Abstract
Hypoxia inducible factor-1 (HIF-1) is the master regulator of metabolic adaptation to hypoxia. It is appreciated that HIF-1α accumulation is achieved under normoxic conditions by e.g., nitric oxide. We determined molecular mechanisms of HIF-1α accumulation under the impact of S-nitrosoglutathione (GSNO). In human embryonic kidney cells GSNO provoked nuclear accumulation of HIF-1α. This appeared unrelated to gene transcription and protein translation, thus pointing to inhibition of HIF-1α degradation. Indeed, GSNO as well as the hypoxia mimic CoCl2 decreased ubiquitination of HIF-1α and GSNO-induced HIF-1α failed to coimmunoprecipitate with pVHL (von Hippel Lindau protein). Considering that HIF-1α-pVHL interactions require prolyl hydroxylation of HIF-1α, we went on to demonstrate inhibition of HIF-1α prolyl hydroxylases (PHDs) by GSNO. In vitro HIF-1α-pVHL interactions revealed that GSNO dose-dependently inhibits PHD activity but not the interaction of a synthetic peptide resembling the hydroxylated oxygen-dependent degradation domain of HIF-1α with pVHL. We conclude that GSNO-attenuated prolyl hydroxylase activity accounts for HIF-1α accumulation under conditions of NO formation during normoxia and that PHD activity is subject to regulation by NO.
INTRODUCTION
The heterodimeric transcription factor hypoxia inducible factor-1 (HIF-1) plays a central role in adaptation to decreased oxygen availability (Semenza, 2002; Wenger, 2002; Brüne and Zhou, 2003). HIF-1 is composed of the two basic helix-loop-helix-Per-Arnt-Sim (bHLH-PAS) proteins HIF-1α and the aryl hydrocarbon receptor nuclear translocator (ARNT), also known as HIF-1β (Wang and Semenza, 1995). In many cell types, the mRNA of both HIF-1α and HIF-1β appear permanently expressed and HIF-1β protein is constitutively present. However, HIF-1α protein is kept at a low or undetectable level under normoxia. In hypoxia, HIF-1α is strikingly induced, translocates to the nucleus, and dimerizes with HIF-1β to form HIF-1, which binds to hypoxia-responsive elements (HRE) in regulatory regions of an impressive array of target genes involved in angiogenesis, erythropoiesis, vasomotor control, and energy metabolism as well as in cell survival decisions (for references, see Maxwell et al., 2001; Wenger, 2002; Zhu et al., 2002). This makes HIF-1 the master regulator of oxygen homeostasis to meet cell and tissue requirements in a situation of oxygen deficiency.
In normoxia HIF-1α is bound to the von Hippel Lindau protein (pVHL) (Maxwell et al., 1999; Hon et al., 2002; Min et al., 2002), which is the substrate recognizing component of an E3 ubiquitin ligase complex (Cockman et al., 2000; Ohh et al., 2000; Tanimoto et al., 2000). Consequently, HIF-1α is polyubiquitinated and degraded by the 26S proteasome system, thus accounting for its low normoxic level of expression (Salceda and Caro, 1997; Huang et al., 1998; Kallio et al., 1999). The requirement of pVHL for HIF-1α degradation is underscored in cells that do not contain a functional pVHL, which leads to high levels of HIF-1α in normoxia (Maxwell et al., 1999; Krieg et al., 2000; Clifford et al., 2001). However, recently, a pVHL- and oxygen-independent HIF-1α degradation pathway has been reported (Isaacs et al., 2002).
In normoxia HIF-1α is hydroxylated at Pro564 and Pro402, which is required for pVHL binding (Ivan et al., 2001; Jaakkola et al., 2001; Masson et al., 2001; Yu et al., 2001). Prolyl hydroxylation is performed by enzymes sharing homology with the EGL-9 protein from Caenorhabditis elegans (Epstein et al., 2001). The human enzymes have been termed prolyl hydroxylase domain containing protein 1, 2, and 3 (PHD1, PHD2, and PHD3; Epstein et al., 2001) or HIF1α prolyl hydroxylase 1, 2, and 3 (Bruick and McKnight, 2001). The existence of a fourth PHD has been described (Oehme etal., 2002). In addition, in normoxia a protein known as factor inhibiting HIF (FIH-1) has been identified (Mahon et al., 2001). The latter enzyme hydroxylates Asn803 of HIF-1α, which attenuates the C-terminal transactivation domain (C-TAD) of the transcription factor by abrogating the recruitment of transcriptional coactivators such as CBP/p300 (Hewitson et al., 2002; Lando et al., 2002a). Elegant work implies that stabilization and transactivation of HIF-1α are two separate processes regulated by hydroxylation at distinct residues (Kaelin, 2002; Semenza, 2002). Because PHDs and FIH-1 hydroxylate HIF-1α in an oxygen-dependent manner, they function as oxygen sensors in vivo. It is now appreciated that hypoxia, transition metals such as CoCl2, and the iron chelator desferroxamine (DFX) directly inhibit PHDs with concomitant HIF-1α stabilization.
Recent evidence suggests that HIF-1α can be accumulated and activated by certain growth factors, cytokines (Hellwig-Bürgel et al., 1999), and hormones during normoxia. In addition, a modulatory role of nitric oxide (NO) emerged (Kimura et al., 2000; Palmer et al., 2000; Sandau et al., 2000, 2001a). Regulation of HIF-1 activity by NO is likely to be of (patho)-physiological relevance but molecular mechanisms have not been defined yet. Initial observations suggested that NO inhibits hypoxia-induced HIF-1α stabilization and HIF-1 transcriptional activation (Liu et al., 1998; Sogawa et al., 1998; Huang et al., 1999). More recent studies indicated that chemically diverse NO donors or enhanced endogenous NO formation by inducible NO-synthase or NO formation in a coculture system under normoxic conditions provoked HIF-1α stabilization, HIF-1 DNA-binding, and activation of downstream target gene expression (Kimura et al., 2001; Sandau et al., 2001a, 2001b; Zhou et al., 2003). Studies performed in several cell systems such as tubular LLC-PK1, human glioblastoma, human hepatoma, or bovine pulmonary artery endothelial cells imply that this is neither species specific nor restricted to certain cell types. Guanylyl cyclase antagonists and lipophilic cGMP analogues did not attenuate/mimic HIF-1α accumulation and thus excluded a role of the soluble guanylyl cyclase-cGMP pathway (Kimura et al., 2000; Palmer et al., 2000; Sandau et al., 2001a), thus leaving molecular mechanisms of NO action unresolved.
Herein, we demonstrate the inhibitory effect of NO on HIF-1α ubiquitination and interaction with pVHL. We present evidence that PHDs are targeted by NO, which suggests attenuation of prolyl hydroxylation as the underlying mechanism of NO-induced HIF-1α accumulation in normoxia. Importantly, we demonstrate regulation of HIF-1α prolyl hydroxylase activity as a mechanism of controlling the stability of HIF-1α.
