Abstract
Interleukin (IL)-4-mediated nuclear signaling by Stat6 has been implicated in lymphoid cell proliferation and the transcriptional activation of genes encoding major histocompatability complex (MHC) class II molecules and Fc receptors. To investigate IL-4-mediated transcriptional events, we cloned two naturally occurring human Stat6 isoforms, Stat6b and Stat6c, that encoded an NH2-terminal truncation or an SH2 domain deletion, respectively. Stat6 variant mRNAs were differentially expressed in many human tissues. To elucidate the biologic role of each isoform, we examined the consequences of overexpression in IL-4-responsive FDC-P2 cells. Stat6 and Stat6b (to a lesser extent) enhanced DNA synthesis, up-regulated endogenous MHC class II and Fcγ receptors, and became tyrosine phosphorylated in response to IL-4 stimulation. In contrast, Stat6c, which lacks functionally critical SH2 domain residues, unexpectedly inhibited IL-4-mediated mitogenesis and cell surface antigen expression and was not tyrosine phosphorylated. Although Stat6c only modestly diminished endogenous Stat6 tyrosine phosphorylation, it abolished endogenous Stat6 FcγRI and Iɛ DNA binding activity and FcγRI–luciferase reporter transcriptional activation. Our results indicate that the molecular mechanism of inhibition by Stat6c was due to suppression of endogenous Stat6 dimer formation. Thus, Stat6b and Stat6c are naturally occurring attenuated and dominant negative Stat6 variants, respectively, that affect IL-4-mediated biologic responses through differential transcriptional regulation.
Interleukin (IL)-4 is a pleiotropic cytokine that plays a prominent role in the regulation of inflammatory and cell-mediated immune responses (1). Numerous cell types proliferate and/or differentiate in response to IL-4 (2). Proliferation of quiescent T cells as well as B cells in the presence of anti-IgM antibodies (3) is induced by IL-4. Lymphocytic expression of major histocompatability complex (MHC) class II molecules (4), CD23 (5) and the IL-4 receptor (6), as well as eosinophilic regulation of Fcγ receptors is intimately associated with IL-4 exposure (7). IL-4 also mediates transcription of unrearranged IgE and IgG1 constant regions, leading to isotype class switching and subsequent biosynthesis (8, 9). Recently, IL-4 has been shown to be the major regulator of the lymphokine-producing phenotype of CD4+ T lymphocytes and to facilitate differentiation to Th2 cells (10).
IL-4 treatment induces tyrosine phosphorylation of the IL-4 receptor, designated IL-4Rα (11, 12), a member of the hematopoietin receptor superfamily (13, 14). Unlike several members of the hematopoietin receptor superfamily, IL-4Rα is ubiquitously expressed on cells of hematopoietic and nonhematopoietic origin. IL-4Rα activation results in tyrosine phosphorylation of multiple substrates including Jak1, Jak3 (15, 16), IRS-1 (17), IRS-2/4PS (18), and Stat6 (13, 14, 19, 20). Distinct regions of IL-4Rα have also been shown to control growth and gene expression (21). Phosphorylation of specific tyrosine residues within the two GYKXF motifs present in the IL-4Rα has been proposed to be crucial for binding to and activation of Stat6 (13, 22).
Selective activation of STATs results in dimerization and translocation to the nucleus, where each interacts with unique DNA response elements and activates transcription (23, 24). Stat6 activation correlates with mitogenic and pleiotropic functional responses induced by IL-4 (22), IL-13 (25), and platelet-derived growth factor (26). Although phenotypic analysis of Stat6−/− mice have elegantly demonstrated a role for Stat6 in IL-4-induced lymphocyte proliferation, Th2 helper T cell differentiation, Ig class switching, and cell surface antigen expression (27–29), the mechanism(s) by which Stat6 induces these effects remain incompletely understood. Here, we have isolated and cloned two novel Stat6 homologs and investigated their biologic function and mechanistic basis for their effects.
MATERIALS AND METHODS
Materials.
Anti-Stat6 peptide sera used for immunoprecipitation or immunoblot analysis were raised against amino acid residues 689–711 (NH2-VPQVYPPHSHSIPPYQGLSPEES-COOH) or 787–804 (NH2-GEDIFPPLLPPTEQDLTK-COOH), respectively. Anti-phosphotyrosine monoclonal antibody was purchased from Upstate Biotechnology (Lake Placid, NY). Murine IL-4 was obtained from PeproTech (Rocky Hill, NJ). Antibodies to CD16/CD32 or I-Ad MHC class II conjugated to fluorescein isothiocyanate (FITC) were obtained from PharMingen. The sequences of one strand of the double-stranded Iɛ and FcγR1 probes used for electrophoretic mobility shift assay (EMSA) were 5′-GATCTAACTTCCCAAGAACAG-3′ and 5′-GTATTTCCCAGAAAAGGAAC-3′, respectively.
cDNA Cloning and Transfection.
Human Stat6 cDNA, cloned in our laboratory (26), was used for Stat6 variant screening. A cDNA library was constructed using oligo(dT)-primed human M426 fibroblast cDNAs packaged into λpCEV29. For library screening, the bacterial strain Y1088 was infected with phage (2 × 104 plaques per 150-mm plate). Nitrocellulose filters were hybridized with 32P-labeled full-length human Stat6 cDNA in Hybrisol-I (Oncor) at 42°C for 20 h, washed under low stringency conditions (3× SSC, 0.1% SDS; once at 25°C for 30 min, three times at 40°C for 30 min), and exposed to x-ray film. The cDNA inserts from plaque-purified clones were sequenced.
pCEV29–Stat6 variant or control pCEV29 cDNAs containing the neomycin gene were electroporated into FDC-P2 cells overexpressing the erythropoietin (EPO) receptor. Stable transfectants were generated by selection in geneticin (750 μg/ml), and clonal transfectants were established by single cell dilution. Transfectants were maintained in RPMI media containing EPO (1 unit/ml)/geneticin (750 μg/ml) and used throughout this study unless otherwise stated.
