Abstract
Mutually catalytic sets of simple organic molecules have been suggested to be capable of self-replication and rudimentary chemical evolution. Previous models for the behavior of such sets have analyzed the global properties of short biopolymer ensembles by using graph theory and a mean field approach. In parallel, experimental studies with the autocatalytic formation of amphiphilic assemblies (e.g., lipid vesicles or micelles) demonstrated self-replication properties resembling those of living cells. Combining these approaches, we analyze here the kinetic behavior of small heterogeneous assemblies of spontaneously aggregating molecules, of the type that could form readily under prebiotic conditions. A statistical formalism for mutual rate enhancement is used to numerically simulate the detailed chemical kinetics within such assemblies. We demonstrate that a straightforward set of assumptions about kinetically enhanced recruitment of simple amphiphilic molecules, as well as about the spontaneous growth and splitting of assemblies, results in a complex population behavior. The assemblies manifest a significant degree of homeostasis, resembling the previously predicted quasi-stationary states of biopolymer ensembles (Dyson, F. J. (1982) J. Mol. Evol. 18, 344–350). Such emergent catalysis-driven, compositionally biased entities may be viewed as having rudimentary “compositional genomes.” Our analysis addresses the question of how mutually catalytic metabolic networks, devoid of sequence-based biopolymers, could exhibit transfer of chemical information and might undergo selection and evolution. This computed behavior may constitute a demonstration of natural selection in populations of molecules without genetic apparatus, suggesting a pathway from random molecular assemblies to a minimal protocell.
The potential prebiotic synthesis of diverse organic compounds has been previously demonstrated by experiments (1–3). Yet, bridging the gap between organosynthesis and the emergence of self-replication and inheritance has remained a major challenge (4, 5). One school of thought centers on individual molecules endowed with a capacity for self-replication, a feat that often necessitates careful engineering of complex chemical structures (6–9). Mathematical analyses (10–12) and experimental testing (13–16) of in vitro evolution focus on nucleic acid polymers, whose de novo abiotic generation is considered by many as improbable (17).
A fundamentally different approach has envisaged primordial self-replication as the collective property of ensembles of relatively simple molecules, interconnected by networks of mutually catalytic interactions (4, 18–26). Within such assemblies, molecules may be held together by noncovalent interactions (23, 27–29). The experimental demonstration that amphiphilic assemblies display self-replication behavior (30–32) has led to increasing theoretical interest in this approach (32–35).
Critics have argued that noncovalent assemblies might lack the capacity of storing and transferring information. Therefore, they could not undergo chemical selection and evolution in the absence of informational biopolymers (13). Yet, concrete models of self-sustaining metabolism without encoding biopolymers have been explored (4, 20–22, 36). One of these (4, 20), a quantitative embodiment of Oparin's prebiotic evolution scenario (37), has analyzed the homeostatic behavior of an ensemble of molecules through a state vector that undergoes step-wise changes. The time-dependent distribution of molecular populations was computed by using a transition probability matrix. The existence of quasi-stationary (homeostatic) states (QSSs) was formally related to the average catalytic properties of the molecular constituents through a mean field approximation.
We use here computer simulations based on the Graded Autocatalysis Replication Domain (GARD) model (38, 39) to analyze the kinetic behavior of mutually catalytic heterogeneous amphiphilic assemblies. Under nonequilibrium conditions, these are shown to spontaneously attain QSSs with high compositional information and a capacity to undergo self-replication and mutation-like changes.
Results
Compositional Assemblies.
The compositional state of a noncovalent molecular assembly is defined by an NG-dimensional vector n, whose components ni are the internal counts of different molecular types (NG is the molecular repertoire size). The time-dependent change of the composition is dictated by
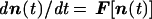 |
1 |
where F is a function governed by the endogenous chemical kinetics, in analogy to previously explored formalisms for multicomponent systems (20, 40, 41).
For two compositional assemblies, p and q, a degree of similarity is defined as the scalar product
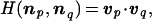 |
2 |
where v = n/|n| is a normalized compositional vector (|n| is the norm of n). For any process leading from np to nq, H represents the time-related compositional change, with H = 1 denoting perfect homeostasis, and H = 0 indicating orthogonality, i.e., a large change of compositional state.
