Abstract
The JB6 mouse epidermal cell system, which includes tumor promotion-sensitive (P+) and tumor promotion-resistant (P−) cells, is a well-established and extensively used cell culture model for studying the mechanism of late-stage tumor promotion. Tumor promoters, such as 12-O-tetradecanoylphorbol 13-acetate (TPA) or epidermal growth factor (EGF), induce high levels of activator protein 1 (AP-1) activity and large, tumorigenic, anchorage-independent colonies in soft agar at a high frequency in JB6 P+ cells, but not in JB6 P− cells. We report here a molecular explanation for the defect in the AP-1 activation and promotion-resistant phenotype of P− cells. We demonstrate that the lack of AP-1 activation and cell transformation responses to TPA and EGF in P− cells appears attributable to the low level of mitogen-activated protein kinase (MAPK) (extracellular signal-regulated protein kinase, Erk) in these cells. TPA and EGF induce transactivation of AP-1 activity in P+ cells but not in P− cells. Nonphosphorylated forms and TPA- or EGF-induced phosphorylated forms of Erks (Erk1 and Erk2) in P− cells were much lower than those in P+ cells. Stable transfection of wild-type MAPK (Erk2) into P− cells restored its response to TPA and EGF for both AP-1 activation and cell transformation. These results suggest that the shortage of MAPK (Erk1 and Erk2) appears to be an important contributor to the tumor promotion-resistant phenotype in JB6 cells.
Chemical carcinogenesis is known to be a multistep process that includes initiation, promotion, and progression (1–4). The availability of the JB6 promotable mouse epidermal cell lines has allowed the extension of tumor promotion studies to an easily manipulated cell culture system (5–10). The promotion-sensitive JB6 cells, originally derived from primary mouse epidermal cells, undergo a response analogous to second stage tumor promotion in mouse skin (5) and are extensively used as an in vitro model for studying the promotion of neoplastic transformation (6–19). This model system includes transformation-sensitive (P+) and transformation-resistant (P−) cells. Exposure of JB6 P+ cells to tumor promoters, such as 12-O-tetradecanoylphorbol 13-acetate (TPA) or to epidermal growth factor (EGF), irreversibly induces anchorage-independent phenotype and tumorigenicity (6–12). In contrast, the P− cells are resistant to tumor promoter induction of anchorage-independence and tumorigenicity (5, 11). The differences between P+ and P− cells and the mechanism of promotion resistance in P− cells have been the subjects of extensive study (5, 6, 12). Of particular interest was the observation that a high level of activator protein 1 (AP-1) transactivating activity measured by reporter constructs was induced by TPA or EGF in P+ but not in P− cells (6). AP-1 is a heterodimeric complex containing products of the jun and fos oncogene families (20). The inquiry as to whether this AP-1 response is required for transformation and might therefore explain the differential sensitivity of P− and P+ cells led to our findings that retinoids and glucocorticoids as well as expression of dominant-negative Jun blocked both induced AP-1 and induced transformation (7). Further evidence implicating AP-1 as required came from the observation that transcription factor-specific retinoids that transrepress AP-1 blocked TPA-induced cell transformation (21). To address the question as to what was limiting the AP-1 response and cell transformation in P− cells, we examined the possibility that the concentration of one or more AP-1 proteins might be insufficient. However, overexpression of c-Jun, the only differentially expressed Jun or Fos family protein (19), failed to restore the AP-1 activation response to the P− cells (15). Knowing that Jun and Fos family proteins interact with mitogen-activated protein kinase (MAPK) family proteins in JB6 cells (14) and that phosphorylation of extracellular signal-regulated protein kinases (Erks) can activate AP-1 activity (22) provided a rationale for examining the possibility that proteins in the Erk family of MAPKs might be limiting. We demonstrate here that P− cells, which show reduced levels of Erk expression and activation, can be converted to promotion sensitive phenotype when wild-type Erk is overexpressed.
MATERIALS AND METHODS
Plasmids and Reagents.
AP-1 luciferase reporter plasmid (−73/+63 collagenase-luciferase) and cytomegalovirus (CMV)-neo vector plasmid were constructed as reported previously (9, 21). Rat wild-type Erk2 was a generous gift from Melanie H. Cobb (University of Texas Southwestern Medical Center) (23). Fetal bovine serum (FBS) was from GIBCO; LipofectAMINE was from GIBCO/BRL; Eagle’s minimal essential medium (MEM) and TPA were from Calbiochem; EGF was from Collaborative Research; luciferase assay substrate was from Promega; and PhosphoPlus MAPK antibody kit was from New England Biolabs.
Cell Culture.
