Abstract
Two ecotypes of the prokaryote Prochlorococcus adapted to distinct light niches in the ocean have been described recently. These ecotypes are characterized by their different (divinyl-) chlorophyll (Chl) a to Chl b ratios and 16S rRNA gene signatures, as well as by their significantly distinct irradiance optima for growth and photosynthesis [Moore, L. R., Rocap, G. & Chisholm, S. W. (1998) Nature (London) 393, 464–467]. However, the molecular basis of their physiological differences remained, so far, unexplained. In this paper, we show that the low-light-adapted Prochlorococcus strain SS120 possesses a gene family of seven transcribed genes encoding different Chl a/b-binding proteins (Pcbs). In contrast, Prochlorococcus sp. MED4, a high-light-adapted ecotype, possesses a single pcb gene. The presence of multiple antenna genes in another low-light ecotype (NATL2a), but not in another high-light ecotype (TAK9803–2), is demonstrated. Thus, the multiplication of pcb genes appears as a key factor in the capacity of deep Prochlorococcus populations to survive at extremely low photon fluxes.
The ecological distribution of photosynthetic organisms is strongly controlled by the availability of solar light. A remarkable example of the role of light in modeling natural communities in the ocean was provided recently by the discovery of two ecotypes of the prokaryote Prochlorococcus living at different depths (1). These ecotypes, characterized by their different (divinyl-) chlorophyll (Chl) a to Chl b ratios and 16S rRNA gene signatures, were shown to have significantly distinct irradiance optima for growth and photosynthesis (2–4). Thus, major differences can be expected between the light-harvesting complexes of these ecotypes. The nature of the antenna genes of Prochlorococcus, as well as the two other prokaryotes that contain Chl b as the major accessory pigment, has been identified recently (5). The encoded proteins, named Pcbs for prochlorophyte Chl b-binding proteins, seem to be phylogenetically and structurally different from the antenna proteins found in photosynthetic eukaryotes (6). Although Prochlorothrix was shown to possess three different pcb genes, a single gene copy was found in Prochloron (5, 7). Up to now, only one pcb gene was reported for each of two Prochlorococcus clones (5), representative of the high-light- and low-light-adapted ecotypes (1, 8), MED4, from surface waters of the Mediterranean Sea, and SS120, isolated from the deep waters (120 m) of the Sargasso Sea, respectively. However, the biochemical characterization of their thylakoid proteins revealed a much more prominent band of antenna proteins extending over a wider range of molecular masses in SS120 (31–38 kDa) than in MED4 (31–32.5 kDa) at low light (9), suggesting that the former strain might in fact possess several proteins. In this study, we reexamined these strains and other representatives of high- and low-light-adapted ecotypes and found that the latter but not the former had multiple antenna genes. The evolutionary and ecological implications of this discovery are discussed.
Materials and Methods
Culturing and Spectrofluorimetry.
Prochlorococcus marinus clone SS120 (CCMP1375; ref. 1), Prochlorococcus sp. clone MED4 (CCMP1378; ref. 1), and the unicyanobacterial Prochlorococcus sp. strains NATL2a (isolated at 30 m in the North Atlantic Ocean, 38° 59′ N, 40° 33′ W) and TAK9803–2 (isolated at 20 m in the Takapoto lagoon, 14° 30′ N, 145° 20′ W) were grown at 24 ± 1°C in PCR-S11 medium (10). To obtain comparable fluorescence excitation spectra using a Perkin–Elmer LS50 spectrofluorimeter, all strains were maintained under blue light at ≈20 μmol Q m−2⋅s−1 (Q is Quanta), a value intermed- iate between the growth irradiance maxima of MED4 and SS120 (11).