In the realm of large assemblies, i.e., N>>NG (N = Σni is the assembly size), all randomly formed assemblies have nearly identical n, and H≈1 is trivial. The analyses below therefore are performed under small assembly and large repertoire conditions (N<NG) (cf. refs. 42 and 43) whereby randomly formed assemblies are nearly orthogonal. In this context it is helpful to quantify an assembly's information content in a way analogous to that used for ensembles of biopolymer sequences (12, 41). We use a measure of compositional bias I, related to the improbability of spontaneous formation, and therefore also to compositional entropy:
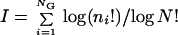 |
3 |
A Kinetic Model.
We consider the behavior of diverse lipid-like amphiphilic molecules of the kinds formed in the laboratory or under simulated prebiotic conditions (2, 3, 35, 44). These will spontaneously aggregate in an aqueous medium to form molecular assemblies governed by hydrophobic interactions (27, 45). A central assumption of the present model is that compounds already present within an assembly may enhance the rate of joining and leaving of new molecular species. For large NG values, this will result in a complex mutually catalytic network (cf. refs. 24, 46, and 47). Catalyzed joining may be akin to catalyzed “flipping” of molecules between two leaflets of a lipid bilayer (48). In future embodiments of the present model, assembly formation could involve catalyzed covalent changes, comparable to those observed for accelerated vesicle formation (31, 49) and other lipid catalysis reactions (50–52).
For computer simulations of the dynamics of such molecular assemblies, we use chemical kinetics rules based on the previously proposed Graded Autocatalysis Replication Domain (GARD) model (38, 39). Accordingly, the time-dependent changes in the composition of an assembly may be described by NG differential equations (Eq. 1). The function F could in principle be deduced by ab initio computations for the interaction within each molecular pair, by using, for example, force field equations (53–55). However, because the modeled system may contain thousands of different compounds, for which a detailed knowledge is lacking, it is more advisable to use a statistical approach (56). Such an analysis may be based on a previously proposed probabilistic formalism for ligand-receptor interactions (57) as described (34, 38, 56, 58).
The minimal kinetic model pursued here assumes that the rate of energetically favorable entry of an extraneous molecular species into a preformed assembly is enhanced to some degree (even very small) in a concentration-dependent way by every type of molecule present inside the assembly. Thus the function F assumes the specific form
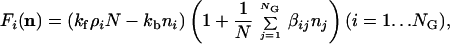 |
4 |
where kf and kb are, respectively, the basal forward and backward reaction rates (with kf>kb signifying spontaneous aggregation). Although more elaborate kinetic models exist for micelle formation (32, 54, 59), we use here a highly simplified formalism, which assumes that a compound i joins an assembly with a probability proportional to its external free concentration ρi and to the total size of the assembly N. The ensuing logistic growth behavior (60) may be shown to be equivalent to that of the original Graded Autocatalysis Replication Domain (GARD) model (39). Mutual rate enhancement exerted by molecule type j on molecule type i is represented by the element βij of an NG × NG matrix. The choice of rate enhancement distribution characteristics is guided by experimental results for lipid catalysis (52).
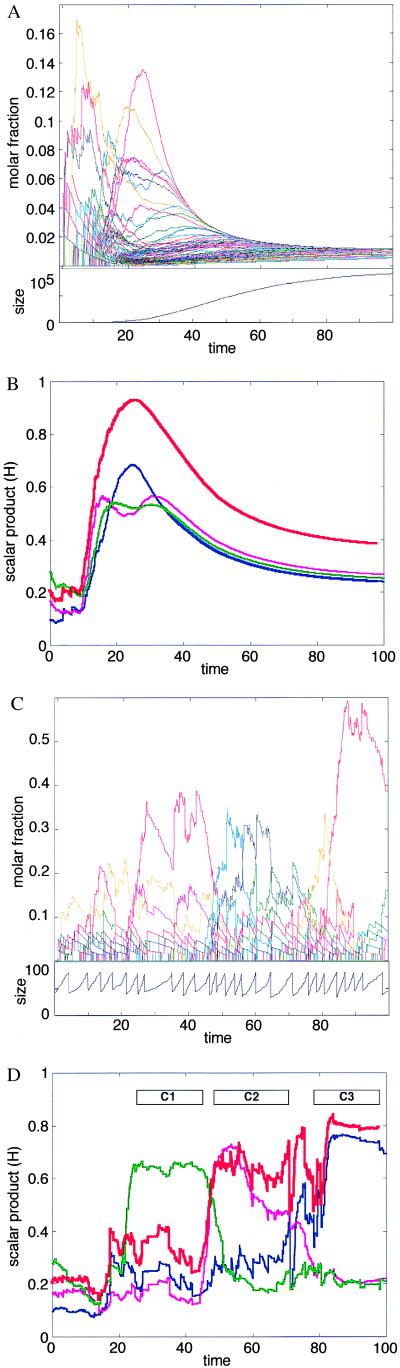
The typical simulated behavior of a molecular assembly, as dictated by Eq. 4, is shown in Fig. 1 A and B. It may be seen that if an assembly is allowed to form and grow in a finite pool of compounds, i.e., in a closed system, the molar fractions of some components increase temporarily. Thus, the compositional vector n as well as the similarity value H trace a complex trajectory, transiently passing through highly idiosyncratic compositions, but finally decaying to an equimolar equilibrium composition. Although the transient composition is kinetically dictated by the values of the mutual rate enhancement factors βij, the final state is related only to the ratio of the basal rate constants, i.e., to thermodynamic equilibrium parameters.
