrations of a GST-Jem1p-J domain fusion protein and measured steady state ATP hydrolysis, as above. The results shown in Figure 6B indicate that the J domain of Jem1p also activated the ATPase activities of each protein identically.
Kar2-1p and Kar2-133p Exhibit a Reduced Peptide Affinity and Compromised Peptide-stimulated ATPase Activity
BiP binds polypeptides in the lumen of the yeast ER to drive protein translocation (Sanders et al., 1992; Matlack et al., 1999), to engineer protein folding (Te Heesen and Aebi, 1994; Simons et al., 1995), and to facilitate ERAD (Gillece et al., 1999; Nishikawa et al., 2001). We therefore wished to define whether the affinities of the wild-type and mutant proteins for a peptide substrate differed. Using a fluorescein-labeled peptide (F-APPY), increasing concentrations of the proteins, and measurements of fluorescence anisotropy to detect F-APPY-BiP complexes (Montgomery et al., 1999), we found that the peptide affinities for wild-type BiP (5.1 ± 0.21 μM), Kar2-1p (13.9 ± 0.60 μM), and Kar2-133p (15.4 ± 0.41 μM) differed by 2.7–3.0-fold (Figure 7A). Because saturation was not achieved for the mutant proteins (yeast BiP aggregates in vitro at concentrations required to achieve saturation; our unpublished data), the calculated KDs for Kar2-1p and Kar2-133p are likely an under-estimate of the true KDs. Therefore, the peptide affinities for the ERAD-defective Kar2 mutants are significantly reduced relative to wild-type BiP. Furthermore, the peptide affinity for wild-type BiP is similar to that reported for the cytoplasmic yeast Hsp70, Ssa1p (∼5 μM; Pfund et al., 2001), which is 63% identical to Kar2p, supporting the efficacy of using this experimental method.
Hsp70-peptide interactions are controlled by interdomain coupling between the ATPase and peptide-binding domains (see for example, Ha et al., 1997; Davis et al., 1999). If the coupling between these domains is compromised, then the regulation of peptide binding and/or release may be altered and the delivery of an ERAD substrate to the cytoplasm would be attenuated. To assess interdomain coupling of wild-type BiP and the Kar2-1 and Kar2-133 proteins, we first needed to identify a more soluble, but related peptide substrate for yeast BiP. The peptide ultimately chosen, known as p5, is a shorter derivative of APPY (Pierpaoli et al., 1998) and when labeled with the fluorescent reporter, AEDANS bound to the wild-type and mutant proteins, as measured by fluorescence anisotropy (our unpublished data). Next, we added saturating concentrations of unlabeled p5 (700 μM peptide to 0.7 μM BiP) and measured steady state ATP hydrolysis overtime. Although the endogenous ATPase activities of wild-type and the mutant Kar2 proteins were similar (Figures 6 and 7B), we observed an ∼twofold reduced p5-mediated activation of ATP hydrolysis for the mutant proteins compared with wild-type BiP (Figure 7B). This second result suggests that the interdomain coupling between peptide interaction and ATP hydrolysis is altered to some degree in the Kar2 mutants.