MATERIALS AND METHODS
Materials
S-nitrosoglutathione (GSNO) was synthesized as described (Hart, 1997). Commercially available chemicals were of the highest grade of purity. Specifically, medium, fetal calf serum (FCS) and supplements were purchased from PAA (Linz, Austria). The membrane-permeable proteasome inhibitor MG132 (Lee and Goldberg, 1998), actinomycin D, cycloheximide (CHX), DAPI, and FITC-labeled anti-mouse secondary antibodies were ordered from Sigma (Taufkirchen, Germany). Protein assay kits were bought from Bio-Rad (Richmond, CA). Luciferase assay systems were delivered by Promega (Heidelberg, Germany). Ni-NTA-agarose was from Qiagen (Hilden, Germany). The anti-HA mAb came from BabCO (Richmond, CA). Anti-mouse-dynal beads were purchased from Dynal (Mannheim, Germany). Protease inhibitor cocktails came from Roche (Mannheim, Germany). Nitrocellulose membrane, ECL detection system and horseradish peroxidase (HRP)-labeled anti-mouse secondary antibodies were delivered by Amersham Life Science (Freiburg, Germany). Multitest slides and cover slips were ordered from ICN Biomedicals (Costa Mesa, CA) and FluorSave mounting medium was bought from Calbiochem (Bad Soden, Germany). The CoolSNAP CCD camera was from Roper Scientific (Tucson, AZ), whereas the MetaMorph software package came from Universal Imaging (West Chester, PA). Peqlab delivered peqGOLD RNAPure kit and primers were ordered from MWG-Biotech (Ebersberg, Germany). HIF-1α antibodies, Advantage RT-for-PCR kit, AdvanTaq PCR kit were purchased from Becton Dickinson (Heidelberg, Germany). The plasmid pGLEPOHRE, provided by Dr. T. Kietzmann (University of Göttingen, Göttingen, Germany), harbors three erythropoietin hypoxia-responsive elements (HRE) in front of the SV40 promoter and was described previously (Sandau et al., 2001b). The pCMV-HIS6-ubiquitin plasmid was a gift from Dr. D. Bohmann (European Molecular Biology Laboratory, Heidelberg, Germany). Plasmids pcDNA3-HIF-1α, pCR3.1-HA-VHL, and pGalluc (O'Rourke et al., 1999) were kindly provided by Dr. P.J. Ratcliffe (Wellcome Trust Centre for Human Genetics, University of Oxford, UK). The pGal-HIF-1α727–826 plasmid was generously provided by Dr. D. Lando and Dr. M.L. Whitelaw (Lando et al., 2002b).
Cell Culture
Human embryonic kidney (HEK293) cells were cultured in DMEM with 4.5 g/l d-glucose. Medium was supplemented with 10% FCS, 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were transferred two times a week, and medium was changed before experiments. Cells were kept in a humidified atmosphere of 5% CO2 in air at 37°C.
Indirect Immunofluorescence and Fluorescent Microscopy
HEK293 cells grown on multitest slides were stimulated with 100 μM CoCl2 or 1 mM GSNO for 2 h. Cells were fixed with 4% paraformaldehyde for 5 min and permeabilized with 4% paraformaldehyde/0.2% Triton X-100 for 5 min at room temperature. To block nonspecific antibody binding, slides were incubated for 30 min at room temperature with 5% milk/PBS. Slides were successively incubated with the HIF-1α antibody (1:100 in 1% milk/PBS) at 4°C overnight and FITC-labeled secondary antibodies (1:100 in 1% milk/PBS) at 37°C for 1 h. Finally, 0.2 μg/ml DAPI/PBS was added at room temperature for 2 min. Slides were washed three times for 5 min each with PBS and briefly rinsed with distilled water. Coverslips were mounted to the multitest slides with intermediate FluorSave mounting medium. Slides were examined by an Axioskop fluorescent microscope (Zeiss, Oberkochen, Germany). Photographs were taken with a CoolSNAP CCD camera and images were created by the MetaMorph software package (Universal Imaging).
Cell Transfection
HEK293 cells, 2 × 106, were plated in 10-cm dishes 1 d before transfection. At a rate of 60% confluence, cells were transfected with plasmids, using the calcium phosphate precipitation method (Sambrook et al., 1989). Briefly, plasmids in the presence of 125 mM CaCl2 and HBS buffer (25 mM HEPES, 140 mM NaCl, 0.75 mM Na2HPO4, pH 7.05) were incubated for 30 min at room temperature and added drop-wise to cells. Sixteen hours later medium was changed and incubations were continued for another 8-h period before cell stimulation.
Reporter Assay
HEK293 cells, 2 × 105, were transfected with 1 μg pGLEPOHRE or cotransfected with 0.5 μg pGal-HIF-1α727–826 and 1 μg pGal-Luc by the calcium phosphate precipitation method. Twenty-four hours after transfection the medium was replaced with medium containing 1 mM GSNO, 100 μM CoCl2, or cells were exposed to 1% hypoxia for an additional 16 h. In all experiments cells were lysed according and luciferase activity was measured using a commercial kit. All data were normalized to controls.
Western Blot Analysis
HIF-1α was quantified by Western analysis. Briefly, cells were stimulated by agonists for indicated times. In case of actinomycin D, cells were preincubated for 30 min. Cells were then scraped off, lysed in 300 μl lysis buffer B, and sonicated, followed by centrifugation (15000 × g, 15 min). Eighty micrograms protein was added to the same volume of 2× SDS-PAGE sample buffer and boiled for 5 min. Proteins were resolved on 7.5% SDS-polyacrylamide gels. Gels were washed with blotting buffer (25 mM Tris, 192 mM glycine, 20% methanol, pH 8.3) for 5 min, proteins were blotted onto nitrocellulose membranes by a semidry transfer, and unspecific binding sites were blocked with 5% milk/TTBS (50 mM Tris/HCl, 140 mM NaCl, 0.05% Tween-20, pH 7.2) for 1 h. The HIF-1α-antibody (1:1000 in 1% milk/TTBS) was added and incubated overnight at 4°C. Afterward, nitrocellulose membranes were washed three times for 5 min each with TTBS. For protein detection, blots were incubated with a HRP-labeled goat anti-mouse secondary antibody (1:2000 in 1% milk/TTBS) for 1 h and washed three times for 5 min each with TTBS, followed by ECL detection.
Semiquantitative RT-PCR
HEK293 cells, 2 × 106, were plated 1 d before experiments. The following day medium was changed, and cells were stimulated with 100 μM CoCl2 or 1 mM GSNO for 2 h. Total RNA was isolated using the peqGOLD RNAPure kit. One microgram total RNA was applied to complete the reverse transcription with an Advantage RT-for-PCR kit using hexamer random primers. PCR was performed with an AdvanTaq PCR kit. The following primer pairs were selected: HIF-1α, 5′-CTCAAAGTCGGACAGCCTCA-3′, 5′-CCCTGCAGTAGGTTTCTGCT-3′; actin, 5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′, 5′-CTAGAAGCATTTGCGGTCGACGATGGAGGG-3′. Amplification program: 95°C, 30 s; 56°C, 30 s; 72°C, 1 min; 20 cycles; 72°C, 10 min. RT-PCR products were separated on 2% agarose gels and visualized with ethidium bromide.
HIF-1α Ubiquitination Assay
HEK293 cells were cotransfected with 0.3 μg of a plasmid encoding full-length HIF-1α (pcDNA3-HIF-1α) and 3 μg pCMV-HIS6-ubiquitin plasmid encoding HIS-tagged ubiquitin. After stimulation with 100 μM CoCl2 or 1 mM GSNO for the indicated time, cells were scraped off and centrifuged. Lysis buffer A, 300 μl (50 mM Tris, 150 mM NaCl, 8 M urea, pH 7.5), was added to each pellet and immediately vortexed three times for 15 s. After centrifugation at 15,000 × g for 30 min lysates were transferred to fresh tubes. Protein of the supernatant, 500 μg, was mixed with 100 μl Ni-NTA-agarose (1:1 resuspended in lysis buffer A) and incubated, while rolling, at room temperature for 1 h. Afterward, beads were pelleted by centrifuging at 1000 × g for 5 min, washed three times with 200 μl lysis buffer A, resuspended in 50 μl 2× sample buffer (125 mM Tris/HCl, 2% SDS, 10% glycerin, 1 mM dithiothreitol (DTT), 0.002% bromophenol blue, pH 6.9) and heated at 95°C for 10 min. Beads were removed by centrifugation. Proteins were electrophoretically separated on 7.5% SDS-gels, followed by Western analysis using HIF-1α antibodies.