Gene Isolation and Sequencing.
pcb fragments were amplified directly from Prochlorococcus cells by using several sets of primers: P-G-pcb-1-a-S-20, 5′-ATCGARACNTAYGGNAAYCC-3′; P-G-pcb-25-a-S-20, 5′-TNACNTAYGGNTGGTGGGC-3′; P-G-pcb-760-a-A-20, 5′-GCNGCNATDATNGCCATCCA-3′; and P-G-pcb-998-a-A-20, 5′-ACNCKYTTRAARTCRAANCC-3′. PCR was performed with the Ready-to-Go PCR Beads kit (Amersham Pharmacia). pcb fragments of 730 to 1014 bp were purified with the Wizard kit (Promega) and cloned by using the TOPO-TA cloning kit (Invitrogen). Complete pcb genes were obtained by screening a genomic library of Prochlorococcus SS120 established in λGEM-12 (Promega), and DNA sequences were determined on a Vistra 725 automated sequencer (Molecular Dynamics). Protein sequences were aligned by using pileup (Wisconsin Package, Version 8; Genetics Computer Group, Madison, WI), followed by manual refinement on the genetic data environment software package (12).
Northern Blotting.
Total RNA was isolated and analyzed as described previously (13). Gene-specific pcb fragments of 271 to 298 bp were amplified by PCR with the cloned genes as templates. From these, [32P]UTP-labeled specific RNA probes were synthesized by using the Lig'nScribe and MAXIscript kits (Ambion, Austin, TX). After hybridization under stringent conditions (61.5°C), detection was performed by using a Storm PhosphorImager (Molecular Dynamics). The absence of cross-hybridization between each specific RNA probe and transcripts of all other pcb genes was verified by RNase protection assay experiments (data not shown).
Results
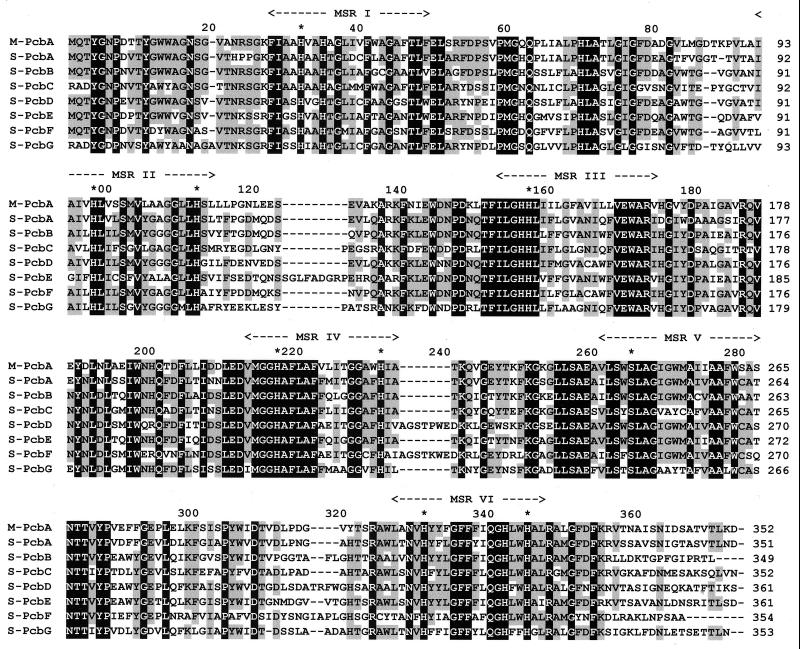
Fragments of the pcb gene were amplified by PCR directly from whole Prochlorococcus MED4 and SS120 cells. We confirmed that the high-light-adapted genotype contained a single pcb gene. Only one PCR product with the expected sequence (5) was obtained, and hybridization of restriction digests of chromosomal DNA with pcb probes at low stringency resulted only in the banding pattern predicted from the reported pcb sequence (data not shown). However, from the low-light strain, seven different pcb genes were recovered. We arbitrarily called them pcbA to pcbG. The deduced proteins have lengths from 351 to 363 aa, with sequence identities ranging from 50% to 75% (Table 1). They all contain the six potential transmembrane helices found in other Pcbs (5), with conserved histidines that can serve as potential ligands for Chl molecules (Fig. 1).
Table 1.