Wild-type BiP Increases the Solubility of an ERAD Substrate in kar2-1 Microsomes But Exacerbates the ERAD Defect
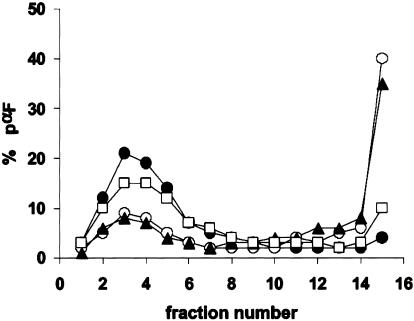
BiP retains ERAD substrates in an extended conformation as they retro-translocate from the ER, and defects in BiP function can lead to substrate aggregation (Nishikawa et al., 2001). To examine whether the solubility of an ERAD substrate was reduced in the ER of the kar2 mutants, we measured the oligomeric state of pαF. Microsomes containing radiolabeled pαF were treated with detergent, the extract was centrifuged on a continuous sucrose gradient, and the amount of pαF in each fraction was measured after SDS-PAGE and phosphorimager analysis (Nishikawa et al., 2001). When the sedimentation of pαF derived from wild-type microsomes was examined, we found that ∼5% of the total pαF resided at the bottom of the gradient, whereas ∼40% of the pαF resided at the bottom of the gradients when kar2-1– and kar2-133–derived microsomes were used, respectively (Figure 8). These data suggest that the propensity of pαF to aggregate in the ER of kar2-1 yeast is enhanced, an effect that would hinder retro-translocation. We then examined pαF solubility in extracts prepared from kar2-1 mutant yeast that had been transformed with a wild-type KAR2 expression vector (pMR109) and observed that pαF was now largely absent from the bottom of the gradient and that the percentage of soluble pαF (fractions 2–5) increased.
Figure 8.
PαF aggregation is enhanced in kar2-1–derived mutant microsomes but can be resolubilized by wild-type BiP. ΔGppαF was translocated into microsomes derived from wild-type (•), kar2-1 (○), and kar2-133 (▴) cells, and kar2-1 yeast transformed with a KAR2 overexpression plasmid (pMR109; □). The microsomes were washed and incubated at 37°C for 30 min before Triton X-100 extraction and sedimentation in a 5–40% sucrose gradient. The percentages of pαF, the in vitro ERAD substrate, in fractions from the top (fraction 1) to the bottom (fraction 14) of the gradient are shown.
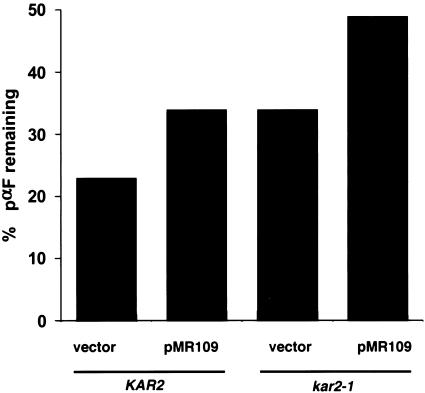
To explore whether increasing pαF solubility enhanced its degradation, we performed the in vitro ERAD assay using microsomes from the wild-type and kar2-1 strains containing either pMR109 or a vector control. The concentration of BiP was greater in microsomes prepared from the wild-type and kar2-1 strains containing pMR109 compared with yeast harboring the vector control: BiP levels were elevated by ∼50% when pMR109 was present in the wild-type strain and by ∼90% when the same plasmid was present in the kar2-1 strain as assessed by quantitative immunoblot analysis (our unpublished data). We then observed that microsomes from the kar2-1 strain degraded pαF less efficiently than microsomes from the isogenic wild-type strain, as shown previously (Brodsky et al., 1999; also see Figure 4B). Surprisingly, ERAD was inhibited by 40–50% in microsomes prepared from either strain containing the wild-type BiP expression vector compared with the vector control (Figure 9). This result suggests that the kar2-1 mutation may be dominant with respect to ERAD because the ERAD defect was not rescued by expression of wild-type protein; however, it is formally possible that wild-type BiP failed to rescue the ERAD defect in kar2-1–derived microsomes because the amount of BiP may be too high in this system. Regardless, these data show that increasing the amount of BiP augments polypeptide solubility in a kar2 mutant but is insufficient to restore ERAD to wild-type levels. Furthermore, increasing the amount of BiP in microsomes prepared from wild-type cells inhibited ERAD. A model for these observations is presented below.
Figure 9.