JB6 P+ mouse epidermal cell line (Cl 41) and its transfectants or P− cell line (Cl 30.7b) and its transfectants were cultured in monolayers at 37°C, 5% CO2 by using Eagle’s MEM containing 5% FBS, 2 mM l-glutamine, and 25 μg of gentamicin per ml (21).
Generation of Stable Cotransfectants.
JB6 P− cells (Cl 30.7b) were cultured in a 6-well plate until they reached 85–90% confluence. We used 2 μg of AP-1 luciferase reporter plasmid and 0.3 μg of CMV-neo vector with or without 12 μg of wild-type Erk2 plasmid DNA and 15 μl of LipofectAMINE reagent to transfect each well in the absence of serum. JB6 P+ cells (Cl 41) were transfected with 2 μg of AP-1 luciferase reporter plasmid and 0.3 μg of CMV-neo plasmid DNA. After 10–12 hr, the medium was replaced by 5% FBS MEM. Approximately 30–36 hr after the beginning of the transfection, the cells were digested with 0.033% trypsin, and cell suspensions were plated into 100-mm dishes and cultured for 24–28 days with G418 selection (300 μg/ml). Stable transfectants were screened by assay of the luciferase activity and PhosphoPlus MAPK antibody kit. Stably transfected 30.7b AP-1 mass3, 30.7b AP-1 mass4, 30.7b MAPK-WT mass2, and C141 AP-1 mass1 were established and cultured in G418-free MEM for at least two passages before each experiment. Clonal transfectants were also established by limited dilution method.
Assay for AP-1 Activity.
Confluent monolayers of JB6 P+ or P− cell stable transfectants were trypsinized and 8 × 103 viable cells suspended in 100 μl 5% FBS MEM were added into each well of a 96-well plate. Plates were incubated at 37°C in a humidified atmosphere of 5% CO2/95% gas air. Twelve to 24 hr later, cells were starved by culturing cells in 0.1% FBS MEM for 12 hr before exposure to TPA or EGF. The cells were exposed to EGF or TPA as indicated for 24 hr in 37°C, 5% CO2 incubation. The cells were extracted with lysis buffer, and luciferase activity was measured by using a luminometer (monolight 2010). The results are expressed as the relative AP-1 activity or relative luciferase units (RLU) (21, 24). The relative AP-1 activity was presented as the luciferase activity relative to the medium control cells.
Anchorage-Independent Transformation Assay.
Cells (1 × 104) in a 60-mm dish were exposed to TPA or EGF in 1 ml of 0.33% BME agar containing 10% FBS over 3.5 ml of 0.5% BME agar containing 10% FBS medium. The cultures were maintained in 37°C, 5% CO2 incubator for 14–16 days, and the induced cell colonies were scored by the methods described (11).
MAPK Analysis.
Erk activation was determine by immunoblotting with phospho-specific antibodies. Cell extracts were analyzed for Erk1 and -2 with antibodies against Erk1 and Erk2 (p44 and P42) and for phosphorylated Erk1 and 2 with phospho-specific antibodies against phosphorylated tyrosine 204 of p44 and p42 MAPKs (25, 26). Antibodies were from the PhosphoPlus MAPK antibody kit (New England Biolabs) and were used according to manufacturer’s recommendations. Antibody bound proteins were detected by chemiluminescence (ECL; Amersham).
RESULTS
Establishment of P− and P+ AP-1 Reporter Cells: Retention of Differential Response Phenotypes.
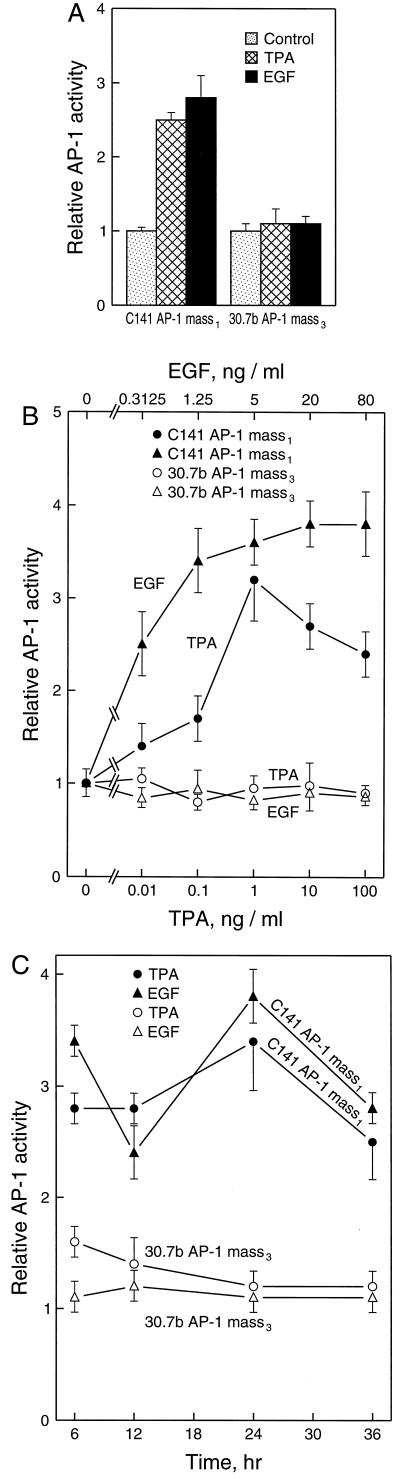
JB6 cell lines Cl 41 and Cl 30.7b represent typical P+ and P− cells, respectively (5–10). To compare the AP-1 induction by different tumor promoters between P+ and P− cells, we established stable AP-1 reporter (−73/+63 collagenase luciferase) transfectants from Cl 41 and Cl 30.7b cells. These stable transfectants were generated by “mass selection” of pooled clones. After transfecting cells with plasmids in 100-mm dishes and selecting by G418 (300 μg/ml), more than 10 colonies per dish were grown. Instead of ring cloning of these cells (21), all of these colonies in the dish were grown