RNase Protection Assay.
Total RNA was isolated from a variety of human tissues or obtained from CLONTECH. A 344-bp fragment from the 5′ end of the human Stat6c cDNA was amplified by PCR and cloned in the pBluescriptII KS(+) vector. The identity of the insert was confirmed by sequencing. The plasmid was linearized at the EcoRI site and a [32P]UTP-labeled 395-bp antisense RNA was synthesized with T7 polymerase. The probe was designed such that Stat6, Stat6b, and Stat6c transcripts would result in 276-, 140-, and 344-bp protected products, respectively. A 125-bp riboprobe synthesized from the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA (PharMingen) was mixed with the Stat6 probe and added as an internal standard to each sample. The size of the protected GAPDH transcript was 97 bp. The RNase protection assay was performed as recommended by the manufacturer (Ambion, Austin, TX). Briefly, the riboprobes were coprecipitated with 50 μg of total RNA from each tissue sample, resuspended in the 20-μl hybridization solution, and incubated at 42°C for 18–20 h. The RNA hybrid digested with RNase A (10 μg) and RNase T1 (100 units) for 30 min at 37°C. Protected products were analyzed on a 6% acrylamide–urea gel and visualized by autoradiography.
Mitogenic Assay.
[3H]Thymidine incorporation into FDC-P2 cells was performed as previously described (12) with the following modifications. FDC-P2 cells or FDC-P2 transfectants stably expressing each Stat6 variant (2 × 105 cells/ml) were washed and resuspended in RPMI 1640 medium with 15% fetal bovine serum containing either IL-3 (5% WEHI) or IL-4 (0.0001–10 ng/ml). After 48 h of stimulation with either cytokine, cells were incubated with [3H]thymidine (2 μCi/ml) for 5 h, washed, and harvested onto glass filters with an automatic harvester (Skatron, Norway). [3H]Thymidine incorporation was measured using a Beckman 5500 scintillation counter. FDC-P2 cells treated with fetal bovine serum alone incorporated less than 0.1% of the counts incorporated in the presence of IL-3. EPO (1 unit/ml) standardization of mitogenic assays showed <5% variation among transfectants.
FACS Analysis of Cell Surface Antigen Expression.
FDC-P2 or FDC-P2 transfectants were untreated or treated with IL-4 (100 ng/ml) for 72 h. 1 × 106 cells were incubated for 60 min on ice with 2 μg of anti-I-Ad, anti-CD23, or anti-CD16/CD32 conjugated to FITC (PharMingen). Cells were washed with 5 ml of ice-cold PBS containing 0.1% sodium azide and resuspended in 100 μl of PBS, 0.1% sodium azide. Flow cytometry was performed and quantitated using a FACScan (Becton Dickinson).
Phosphotyrosine Analysis.
FDC-P2–Stat6 variant transfectants were starved in DMEM, 25 μM sodium orthovanadate for 3 h, stimulated with IL-4 (500 ng/ml) for 20 min, and washed once with cold PBS, 100 μM sodium orthovanadate. Whole cell lysates were prepared by solubilization in RIPA buffer, immunoprecipitated, and immunoblotted as described (26).
Electrophoretic Mobility Shift and Supershift Analysis.
FDC-P2 cells or FDC-P2–Stat6 transfectants were starved for 3 h. Cells were treated for the indicated time period with 500 ng/ml IL-4, washed once with cold PBS, 100 μM sodium orthovanadate, and solubilized in gel shift lysis buffer (26). For EMSA, 5 μg of whole cell lysate was incubated with the 32P-oligonucleotide ([32P]Iɛ) probe in 20 mM Hepes, pH 7.9, 40 mM KCl, 1 mM MgCl2, 100 μM EDTA, 500 μM DTT, 6.0% glycerol, 1 mg/ml BSA, and 100 μg/ml poly(dIdC) for 15 min and then fractionated on 0.22× TBE (100 mM Tris borate, pH 8.0, 2 mM EDTA), 4.5% acrylamide gels.
Luciferase Reporter Analysis.
Luciferase reporter plasmids were constructed using a 4 × FcγRI site (5′-GTATTTCCCAGAAAAGGAAC-3′) cloned into the NheI to BglII sites of pGL2-Basic (Promega) containing a TATA-box and minimal c-fos promoter (30). NIH 3T3 cells (1 × 106 cells/plate) overexpressing IL-4Rα were transiently transfected by calcium phosphate precipitation with 1.0–10.0 μg of each Stat6 variant and 5 μg of reporter plasmid. After 24 h, cells were starved overnight in serum-free DMEM and treated with or without IL-4 (500 ng/ml) for 6 h. Cell lysates were prepared, and luciferase activity was measured using a Lumat-LB luminometer (Berthold, Nashua, NH). Relative light units for each sample were normalized to protein concentration as measured by the method of Bradford.
RESULTS
Isolation of Human Stat6 Variant cDNAs and Comparison of Deduced Amino Acid Sequences.