HIF-1α-pVHL Coimmunoprecipitation
HEK293 cells were cotransfected with 1 μg pcDNA3-HIF-1α and 1 μg pCR3.1-HA-VHL encoding HA-tagged full-length pVHL. Cells were subjected to 1% hypoxia or stimulated with 100 μM CoCl2 or 1 mM GSNO for 4 h and subsequently exposed to 10 μM MG132 for 1 h. Cells were scraped off the dishes and collected. To each cell pellet 300 μl lysis buffer B (50 mM Tris, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1 mM PMSF, protease inhibitor cocktail, pH 7.5) was added, followed by immediate vortexing (three times, 15 s). After centrifugation (15000 × g for 30 min) supernatants were transferred to fresh tubes. Any supernatant (1 mg protein) was supplied with 1 μg anti-HA antibody and incubated at 4°C for 1 h. Thereafter, 20 μl anti-mouse-dynal beads were added, and incubations were continued at 4°C overnight. Beads were collected, washed three times with 100 μl lysis buffer B, finally supplemented with 50 μl 2× sample buffer, and boiled at 95°C for 10 min. Beads were removed by centrifugation, and supernatants were loaded on 7.5% SDS-gels. Western analysis was performed using HIF-1α antibodies.
In Vitro Protein Interaction Assay
The impact of GSNO on recombinant HIF-1α prolyl hydroxylases was investigated by in vitro protein interaction assays. The assay makes use of the interaction between the C-terminal ODD and 35S-pVHL, which requires the presence of hydroxyproline in the ODD. To test for hydroxylase independent effects of GSNO during protein interactions, we incubated a synthetic peptide resembling the biotinylated hydroxyproline ODD (Biosyntan, Berlin, Germany) and 35S-pVHL expressed in a T7 coupled rabbit reticulocyte lysate in vitro transcription/translation system (Promega) in the presence and absence of GSNO. Streptavidine-agarose was added and incubations went on for 16 h on an end-over-end rotator at 4°C. Unbound 35S-pVHL was removed by washing, beads were boiled in SDS-PAGE loading buffer, and the supernatant was subjected to 15% SDS-PAGE. pVHL was detected by autoradiography. The VHL gene contains an internal in-frame methionine at codon 54, thus two pVHL isoforms are expressed, which are composed of 213 and 160 amino acid residues (for references, see Ivan and Kaelin, 2001). As both isoforms behave similarly they are found in all our interaction assays. The impact of GSNO on PHD activity was tested in a GalDBD-HIF-1α549–582-pVHL in vitro interaction assay, performed as described previously (Jaakkola et al., 2001). Briefly, we produced GalDBD-HIF-1α549–582, PHD, and 35S-labeled pVHL in rabbit reticulocyte or wheat germ lysate (Promega). GalDBD-HIF-1α549–582 was purified with GalDBD-antibodies conjugated to agarose beads (Santa Cruz Biotechnology, Heidelberg, Germany) and suspended in a reaction buffer (20 mM Tris, 5 mM KCl, 1.5 mM MgCl2, pH 7.5) supplemented with 1 mM ascorbate, 1 mM α-ketoglutarate, and 20 μM FeCl2. GalDBD-HIF-1α549–582 was then incubated for 30 min with GSNO before recombinant PHD expressed in reticulocyte lysate or wheat germ extract was added. In all experiments unprogrammed reticulocyte or wheat germ lysate served as negative controls. The reaction was terminated after 30 min by addition of desferroxamine (final concentration, 100 μM). The beads were washed several times before 35S-pVHL was added. The binding reaction and detection of 35S-pVHL were done as described above. The effect of GSNO on PHD function was also tested in a hypoxia workstation (In Vivo2 400, Ruskinn Technologies, Leeds, UK). To this end GalDBD-HIF-1α549–582 was diluted in reaction buffer, GSNO was added in a 1% oxygen atmosphere. The solution was equilibrated in hypoxia for 30 min. Recombinant PHD expressed in rabbit reticulocyte lysate was equilibrated separately for 30 min. PHD was then added to the substrate solution. After 30 min the reaction was stopped in the hypoxic atmosphere by addition of DFX. pVHL binding and detection were done in normoxia as described above.
RESULTS
GSNO Provoked Accumulation, Nuclear Translocation, and Transactivation of HIF-1α
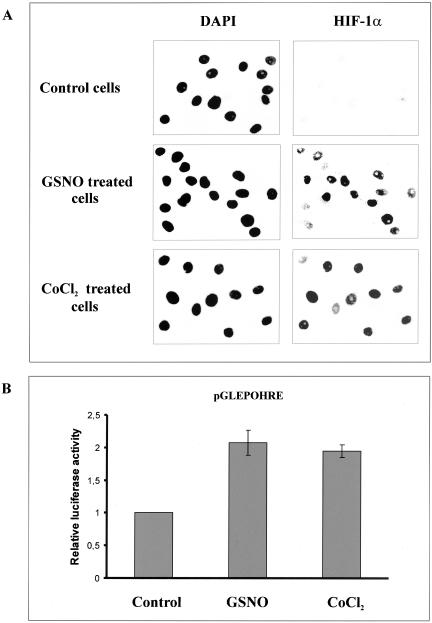
There is considerable evidence that NO or NO-derived species induce HIF-1α under normoxia. These results have been obtained in several cellular systems, among others in human embryonic kidney (HEK293) cells. Therefore, we used this cell line to examine mechanisms of NO-evoked HIF-1α accumulation. In a first set of experiments we incubated HEK293 cells with 1 mM GSNO or the hypoxia mimic CoCl2 (100 μM) for 2 h to analyze induction and subcellular localization of HIF-1α (Figure 1A). Immunofluorescence analysis highlighted HIF-1α immunoreactivity predominantly in the nucleus based on DAPI counterstaining. As expected, HIF-1α was absent, i.e., below the detection limit in untreated control cells.
Figure 1.
GSNO-evoked nuclear accumulation and transactivation of HIF-1α. (A) HEK293 cells were stimulated for 2 h with 1 mM GSNO or 100 μM CoCl2 or remained untreated. Nuclei were visualized by DAPI staining. HIF-1α was detected by an anti-HIF-1α mAb and FITC-labeled anti-mouse secondary antibody. Each experiment was performed at least three times and representative data are shown. (B) HEK293 cells, 2 × 105, were transfected with the pGLEPOHRE plasmid and stimulated for 16 h with 1 mM GSNO or 100 μM CoCl2, or left untreated (control). After cell lysis luciferase activity was measured and normalized compared with controls. Data are the mean ± SD (n = 3).
In line with previous reports obtained in A-172 cells (Kimura et al., 2000) or HepG2 cells (Sandau et al., 2001b), we demonstrated induction of a hypoxia-inducible luciferase reporter gene construct by GSNO and CoCl2 in HEK293 cells (Figure 1B), which suggests that HIF-1 induced by GSNO is transcriptionally active.
GSNO Induced HIF-1α Protein in a Dose- and Time-dependent Manner
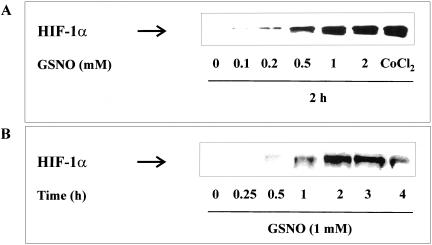
First experiments determined the dose- and time-dependent impact of GSNO on HIF-1α accumulation in HEK293 cells. GSNO at concentrations ranging from 0.1 mM to 2 mM were used to stimulate cells for 2 h. Maximal effects were achieved with 1 mM of the NO donor (Figure 2A). Interestingly, GSNO was equipotent compared with the hypoxia mimic CoCl2.
Figure 2.
Concentration- and time-dependent HIF-1α accumulation. (A) HEK293 cells were stimulated with GSNO (0.1–2 mM) for 2 h or (B) with 1 mM GSNO for 15 min up to 4 h. Western analysis with an anti-HIF-1α mAb was performed as described in MATERIALS AND METHODS. Each experiment was performed at least three times and representative data are shown.
The time response toward 1 mM GSNO revealed a maximal HIF-1α accumulation at 2 h, with a declining response afterward (Figure 2B).