as a “mass stable culture.” To be sure that the original tumor promoter sensitivity in the reporter gene transfectants was not altered by the process of gene transfection, the transformation response to TPA and EGF was measured by using anchorage-independent transformation in soft agar. The JB6 P+ cell transfectant, Cl 41 AP-1 mass1 and P− cell transfectant, Cl 30.7b AP-1 mass3, showed the same characteristic transformation response to TPA and EGF as seen with their corresponding parental P+ Cl 41 or P− Cl 30.7b (data not shown). TPA and EGF induced high levels of AP-1 activation in C1 41 AP-1 mass1 at all the time points and doses studied (Fig. 1), but no significant AP-1 transactivation by TPA or EGF was observed in Cl 30.7b AP-1 mass3 (Fig. 1A). These results were further confirmed by dose response observation (Fig. 1B). During the time course study, only a low level (1.6-fold) of AP-1 activation was observed at 6 hr in the Cl 30.7b AP-1 mass3 cells that were exposed to TPA (Fig. 1C). These data are consistent with previous findings of Bernstein and Colburn (6) in a transient transfection study.
Figure 1.
TPA- and EGF-induced AP-1 activation in JB6 P+ cells but not P− cells. A total of 8 × 103 of JB6 P+, C1 41 AP-1 mass1 or P−, Cl 30.7b AP-1 mass3 suspended in 5% FBS MEM was seeded into each well of 96-well plates. After overnight culture at 37°C, the cells were starved for 24 hr by changing medium with 0.1% FBS MEM. Then, (A) the cells were treated with TPA (10 ng/ml) or EGF (10 ng/ml) for 24 hr before assaying for luciferase activity; (B) for dose response study, the cells were treated with the indicated dose of TPA or EGF and incubated for 24 hr, then luciferase activity was determined; and (C) for the time course study, the cells were exposed to TPA (10 ng/ml) or EGF (10 ng/ml). The luciferase activity was measured at time points as indicated. The results were expressed as relative AP-1 activity. The relative AP-1 activity was presented as the luciferase activity relative to the medium control.
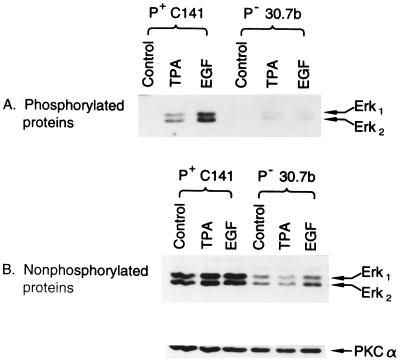
AP-1 Nonresponsiveness in JB6 P− Cells Is Accompanied by Low Levels of MAP Kinase.
Previous studies showed that neither protein kinase C (PKC) (27) nor c-Jun (15) was the factor limiting the AP-1 response in P− cells. For these reasons and because others have demonstrated a requirement for Erk1 and -2 in AP-1 transactivation (22), we hypothesized that the lack of AP-1 transactivation response in P− cells may be caused by a low level of functioning Erks. To test this hypothesis, we analyzed the Erk1 and Erk2 proteins and their TPA- or EGF-induced phosphorylated (activated) forms in P+ Cl 41 and P− Cl 30.7b cells. Because PKCα is invariant in JB6 variants with or without tumor promoter (28), we used it as control for the loaded sample protein. The results showed that the levels of Erk1 and Erk2 proteins and TPA- or EGF-induced phosphorylated Erk1 and Erk2 proteins in the P− cells were much lower than those in P+ cells (Fig. 2). Sequence analysis of a 193-bp fragment (bases 507–699) of the mRNA coding sequence generated by reverse transcription–PCR of RNA from JB6 Cl 30.7b and cloned into a TA cloning vector indicated that the conserved phosphorylation domain of Erk2 to which the phospho-specific antibody was raised had not been mutated. Thus, the deficiency in activated (phosphorylated) Erk levels in Cl 30.7b cannot be attributed to a lack of antibody recognition of this domain. Similarly, because this unmutated domain is a suitable substrate for phosphorylation, the phosphorylation of Erk is not limiting. In fact, the low level of Erk proteins that are present in 30.7b appear to be phosphorylated efficiently (see Fig. 3B, 30.7b AP1 mass4). Thus Erk protein, not its phosphorylation site, is deficient.