Percentages of sequence identities (upper right triangle) and sequence similarities (lower left triangle) between the different Pcb proteins from Prochlorococcus MED4 (M-PcbA) and SS120 (S-PcbA to S-PcbG)
| M-PcbA | S-PcbA | S-PcbB | S-PcbC | S-PcbD | S-PcbE | S-PcbF | S-PcbG | |
|---|---|---|---|---|---|---|---|---|
| M-PcbA | — | 75 | 64 | 63 | 63 | 63 | 57 | 54 |
| S-PcbA | 87 | — | 71 | 66 | 66 | 66 | 61 | 58 |
| S-PcbB | 77 | 80 | — | 61 | 70 | 71 | 65 | 56 |
| S-PcbC | 78 | 79 | 73 | — | 56 | 57 | 53 | 66 |
| S-PcbD | 77 | 80 | 80 | 70 | — | 66 | 67 | 55 |
| S-PcbE | 78 | 80 | 79 | 71 | 78 | — | 57 | 55 |
| S-PcbF | 74 | 76 | 76 | 68 | 80 | 71 | — | 50 |
| S-PcbG | 72 | 75 | 70 | 80 | 71 | 71 | 68 | — |
Sequence similarities comprise identical sequences plus conservative substitutions within the groups DN, EQ, ST, KR, FYW, and LIVM.
Figure 1.
Alignment of the deduced protein sequences from the Prochlorococcus MED4 pcbA gene (M-PcbA) and from Prochlorococcus SS120 pcbA to pcbG genes (S-PcbA to S-PcbG). Identical residues are shown in white type on a black background. Black type on gray squares indicates that the percentage of conserved residues is >60% (i.e., at least 5 of 8 sequences). Predicted membrane-spanning regions (MSR) are indicated by arrows, and stars mark putative Chl-binding residues.
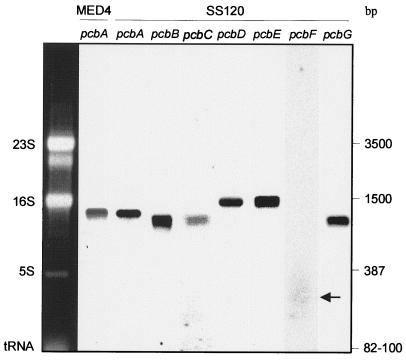
All genes are expressed (Fig. 2) as monocistronic transcripts (1.35–1.5 kb). The transcript of pcbF most likely has a much faster turnover than those of the six other genes, because it is detected only as a degraded product on Northern blots, whereas rehybridization of the membrane with another probe (pcbG) attested the good quality of the mRNA. A partial physical mapping of the Prochlorococcus SS120 genome by pulse-field gel electrophoresis, sequence analysis, and Southern hybridization revealed that these genes are not clustered, but randomly spread throughout the genome (data not shown). The fairly wide distance between these genes suggests that different promoters most likely control their transcription. The independent regulation of their expression might provide a higher level of plasticity to the antenna system of this organism.
Figure 2.
Detection of antenna gene transcripts by Northern blotting from Prochlorococcus MED4 (pcbA) and SS120 (pcbA to pcbG) strains. Transcripts of pcbF have been indicated by an arrow. The sizes of major RNA molecules are indicated.
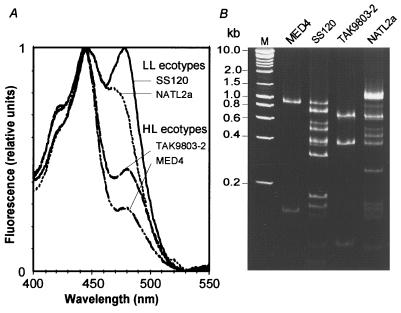
To generalize these observations, two other Prochlorococcus strains, TAK9803–2 and NATL2a, were studied. The phylogenetic analysis of the 16S rRNA genes of these strains shows that the former strain belongs to the high-light-adapted clade (14), as does MED4 (8), whereas NATL2a groups with SS120, outside of this cluster (2, 8). Furthermore, spectrofluorimetry (Fig. 3A) shows that the TAK9803–2 strain has a high ratio of divinyl-Chl a to b, which is characteristic of all high-light-adapted Prochlorococcus strains, whereas NATL2a displays a low Chl a to b ratio, typical for a low-light-adapted strain (2, 3). PCR fragments of ≈1,000 bp were amplified from the pcb genes of these two strains. The amplification products subsequently were cleaved with the restriction endonuclease HaeIII, an enzyme that cuts all eight known pcb genes from Prochlorococcus between one and three times. Although the exact number of pcb genes cannot be determined with certainty by this method, summation of the sizes of the different bands shown in Fig. 3B suggests that there is a single gene in TAK9803–2 as there is in MED4, and there are at least five genes in NATL2a. In the latter, there could be even more pcb genes, as some of the PCR-products apparently were not cleavable by HaeIII (Fig. 3B, top band of lane 5).