Increasing the concentration of wild-type BiP exacerbates the kar2-1 ERAD defect and reduces ERAD efficiency in wild-type microsomes. Wild-type (KAR2) and kar2-1 mutant microsomes prepared from strains harboring a control vector (vector) or overexpressing BiP (pMR109) were prepared and an in vitro ERAD assay was performed as described in MATERIALS AND METHODS. Data represent the means of three independent experiments and standard deviations are <10% of the means.
DISCUSSION
To better define the role of BiP during ERAD, we devised a genetic screen to obtain ERAD-specific kar2 mutants and isolated the kar2-1 allele, which was previously coopted to show that BiP is required for the degradation of soluble (ER lumenal) ERAD substrates but not for the proteolysis of integral ER membrane proteins (Brodsky et al., 1999). The position of the mutated residue in Kar2-1p suggested that another previously isolated kar2 mutant, known as kar2-133, would exhibit similar properties. Indeed, kar2-1 and kar2-133 yeast are defective for the ERAD of three soluble substrates (CPY*: Zhang et al., 2001; pαF and AiPiZ: Brodsky et al., 1999), and we report here that expression of Kar2-1p or Kar2-133p induces the UPR and leads to the aggregation of one of these substrates. Furthermore, the Kar2-1 and Kar2-133 proteins exhibit reduced peptide affinities compared with wild-type BiP, and the addition of peptide fails to enhance ATP hydrolysis by the mutant proteins to the same degree as for wild-type BiP. This effect, although partial, most likely results from interdomain communication defects in the mutant proteins because an equal degree of J domain–stimulated ATPase activity was observed for Kar2-1/133p and wild-type BiP. Although we cannot formally exclude the possibility that the reduced peptide affinity of the mutants was the cause of lower peptide-stimulated ATPase activity, we note that the concentration of peptide used in this latter experiment was 1000-fold greater than the concentration of BiP, and—if the affinities of BiP for APPY and p5 are comparable—was >40-fold higher than the predicted KD.
In light of these data, why are Kar2-1p– and Kar2-133p–expressing cells and microsomes derived from these strains translocation-proficient? The ability of BiP to bind a translocating polypeptide is essential to drive protein import, whether through its action as a “motor” or as a “ratchet” (Sanders et al., 1992; Matlack et al., 1999). However, it was previously noted by Rapoport and colleagues that 6–7 BiPs bind ppαF, and a BiP mutant lacking the peptide binding domain “lid”—and that exhibits a faster off-rate for peptide when ADP-bound (Misselwitz et al., 1998)—supported ratcheting, but higher concentrations of the protein were required (Matlack et al., 1999). Thus, a BiP mutant with a lower peptide affinity should catalyze protein translocation as long as greater amounts of the protein are present. In fact, this may be the case in kar2-1 and kar2-133 yeast and in Kar2-1/133p-containing microsomes (Table 1). We further note that the positions of the kar2-1 and kar2-133 mutations are likely to weaken interactions between the lid and the loops in the peptide-binding domain, resulting in proteins with similar properties as the “lid-less” BiP mutants used by Matlack et al. (1999); as expected, Hsc70 mutants with predicted weakened interactions between the loops and the lid exhibit an increase in peptide off-rates (Hu et al., 2002; see below). In contrast, a BiP mutant in which peptide binding is completely abolished should be lethal and would not be isolated in our screen: Mutations in DnaK that significantly reduce the chaperone's peptide affinity cannot complement the ΔdnaK phenotype (for example, see Burkholder et al., 1996). Therefore, we propose that kar2-1 and kar2-133 yeast are viable and translocation-proficient because they can balance the reduced affinity between BiP and peptides by synthesizing greater amounts of BiP via the UPR.