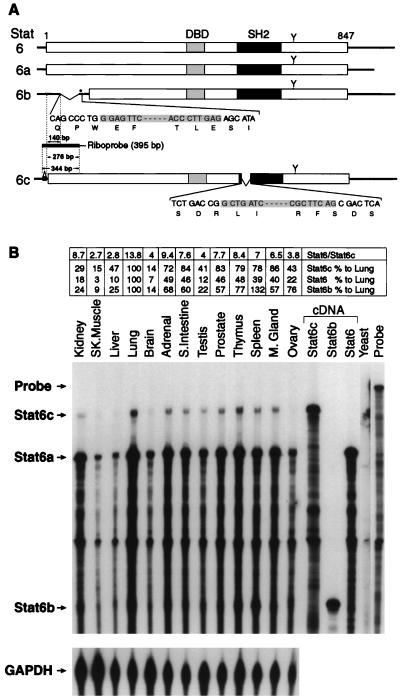
To investigate IL-4Rα-mediated signal transduction and transcriptional activation, we cloned wild-type human Stat6 and three Stat6 variant cDNAs (Stat6a, Stat6b, and Stat6c) from a human M426 embryonic lung fibroblast cDNA library. In comparison with Stat6 cDNA, Stat6a possessed a dramatically shorter 3′ noncoding region and a polyadenylation sequence juxtaposed to the termination codon (Fig. 1A). Differences among the Stat6, Stat6b, and Stat6c cDNA noncoding regions were noted primarily near the polyadenylation sequences. Stat6c also contained a 68-bp insertion upstream of the initiation codon. The Stat6 and Stat6a cDNA coding regions were identical, whereas Stat6b possessed a 139-bp deletion encompassing the last bp of codon 39 up to and including codon 86, resulting in the introduction of a stop codon. Stat6c contained an 84-bp deletion comprising the last bp of codon 536 up to and including the first 2 bp of codon 564.
Figure 1.
(A) Schematic diagram of Stat6, Stat6a, Stat6b, and Stat6c cDNAs. The Stat6-encoded cDNA (35) is illustrated as an open box. The putative DNA binding domain (DBD), SH2 domain, and a known tyrosine residue phosphorylated upon IL-4 treatment (Y) are indicated. Bases deleted in Stat6b and Stat6c are inclusively shaded. In Stat6b, an in-frame stop codon is designated by an asterisk. The riboprobe spanning the Stat6c insertion/Stat6b deletion used in the RNase protection assay and designed to protect hybrid products of 276, 140, and 344 bp for Stat6, Stat6b, and Stat6c, respectively, as described in Materials and Methods is depicted. (B) RNase protection assay of Stat6 variant RNAs isolated from human tissues. Total RNA prepared from several human tissues was used in the RNase protection assay. The protected products were analyzed on 6% acrylamide–urea gels, and the dried gel was exposed to x-ray film for 24 h with intensifying screens. The GAPDH antisense probe (97 bp) was run as an internal standard for each tissue and exposed for 3 h. Stat6, Stat6b, and Stat6c cDNAs were also subject to RNase protection and shown as controls. Quantitative comparison of Stat6 transcripts was performed by excision of each fragment, liquid scintillation counting, and normalization to the internal GAPDH antisense probe. The relative amount of each Stat6 variant transcript was quantitated relative to lung tissue (100%) for the same variant. Additionally, the relative ratio of Stat6/Stat6c transcript is presented.
The deduced amino acid sequence of each Stat6 variant was compared (Fig. 1A). The encoded gene products of Stat6 and Stat6a were identical, and because no significant biologic differences have been observed when these cDNAs were expressed in FDC-P2 or NIH 3T3 cells (data not shown), Stat6a herein will be referred to as Stat6. Stat6b possessed an NH2-terminal truncation of at least 110 amino acids due to the introduction of a stop codon and utilization of an internal initiation site, presumably Met111 (Fig. 1A). The deduced Stat6c amino acid sequence was identical to that of Stat6 except for a deletion of amino acid residues 537–564 within the SH2 domain of the molecule.
Detection and Quantitative Expression of Stat6 Variant mRNA in Human Tissues.
To determine whether the Stat6b and Stat6c cDNAs were authentic copies of mRNAs, reverse transcriptase (RT)-PCR analysis utilizing oligonucleotide primers designed to detect each variant was performed on RNA isolated from various human tissues. Primers proximal but upstream of the Stat6c noncoding insertion and adjacent but downstream of the Stat6b deletion amplified Stat6, Stat6b, or Stat6c as unique amplicons in multiple tissue samples (data not shown). A second RT-PCR analysis using primers flanking the SH2 domain further verified the existence of the Stat6c SH2 domain deletion. The identity of each amplicon was confirmed by cDNA sequencing (data not shown).
To investigate quantitative differences in the expression of each Stat6 variant transcript among the various human tissues, we performed a ribonuclease (RNase) protection assay. Individual Stat6 variant mRNAs were normalized to GAPDH mRNA for each tissue sample. As shown in Fig. 1B, transcripts encoding Stat6, Stat6b, or Stat6c were detected at varying levels in all tissues studied. Stat6b and Stat6c transcripts were expressed to the greatest extent in spleen and lung, respectively. Among the variants, Stat6 mRNA was consistently quantitated at two to four times the level of Stat6b mRNA depending on the tissue analyzed. Interestingly, the Stat6 transcript was expressed at 2.7 to 13.8 times the amount of the Stat6c transcript in the various tissues. We conclude that Stat6 variant mRNAs are differentially expressed in a variety of human tissues.
Effect of Stat6 Isoform Expression on IL-4-Induced [3H]Thymidine Uptake and Cell Surface Antigen Expression in FDC-P2 Cells.