HIF-1α mRNA Transcription and Protein Translation Is Not Affected by GSNO
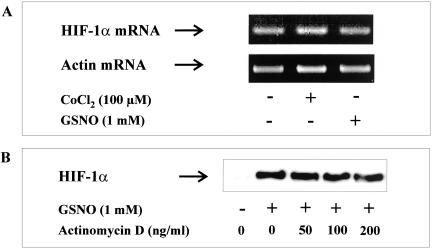
We started to determine alteration of HIF-1α mRNA levels in response to GSNO-treatment. After the addition of 1 mM GSNO for 2 h, total RNA was isolated and HIF-1α mRNA was quantified by RT-PCR using actin as an internal control (Figure 3A). Visualization and quantification of RT-PCR products revealed no significant changes, which indicated that the accumulation of HIF-1α protein is not caused by enhanced transcription of HIF-1α. Application of 100 μM CoCl2 for 2 h had no effect on HIF-1α gene transcription either.
Figure 3.
HIF-1α mRNA transcription under the impact of CoCl2 or GSNO. (A) HEK293 cells were stimulated with 1 mM GSNO or 100 μM CoCl2 for 2 h. HIF-1α mRNA was quantified by reverse transcription and PCR (RT-PCR). DNA products were visualized with ethidium bromide on 2% agarose gels. (B) Cells were preincubated for 30 min with the indicated concentrations of actinomycin D followed by incubations with 1 mM GSNO for 4 h. Western analysis with an anti–HIF-1α mAb was used to detect HIF-1α protein. Each experiment was performed at least three times and representative data are shown.
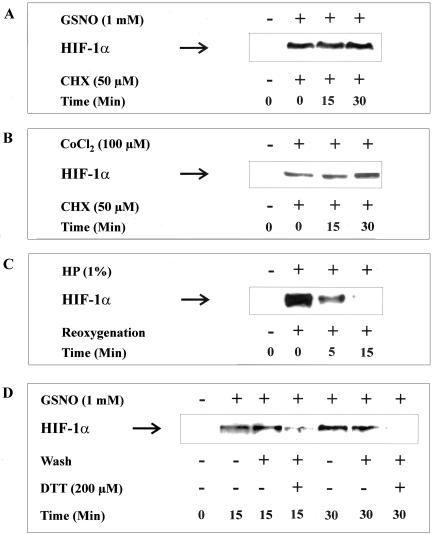
We then used the general inhibitor of mRNA synthesis, actinomycin D, to check if a transcriptional process is required for HIF-1α accumulation (Figure 3B). HEK cells were preincubated with actinomycin D for 30 min, followed by stimulation with GSNO for 4 h. Actinomycin D had no effect on GSNO-evoked HIF-1α accumulation at concentrations ranging from 50 to 200 ng/ml. Ruling out transcriptional regulation, we went on to determine HIF-1α translation under the influence of GSNO and performed a set of experiments using cycloheximide (CHX) to block protein synthesis (Figure 4A). HIF-1α accumulation was achieved with 1 mM GSNO for 2 h followed by the addition of CHX for 15–30 min. HIF-1α protein levels remained constant >30 min although protein synthesis was blocked. The same observation was made in cells exposed to 100 μM CoCl2 for 2 h and subsequently treated with CHX (Figure 4B). To identify the onset of HIF-1α degradation after hypoxic accumulation, we performed hypoxia/reoxygenation studies (Figure 4C). Incubations of HEK293 cells in a 1% oxygen atmosphere for 2 h evoked massive HIF-1α accumulation as determined by Western analysis. Reoxygenation for 5 min reduced HIF-1α significantly, whereas at 15 min of reoxygenation HIF-1α had disappeared completely.
Figure 4.
HIF-1α protein translation under the impact of CoCl2, hypoxia, or GSNO. HEK293 cells were stimulated with 1 mM GSNO (A and D) or 100 μM CoCl2 (B) or subjected to 1% hypoxia (C). Specifically, after stimulation with/without 1 mM GSNO for 2 h (A) or 100 μM CoCl2 for 2 h (B), 50 μM cycloheximide (CHX) was added for 15 and 30 min; after a 2-h period of hypoxia (1% oxygen) reoxygenation was done for 5 and 15 min (C); after a 1-h stimulation with 1 mM GSNO, medium was changed and DTT was added as indicated followed by a 15- or 30-min lasting incubation (D). Western analysis with an anti-HIF-1α mAb was used to detect HIF-1α protein. Each experiment was performed at least three times and representative data are shown.
A similar approach was followed with GSNO as an agonist (Figure 4D). HEK293 cells were stimulated with 1 mM GSNO for 1 h. Thereafter, medium was changed (wash), and some samples additionally received 200 μM of the reducing agent DTT. Incubations continued for 15 or 30 min before Western analysis of HIF-1α. Whereas simply changing the medium only partially reversed HIF-1α accumulation, the addition of DTT most effectively decreased the appearance of HIF-1α within 15–30 min. This is in some analogy to the recent observation that DTT attenuates (Z)-1–1[2-(aminoethyl-amino]diazen-1-ium-1,2-diolate (NO donor known as NOC-18) evoked HIF-1 DNA-binding (Palmer et al., 2000). The requirement of DTT can be rationalized based on the assumption that intracellular S-nitrosothiols that are formed in response to exogenously supplied GSNO are not removed by washing cells but are subjected to degradation in the presence of the DTT. Taken together these results demonstrate that the half-life of HIF-1α in a hypoxia/reoxygenation or GSNO/wash plus DTT neutralization setting is substantially shorter than the half-life of HIF-1α induced by GSNO or CoCl2. Taking into consideration that CoCl2 is reported to block HIF-1α degradation these results suggest that GSNO may lead to accumulation of HIF-1α by attenuating its degradation.
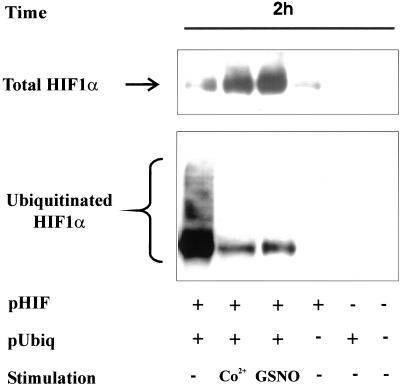
GSNO Attenuated Ubiquitination of HIF-1α
Considering that ubiquitination of HIF-1α is a prerequisite for its degradation, we determined the impact of GSNO on this critical step leading to proteasomal proteolysis and compared the action of NO to the hypoxia mimicking reagent CoCl2. To follow HIF-1α ubiquitination, we cotransfected HEK293 cells with 0.3 μg pcDNA3-HIF-1α and 3 μg pCMV-HIS6-ubiquitin expression plasmids. After the addition of 100 μM CoCl2 or 1 mM GSNO we noticed marked HIF-1α protein accumulation at 2 h as determined by Western analysis (Figure 5). As shown by Ni-NTA-agarose affinity purification of ubiquitinated HIF-1α and subsequent Western blot analysis, GSNO as well as CoCl2 dramatically down-regulated HIF-1α ubiquitination compared with controls.
Figure 5.
HIF-1α accumulation and ubiquitination after GSNO treatment. HEK293 cells were cotransfected with 3 μg pCMV-HIS6-ubiquitin and 0.3 μg pcDNA3-HIF-1α expression plasmids for 16 h. As controls, cells were transfected with either of the two plasmids or received vehicle only. Medium was changed, and incubations continued for another 8-h period before stimulation with 100 μM CoCl2 or 1 mM GSNO for 2 h. HIF-1α and ubiquitinated HIF-1α were determined by Western analysis as described in MATERIALS AND METHODS. Each experiment was performed at least three times and representative data are shown.
Evidently, accumulation of HIF-1α and decreased ubiquitination of the protein are in line with our hypothesis that HIF-1α accumulation results from impaired degradation. These observations are in close analogy to the well-established action of the hypoxia mimic CoCl2 used for comparison in our study. Of note, expression of HIS-tagged proteins, i.e., HIS6-ubiquitin, did not cause HIF-1α accumulation, which precludes unspecific chelation of Fe2+ by the HIS-tag. Considering that ubiquitination of HIF-1α requires association with pVHL, we looked into the HIF-1α-pVHL interaction.