Figure 2.
Low level of MAPK (Erk1 or Erk2) expression and activation in JB6 P− Cl 30.7b cells. A total of 8 × 104 of JB6 P+ C1 41 or P− Cl 30.7b cells was seeded into each well of 6-well plates. After culture at 37°C for 24 hr, the cells were starved for 48 hr by replacing medium with 0.1% FBS MEM. The medium was changed with fresh 0.1% FBS MEM and cultured for 4 hr. The cells were exposed to TPA (10 ng/ml) or EGF (10 ng/ml) for 30 min, then extracted and analyzed by Western blot analysis by using the PhosphoPlus MAPK antibody kit by New England Biolabs and visualized by chemiluminescence (ECL, Amersham). (A) Phosphorylated Erk proteins detected with phospho-specific MAPK antibody. (B) Erk1 and Erk2 proteins detected with MAPK antibody.
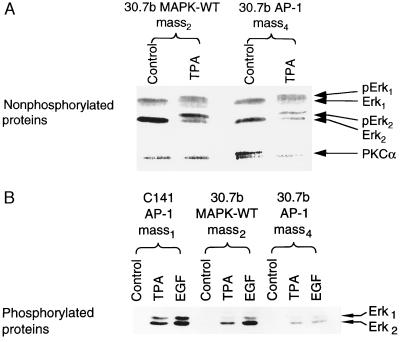
Figure 3.
Overexpression of Erk2 in JB6 P− Cl 30.7b cells. A total of 8 × 104 of Cl 30.7b AP-1 mass4, Cl 30.7b MAPK-WT mass2 or Cl 41 AP-1 mass1 was treated, extracted, and analyzed as described in Fig. 2. (A) Erk1 and Erk2 proteins detected with MAPK antibody. To visualize the conversion of unphosphorylated Erk to phosphorylated Erk in these transfectants, extracts were separated on an 8% polyacrylamide gel, transferred, and blotted as above. Under these conditions the gel mobility shift due to phosphorylation of Erks could be visualized. (B) Phosphorylated Erk proteins detected with phospho-specific MAPK antibody.
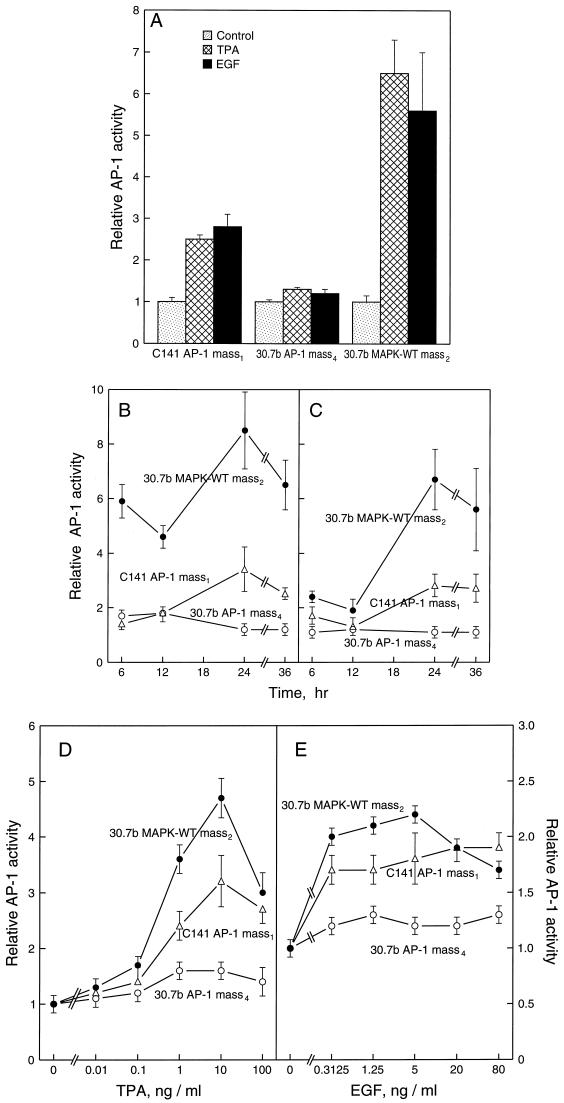
Expression of Wild-Type Erk Restores AP-1 Transactivation Response to P− cells.