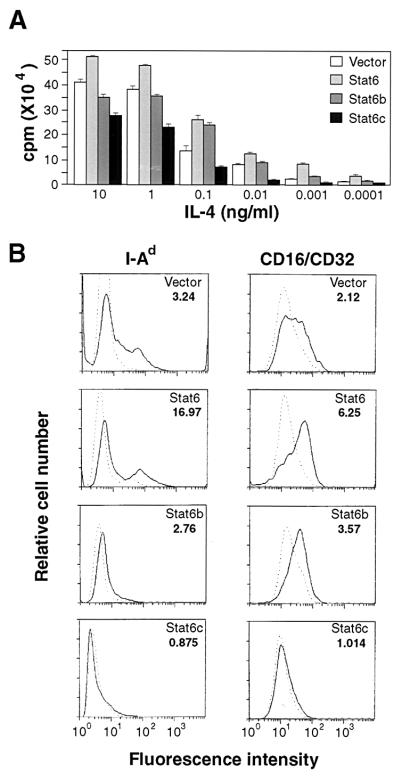
We next investigated the effect of each Stat6 isoform on IL-4-mediated proliferation by expressing each gene product in FDC-P2 cells and examining IL-4-induced DNA synthesis. As shown in Fig. 2A, IL-4 (10 ng/ml) induced 25% greater [3H]thymidine incorporation in FDC-P2 cells overexpressing Stat6 (FDC-P2–Stat6) than similarly treated FDC-P2 empty vector transfectants. DNA synthesis induced by IL-4 in FDC-P2 cells overexpressing Stat6b (FDC-P2–Stat6b) was similar to that observed in control FDC-P2 cells. In contrast, treatment with saturating concentrations of IL-4 (10 ng/ml) resulted in reduced [3H]thymidine incorporation by at least 30% in FDC-P2–Stat6c. Expression of Stat6c inhibited IL-4-mediated [3H]thymidine incorporation by 50–70% at lower IL-4 concentrations when compared with empty vector transfected cells. Thus, expression of Stat6 enhances, whereas Stat6c inhibits, IL-4-induced DNA synthesis in FDC-P2 transfectants.
Figure 2.
(A) Effect of Stat6 isoform expression on IL-4-induced mitogenesis in FDC-P2 cells. FDC-P2–Stat6 (light gray bar), FDC-P2–Stat6b (medium gray bar), FDC-P2–Stat6c (black bar), or control FDC-P2 (open bar) transfectants were treated with IL-4 at the indicated concentration. Each bar represents the average of three determinations ± SEM. Background uptake was typically less than 2000 cpm for each transfectant. (B) IL-4 induction of cell surface antigen expression on FDC-P2 or FDC-P2–Stat6 isoform transfectants. Cell surface staining of FDC-P2–Stat6, FDC-P2–Stat6b, FDC-P2–Stat6c, or control FDC-P2 transfectants was performed with FITC-conjugated anti-I-Ad or anti-CD16/32 after 72 h in the presence (solid line) or absence (dotted line) of IL-4. Fold induction (fluorescence intensity in the presence of IL-4/fluoresence intensity in the absence of IL-4) is presented in the upper right corner of each histogram.
IL-4 has pronounced effects on the cell surface expression of I-Ad (MHC class II) molecules and Fc receptors (4, 5). In human monocytes, IL-4 has been shown to induce Stat6 binding to the FcγRI promoter (13, 14). Therefore, we analyzed whether expression of the different Stat6 isoforms had any effect on the levels of IL-4-inducible cell surface antigens in FDC-P2 cells by flow cytometry. As expected, I-Ad and CD16/CD32 cell surface staining was increased in IL-4-treated FDC-P2 cells (Fig. 2B). Enhanced I-Ad and CD16/CD32 staining was observed in FDC-P2–Stat6 transfectants. FDC-P2–Stat6b transfectants also showed up-regulation of IL-4-induced I-Ad and CD16/CD32 expression but to a much lesser extent. In contrast, the ability of IL-4 to induce I-Ad and CD16/CD32 molecules was abolished in FDC-P2–Stat6c transfectants. Similar effects on CD23 molecules were also observed (data not shown). These results strongly suggest that Stat6 plays a significant role in mediating IL-4-induced I-Ad, CD16/CD32, and CD23 cell surface expression in FDC-P2 cells. Moreover, Stat6c has potent dominant inhibitory effects on the ability of IL-4 to mediate up-regulation of these cell surface antigens.
Effects of IL-4 Stimulation on Tyrosine Phosphorylation of Stat6 Isoforms Expressed in FDC-P2 Cells.
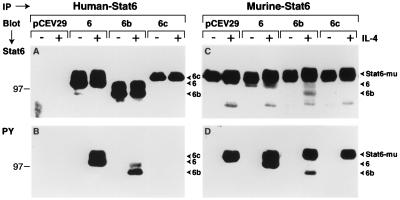
To gain insight into the mechanistic basis by which Stat6b and Stat6c might be exerting effects on IL-4-mediated proliferation and functional responses, we analyzed the expression and tyrosine phosphorylation of each Stat6 isoform in the FDC-P2 transfectants. Expression was first examined using anti-human Stat6 serum that does not recognize murine Stat6. Whole cell lysates from untreated or IL-4-treated FDC-P2 or FDC-P2–Stat6 isoform transfectants were immunoprecipitated with the anti-human Stat6 serum and subjected to SDS/PAGE; resolved proteins were subsequently immunoblotted with anti-human Stat6 serum. As shown in Fig. 3A, a 100-kDa species was readily observed in immunoprecipitates from FDC-P2 cell lysates overexpressing human Stat6. Stat6b and Stat6c were detected as 95- and 102-kDa species in FDC-P2–Stat6b or FDC-P2–Stat6c immunoprecipitates, respectively. Stat6 and Stat6b were expressed at similar levels, whereas Stat6c was expressed at levels 3-fold lower than that of either of the other isoforms. No human Stat6 was detected in immunoprecipitates from FDC-P2 cells transfected with the pCEV29 vector alone (Fig. 3A).