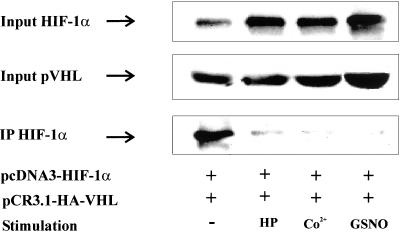
GSNO Attenuated the HIF-1α-pVHL Interaction
To analyze the HIF-1α-pVHL interaction in vivo, we used a coimmunoprecipitation approach and therefore transfected HEK293 cells with 1 μg pCR3.1-HA-VHL and 3 μg pcDNA3-HIF-1α expression plasmids. Twenty-four hours posttransfection, HEK293 cells were subjected to 1% hypoxia and stimulated with 100 μM CoCl2 or 1 mM GSNO for 4 h, followed by the addition of 10 μM MG132 for 1 h. These amounts of MG132 caused some accumulation of HIF-1α in otherwise untreated controls but did not impair further accumulation of HIF-1α under hypoxia, CoCl2, or GSNO compared with controls. The HIF-1α-pVHL interaction was assessed by coimmunoprecipitation (Figure 6). Accumulation of HIF-1α in the cell lysate that was used for immunoprecipitation was followed by Western analysis (input) and revealed increasing amounts of HIF-1α after hypoxia or CoCl2 or GSNO treatment.
Figure 6.
In vivo interaction between HIF-1α and pVHL. HEK293 cells were cotransfected with 1 μg pCR3.1-HA-VHL and 3 μg pcDNA3-HIF-1α expression plasmids. After changing medium at 16 h, incubations continued for 8 h before stimulation with 1% hypoxia (HP), 100 μM CoCl2, or 1 mM GSNO for 4 h, followed by MG132 for 1 h as described in MATERIALS AND METHODS. Coimmunoprecipitation was performed with an anti-HA mAb, which recognized heme agglutinin-tagged pVHL and anti-mouse antibody-coated magnetic beads. Expression of pVHL (input) and accumulation of HIF-1α in cell lysates (input) as well as immunoprecipitates (IP) was determined by Western analysis using an anti–HIF-1α mAb as described in MATERIALS AND METHODS. Each experiment was performed at least three times and representative data are shown.
When pVHL was immunoprecipitated followed by HIF-1α Western analysis (IP), we noticed a significant pVHL-HIF-1α interaction in normoxic controls. In response to GSNO, hypoxia, or CoCl2, however, we observed a dramatically decreased amount of HIF-1α that coprecipitated with pVHL despite high levels of HIF-1α. As a further control we assured that the amount of pVHL expression between samples was equivalent (pVHL input). These studies suggest that accumulation of HIF-1α in response to GSNO is associated with an impaired HIF-1α-pVHL interaction. This effect may be the result of a chemical modification of the ODD and/or pVHL by GSNO or result from attenuated prolyl hydroxylation of the ODD, which is required for pVHL binding.
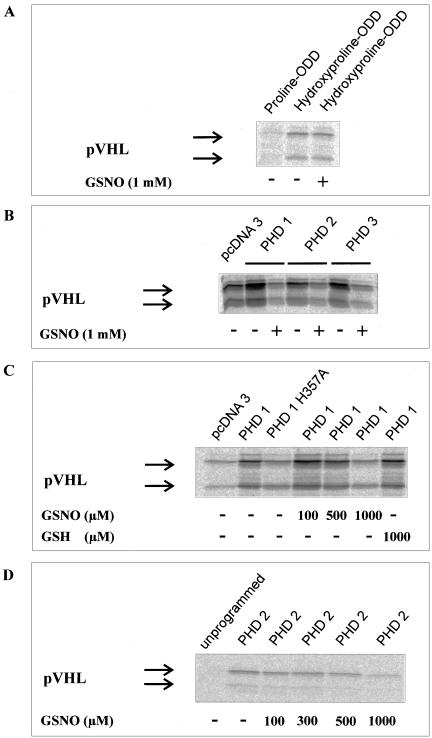
In Vitro HIF-1α-pVHL Interactions under the Impact of GSNO
It is established that NO or NO-derived species may cause posttranslational protein modifications (Stamler et al., 1992; Brüne et al., 1996). Therefore, it seemed conceivable that a chemical reaction involving NO impinges on the interaction of the hydroxylated ODD with pVHL. We tested this hypothesis by incubating a synthetic 19mer hydroxyproline564-ODD with pVHL. GSNO (1 mM) had no effect on this interaction when assaying for a direct interaction between the hydroxylated-ODD and pVHL (Figure 7A). This makes a chemical modification of HIF-1α-ODD and/or pVHL unlikely, which would abrogate binding of HIF-1α-ODD to pVHL.
Figure 7.
In vitro interaction between HIF-1α-ODD and pVHL. To test direct chemical effects of GSNO on the interaction of HIF-1α with pVHL, a synthetic biotinylated 19mer peptide resembling hydroxyproline564-HIF-1α556–574 was incubated with 35S-labeled pVHL in the absence (lane 2) or presence (lane 3) of 1 mM GSNO. Wild-type proline564-HIF-1α556–574 served as negative control (A). GalDBD-HIF-1α549–582 was incubated with reticulocyte lysate programmed with empty vector, PHD1, PHD2, or PHD3 in the absence or presence of 1 mM GSNO before the addition of 35S-pVHL (B). Detection of pVHL after precipitation with a Gal antibody indicates hydroxylation of the ODD and thus activity of hydroxylating enzymes. GalDBD-HIF-1α549–582 was incubated with PHD1, an inactive PHD 1 mutant (PHD1 H357A), or PHD1 in the presence of increasing concentrations of GSNO. Reduced glutathione served as a negative control (C). GalDBD-HIF-1α549–582 and PHD2 were expressed in wheat germ lysate to exclude heme from the assay. The effect of increasing GSNO concentrations was tested as detailed above (D).
In a further step we analyzed the impact of GSNO on the activity of the human enzymes known to hydroxylate HIF-1α. To this end we incubated GalDBD-HIF-1α549–582 with 1 mM GSNO for 30 min and then added recombinant PHD1, PHD2, or PHD3 to the reaction mixture. The activity of all enzymes was strongly inhibited by GSNO under aerobic conditions when compared with untreated controls, based on the largely attenuated pVHL-GalDBD-HIF-1α549–582 interaction (Figure 7B).
Interestingly, the activity of PHDs in GSNO-treated samples was below the activity noticed in unprogrammed reticulocyte lysate, which has been reported to contain low levels of PHD2 (Ivan et al., 2002). The inhibitory effect of GSNO on PHD1 was shown to be dose dependent (Figure 7C). GSNO (1 mM) reduced the activity to a level comparable to an inactive PHD1 mutant (Epstein et al., 2001). Importantly, reduced glutathione had no effect on PHD1 activity, confirming that the NO moiety of the NO donor accounts for GSNO action. The dose response experiment was repeated using PHD2 and GalDBD-HIF-1α549–582 expressed in wheat germ lysate (Figure 7D) to test whether the NO-scavenging effect of reticulocyte heme impinges on the experimental outcome, assuming that heme will lower the steady state concentration of NO during incubations. Although not directly comparable because of the use of PHD1 vs. PHD2, the dose response was shifted to lower doses in the wheat germ assay, which may suggest that heme interferes with the NO effect in the reticulocyte lysate.
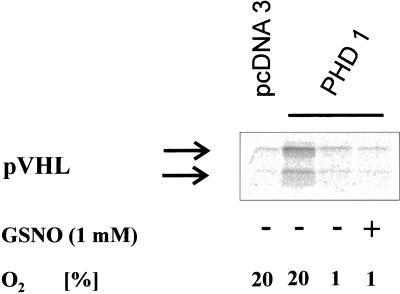
Finally, we tested the effect of GSNO on PHD activity in a hypoxic atmosphere. Our interest in these experiments was sparked by a report (Wang et al., 2002) postulating activation of HIF-1α prolyl hydroxylase activity in cytoplasmic extracts prepared from hypoxic cells.
However, we were unable to demonstrate activation of recombinant PHD1 (Figure 8), PHD2, or PHD3 (our unpublished results) by 1 mM GSNO in an atmosphere of 1% O2.
Figure 8.