Figure 1.
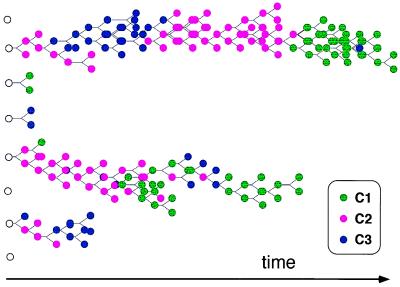
Results of computer simulations for the kinetics of spontaneous aggregation in amphiphilic assemblies. An initial assembly was seeded randomly by choosing Nmin individual molecules out of a pool containing nTOT molecules of each of NG possible types. Here NG = 100, nTOT = 1,000, Nmin = 40. A Monte Carlo type method was used for performing discrete stochastic changes in the assembly, as dictated by Eq. 4, and by using methods as described (68). At each time step, the change in the count of molecules of each species (Δni) was calculated by random sampling from a Poisson distribution with average Fi(n)Δt, where Δt defines the time scale of the process. In the results presented here Δt = 0.05 sec. The logarithms of the rate enhancement factors βij were sampled from a normal distribution [used as a continuous approximation for the binomial distribution (57)] with an average μ = −4 and a SD σ = 4. These numerical parameter values were selected so that the resultant rate enhancement would conform with an experimentally based distribution derived from multiple reported data sets for lipid micelle catalysis (52). The molar fractions of free molecules of kind i is defined as ρi = (nTOT − ni)/NG. For the forward and backward reaction rates we use the values kf = 10-2 and kb = 10-5 sec-1 (for simplicity, all molecules are assumed to have identical uncatalyzed rate constants, and to differ only in their mutual rate enhancement properties). The program was written for matlab, version 5 (Mathworks, Natick, MA). The detailed results of the computer simulations are available on line at http://ool.weizmann.ac.il/PNAS2000. (A) The time dependence of the molar fraction ni/N for each species i, in an assembly whose growth is limited only by the finite supply of molecules (Upper). Because all of the species are thermodynamically equivalent, they reach the same molar fraction after a certain time. In the transient the random network of mutual rate enhancements determines significant and nontrivial differences among the various molecular types. The change of assembly size N with time is shown (Lower): the initial increase is nearly exponential, leveling off to zero growth as the external molecular supply is exhausted and equilibrium is reached. (B) The change of the similarity values H (Eq. 2) with time. The red line depicts H values relative to the asymptotic composition n* reached by an assembly that forms and expands indefinitely with unlimited supply of all molecular species ni. n* represents the asymptotic steady-state solution of Eq. 4. The three thin lines measure the similarity to the three main composomes of Fig. 3. (C) The time-dependent behavior of a system similar to that in A and B, but with an added process of splitting the assembly when its size reaches 2Nmin. The splitting is performed by randomly dividing such an assembly into two daughter assemblies of size ≈Nmin each. The count of each molecular species in a daughter assembly is sampled from a binomial distribution with ni trials and probability 0.5. In addition, a constant population condition (11) is implemented by decomposing one randomly selected assembly after each splitting event. This is done by breaking the chosen assembly into its monomeric constituents and replenishing the external species concentrations ρi. The change of assembly size N with time, indicating the periodical splits, is shown (Lower). (D) The analysis of H relative to specific compositions for the same simulation shown in C. The initial random assembly proceeds through relatively abrupt transitions from one composome to another. Composomes are marked as C1 (green), C2 (magenta), and C3 (blue) (see Fig. 3).