Figure 3.
Characterization of different low-light (LL)- and high-light (HL)-adapted Prochlorococcus ecotypes grown at 20 μmol Q m−2⋅s−1. (A) Excitation fluorescence spectra of whole cells normalized to the divinyl-Chl a peak (444 nm). The variable height of the second peak, mainly monovinyl-Chl b (468 nm) for NATL2a and divinyl-Chl b (478 nm) for the other strains, indicates the widely different Chl b to Chl a ratios between strains. (B) Pattern of amplified pcb fragments obtained on polyacrylamide gels after cleavage by HaeIII. Lane M, size markers.
Discussion
Our results show that part of the genetic and likely ecophysiological differentiation between high-light- and low-light-adapted Prochlorococcus strains can be explained by a multiplication of pcb genes in the latter strains. Because these genes are all expressed and show considerable sequence variation, these findings result in an extraordinary diversity of light-harvesting complexes of low-light ecotypes, probably associated with a diversification of their properties. A reexamination of thylakoid profiles obtained by Partensky et al. (9), which showed a thickening of the antenna band of SS120 from high to low light, suggests that the expression of some of the pcb genes might be specifically induced under low-light conditions. This phenomenon results in a relative increase in the size of antenna complexes, which could enable such a low-light-adapted strain to efficiently use the scarce photons reaching natural oceanic populations at depths below 100 m. Nevertheless, it is worth noting that despite the apparently higher plasticity of the antenna of SS120 as compared with MED4, its photosynthetic apparatus is more sensitive to photodamage at high light, as shown by the strong photoinhibition of these cells above 100 μmol Q m−2⋅s−1 (2, 11).
All higher plants studied to date possess 6 to 15 Chl a/b binding proteins encoded by members of the Lhc gene family and having distinct functions within the photosynthetic apparatus (6). During acclimation to changes in photon fluxes, one of the major alterations exhibited by plants is a variation in their antennae size. This adaptation is attributable mainly to changes in the relative abundance of the individual proteins regulated at the transcriptional or translational level. However, there is no evidence, either from the literature or from the available sequence data, for a lower number of antenna genes in sun plants than there are in shade plants, nor is there evidence for a comparable degree of sequence variation within a given type of eukaryotic antenna gene (e.g., Lhca or Lhcb). Thus, multiplying the number of antenna genes is a specific adaptation mechanism to low light. This phenomenon seems to be unique to Prochlorococcus, but its further analysis may provide fundamental insights in the evolution and adaptation of light-harvesting systems that may also be relevant to higher plants. Such a multiplication is even more surprising in the case of this organism, because it has the smallest genome of all oxyphototrophic organisms and displays many examples of genome compactness (15). For instance, Synechocystis PCC6803, a cyanobacterium with a twice bigger genome (3.6 Mbp) as Prochlorococcus (15), has no photosynthetic genes in more than four copies (e.g., petF, encoding ferredoxin; ref. 16).
One intriguing question that remains to be solved is whether the multiple pcb gene copies are expressed differentially depending on the light conditions, as was previously shown for the different copies of psbA (17) and psbD (18) in Synechococcus PCC7942.