To explain the fact that kar2-1 and kar2-133 cells and microsomes obtained from these strains are ERAD-defective, we note that pαF aggregates in kar2-1– and kar2-133–derived microsomes (Figure 8), a phenomenon that would preclude retro-translocation through the pore formed by the Sec61p complex in the ER membrane. We suggest further that nucleotide and/or cochaperone-mediated pαF release from Kar2-1/133p is altered. For example, if pαF is released prematurely, then the polypeptide may aggregate before it can be retro-translocated. Several lines of evidence support this view. First, expression of wild-type BiP restores pαF solubility (Figure 8), suggesting that it binds Kar2-1/133p–released polypeptides. Second, mutations in bovine Hsc70 in which interdomain coupling is altered exhibit an increase in peptide off-rate (Ha et al., 1997). Third, conversion of glycine-468 to aspartic acid in DnaK was proposed to perturb the conformation of loop 5,6 in the peptide-binding domain, and when combined with a second mutation (G455D), the peptide off-rate increased (Buchberger et al., 1999). Based on their positions (Figure 5), the kar2-1 and kar2-133 gene products may exhibit a similar kinetic defect; however, the contribution of single mutations, ATP, and/or Hsc70/DnaK cochaperones on peptide release was not examined in these other studies. Thus, it will be vital in future work to assess the peptide affinities and on- and off-rates for wild-type BiP and Kar2-1/133p in the presence and absence of each of the growing number of known BiP cochaperones, which include activators of ATP hydrolysis (e.g., Sec63p, Scj1p, and Jem1p) and nucleotide exchange factors (reviewed in Fewell et al., 2001); for example, the Sls1p nucleotide exchange factor catalyzes nucleotide release from yeast BiP, and the introduction of the kar2-1 and kar2-133 alleles into sls1Δ yeast results in synthetic effects on cell growth (Kabani et al., 2000b).
Another, nonmutually exclusive scenario is that the Kar2-1p– and Kar2-133p–mediated ERAD defects arise at least in part from defective “gating” of the translocation channel during retro-translocation. Experiments from Johnson and colleagues (Hamman et al., 1998; Haigh and Johnson, 2002) indicate that BiP helps seal the translocation channel in the mammalian ER during translocation, but it is not clear how BiP facilitates channel reopening during retro-translocation (Johnson and Haigh, 2000). Therefore, it is formally possible that the Kar2p mutants are able to gate the translocon during protein import, but cannot do so during ERAD; this hypothesis is currently being examined. BiP-dependent gating might also require the participation of cochaperones, and the differential interaction between Kar2-1p and translocation/retro-translocation-specific factors might also explain why kar2-1 yeast are selectively defective for ERAD.
Finally, as noted above, we have shown that increasing the amount of BiP in the ER does not facilitate ERAD in wild-type yeast but instead slows retro-translocation and degradation. In the kar2-1 background, the introduction of wild-type BiP helps resolubilize pαF but ERAD efficiency is also reduced. These data may be explained by the fact that prevention of polypeptide aggregation requires only BiP binding, whereas ERAD requires both polypeptide binding and release, which might be regulated by cochaperones. PαF might have become bound to and trapped in the ER by exaggerated amounts of BiP (Figure 9), but the concentrations and/or activities of cochaperones required for release might not have risen in parallel. Consistent with our work, increasing the level of BiP slows the ERAD of mutant ribophorin in mammalian cells (deVirgilio et al., 1999) and the concept that ERAD can proceed only after BiP release is well established (Knittler et al., 1995; Beggah et al., 1996; Skowronek et al., 1998; Chillaron and Haas, 2000). It is possible that under conditions of ER stress, when the concentration of BiP rises via the UPR, ERAD efficiency may decline, but the solubility of putative ERAD substrates is maintained. In turn, if ER stress is removed and/or the level of BiP can return to that in the unstressed state, BiP-bound, aggregation-prone substrates may be given a “second-chance” to fold or may then be targeted for degradation. To test this hypothesis, it will be important in the future to correlate how different levels of ER stress affect the concentration of BiP, ERAD efficiency, and the solubility of ERAD substrates.