QSSs.
Next, we explored the behavior of the system when it is kept far from thermodynamic equilibrium. Under conditions of unlimited supply and unlimited growth, the molar fractions ni/N reach a single nontrivial asymptotic stationary state n* (see Fig. 1 B and D). This is observed also for the linear equation dn/dt = Bn, with Bij = kfρi(1 + βij), obtained as an approximation from Eq. 4 by assuming kb = 0 and ρi = constant. This equation has a single attractor, which corresponds to the eigenvector with the highest (real and positive) eigenvalue, λmax (cf. ref. 47).
A more interesting nonequilibrium behavior is observed when the growing assemblies undergo disruption by processes akin to those experimentally imposed by surface tension or turbulence (31, 35, 61, 62). This perturbation serves as an external free energy input, as it regenerates high free energy water-dispersed molecules from the thermodynamically favorable assemblies. In the computer simulations, when an assembly reaches a maximal size, it undergoes splitting, by randomly dividing the molecular components between two daughter assemblies. Assembly population growth is regulated according to a constant population rule (Fig. 1 C and D and refs. 10 and 11).
Assembly splitting results in a simple form of compositional inheritance (cf. ref. 42), whereby molecular compositions generally are preserved from one “generation” to another. There are also mutation-like compositional changes inherent to the underlying kinetics of a molecular joining process in complex assemblies. It is demonstrated that inheritance is more accurate in cases where the parent assembly tends to grow homeostatically and have a high information content I (cf. ref. 56). The graded replication fidelity is quantified here by computing an average H value for a parent assembly versus both daughter assemblies (see Fig. 4A).
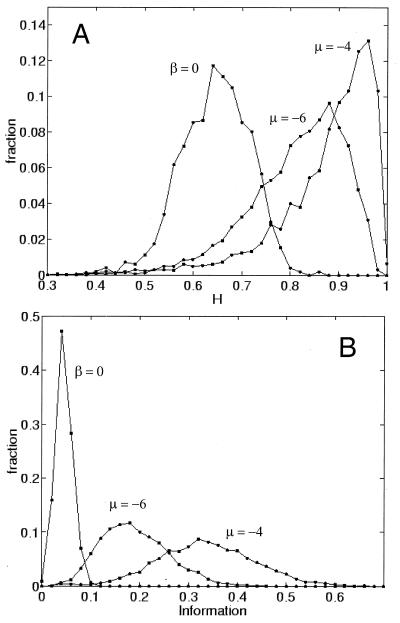
Figure 4.
Probability distributions for assembly characteristics as a function of the degree of mutual rate enhancement. β = 0 indicates no catalysis; medium and high catalysis (μ = −6 and μ = −4, respectively) represent β matrices sampled from normal distributions of the rate enhancement factors with the indicated mean and a SD of 4 (see legend to Fig. 1). Each distribution is computed for 4,000 assemblies, and values are sampled immediately after split events. (A) The homeostasis parameter H computed for pairs of parent and progeny. As the mutual rate enhancement is increased, H values shift form 0.65 ± 0.07, corresponding to random similarity values, through 0.81 ± 0.10 for low catalysis and 0.88 ± 0.10 for high catalysis, showing that the denser catalytic networks also are characterized by a higher level of average homeostasis. This finding indicates that a larger fraction of the assemblies tend to transmit their unique composition to their progeny. (B) The information or compositional bias parameter I. When there is no catalysis, I assumes very low values (0.05 ± 0.02), corresponding to low-information assemblies near thermodynamic equilibrium. The introduction of mutual rate enhancement leads to an appreciable increase in the information content to 0.19 ± 0.07 (μ = −6) and 0.34 ± 0.10 (μ = −4) (the maximal value of I is 1).