In vitro effect of GSNO on PHD1 in hypoxia. GalDBD-HIF-1α549–582 was incubated with PHD1 in normoxia (lane 2) or in hypoxia in the absence (lane 3) or presence (lane 4) of 1 mM GSNO. The reaction was stopped by addition of 200 μM DFX. GalDBD-HIF-1α549–582 was precipitated with a Gal antibody, and 35S-pVHL was added. Detection of pVHL indicates enzymatic hydroxylation of the HIF-1α-ODD.
GSNO Stimulated the C-terminal HIF-1α Transactivation
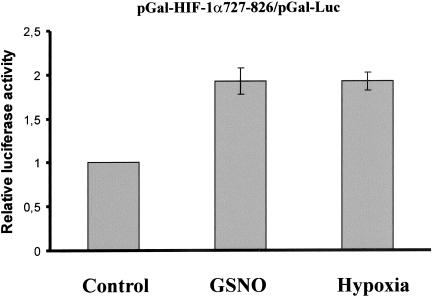
FIH-1 is known to hydroxylate asparagine803 of HIF-1α and thus to attenuate HIF-1 transactivation. Considering that FIH-1 and PHDs share common regulatory features, we were interested to study the impact of GSNO on FIH-1, expecting its inhibition. For these experiments HEK293 cells were cotransfected with pGal-HIF-1α727–826 and pGal-Luc. Luciferase activity was determined under the impact of 1 mM GSNO and 1% hypoxia, compared with controls (Figure 9).
Figure 9.
C-terminal HIF-1α transactivation after GSNO-treatment. HEK293 cells, 2 × 105, were cotransfected with 0.5 μg pGal-HIF-1α727–826 and 1 μg pGal-Luc plasmids and stimulated for 16 h with 1 mM GSNO, exposed to 1% hypoxia, or left untreated (control). After cell lysis, luciferase activity was measured and normalized compared with controls. Each experiment was performed at least three times and representative data are shown. Data are the mean ± SD (n = 3).
Although the assay is not directly proving inhibition of FIH-1 by NO, GSNO and hypoxia stimulated luciferase activity to a similar extent, implying that GSNO impaired FIH-1 activity comparable to hypoxia.
DISCUSSION
It is well established that HIF-1 can be activated by conditions other than hypoxia. Recently, NO emerged as one agonist that provokes accumulation of HIF-1α under normoxia. Here we provide evidence on molecular mechanisms used by NO to stabilize HIF-1α and to cause HIF-1 transactivation. Accumulation of active HIF-1α results from inhibition of the HIF-1α hydroxylases PHD1, PHD2, PHD3, and FIH-1. Consistently, the HIF-1α-pVHL interaction is lost, and ubiquitination of HIF-1α is largely attenuated under these conditions. Our data suggest that the oxygen-sensing enzymes, known as HIF-1α prolyl hydroxylases, are subject to regulation by the (patho-) physiological effector molecule NO.
HIF-1 is composed of an α-subunit, which is continuously degraded under normoxia but stabilized under hypoxic conditions, and a constitutive β-subunit. It has been observed that active HIF-1 can be induced by the oncogene pp60(c-Src) in normoxia by enhanced HIF-1α synthesis (Karni et al., 2002). Because we have shown that NO does not affect HIF-1α production, these two mechanisms of regulation are clearly distinct. In hypoxia, formation of active HIF-1 occurs through inhibition of the human oxygen-dependent HIF-1α prolyl hydroxylases (PHD1, PHD2, PHD3) by lack of oxygen (Semenza, 2001; Oehme et al., 2002). PHDs regulate HIF-1α stability by oxygen-dependent hydroxylation of the conserved proline residues Pro564 and/or Pro402. Hydroxylated proline residues allow pVHL, the substrate recognizing subunit of an E3 ubiquitin ligase, to bind HIF-1α. The multiprotein ubiquitin-ligase complex adds ubiquitin to HIF-1α, which is then rapidly degraded by the 26S-proteasomal system. It is proposed that hypoxia blocks prolyl hydroxylation, which results in the failure of pVHL binding, decreased ubiquitination, and concomitant HIF-1α accumulation. This situation is closely resembled under conditions of GSNO treatment. In our experiments GSNO evoked nuclear HIF-1α accumulation and hypoxia-responsive reporter gene activation. The action of GSNO is shared by several other, chemically distinct NO donors such as NOC-18 (Z-1–1[2-aminoethyl-amino]diazen-1-ium-1,2-diolate), SNAP (S-nitroso-N-acetyl-d,l-penicillamine), or SNP (sodium nitroprusside) and is evident in distinct cells such as pig LLC-PK1 cells, human glioblastoma cells, human hepatoma cells, bovine pulmonary artery endothelial cells, or HEK293 cells, thus arguing against a NO donor or cell type–specific response (for references, see Brüne and Zhou, 2003). Consistent with HIF-1α accumulation, ubiquitination of the α-subunit was decreased, and HIF-1α was present under normoxia in a form that does not bind pVHL, implying that NO and hypoxia share overlapping signaling components.
NO is known to interact with iron (II) heme- or nonheme-containing proteins (Grisham et al., 1999). This may be exemplified by spectroscopic studies when NO directly binds to the ferrous ion in protocatechuate 4,5-dioxygenase and catechol 2,3-dioxygenase (Arciero et al., 1985) or to isopenicillin N synthase (Roach et al., 1995). These enzymes coordinate Fe2+ in their catalytic domain in a 2-histidine-1-carboxylate facial triad that is the defining structural motif of mononuclear nonheme iron(II) enzymes (Hegg and Que, 1997). Interestingly, HIF-1α prolyl hydroxylases belong to a nonheme Fe2+-containing family of enzymes. Therefore, it seems reasonable to assume Fe2+-coordination by NO in the catalytic site of PHD and thus competition with dioxygen. Replacement of oxygen by NO would give a molecular explanation for inhibition of enzyme function. This hypothesis is supported by our observations. We cannot exclude, however, that a chemical reaction of NO or one of its metabolites with PHD causes enzyme inactivation at a moiety other than Fe2+. A number of reports from independent groups suggested that treatment of cells with NO donors under hypoxic conditions inhibited HIF-1α accumulation and transactivation of HIF-1 (Liu et al., 1998; Sogawa et al., 1998; Huang et al., 1999). At present we do not have an explanation for the difference between normoxic and hypoxic NO effects. Although our study was in progress, it was reported that NO-evoked downregulation of HIF-1α in hypoxia is mediated by activation of PHDs (Wang et al., 2002) in cytoplasmic extracts containing partially purified PHDs. In our in vitro assay we failed to detect an activating impact of GSNO on recombinant PHDs in hypoxia. This discrepancy may suggest that the activator function of NO on PHD is caused by a cytoplasmic component, which is lacking in our assay.
There is still a lively debate on the formation of reactive oxygen species (ROS) in hypoxia. Although fluorescent dyes, e.g., dihydrofluorescein, seem to indicate enhanced ROS generation in hypoxia (Chandel et al., 2000), other methods such as the detection of hydrogen peroxide using the chemiluminescent dye luminol suggest that ROS production is rather decreased in hypoxia (Fandrey et al., 1994; Vaux et al., 2001). If hypoxia favors increased ROS production and if a chemical reaction of NO with ROS occurs, then NO would antagonize the stabilizing effect of ROS on HIF-1α that has been postulated. Altogether, the regulatory effect of ROS on prolyl hydroxylases in hypoxia and the interplay between ROS and NO in this context remain unresolved at present.