Acknowledgments
We thank Peter Walter, Linda Rotondi, Michael Cascio, Amie McClellan, James Endres, Ricky Rivers, Diane Hoeffelder, Jen Goeckeler, Sheara Fewell, Tom Harper, and Jason Weil for reagents and technical assistance. This work was supported by Grant MCB-0110331 from the National Science Foundation and by grants from the National Institutes of Health (NIH) to L.L.M. and M.D.R. We also acknowledge shared instrumentation Grant 1S10RR 11998-01A1 from the NIH to Dr. Michael Cascio (University of Pittsburgh Medical School).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-12-0847. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-12-0847.
References
- Aridor, M., and Hannan, L.A. (2000). Traffic jam: a compendium of human diseases that affect intracellular transport processes. Traffic 1, 836–851. [DOI] [PubMed] [Google Scholar]
- Beggah, A., Mathews, P., Beguin, P., and Geering, K. (1996). Degradation and endoplasmic reticulum retention of unassembled alpha- and beta-subunits of Na, K-ATPase correlate with interaction of BiP. J. Biol. Chem. 271, 20895–20902. [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi, S., Cwirla, S.E., Dower, W.J., Lipshutz, R.J., Sprang, S.R., Sambrook, J.F., and Gething, M.J. (1993). Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75, 717–728. [DOI] [PubMed] [Google Scholar]
- Brodsky, J.L., and Schekman, R. (1993). A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J. Cell Biol. 123, 1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, J.L., Goeckeler, J., and Schekman, R. (1995). Sec63p and BiP are required for both co- and post-translational protein translocation into yeast microsomes. Proc. Natl. Acad. Sci. USA 92, 9643–9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, J.L., and McCracken, A.A. (1999). ER protein quality control and proteasome-mediated protein degradation. Semin. Cell Dev. Biol. 10, 507–513. [DOI] [PubMed] [Google Scholar]
- Brodsky, J.L., Werner, E.D., Dubas, M.E., Goeckeler, J.L., Kruse, K.B., and McCracken, A.A. (1999). The requirement for molecular chaperones during ER associated protein degradation (ERAD) demonstrates that protein import and export are mechanistically distinct. J. Biol. Chem. 274, 3453–3460. [DOI] [PubMed] [Google Scholar]
- Buchberger, A., Gassler, C.S., Buttner, M., McMacken, R., and Bukau, B. (1999). Functional defects of the DnaK756 mutant chaperone of Escherichia coli indicate distinct roles for amino and carboxyl-terminal residues in substrate and co-chaperone interaction and interdomain communication. J. Biol. Chem. 274, 38017–38026. [DOI] [PubMed] [Google Scholar]
- Burkholder, W.F., Zhao, X., Zhu, X., Hendrickson, W.A., Gragerov, A., and Gottesman, M.E. (1996). Mutations in the C-terminal fragment of DnaK affecting peptide binding. Proc. Natl. Acad. Sci. USA 93, 10632–10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassagrande, R., Stern, P., Diehn, M., Shamu, C., Osario, M., Zuniga, M., Brown, P.O., and Ploegh, H. (2000). Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell 5, 729–735. [DOI] [PubMed] [Google Scholar]
- Chevalier, M., King, L., Wang, C., Gething, M.J., Elguindi, E., and Blond, S.Y. (1998). Substrate binding induces depolymerization of the C terminal peptide binding domain of murine Grp78/BiP. J. Biol. Chem. 273, 26827–26835. [DOI] [PubMed] [Google Scholar]
- Chillaron, J., and Haas, I.G. (2000). Dissociation from BiP and retrotranslocation of unassembled immunoglobulin light chains are tightly coupled to proteasome activity. Mol. Biol. Cell 11, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi, A., and Schekman, R. (1997). The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J. Cell Biol. 137, 1483–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J.S., Shamu, C.E., and Walter, P. (1993). Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197–1206. [DOI] [PubMed] [Google Scholar]