To test whether the shortage of Erk1 and Erk2 in P− cells is responsible for the lack of AP-1 activation response to TPA or EGF, we cotransfected AP-1 luciferase reporter plasmid and CMV-neo vector with or without rat wild-type Erk2 into JB6 P− cells Cl 30.7b. After G418 selection, we established stable Cl 30.7b AP-1 mass4 and 30.7b AP-1 MAPK-WT mass2. High levels of introduced Erk2 protein were found in Cl 30.7b MAPK-WT mass2 as compared with these in 30.7b AP-1 mass4 (Fig. 3A). After stimulation with TPA or EGF, the phosphorylated protein of Erk2 in 30.7b MAPK-WT mass2 was much higher than that in 30.7b mass4 (Fig. 3B). However, the activation of Erk1 in 30.7b MAPK-WT mass2 was still lower than that in C1 41 AP-1 mass1 cells (Fig. 3B). To determine whether tumor promoter-induced AP-1 activation response in P− cells was restored by the introduction of wild-type Erk2, we incubated the stable transfectant that expressed the wild-type Erk2 with TPA or EGF. High levels of AP-1 activity were observed in Cl 30.7b MAPK-WT mass2 at all time points and doses studied (Fig. 4). The fold AP-1 induction by TPA or EGF in Cl 30.7b MAPK-WT mass2 was even higher than that in P+ cells (Fig. 4). Restoration of AP-1 transactivation response was also observed in two independent clonal wild-type Erk2 transfectants (see Table 1). In contrast, in Cl 30.7b cells transiently overexpressing PKCα there was no significant gain of AP-1 transactivation. Fold induction by TPA in parental P− cells was 0.9–1.3, whereas fold induction in P−/PKCα transfectants was 1.0- to 1.1-fold. Thus, the rescue by Erk2 of AP-1 response is relatively specific.
Figure 4.
Restoration of AP-1 response to TPA or EGF stimulation in wild-type Erk2-transfected JB6 P− cells. A total of 8 × 103 of Cl 30.7b AP-1 mass4, Cl 30.7b MAPK-WT mass2, or Cl 41 AP-1 mass1 was seeded into each well of 96-well plates. After overnight culture at 37°C, the cells were starved for 24 hr by replacing medium with 0.1% FBS MEM. Then, (A) the cells were treated with TPA (10 ng/ml) or EGF (10 ng/ml) for 24 h, the luciferase activity was measured as described (9, 24). (B and C) For time course study, the cells were exposed to TPA (10 ng/ml) (B) or EGF (10 ng/ml) (C) for the times as indicated. (D and E) For the dose response study, the cells were treated with the indicated doses of TPA or EGF and incubated for 24 hr before assaying luciferase activity. The luciferase activity was measured and the results were presented as relative AP-1 activity.
Table 1.
Clonal wild-type Erk2 transfectants show increased Erk2 and a gain of AP-1 transactivation and transformation responses to TPA and EGF
| Cells | Relative levels of Erk2 expression* | Fold AP-1 induction†
|
Transformation response‡, soft agar colonies/104 cells
|
|||
|---|---|---|---|---|---|---|
| TPA 10 ng/ml | EGF 10 ng/ml | Control | TPA 10 ng/ml | EGF 10 ng/ml | ||
| 30.7b AP-1 mass3 | 1.0 | 1.1 ± 0.1 | 1.1 ± 0.1 | 0 | 75 | 5 |
| 30.7b MAPK-WT C4 | 4.8 | 2.1 ± 0.1 | 2.2 ± 0.2 | 15 | 750 | 905 |
| 30.7b MAPK-WT C5 | 2.8 | 2.6 ± 0.0 | 2.5 ± 0.1 | 10 | 725 | 900 |
Relative levels of Erk2 were determined by Western blot analysis by using Erk-specific antibody as described in Fig. 3. Exposures of the x-ray film were scanned and quantitated on a Molecular Dynamics personal scanner.
AP-1 transactivation assay was as described.
Anchorage-independent tranformation assays were as described in Fig. 5. Two independent assays were done, and mean results are reported.
Introduction of Wild-Type Erk2 Converts P− Cells to P+ Phenotype.