Using a hydroxyproline-specific HIF-α antibody, it has been demonstrated very recently that oncogenes such as v-Src and RasV12 stabilize HIF-1α in normoxia by attenuating prolyl hydroxylation (Chan et al., 2002). Our experiments demonstrate PHD inhibition by NO and thus suggest that not only oncogenes but also the physiological signaling molecule NO participates in the regulation of PHD activity. Thus, NO evolves as a most interesting mediator that provokes HIF-1 responses under normoxia, whereas it attenuates HIF-1 responses under hypoxia. The (patho-)physiological implications of these findings are intriguing. One may speculate whether inflammatory conditions, characterized by massive NO generation, use the HIF system to meet demands of increased metabolic/energy supply. Importantly, coculture and transwell experiments with NO-producing macrophages showed that NO functions as an intracellular as well as intercellular mediator that provoked HIF-1α accumulation in NO-sensitive detector cells (Zhou et al., 2003). This implies that effects of NO generated by GSNO are reflected by NO produced by the inducible NO-synthase (iNOS) in cells, although it remains to be determined whether comparable amounts of NO are generated under both experimental conditions. Inhibition of HIF hydroxylases provides an explanation why NO produced by the inducible NO-synthase is sufficient to induce accumulation of HIF-1α in normoxia. On the other side, in hypoxia the formation of NO has been reported to suppress HIF-1α. This may be relevant to limit a hypoxic response evoked by HIF-1.
In summary, our data provide evidence that NO impairs the activity of HIF-1α prolyl hydroxylases under normoxia, which provokes HIF-1α accumulation and HIF-1 activation. We suggest that the regulation of PHD and FIH-1 activity by NO is an important modulatory component in controlling regulation of stability and transactivating function of HIF-1α in normoxia.
Acknowledgments
We are grateful to Dr. D. Bohmann (European Molecular Biology Laboratory, Heidelberg, Germany), Dr. P.J. Ratcliffe (Wellcome Trust Centre for Human Genetics, University of Oxford, UK), and Dr. T. Kietzmann (University of Göttingen, Göttingen, Germany) for providing reagents. The technical assistance of Sandra Christmann, Andrea Trinkaus, and T. Svensson is highly appreciated. The work was supported by grants from Deutsche Forschungsgemeinschaft (BR999), Deutsche Krebshilfe (10–2008-Br2), and EC (QLK6-CT-2000–00064) to B.B. and by Deutsche Forschungsgemeinschaft (SFB367) to W.J. and E.M.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02-12-0791. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-12-0791.
References
- Arciero, D.M., Orville, A.M., and Lipscomb, J.D. (1985). [17O]Water and nitric oxide binding by protocatechuate 4, 5-dioxygenase and catechol 2,3-dioxygenase. Evidence for binding of exogenous ligands to the active site Fe2+ of extradiol dioxygenases. J. Biol. Chem. 260, 14035–14044. [PubMed] [Google Scholar]
- Bruick, R.K., and McKnight, S.L. (2001). A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294, 1337–1340. [DOI] [PubMed] [Google Scholar]
- Brüne, B., Mohr, S., and Messmer, U.K. (1996). Protein thiol modification and apoptotic cell death as cGMP-independent nitric oxide (NO) signaling pathways. Rev. Physiol. Biochem. Pharmacol. 127, 1–30. [DOI] [PubMed] [Google Scholar]
- Brüne, B., and Zhou, J. (2003). The role of nitric oxide (NO) in stability regulation of hypoxia inducible factor-1alpha (HIF-1alpha). Curr. Med. Chem. 10, 845–855. [DOI] [PubMed] [Google Scholar]
- Chan, D.A., Sutphin, P.D., Denko, N.C., and Giaccia, A.J. (2002). Role of prolyl hydroxylation in oncogenically stabilized hypoxia-inducible factor-1alpha. J. Biol. Chem. 277, 40112–40117. [DOI] [PubMed] [Google Scholar]
- Chandel, N.S., McClintock, D.S., Feliciano, C.E., Wood, T.M., Melendez, J.A., Rodriguez, A.M., and Schumacker, P.T. (2000). Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J. Biol. Chem. 275, 25130–25138. [DOI] [PubMed] [Google Scholar]
- Clifford, S.C., Cockman, M.E., Smallwood, A.C., Mole, D.R., Woodward, E.R., Maxwell, P.H., Ratcliffe, P.J., and Maher, E.R. (2001). Contrasting effects on HIF-1alpha regulation by disease-causing pVHL mutations correlate with patterns of tumourigenesis in von Hippel-Lindau disease. Hum. Mol. Genet. 10, 1029–1038. [DOI] [PubMed] [Google Scholar]
- Cockman, M.E. et al. (2000). Hypoxia inducible factor-alpha binding and ubiquitylation by the von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 275, 25733–25741. [DOI] [PubMed] [Google Scholar]
- Epstein, A.C. et al. (2001). C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54. [DOI] [PubMed] [Google Scholar]
- Fandrey, J., Frede, S., and Jelkmann, W. (1994). Role of hydrogen peroxide in hypoxia-induced erythropoietin production. Biochem. J. 303, 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisham, M.B., Jourd'Heuil, D., and Wink, D.A. (1999). Nitric oxide. I. Physiological chemistry of nitric oxide and its metabolites: implications in inflammation. Am. J. Physiol. 276, G315–321. [DOI] [PubMed] [Google Scholar]
- Hart, T.W. (1997). Some observation concerning the S-nitroso and S-phenylsulfonyl derivatives of l-cysteine and glutathione. Tetrahedron Lett. 26, 2013–2016. [Google Scholar]
- Hegg, E.L., and Que, L., Jr. (1997). The 2-His-1-carboxylate facial triad—an emerging structural motif in mononuclear non-heme iron(II) enzymes. Eur. J. Biochem. 250, 625–629. [DOI] [PubMed] [Google Scholar]
- Hellwig-Bürgel, T., Rutkowski, K., Metzen, E., Fandrey, J., and Jelkmann, W. (1999). Interleukin-1beta and tumor necrosis factor-alpha stimulate DNA binding of hypoxia-inducible factor-1. Blood 94, 1561–1567. [PubMed] [Google Scholar]
- Hewitson, K.S. et al. (2002). Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 277, 26351–26355. [DOI] [PubMed] [Google Scholar]
- Hon, W.C. et al. (2002). Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature 417, 975–978. [DOI] [PubMed] [Google Scholar]
- Huang, L.E., Gu, J., Schau, M., and Bunn, H.F. (1998). Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 95, 7987–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L.E., Willmore, W.G., Gu, J., Goldberg, M.A., and Bunn, H.F. (1999). Inhibition of hypoxia-inducible factor 1 activation by carbon monoxide and nitric oxide. J. Biol. Chem. 274, 9038–9044. [DOI] [PubMed] [Google Scholar]
- Isaacs, J.S., Jung, Y.J., Mimnaugh, E.G., Martinez, A., Cuttitta, F., and Neckers, L.M. (2002). Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 alpha-degradative pathway. J. Biol. Chem. 277, 29936–29944. [DOI] [PubMed] [Google Scholar]
- Ivan, M. et al. (2002). Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc. Natl. Acad. Sci. USA 99, 13459–13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan, M., and Kaelin, W.G., Jr. (2001). The von Hippel-Lindau tumor suppressor protein. Curr. Opin. Genet. Dev. 11, 27–34. [DOI] [PubMed] [Google Scholar]
- Ivan, M. et al. (2001). HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468. [DOI] [PubMed] [Google Scholar]
- Jaakkola, P. et al. (2001). Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472. [DOI] [PubMed] [Google Scholar]
- Kaelin, W.G., Jr. (2002). How oxygen makes its presence felt. Genes Dev. 16, 1441–1445. [DOI] [PubMed] [Google Scholar]
- Kallio, P.J., Wilson, W.J., O'Brien, S., Makino, Y., and Poellinger, L. (1999). Regulation of the hypoxia-inducible transcription factor 1alpha by the ubiquitin-proteasome pathway. J. Biol. Chem. 274, 6519–6525. [DOI] [PubMed] [Google Scholar]