- Davis, J.E., Voisine, C., and Craig, E.A. (1999). Intragenic suppressors of Hsp70 mutants: interplay between the ATPase- and peptide-binding domains. Proc. Natl. Acad. Sci. USA 96, 9269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deVirgilio, M., Kitzmuller, C., Schwaiger, E., Klein, M., Kreibich, G., and Ivessa, N.E. (1999). Degradation of a short-lived glycoprotein from the lumen of the endoplasmic reticulum: the role of N-linked glycans and the unfolded protein response. Mol. Biol. Cell 10, 4059–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard, L., Molinari, M., and Helenius, A. (1999). Setting the standards: quality control in the secretory pathway. Science 286, 1882–1888. [DOI] [PubMed] [Google Scholar]
- Fewell, S.W., Travers, K.J., Weissman, J.S., and Brodsky, J.L. (2001). The action of molecular chaperones in the early secretory pathway. Annu. Rev. Genet. 35, 149–91. [DOI] [PubMed] [Google Scholar]
- Flynn, G.C., Pohl, J., Flocco, M.T., and Rothman, J.E. (1991). Peptide binding specificity of the molecular chaperone BiP. Nature 353, 726–730. [DOI] [PubMed] [Google Scholar]
- Friedlander, R., Jarosch, E., Urban, J., Volkwein, C., and Sommer, T. (2000). A regulatory link between ER-associated protein degradation and the unfolded protein response. Nat. Cell. Biol. 2, 379–84. [DOI] [PubMed] [Google Scholar]
- Fujiki, Y., Hubbard, A.L., Fowler, S., and Lazarow, P.B. (1982). Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillece, P., Luz, J.M., Lennarz, W.J., de La Cruz, F.J., and Römisch, K. (1. 999). Export of a cysteine-free misfolded secretory protein from the endoplasmic reticulum for degradation requires interaction with protein disulfide isomerase. J. Cell Biol. 147, 1443–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, J.H., Hellman, U., Johnson, E.R., Li, L., McKay, D.B., Sousa, M.C., Takeda, S., Wernstedt, C., and Wilbanks, S.M. (1997). Destabilization of peptide binding and interdomain communication by an E543K mutation in the bovine 70-kDa heat shock cognate protein, a molecular chaperone. J. Biol. Chem. 272, 27796–27803. [DOI] [PubMed] [Google Scholar]
- Haigh, N.G., and Johnson, A.E. (2002). A new role for BiP: closing the aqueous translocon pore during protein integration into the ER membrane. J. Cell Biol. 156, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman, B.D., Hendershot, L.M., and Johnson, A.E. (1998). BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocation pore before and early in translocation. Cell 92, 747–758. [DOI] [PubMed] [Google Scholar]
- Hartl, F.U. (1996). Molecular chaperones in cellular protein folding. Nature 381, 571–580. [DOI] [PubMed] [Google Scholar]
- Hayes, S.A., and Dice, J.F. (1996). Role of molecular chaperones in protein degradation. J. Cell Biol. 132, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhfeld, J., Cyr, D.M., and Patterson, C. (2001). From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Rep. 2, 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich, A.L., Weber-Ban, E.U., and Finley, D. (1999). Chaperone rings in protein folding and degradation. Proc. Natl. Acad. Sci. USA 96, 11033–11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, S.M., Liang, P.H., Hsiao, C.D., and Wang, C. (2002). Characterization of the L399P and R447G mutants of hsc70: the decrease in refolding activity is correlated with an increase in the rate of substrate dissociation. Arch. Biochem. Biophys. 407, 135–141. [DOI] [PubMed] [Google Scholar]
- Johnson, A.E., and Haigh, N.G. (2000). The ER translocon and retrotranslocation: is the shift into reverse manual or automatic? Cell 102, 709–712. [DOI] [PubMed] [Google Scholar]
- Kabani, M., Boisrame, A., Beckerich, J-M., and Gaillardin, C. (2000a). A highly representative two-hybrid library for the yeast Yarrowia lipolytica. Gene 241, 309–315. [DOI] [PubMed] [Google Scholar]