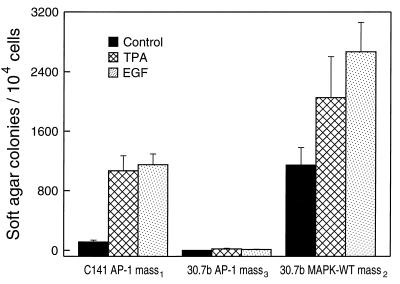
Our previous results demonstrated that induced AP-1 activity is important and required for cell transformation (7). If AP-1 nonresponsiveness is the only factor limiting transformation response in the P− cells, then wild-type Erk expression should render the P− cells promotion-sensitive. Thus, we tested the P−/Erk2 transfectants for tumor promoter-induced transformation response in soft agar. The results indicate that a high frequency of cell transformation was observed in Cl 30.7b MAPK-WT mass2, whereas no significant anchorage-independent transformation was observed in the reporter only Cl 30.7b AP-1 mass4 (Fig. 5). The overexpression of wild-type Erk2 gave rise to a significant frequency of Cl 30.7b cell transformation without added tumor promoter (Fig. 5) and anchorage-independent colonies that were smaller than those induced by TPA or EGF. The transformation frequency induced by TPA or EGF in P−/Erk transfectants was higher than that in P+ reporter only cells (Fig. 5), paralleling the elevated AP-1 transactivation induced by TPA or EGF in these Erk2 transfectants (Fig. 4). Moreover, a gain of transformation response to TPA or EGF occurred in two independent clonal wild-type Erk2 transfectants (see Table 1). This restoration of AP-1 response occurred without transformation by Erk2 alone. In summary, expression of Erk2 in P− cells restores not only AP-1 response but also transformation response.
Figure 5.
Overexpression of wild-type Erk2 converts the P− cell to P+ phenotype. A total of 1 × 104 of C1 41 AP-1 mass1, Cl 30.7b AP-1 mass3 or Cl 30.7b MAPK-WT mass2 was or was not exposed to TPA (10 ng/ml) or EGF (10 ng/ml) in 1 ml of 0.33% BME agar containing 10% FBS laid over 3.5 ml of 0.5% BME agar containing 10% FBS in each well of a 60-mm-diameter dish. The cultures are maintained in a 37°C, 5% CO2 incubator for 14–16 days, and the cell colonies were scored as described (11). The results were presented as soft agar colonies per 104 cells.
DISCUSSION
This report provides a molecular explanation for the defect in tumor promoter-induced AP-1 activation in promotion-resistant JB6 cells. That is, the P− cells show insufficient levels and consequently insufficient amounts of activated forms, of the MAPKs (Erks) needed to activate AP-1. Protein analysis revealed low levels of Erks 1 and 2 in P− Cl 30.7b cells relative to those in the P+ cells. That Erk protein is limiting for AP-1 activation and cell transformation as suggested by the finding that overexpression of Erk2 restores the capacity to induce AP-1 activity and cell transformation in response to tumor promoters TPA and EGF. This rescue of AP-1 transactivation and cell transformation by expression of wild-type Erk2 was verified in multiple clonal Cl 30.7b/Erk2 transfectants. The Erk rescue is relatively specific. Expressing another kinase, PKCα, does not restore AP-1 transactivation. Elucidating the Erk-dependent events required for AP-1 transactivation will be important.
It should be noted that promotion-resistant JB6 cells are not generally unresponsive to tumor promoters. P− cells respond to TPA or EGF with activation of PKC and EGF receptor kinase, respectively (27, 29, 30), with induction of immediate early gene transcription such as that of TPA-inducible sequences (31) and tissue inhibitor of metalloproteinases (32). Thus, many of the molecular apparati needed to signal cellular responses initiated by PKC or EGF receptor kinase activation are intact. The molecular responses impaired in P− cells appear to be limited to those required for tumor promoter-induced transformation.
What limits the Erk protein level in P− cells? Apparently one or more steps downstream or independent of PKC or EGF receptor kinase that indirectly or directly regulate Erk1 and -2 protein levels is deficient. The possibility that Erk was present at a high level but was mutationally inactivated and not recognized by antibody was excluded by sequence analysis of P− Erk in the conserved phosphorylation domain. Studies of the stability of the Erk protein and its mRNA should be informative. It is noteworthy that the phosphorylation of Erk, presumably by MAPK kinase (MEK) (33), appears not to be limiting in P− cells. MEK has been shown to be the only enzyme responsible for activating Erk1 and -2 by phosphorylation on Thr-183 and Tyr-185 (34, 35). In addition to being activated by MEK, phospho-Erk is inactivated by a specific phosphatase, PAC1 (36). It is unlikely that increased levels of PAC1 could contribute to the lack of phospho-Erk in P− cells.