- Karni, R., Dor, Y., Keshet, E., Meyuhas, O., and Levitzki, A. (2002). Activated pp60c-Src leads to elevated hypoxia-inducible factor (HIF)-1alpha expression under normoxia. J. Biol. Chem. 277, 42919–42925. [DOI] [PubMed] [Google Scholar]
- Kimura, H., Weisz, A., Kurashima, Y., Hashimoto, K., Ogura, T., D'Acquisto, F., Addeo, R., Makuuchi, M., and Esumi, H. (2000). Hypoxia response element of the human vascular endothelial growth factor gene mediates transcriptional regulation by nitric oxide: control of hypoxia-inducible factor-1 activity by nitric oxide. Blood 95, 189–197. [PubMed] [Google Scholar]
- Kimura, H., Weisz, A., Ogura, T., Hitomi, Y., Kurashima, Y., Hashimoto, K., D'Acquisto, F., Makuuchi, M., and Esumi, H. (2001). Identification of hypoxia-inducible factor 1 ancillary sequence and its function in vascular endothelial growth factor gene induction by hypoxia and nitric oxide. J. Biol. Chem. 276, 2292–2298. [DOI] [PubMed] [Google Scholar]
- Krieg, M., Haas, R., Brauch, H., Acker, T., Flamme, I., and Plate, K.H. (2000). Up-regulation of hypoxia-inducible factors HIF-1alpha and HIF-2alpha under normoxic conditions in renal carcinoma cells by von Hippel-Lindau tumor suppressor gene loss of function. Oncogene 19, 5435–5443. [DOI] [PubMed] [Google Scholar]
- Lando, D., Peet, D.J., Gorman, J.J., Whelan, D.A., Whitelaw, M.L., and Bruick, R.K. (2002a). FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16, 1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando, D., Peet, D.J., Whelan, D.A., Gorman, J.J., and Whitelaw, M.L. (2002b). Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 295, 858–861. [DOI] [PubMed] [Google Scholar]
- Lee, D.H., and Goldberg, A.L. (1998). Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8, 397–403. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Christou, H., Morita, T., Laughner, E., Semenza, G.L., and Kourembanas, S. (1998). Carbon monoxide and nitric oxide suppress the hypoxic induction of vascular endothelial growth factor gene via the 5′ enhancer. J. Biol. Chem. 273, 15257–15262. [DOI] [PubMed] [Google Scholar]
- Mahon, P.C., Hirota, K., and Semenza, G.L. (2001). FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 15, 2675–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson, N., Willam, C., Maxwell, P.H., Pugh, C.W., and Ratcliffe, P.J. (2001). Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 20, 5197–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell, P.H., Pugh, C.W., and Ratcliffe, P.J. (2001). The pVHL-hIF-1 system. A key mediator of oxygen homeostasis. Adv. Exp. Med. Biol. 502, 365–376. [PubMed] [Google Scholar]
- Maxwell, P.H. et al. (1999). The tumour suppressor protein VHL targets hypoxia-inducible facto4rs for oxygen-dependent proteolysis. Nat. Lett. 399, 271–275. [DOI] [PubMed] [Google Scholar]
- Min, J.H., Yang, H., Ivan, M., Gertler, F., Kaelin, W.G., Jr., and Pavletich, N.P. (2002). Structure of an HIF-1alpha-pVHL complex: hydroxyproline recognition in signaling. Science 296, 1886–1889. [DOI] [PubMed] [Google Scholar]
- Oehme, F., Ellinghaus, P., Kolkhof, P., Smith, T.J., Ramakrishnan, S., Hutter, J., Schramm, M., and Flamme, I. (2002). Overexpression of PH-4, a novel putative proline 4-hydroxylase, modulates activity of hypoxia-inducible transcription factors. Biochem. Biophys. Res. Commun. 296, 343–349. [DOI] [PubMed] [Google Scholar]
- Ohh, M., Park, C.W., Ivan, M., Hoffman, M.A., Kim, T.Y., Huang, L.E., Pavletich, N., Chau, V., and Kaelin, W.G. (2000). Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2, 423–427. [DOI] [PubMed] [Google Scholar]
- O'Rourke, J.F., Tian, Y.M., Ratcliffe, P.J., and Pugh, C.W. (1999). Oxygen-regulated and transactivating domains in endothelial PAS protein 1: comparison with hypoxia-inducible factor-1alpha. J. Biol. Chem. 274, 2060–2071. [DOI] [PubMed] [Google Scholar]
- Palmer, L.A., Gaston, B., and Johns, R.A. (2000). Normoxic stabilization of hypoxia-inducible factor-1 expression and activity: redox-dependent effect of nitrogen oxides. Mol. Pharm. 58, 1197–1203. [DOI] [PubMed] [Google Scholar]
- Roach, P.L., Clifton, I.J., Fulop, V., Harlos, K., Barton, G.J., Hajdu, J., Andersson, I., Schofield, C.J., and Baldwin, J.E. (1995). Crystal structure of isopenicillin N synthase is the first from a new structural family of enzymes. Nature 375, 700–704. [DOI] [PubMed] [Google Scholar]
- Salceda, S., and Caro, J. (1997). Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. J. Biol. Chem. 272, 22642–22647. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Sandau, K.B., Fandrey, J., and Brune, B. (2001a). Accumulation of HIF-1alpha under the influence of nitric oxide. Blood 97, 1009–1015. [DOI] [PubMed] [Google Scholar]
- Sandau, K.B., Faus, H.G., and Brune, B. (2000). Induction of hypoxia-inducible-factor 1 by nitric oxide is mediated via the PI 3K pathway. Biochem. Biophys. Res. Commun. 278, 263–267. [DOI] [PubMed] [Google Scholar]
- Sandau, K.B., Zhou, J., Kietzmann, T., and Brune, B. (2001b). Regulation of the hypoxia-inducible factor 1alpha by the inflammatory mediators nitric oxide and tumor necrosis factor-alpha in contrast to desferroxamine and phenylarsine oxide. J. Biol. Chem. 276, 39805–39811. [DOI] [PubMed] [Google Scholar]
- Semenza, G.L. (2001). Hif-1, O(2), and the 3 phds. How animal cells signal hypoxia to the nucleus. Cell 107, 1–3. [DOI] [PubMed] [Google Scholar]
- Semenza, G.L. (2002). HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol. Med. 8, S62–67. [DOI] [PubMed] [Google Scholar]
- Sogawa, K., Numayama-Tsuruta, K., Ema, M., Abe, M., Abe, H., and Fujii-Kuriyama, Y. (1998). Inhibition of hypoxia-inducible factor 1 activity by nitric oxide donors in hypoxia. Proc. Natl. Acad. Sci. USA 95, 7368–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler, J.S., Singel, D.J., and Loscalzo, J. (1992). Biochemistry of nitric oxide and its redox-activated forms. Science 258, 1898–1902. [DOI] [PubMed] [Google Scholar]
- Tanimoto, K., Makino, Y., Pereira, T., and Poellinger, L. (2000). Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. EMBO J. 19, 4298–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux, E.C., Metzen, E., Yeates, K.M., and Ratcliffe, P.J. (2001). Regulation of hypoxia-inducible factor is preserved in the absence of a functioning mitochondrial respiratory chain. Blood 98, 296–302. [DOI] [PubMed] [Google Scholar]
- Wang, F., Sekine, H., Kikuchi, Y., Takasaki, C., Miura, C., Heiwa, O., Shuin, T., Fujii-Kuriyama, Y., and Sogawa, K. (2002). HIF-1alpha-prolyl hydroxylase: molecular target of nitric oxide in the hypoxic signal transduction pathway. Biochem. Biophys. Res. Commun. 295, 657–662. [DOI] [PubMed] [Google Scholar]
- Wang, G.L., and Semenza, G.L. (1995). Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 270, 1230–1237. [DOI] [PubMed] [Google Scholar]
- Wenger, R.H. (2002). Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J. 16, 1151–1162. [DOI] [PubMed] [Google Scholar]
- Yu, F., White, S.B., Zhao, Q., and Lee, F.S. (2001). HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. USA 98, 9630–9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Fandrey, J., Schumann, J., Tiegs, G., and Brune, B. (2003). NO and TNF-alpha released from activated macrophages stabilize HIF-1alpha in resting tubular LLC-PK1 cells. Am. J. Physiol. Cell Physiol. 284, C439–C446. [DOI] [PubMed] [Google Scholar]
- Zhu, H., Jackson, T., and Bunn, H.F. (2002). Detecting and responding to hypoxia. Nephrol. Dial. Transplant 17(Suppl 1), 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]