Governed by the processes of splitting and decomposition, the system is observed to pass through a set of QSSs (4, 20, 61–64) (Figs. 1 C and D and 2). Such states are stable for time intervals that encompass numerous growth/splitting cycles, constituting local attractors in compositional space. Such persistent increases in the molar fractions of certain components are in contrast to the transient increases seen without splitting. However, because of the mutation-like fluctuations introduced through the stochastic splitting of small assemblies, abrupt transition from one QSS to another may occur. Under the above-mentioned linear approximation of Eq. 4, these multiple QSSs may be shown to be related to eigenvalues of the matrix B, which have a positive real part in the vicinity of λmax.
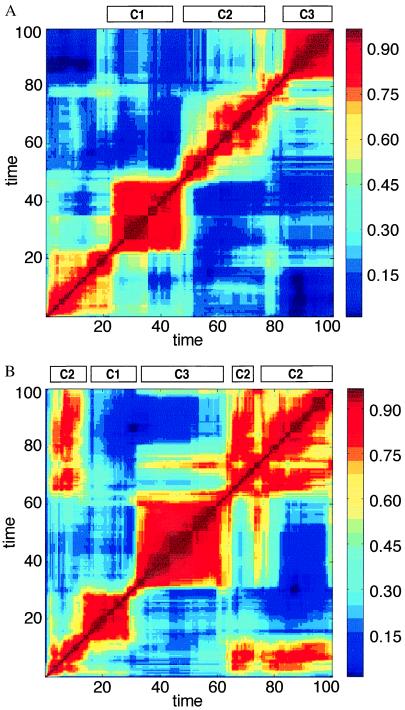
Figure 2.
A time correlation matrix for H values (Eq. 2), where the ordinate and the abscissa represent np and nq, compositional vectors at different points in the time-dependent evolution of a particular assembly. In this case H for nearly disposed time steps assumes the meaning of degree of homeostasis. Red colors signify higher H value (bar on right). (A) The H matrix for the run shown in Fig. 1 C and D. The four main red squares around the diagonal signify time intervals in which the composition does not undergo major changes (QSSs). The bars labeled C1, C2, and C3 are defined by the maximal similarity between the composition at a given time point and one of the three major composomes of Fig. 3. Intercomposome dissimilarity is displayed as blue off-diagonal areas (low H values). A statistical analysis of the lifetimes of different composomes revealed an exponential distribution, probably related to the Poissonian nature of the mutation-like compositional changes. In computer simulation that encompass 20,000 growth and split steps, the following prevalences were observed: C1, 0.13; C2, 0.37; C3, 0.50. (B) The influence of different initial conditions is seen in a different run with the same kinetic parameters but with a different initial composition. The H matrix displays off-diagonal red rectangles, representing compositions that emerge more than once.
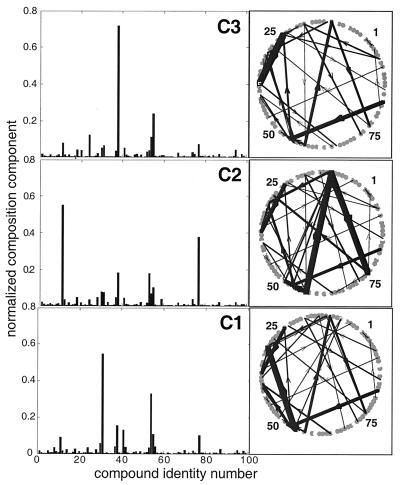
Each of the QSSs is characterized by a different, highly unusual “compositional genome,” or “composome” (Fig. 3). They may be regarded as different mutually catalytic networks, or metabolic pathways, encompassing different subsets of compounds derived from the global chemistry (Fig. 3 Right). The red square patches in the H correlation matrix (Fig. 2) represent QSSs, i.e., time spans in which the normalized compositional vector ν remains rather constant, corresponding to plateaus of high H values (Fig. 1D), a hallmark of homeostatic behavior. Different runs with the same βij parameters, but with different initial composition yield different time courses and H correlation matrices (compare Fig. 2 A and B). However, in numerous different runs, the same composomes are observed with specific time-averaged fractional incidences (Fig. 2A, legend).
Figure 3.
The compositions and “metabolic” networks for the three composomes of the previous figures. A fuzzy c-means clustering algorithm of matlab was applied to a data set of 1,000 compositions sampled immediately after split events. (Left) Histograms represent normalized molar fractions at the cluster centers. (Right) The respective “metabolic” networks, where the width of each arrow represents the effective strength of the catalytic enhancement, calculated as nj⋅βij (arrows with nj⋅βij<20 are omitted).