- Kabani, M., Beckerich, J.M., and Gaillardin, C. (2000b). Sls1p stimulates Sec63p-mediated activation of Kar2p in a conformation-dependent manner in the yeast endoplasmic reticulum. Mol. Cell. Biol. 20, 6923–6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani, M., Beckerich, J.M., and Brodsky, J.L. (2002). Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 22, 4677–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittler, M.R., Dirks, S., and Haas, I.G. (1995). Molecular chaperones involved in protein degradation in the endoplasmic reticulum: quantitative interaction of the heat shock cognate protein BiP with partially folded immunoglobulin light chains that are degraded in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 92, 1764–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman, S.K., and Schekman, R. (1995). Interaction between BiP and Sec63p is required for the completion of protein translocation into the ER of S. cerevisiae. J. Cell Biol. 131, 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack, K.E.S., Plath, K., Misselwitz, B., and Rapoport, T.A. (1997). Protein transport by purified yeast Sec complex and Kar2p without membranes. Science 277, 938–941. [DOI] [PubMed] [Google Scholar]
- Matlack, K.E., Misselwitz, B., Plath, K., and Rapoport, T.A. (1999). BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell 97, 553–564. [DOI] [PubMed] [Google Scholar]
- McClellan, A.J., Endres, J., Vogel, J.P., Palazzi, D., Rose, M.D., and Brodsky, J.L. (1998). Specific molecular chaperone interactions and an ATP-dependent conformational change are required during posttranslational protein translocation into the yeast ER. Mol. Biol. Cell 9, 3533–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken, A.A., and Brodsky, J.L. (1996). Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J. Cell Biol. 132, 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz, B., Staeck, O., and Rapoport, T.A. (1998). J proteins catalytically activate Hsp70 molecules to trap a wide range of peptide sequences. Mol. Cell 2, 593–603. [DOI] [PubMed] [Google Scholar]
- Montgomery, D.L., Morimoto, R.I., and Gierasch, L.M. (1999). Mutations in the substrate binding domain of the Escherichia coli 70 kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling. J. Mol. Biol. 286, 915–932. [DOI] [PubMed] [Google Scholar]
- Mori, K., Sant, A., Kohno, K., Normington, K., Gething, M.J., and Sambrook, J. (1992). A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 7, 2583–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, M.W., and Brodsky, J.L. (2001). Yeast ribosomes bind to highly purified reconstituted Sec61p complex and to mammalian p180. Traffic 2, 705–716. [DOI] [PubMed] [Google Scholar]
- Nishikawa, S.I., Fewell, S., Kato, Y., Brodsky, J.L., and Endo, T. (2001). Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 153, 1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, D.T., Brown, J.D., and Walter, P. (1996). Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 134, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, D.T., Spear, E.D., and Walter, P. (2000). The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 150, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpaoli, E.V., Gisler, S.M., and Christen, P. (1998). Sequence-specific rates of interaction of target peptides with the molecular chaperones DnaK and DnaJ. Biochemistry 37, 16741–16748. [DOI] [PubMed] [Google Scholar]
- Pfund, C., Huang, P., Lopez-Hoyo, N., and Craig, E.A. (2001). Divergent functional properties of the ribosome-associated chaperone Ssb compared with other Hsp70s. Mol. Biol. Cell 12, 3773–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon, M., Schekman, R., and Römisch, K. (1997). Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 16, 4540–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper, R.K., Böhmler, S., Bordallo, J., Sommer, T., and Wolf, D.H. (1997). Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388, 891–895. [DOI] [PubMed] [Google Scholar]
- Plemper, R.K., and Wolf, D.H. (1999). Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem. Sci. 24, 266–270. [DOI] [PubMed] [Google Scholar]
- Polaina, J., and Conde, J. (1982). Genes involved in the control of nuclear fusion during the sexual cycle of Saccharomyces cerevisiae. Mol. Gen. Genet. 186, 253–258. [DOI] [PubMed] [Google Scholar]