Acknowledgments
We thank Prof. S. W. Chisholm and Dr. L. Moore for providing culture material for use in this study and Ms. F. Legall for taking care of the cultures. We also thank Drs. D. Vaulot and S. Loiseaux-de-Goër for critically reading the manuscript. This study was funded by the European Community program MAS3-CT97–0128 (Promolec program) and supported by the Deutsche Forschungsgemeinschaft to W.R.H. (SFB 429). L.G. was supported by a Ph.D. grant cofunded by the Centre National de la Recherche Scientifique and the Région Bretagne, and G.W.M.v.d.S. was supported by the Centre National de la Recherche Scientifique–Institut National des Sciences de l'Univers.
Abbreviations
- Chl
chlorophyll
- Pcb
prochlorophyte Chl b-binding protein
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AF198526 (pcbB), AF198527 (pcbC), AF198528 (pcbD), AF198529 (pcbE), AF198530 (pcbF), and AF198531 (pcbG)].
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.070040897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.070040897
References
- 1.Chisholm S W, Frankel S L, Goericke R, Olson R J, Palenik B, Waterbury J B, West-Johnsrud L, Zettler E R. Arch Microbiol. 1992;157:297–300. [Google Scholar]
- 2.Moore L R, Rocap G, Chisholm S W. Nature (London) 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 3.Moore L R, Chisholm S W. Limnol Oceanogr. 1999;44:628–638. [Google Scholar]
- 4.West N J, Scanlan D J. Appl Env Microbiol. 1999;65:2585–2591. doi: 10.1128/aem.65.6.2585-2591.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.La Roche J, van der Staay G W, Partensky F, Ducret A, Aebersold R, Li R, Golden S S, Hiller R G, Wrench P M, Larkum A W, Green B R. Proc Natl Acad Sci USA. 1996;93:15244–15248. doi: 10.1073/pnas.93.26.15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durnford D G, Deane J A, Tan S, McFadden G I, Gantt E, Green B R. J Mol Evol. 1999;48:59–68. doi: 10.1007/pl00006445. [DOI] [PubMed] [Google Scholar]
- 7.van der Staay G W M, Yurkova N, Green B R. Plant Mol Biol. 1998;36:709–716. doi: 10.1023/a:1005930210515. [DOI] [PubMed] [Google Scholar]
- 8.Urbach E, Scanlan D J, Distel D L, Waterbury J B, Chisholm S W. J Mol Evol. 1998;46:188–201. doi: 10.1007/pl00006294. [DOI] [PubMed] [Google Scholar]
- 9.Partensky F, LaRoche J, Wyman K, Falkowski P G. Photosynth Res. 1997;51:209–222. [Google Scholar]
- 10.Partensky F, Hess W R, Vaulot D. Microbiol Mol Biol Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore L R, Goericke R, Chisholm S W. Mar Ecol Prog Ser. 1995;116:259–275. [Google Scholar]
- 12.Smith S W, Overbeek R, Woese C R, Gilbert W. Comput Appl Biosci. 1994;10:671–675. doi: 10.1093/bioinformatics/10.6.671. [DOI] [PubMed] [Google Scholar]
- 13.Hess W R, Weihe A, Loiseaux-de Goër S, Partensky F, Vaulot D. Plant Mol Biol. 1995;27:1189–1196. doi: 10.1007/BF00020892. [DOI] [PubMed] [Google Scholar]
- 14.West, N., Schönhuber, W. A., Amann, R. I., Rippka, R. & Scanlan, D. J. (2000) Appl. Environ. Microbiol., in press.
- 15.Strehl B, Holtzendorff J, Partensky F, Hess W R. FEMS Microbiol Lett. 1999;181:261–266. doi: 10.1111/j.1574-6968.1999.tb08853.x. [DOI] [PubMed] [Google Scholar]
- 16.Kotani H, Tabata S. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:151–171. doi: 10.1146/annurev.arplant.49.1.151. [DOI] [PubMed] [Google Scholar]
- 17.Schaefer M R, Golden S S. J Bacteriol. 1989;171:3973–3981. doi: 10.1128/jb.171.7.3973-3981.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bustos S A, Golden S S. Mol Gen Genet. 1992;232:221–230. doi: 10.1007/BF00280000. [DOI] [PubMed] [Google Scholar]