Figure 3.
Expression and IL-4-induced tyrosine phosphorylation of Stat6 isoforms in FDC-P2 cells. (A and B) Detection of human Stat6. FDC-P2 transfectants were starved as indicated and then stimulated with IL-4 (500 ng/ml) for 20 min. Whole cell lysates containing equivalent amounts of protein were then immunoprecipitated (IP) with anti-human Stat6 serum and subjected to SDS/PAGE. Resolved proteins were transferred to Immobilon-P membranes and immunoblotted (Blot) with anti-human Stat6 serum (A) or anti-phosphotyrosine (PY) (B) (26). (C and D) Detection of mouse/human Stat6. FDC-P2 or FDC-P2–Stat6 isoform transfectants were treated with IL-4, immunoprecipitated with anti-Stat6 serum, and subjected to SDS/PAGE. Immobilon-P membranes were immunoblotted with anti-Stat6 serum (C) or anti-phosphotyrosine (D). Bound primary antibody was detected by anti-rabbit or anti-mouse antibody conjugated to horseradish peroxidase followed by enhanced chemiluminescence (Amersham).
To determine whether each Stat6 isoform could be activated by IL-4, we first examined whether these Stat6 species became tyrosine phosphorylated in response to IL-4 treatment. Whole cell lysates from untreated or IL-4-treated FDC-P2–Stat6 transfectants were immunoprecipitated with anti-human Stat6 serum and subjected to SDS/PAGE; resolved proteins were immunoblotted with antiphosphotyrosine antibody. As shown in Fig. 3B, 100- and 95-kDa tyrosine-phosphorylated species were readily detected in IL-4-treated FDC-P2–Stat6 and FDC-P2–Stat6b transfectants, respectively. Stat6 tyrosine phosphorylation was greater than that of Stat6b, and no Stat6c tyrosine phosphorylation was detected.
We next asked whether Stat6 isoform overexpression would affect endogenous murine Stat6 phosphorylation. To assay endogenous Stat6 activation, lysates were immunoprecipitated with an anti-Stat6 serum that recognizes both murine and human Stat6. As shown in Fig. 3C, similar levels of murine Stat6 were observed in FDC-P2 cells and FDC-P2 isoform transfectants. Human Stat6 isoform expression was detected in a manner consistent with that observed utilizing anti-human Stat6 serum. Similar levels of endogenous murine Stat6 tyrosine phosphorylation were detected in native FDC-P2 cells as well as in Stat6 and Stat6b isoform transfectants in response to IL-4 treatment (Fig. 3D). However, Stat6c expression slightly, but consistently, diminished (14.7 ± 2.1%) IL-4-induced endogenous murine Stat6 tyrosine phosphorylation (Fig. 3D). Human Stat6 and Stat6b, but not Stat6c, tyrosine phosphorylation was also detected utilizing this antiserum, confirming our previous results (see Fig. 3B). These results argue that Stat6 and Stat6b, but not Stat6c, are tyrosine phosphorylated in response to IL-4 and that IL-4-mediated tyrosine phosphorylation of endogenous murine Stat6 is only partially reduced by the expression of the human Stat6c isoform.
Differential DNA Binding Activity and Transcriptional Activation of Stat6 Isoforms.
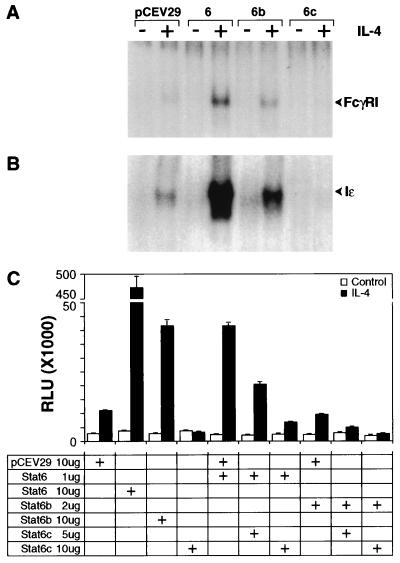
Stat6 has been shown to bind with high affinity to a region within the FcγRI promoter (13, 14). Its DNA binding capacity can be readily distinguished from that of the other STATs by its ability to bind a GAS-like element found in the Ig germ line ɛ promoter of the IL-4-responsive human Cɛ gene (Iɛ). To determine whether expression of the three human Stat6 isoforms affected IL-4-induced FcγRI and Iɛ promoter binding activity, FDC-P2 transfectants were stimulated for 20 min. Whole cell extracts were prepared and assayed for the induction of [32P]FcγRI or [32P]Iɛ DNA binding activity by EMSA (Fig. 4 A and B). Although extracts from untreated FDC-P2 did not contain any FcγRI or Iɛ binding activity, IL-4 treatment led to rapid induction of FcγRI and Iɛ binding. DNA binding activity was confirmed by promoter competition studies and supershift analysis utilizing two independent Stat6 antisera (data not shown). IL-4-induced [32P]FcγRI or [32P]Iɛ binding activity observed in lysates from FDC-P2 transfectants overexpressing human Stat6 was identical to that detected in lysates from IL-4-stimulated parental FDC-P2 cells, albeit at greatly increased levels. Overexpression of Stat6b also led to enhanced DNA binding but to a lesser extent than that observed in FDC-P2–Stat6 lysates. Stat6c did not possess detectable DNA binding activity. In contrast, it inhibited IL-4-induced endogenous murine Stat6 FcγRI and Iɛ DNA binding activity by greater than 80% (Fig. 4 A and B).