- Römisch, K. (1999). Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane. J. Cell Sci. 112, 4185–4191. [DOI] [PubMed] [Google Scholar]
- Rose, M., and Botstein, D. (1983). Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 101, 167–180. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., Misra, L.M., and Vogel, J.P. (1989). KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57, 1211–1221. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., and Hieter, P. (1990). Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Rüdiger, S., Germeroth, L., Schneider-Mergener, J., and Bukau, B. (1997). Substrate specificity of the DnaK chaperone determined by scanning cellulose-bound peptide libraries. EMBO J. 16, 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, S.L., Whitfield, K.M., Vogel, J.P., Rose, M.D., and Schekman, R.W. (1992). Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell 69, 353–365. [DOI] [PubMed] [Google Scholar]
- Schmitz, A., Maintz, M., Kehle, T., and Herzog, V. (1995). In vivo iodination of a misfolded proinsulin reveals co-localized signals for Bip binding and for degradation in the ER. EMBO J. 14, 1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, J.F., Ferro-Novick, S., Rose, M.D., and Helenius, A. (1995). BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J. Cell Biol. 130, 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronek, M.H., Hendershot, L.M., and Haas, I.G. (1998). The variable domain of nonassembled Ig light chains determines both their half-life and binding to the chaperone BiP. Proc. Natl. Acad. Sci. USA 95, 1574–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling, C.J., Rothblatt, J., Hosobuchi, M., Deshaies, R., and Schekman, R. (1992). Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell 3, 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, C.S., Tremblay, J.D., Fewell, S.W., Lewis, J.A., Brodsky, J.L., and Pipas, J.M. (2000). Species-specific elements in the large T-antigen J domain are required for cellular transformation and DNA replication by simian virus 40. Mol. Cell. Biol. 20, 5749–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Heesen, S., and Aebi, M. (1994). The genetic interaction of kar2 and wbp1 mutations. Eur. J. Biochem. 222, 631–637. [DOI] [PubMed] [Google Scholar]
- Thomas, P.J., Qu, B.H., and Pedersen, P.L. (1995). Defective protein folding as a basis of human disease. Trends Biochem. Sci. 20, 456–459. [DOI] [PubMed] [Google Scholar]
- Tsai, B., Ye, Y., and Rapoport, T.A. (2002). Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell. Biol. 3, 246–255. [DOI] [PubMed] [Google Scholar]
- Travers, K.J., Patil, C.K., Wodicka, L., Lockhart, D.J., Weissman, J.S., and Walter, P. (2000). Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249–258. [DOI] [PubMed] [Google Scholar]
- Vogel, J.P., Misra, L.M., and Rose, M.D. (1990). Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J. Cell Biol. 110, 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner, S., Maurizi, M.R., and Gottesman, S. (1999). Posttranslational quality control: folding, refolding, and degrading proteins. Science 286, 1888–1893. [DOI] [PubMed] [Google Scholar]
- Wiertz, E.J., Tortorella, D., Bogyo, M., Yu, J., Mothes, W., Jones, T.R., Rapoport, T.A., and Ploegh, H.L. (1996). Sec61p-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384, 432–438. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Nijbroek, G., Sullivan, M.L., McCracken, A.A., Watkins, S.C., Michaelis, S., and Brodsky, J.L. (2001). Hsp70 Molecular chaperone facilitates endoplasmic reticulum associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell 12, 1303–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, M., and Schekman, R. (1999). The engagement of Sec61p in the ER dislocation process. Mol. Cell 4, 925–934. [DOI] [PubMed] [Google Scholar]
- Zhu, X., Zhao, X., Burkholder, W.F., Gragerov, A., Ogata, C.M., Gottesman, M.E., and Hendrickson, W.A. (1996). Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]