AP-1 is a transcription factor comprised principally of Jun and Fos family heterodimers that binds to a consensus cis element found on the transcriptional promoters of a number of genes whose expression is induced by tumor promoters (20, 37, 38). Evidence for the critical importance of AP-1 activity in transformation by tumor promoters or by oncogenes has been reported (6, 7, 19, 20, 21, 39, 40). Our previous results demonstrated a requirement for AP-1 activation in tumor promoter-induced neoplastic transformation (7, 21, 24). Recent findings with a mouse papilloma keratinocyte model have shown that dominant-negative Jun inhibits both AP-1 activation and progression as measured by matrigel invasion (41). High constitutive levels of AP-1 activity appear to be important for maintenance of tumor phenotype in the JB6 transformed cell line RT101 (24). In the current study, we found that restoring the TPA- or EGF-induced AP-1 activation by introduction of wild-type Erk2 into a P− cell also converts the P− cell to promotion-sensitive phenotype. This result suggests that the promotion-resistant phenotype in this P− cell line is caused by a shortage of Erks and supports the notion that Erk activity is required for tumor promoter-induced cell transformation. This result predicts that blocking Erk activity may be useful as a means of preventing carcinogenesis.
Acknowledgments
We thank Dr. H. H. O. Schmid for critical reading, Dr. Melanie H. Cobb for the generous gift of rat wild-type Erk2, and Ms. Jeanne Ruble for secretarial assistance. This work was supported in part by the Hormel Foundation and National Institutes of Health Grant CA74916.
ABBREVIATIONS
- AP-1
activator protein-1
- EGF
epidermal growth factor
- Erk
extracellular signal-regulated protein kinase
- MAPK
mitogen-activated protein kinase
- P+
tumor promoter-sensitive
- P−
tumor promoter-resistant
- TPA
12-O-tetradecanoyl phorbol-13-acetate
- CMV
cytomegalovirus
- FBS
fetal bovine serum
- MEM
minimal essential medium
- PKC
protein kinase C
- BME
basal medium Eagle
References
- 1.Boutwell R K. Prog Exp Tumor Res. 1964;4:207–250. doi: 10.1159/000385978. [DOI] [PubMed] [Google Scholar]
- 2.Drinkwater N R. In: Genes and Signal Transduction in Multistage Carcinogenesis. Colburn N, editor. New York: Dekker; 1989. pp. 3–17. [Google Scholar]
- 3.Dong Z, Jeffrey A M. Cancer Invest. 1990;8:523–533. doi: 10.3109/07357909009012077. [DOI] [PubMed] [Google Scholar]
- 4.Weinstein I B. Cancer Res. 1988;48:4135–4143. [PubMed] [Google Scholar]
- 5.Colburn N H, Former B F, Nelson K A, Yuspa S H. Nature (London) 1979;281:589–591. doi: 10.1038/281589a0. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein L R, Colburn N H. Science. 1989;244:567–569. doi: 10.1126/science.2541502. [DOI] [PubMed] [Google Scholar]
- 7.Dong Z, Birrer M J, Watts R G, Matrisian L M, Colburn N H. Proc Natl Acad Sci USA. 1994;91:609–613. doi: 10.1073/pnas.91.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simek S L, Kligman D, Patel J, Colburn N H. Proc Natl Acad Sci USA. 1989;86:7410–7414. doi: 10.1073/pnas.86.19.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C S, Ma W-Y, Dong Z. Mol Cell Biol. 1996;16:6427–6435. doi: 10.1128/mcb.16.11.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong Z, Huang C, Ma W-Y. J Biol Chem. 1997;272:9962–9970. doi: 10.1074/jbc.272.15.9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colburn N H, Wendel E J, Abruzzo G. Proc Natl Acad Sci USA. 1981;78:6912–6916. doi: 10.1073/pnas.78.11.6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sobel M E, Dion L D, Vuust J, Colburn N H. Mol Cell Biol. 1983;3:1527–1532. doi: 10.1128/mcb.3.8.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilder P J, Rizzino A. Cancer Res. 1991;51:5898–5902. [PubMed] [Google Scholar]
- 14.Bernstein L R, Ferris D K, Colburn N H, Sobel M E. J Biol Chem. 1994;269:9401–9404. [PubMed] [Google Scholar]