Figure 4.
(A and B) IL-4-induced [32P]FcγRI and [32P]Iɛ binding to Stat6 isoforms. FDC-P2 or FDC-P2–Stat6 isoform transfectants were starved as indicated and then untreated or treated with IL-4 (500 ng/ml) for 20 min. Whole cell lysates were incubated with [32P]FcγRI or [32P]Iɛ for 15 min and assayed by EMSA (26). Binding activity was visualized by autoradiography. (C) Effect of Stat6 isoforms on FcγRI luciferase reporter transcriptional activation in the presence (solid bar) or absence (open bar) of IL-4. The relative luciferase activity of IL-4-stimulated NIH 3T3 cells (solid bar) is compared with unstimulated cells (open bar). The amount of transfected cDNA is indicated below. Endogenous Stat6 activation was identical in control pCEV29 transfected (10 μg) or nontransfected NIH 3T3 cells. Values shown are the mean ± SEM of three determinations.
To gain further insights concerning the role of each Stat6 variant in mediating IL-4-induced transcriptional activation, we utilized the FcγRI promoter coupled to a luciferase reporter containing the minimal fos promoter. As shown in Fig. 4C, IL-4 treatment of NIH 3T3–Stat6 transfectants resulted in a 220-fold induction of the FcγRI luciferase reporter compared with a 5-fold induction observed in IL-4-treated control NIH 3T3–pCEV29 empty vector transfectants. NIH 3T3–Stat6b transfectants exhibited a 20-fold induction.
The mechanism by which Stat6c exerted inhibitory effects was also investigated using the FcγRI–luciferase reporter (Fig. 4C). In contrast to the enhanced transcription observed with Stat6, IL-4-induced luciferase activity was completely abolished in NIH 3T3–Stat6c transfectants. Indeed, Stat6c exerted a dominant negative effect on transcriptional activation even when transfected at a concentration much less than that of Stat6 (data not shown). The dose-dependent inhibition of Stat6-induced transcriptional activation by Stat6c further suggests that Stat6c expression levels predicate transcriptional outcome (Fig. 4C).
Effect of Stat6c on Endogenous Stat6 Dimerization.
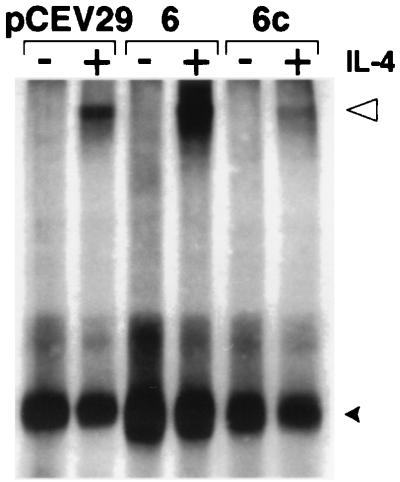
To elucidate the molecular mechanism of Stat6c’s potent transcriptional inactivation, we considered several possibilities. Because endogenous Stat6 tyrosine phosphorylation is only partly diminished by Stat6c, inhibition of IL-4-induced endogenous Stat6 association with IL-4Rα or JAK activation seemed unlikely. Moreover, Stat6c does not directly bind FcγRI or Iɛ promoter elements, making competitive transcriptional inactivation improbable. Therefore, we performed cross-linking studies to investigate whether Stat6c might effect endogenous Stat6 dimerization. Whole cell lysates from IL-4-treated FDC-P2 cells or Stat6 and Stat6c transfectants were incubated with disuccinimidyl glutarate. Immunoprecipitation followed by immunoblotting with anti-Stat6 serum revealed the presence of a Stat6 dimer in IL-4-treated lysates (Fig. 5). FDC-P2–Stat6 transfectants treated with IL-4 exhibited greatly increased levels of the Stat6 dimer when compared with the endogenous Stat6 in the FDC-P2 control cells. Strikingly, endogenous Stat6 dimerization was reduced by greater than 60% in IL-4-treated FDC-P2–Stat6c transfectants in comparison with the FDC-P2 control cells (Fig. 5). Thus, the molecular basis of transcriptional inactivation by Stat6c seems to be due to the suppression of endogenous Stat6 dimer formation.
Figure 5.
Effect of Stat6c on IL-4-induced dimerization of murine Stat6. FDC-P2, FDC-P2–Stat6, or FDC-P2–Stat6c transfectants were incubated in the presence or absence of IL-4. Disuccinimidyl glutarate cross-linking of Stat6 dimers in whole cell lysates was assayed by immunoprecipitation followed by immunoblotting with anti-Stat6 serum (13). Arrows indicate each Stat6 variant. An open arrowhead marks the position of the apparent Stat6 dimer.
DISCUSSION
In the present study, we have investigated the biologic properties of two Stat6 homologs, Stat6b and Stat6c, that were isolated from a human embryonic lung fibroblast cDNA library. Stat6b and Stat6c possessed an NH2-terminal truncation or a SH2 domain deletion, respectively. Although equivalent expression levels of human Stat6 and Stat6b were detected after stable transfection of murine FDC-P2 cells, IL-4-induced MHC class II and Fc receptor cell surface expression, promoter binding, and transcriptional activation were attenuated in Stat6b transfectants relative to those overexpressing Stat6. Furthermore, expression of human Stat6c unexpectedly abolished or dramatically reduced these IL-4-mediated events, most likely through the inhibition of endogenous Stat6 dimerization. Thus, each Stat6 isoform apparently acts to differentially control the extent of IL-4-induced functional responses and cellular proliferation.
Stat6b and Stat6c are naturally occurring isoforms of Stat6. However, differential splicing is not unprecedented within the STAT family (31). Stat1, -3, -5A, and -5B genes have been demonstrated to encode variants that arise from the differential mRNA splicing of exon(s) encoding COOH-terminal domains (31–33). COOH-terminally processed Stat1β failed to activate transcription (32). COOH-terminally truncated Stat5A and B homologs and, in some instances, Stat3β were found to be dominant negative regulators of transcription (31, 33). However, no dominant negative STAT molecule other than Stat6c has been isolated that contains a deletion of critical amino acids within the SH2 domain. Furthermore, STAT variants have yet to be reported that, like Stat6b, possess deletion of exon(s) encoding NH2-terminal domains.
Recent structure–function analyses of Stat1 and Stat4 suggest at least two potential functions for STAT NH2-terminal domains. Two studies indicated that the NH2-terminal domain was necessary for cooperative binding (34) and activation of the interferon promoter (35). However, a third report provided evidence that the NH2-terminal domain of Stat1 was required for its inactivation by an unknown phosphatase and that lack of phosphatase regulation resulted in enhanced activity (36). The reduction in promoter binding and transcriptional activation of Stat6b observed in the present study is consistent with a role for the NH2-terminal domain of Stat6 in cooperative DNA binding and transcriptional activation.
We have demonstrated that a naturally occurring STAT molecule containing an SH2 domain deletion can function in a dominant negative manner. Previous findings would predict that deletion of this critical region would block dimer formation, resulting in the generation of an inactive molecule. We were also unable to detect tyrosine phosphorylation of Stat6c in response to IL-4 stimulation, further implying that this protein should be nonfunctional. However, Stat6c was shown to consistently behave as a dominant negative species. In this regard, expression of human Stat6c was demonstrated to inhibit IL-4-mediated dimerization of endogenous murine Stat6. Although the precise mechanism by which Stat6c inhibits dimerization remains elusive, it does not seem to dramatically decrease endogenous Stat6 tyrosine phosphorylation in response to IL-4 stimulation. Therefore, inhibition of IL-4Rα/Stat6 association or JAK activity seems to be unlikely. Moreover, Stat6c did not bind Iɛ or FcγRI promoter elements, making transcriptional inactivation by a direct competitive mechanism improbable. In this regard, we have yet to detect direct association of Stat6c with endogenous Stat6 in a dimeric complex using epitope-tagged Stat6c (data not shown).
The presence of identical amino acid residues within each isoform except for the deleted regions precludes development of antisera specific for any one isoform. However, RNase protection and RT-PCR analysis that distinguishes among the multiple Stat6 variants at the mRNA level has suggested that each variant may be differentially regulated in a tissue-specific manner. In addition, expression of each Stat6 isoform may result in varying degrees of IL-4-mediated transcriptional regulation and/or interaction with other signaling molecules. The latter possibility is likely for Stat6c, which deletes a critical arginine residue implicated by x-ray crystallographic data in phosphotyrosine–SH2 domain interactions (37). In any case, the attenuated and dominant negative Stat6 isoforms are likely to play important roles in differential responsiveness to IL-4 by governing the spatial and temporal expression of transcriptionally activated target molecules.
Acknowledgments
We thank Drs. Matt Lorenzi, Marcio Chedid, and Ling-Mei Wang for helpful discussions and Dr. Toru Miki for providing the human fibroblast cDNA library. We also acknowledge Paul Kriebel, Nelson Ellmore, Veena Kapoor, Mary May, and Ryan O’Leary for technical assistance.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: IL, interleukin; FITC, fluorescein isothiocyanate; MHC, major histocompatibility complex; EMSA, electrophoretic mobility shift assay; EPO, erythropoietin; RT, reverse transcriptase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
References
- 1.Paul W E. Blood. 1991;77:1859–1870. [PubMed] [Google Scholar]
- 2.Keegan A D, Nelms K, Wang L-M, Pierce J H, Paul W E. Immunol Today. 1994;15:423–432. doi: 10.1016/0167-5699(94)90272-0. [DOI] [PubMed] [Google Scholar]
- 3.Defrance T, Vanbervliet B, Aubry J P, Takebe Y, Arai N, Miyajima A, Yokata T, Lee T, Arai K, de Vries J E, Banchereau J. J Immunol. 1987;139:1135–1141. [PubMed] [Google Scholar]
- 4.Noelle R, Krammer P H, Ohara J, Uhr J W, Vitetta E S. Proc Natl Acad Sci USA. 1984;81:6149–6153. doi: 10.1073/pnas.81.19.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conrad D H, Waldschmidt T, Lee W T, Rao M, Keegan A D, Noelle R J, Lynch R G, Kehry M R. J Immunol. 1987;139:2290–2296. [PubMed] [Google Scholar]