- 15.Watts R G, Ben-Ari E T, Bernstein L R, Birrer M J, Winterstein D, Wendel E, Colburn N H. Mol Carcinog. 1995;13:27–36. doi: 10.1002/mc.2940130106. [DOI] [PubMed] [Google Scholar]
- 16.Lu Y P, Chang R L, Lou Y R, Huang M T, Newmark H L, Reuhl K R, Conney A H. Carcinogenesis. 1994;15:2363–2370. doi: 10.1093/carcin/15.10.2363. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh R, Amstad P, Cerutti P. Mol Cell Biol. 1993;13:6992–6999. doi: 10.1128/mcb.13.11.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su L C, Mukherjee A B, Mukherjee B B. Oncogene. 1995;10:2163–2169. [PubMed] [Google Scholar]
- 19.Ben-Ari E T, Bernstein L R, Colburn N H. Mol Carcinog. 1992;5:62–74. doi: 10.1002/mc.2940050111. [DOI] [PubMed] [Google Scholar]
- 20.Angel P E, Herrlich P A, editors. The FOS and JUN Families of Transcription. Boca Raton, FL: CRC; 1994. [Google Scholar]
- 21.Li J J, Dong Z, Hegamyer G, Dawson M, Colburn N. Cancer Res. 1996;56:483–489. [PubMed] [Google Scholar]
- 22.Frost J A, Geppert T D, Cobb M H, Feramisco J R. Proc Natl Acad Sci USA. 1994;91:3844–3848. doi: 10.1073/pnas.91.9.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robbins D J, Zhen E, Owaki H, Vanderbilt C A, Ebert D, Geppert T D, Cobb M H. J Biol Chem. 1993;268:5097–5106. [PubMed] [Google Scholar]
- 24.Dong Z G, Lavrovsky V, Colburn N H. Carcinogenesis. 1995;16:749–756. doi: 10.1093/carcin/16.4.749. [DOI] [PubMed] [Google Scholar]
- 25.Sturgill T W, Ray L B, Erikson E, Maller J L. Nature (London) 1988;334:715–718. doi: 10.1038/334715a0. [DOI] [PubMed] [Google Scholar]
- 26.Payne D M, Rossomando A J, Martino P, Erickson A K, Her J-H, Shabanowitz J, Hunt D F, Weber M J, Sturgill T W. EMBO J. 1991;10:885–892. doi: 10.1002/j.1460-2075.1991.tb08021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith B M, Colburn N H. J Biol Chem. 1988;263:6424–6431. [PubMed] [Google Scholar]
- 28.Singh N, Aggarwal S. Int J Cancer. 1995;62:107–114. doi: 10.1002/ijc.2910620120. [DOI] [PubMed] [Google Scholar]
- 29.Colburn N H, Wendel E, Srinivas L. J Cell Biochem. 1982;18:261–270. doi: 10.1002/jcb.1982.240180302. [DOI] [PubMed] [Google Scholar]
- 30.Colburn N H, Gindhart T D, Hegamyer G A, Blumberg P M, Delclos K B, Magun B E, Lockyer J. Cancer Res. 1982;18:261–270. [PubMed] [Google Scholar]
- 31.Cmarik J L, Herschman H, Colburn N H. Mol Carcinog. 1994;11:115–124. doi: 10.1002/mc.2940110209. [DOI] [PubMed] [Google Scholar]
- 32.Sun Y, Hegamyer G, Kim H, Sithanandam K, Li H, Watts R, Colburn N H. J Biol Chem. 1995;270:19312–19319. doi: 10.1074/jbc.270.33.19312. [DOI] [PubMed] [Google Scholar]
- 33.Seger R, Ahn N G, Posada J, Munar E S, Jensen A M, Cooper J A, Cobb M H, Krebs E G. J Biol Chem. 1992;267:14373–81. [PubMed] [Google Scholar]
- 34.Lin A, Minden A, Martinetto H, Claret F X, Lange-Carter C, Mercurio F, Johnson G L, Karin M. Science. 1995;268:286–90. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 35.Cobb M H, Goldsmith E J. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 36.Chu Y, Solski P A, Khosravi-Far R, Der C J, Kelly K. J Biol Chem. 1996;271:6497–6501. doi: 10.1074/jbc.271.11.6497. [DOI] [PubMed] [Google Scholar]
- 37.Adler V, Schaffer A, Kim J, Dolan L, Ronai Z. J Biol Chem. 1995;270:26071–26077. doi: 10.1074/jbc.270.44.26071. [DOI] [PubMed] [Google Scholar]
- 38.Dong Z, Colburn N H. In: Early Detection of Cancer: Molecular Markers. Srivastava S, Lippman S M, Hong W K, Mulshine J L, editors. Armonk, NY: Futura Publishing; 1994. pp. 121–128. [Google Scholar]
- 39.Alani R, Brown P, Binetruy B, Dosaker H, Rosenberg R K, Angel P, Karin M, Birrer M J. Mol Cell Biol. 1991;11:6286–6295. doi: 10.1128/mcb.11.12.6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domann F E, Levy J P, Birrer M J, Bowden G T. Cell Growth Differ. 1994;5:9–16. [PubMed] [Google Scholar]
- 41.Dong Z, Crawford H C, Lavrovsky V, Taub D, Watts R, Matrisian L M, Colburn N H. Mol Carcinog. 1997;19:204–212. doi: 10.1002/(sici)1098-2744(199707)19:3<204::aid-mc8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]