- 6.Ohara J, Paul W E. Proc Natl Acad Sci USA. 1988;85:8221–8225. doi: 10.1073/pnas.85.21.8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Andres B, Cardaba B, del Pozo V, Martin-Orozco E, Gallardo S, Tramon P. Immunology. 1994;83:155–160. [PMC free article] [PubMed] [Google Scholar]
- 8.Coffman R L, Ohara J, Bond M W, Carty J, Zlotnick E, Paul W E. J Immunol. 1986;136:4538–4541. [PubMed] [Google Scholar]
- 9.Vitetta E S, Ohara J, Myers C, Layton J, Krammer P H, Paul W E. J Exp Med. 1985;162:1726–1732. doi: 10.1084/jem.162.5.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seder R A, Paul W E, Davis M M, Fazekas de St G. Proc Natl Acad Sci USA. 1991;88:2835–2839. doi: 10.1073/pnas.88.7.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Izuhara K, Harada N. J Biol Chem. 1993;268:13097–13102. [PubMed] [Google Scholar]
- 12.Wang L-M, Keegan A D, Paul W E, Heidaran M A, Gutkind J S, Pierce J H. EMBO J. 1992;11:4899–4908. doi: 10.1002/j.1460-2075.1992.tb05596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou J, Schindler U, Henzel W J, Ho T C, Brasseur M, McKnight S L. Science. 1994;165:1701–1706. doi: 10.1126/science.8085155. [DOI] [PubMed] [Google Scholar]
- 14.Kotanides H, Reich N C. Science. 1993;262:1265–1267. doi: 10.1126/science.7694370. [DOI] [PubMed] [Google Scholar]
- 15.Johnston J A, Kawamura M, Kirken R A, Chen Y Q, Blake T B, Shibuya K, Ortaldo J R, McVicar D W, O’Shea J J. Nature (London) 1994;370:151–153. doi: 10.1038/370151a0. [DOI] [PubMed] [Google Scholar]
- 16.Witthuhn B A, Silvennoinen O, Miura O, Lai K S, Cwik C, Liu E T, Ihle J N. Nature (London) 1994;370:153–157. doi: 10.1038/370153a0. [DOI] [PubMed] [Google Scholar]
- 17.Wang L M, Myers M G, Sun X J, Aaronson S A, White M, Pierce J H. Science. 1993;261:1591–1594. doi: 10.1126/science.8372354. [DOI] [PubMed] [Google Scholar]
- 18.Wang L M, Keegan A D, Li W, Lienhard G E, Pacini S, Gutkind J S, Myers M G, Sun X J, White M F, Aaronson S A, Paul W E, Pierce J H. Proc Natl Acad Sci USA. 1993;90:4032–4036. doi: 10.1073/pnas.90.9.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quelle F W, Shimoda K, Thierfelder W, Fischer C, Kim A, Ruben S M, Cleveland J L, Pierce J H, Keegan A D, Nelms K, Paul W E, Ihle J N. Mol Cell Biol. 1995;15:3336–3343. doi: 10.1128/mcb.15.6.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schindler C, Kashleva H, Pernis A, Pine R, Rothman P. EMBO J. 1994;13:1350–1356. doi: 10.1002/j.1460-2075.1994.tb06388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan J J, McReynolds L J, Keegan A, Wang L H, Garfein E, Rothman P, Nelms K, Paul W E. Immunity. 1996;4:123–132. doi: 10.1016/s1074-7613(00)80677-9. [DOI] [PubMed] [Google Scholar]
- 22.Pernis A, Witthuhn B, Keegan A D, Nelms K, Garfein E, Ihle J N, Paul W E, Pierce J H, Rothman P. Proc Natl Acad Sci USA. 1995;92:7971–7975. doi: 10.1073/pnas.92.17.7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schindler U, Wu P, Rothe M, Brasseur M, McKnight S L. Immunity. 1995;2:689–697. doi: 10.1016/1074-7613(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 24.Darnell J J E, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 25.Malabarba M G, Rui H, Deutsch H H, Chung J, Kalthoff F S, Farrar W L, Kirken R A. Biochem J. 1996;319:865–872. doi: 10.1042/bj3190865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel B K R, Wang L M, Lee C C, Taylor W G, Pierce J H, LaRochelle W J. J Biol Chem. 1996;271:22175–22182. doi: 10.1074/jbc.271.36.22175. [DOI] [PubMed] [Google Scholar]
- 27.Kaplan M H, Schindler U, Smiley S T, Grusby M J. Immunity. 1996;4:1–20. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 28.Shimoda K, Deursen J, Sangster M Y, Sarawar S R, Carson R T, Tripp R A, Chu C, Quelle F W, Nosaka T, Vignali D A A, Doherty P C, Grosveld G, Paul W E, Ihle J N. Nature (London) 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 29.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Nature (London) 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 30.Michieli P, Li W, Lorenzi M V, Miki T, Zakut R, Givol D, Pierce J H. Oncogene. 1996;12:775–784. [PubMed] [Google Scholar]
- 31.Caldenhoven E, Dijk T, Solari R, Armstrong J, Raaijmakers J A M, Lammers J J, Koenderman L, Groot R P. J Biol Chem. 1996;271:13221–13227. doi: 10.1074/jbc.271.22.13221. [DOI] [PubMed] [Google Scholar]
- 32.Kuang A A, Novak K D, Kang S M, Bruhn K, Lenardo M J. Mol Cell Biol. 1993;13:2536–2545. doi: 10.1128/mcb.13.4.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D, Stravopodis D, Teglund S, Kitazawa J, Ihle J N. Mol Cell Biol. 1996;16:6141–6148. doi: 10.1128/mcb.16.11.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vinkemeier U, Cohen S L, Moarefi I, Chait B T, Kuriyan J, Darnell J E., Jr EMBO J. 1996;15:5616–5626. [PMC free article] [PubMed] [Google Scholar]
- 35.Xu X, Sun Y, Hoey T. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 36.Shuai K, Liao J, Song M M. Mol Cell Biol. 1996;16:4932–4941. doi: 10.1128/mcb.16.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waksman G, Shoelson S E, Pant N, Cowburn D, Kuriyan J. Cell. 1993;72:779–790. doi: 10.1016/0092-8674(93)90405-f. [DOI] [PubMed] [Google Scholar]