When different values of the rate enhancement matrix βij or of the basal kinetic parameter are used, each parameter set results in a different set of composomes. This behavior, however, seems to depend critically on the distribution for the βij values. No composomal QSSs are observed, for example, when a normal probability density is used instead of a lognormal one (D.S., unpublished work). As the average values of βij are augmented, the assemblies statistically show higher parent-progeny similarity as estimated by the parameter H (Fig. 4A), as well as increased values of the information parameter I (Eq. 3, Fig. 4B). This is a quantitative demonstration that networks of mutual rate enhancement propagate their high information content, i.e., manifest replication-like properties. For βij = 0 the simulated assembly decays to an equilibrium composition equal to that of the external medium. This finding is consistent with the notion that nonequilibrium conditions are a prerequisite for obtaining an intricate chemical behavior reminiscent of life phenomena (65, 66).
Assembly Population Dynamics.
A question pertinent to the relevance of compositional assemblies to prebiotic scenarios is whether they may potentially undergo natural selection. Three relevant properties already have been indicated: (i) that such assemblies are capable of storing information; (ii) that they are capable of undergoing compositional transitions resembling the accumulation of mutational changes in a sequential genome; and (iii) that the assemblies may generate progeny by undergoing homeostatic expansion and splitting, partially preserving their compositional constitution. It remains to explore the behavior of compositional assemblies under conditions that allow the competitive coexistence of numerous noncovalent aggregates in a given system.
For the population behavior simulations, several assemblies are seeded and are allowed to undergo the same growth and splitting processes described above, under a constant population constraint (11). Seeding an initial set of random assemblies, the emerging lineages manifest different levels of “viability” (Fig. 5). Some disappear right away, whereas others continue to be present for many generations. Within segments of some lineages, specific composomes show a capacity to temporarily “breed true,” but eventually accumulate compositional changes, giving way to alternative QSSs.
Figure 5.
An evolutionary tree for a population of assemblies. The total number of assemblies is kept constant at a population size W = 8. The color coding is according to the three clusters presented in Fig. 3, as indicated in the Inset. Open circles represent the random initial compositions used to seed the population. The length of the lines from a parent assembly to its progeny is proportional to the time taken to reach splitting size. Circles without progeny depict assemblies that were destroyed. The simulation parameters and the time scale are as in Fig. 1.
Discussion
The computer simulation analyses presented here illustrate how spontaneously forming noncovalent molecular assemblies, when endowed with internal mutual rate enhancement, may exist in numerous different compositional QSSs or composomes. These are homeostatic, namely often capable of conserving their compositional integrity over periods of time, through consecutive events of growth and splitting. Homeostasis is rationalized by the formation of complex feedback loops, resembling metabolic pathways, in which many of the molecules within a subset end up collectively catalyzing the joining of their kind. The assemblies undergo mutation-like compositional changes that lead to a transition from one QSS to another in a process that bears some similarity to speciation. Such transitions result from events in which single molecules with advantageous rate enhancement capacities are randomly inserted into an assembly. Finally, it is shown that some lineages of assemblies may be more successful in selectively populating an environment. In this, sets of compositional assemblies bear formal resemblance to quasi-species of biopolymers (10, 11), providing a bridge between the “genome first” and “metabolism first” paradigms (67).
Our approach extends and complements a previously proposed model (4, 20), which uses a mean-field parameter for rate enhancement to describe a transition between disordered and ordered QSSs. A novel attribute of the present approach is an ability to simulate the detailed kinetic behavior of the system by the use of a physicochemically based probabilistic model.
In previous analyses (21, 22, 47), mutual catalysis was characterized by what amounts to a β matrix in which a fraction p of the elements have a constant value β* and the rest are equal to 0. An ever-increasing number of different compounds was shown to result in a highly connected “catalytically closed” network (21, 22). In contrast, our model assumes a graded β matrix and a constant repertoire size, whereby a spontaneous selection process leads to a local decreased molecular diversity, associated with highly connected networks.
The progression toward reduced diversity of low molecular weight monomers constitutes a prerequisite for the subsequent appearance of “alphabet-based” biopolymers, which typically are composed of a restricted number of monomer types (12, 24, 46, 56). Future extensions of the present analyses, with the inclusion of cooperative, nonlinear rate-enhancement kinetics and controlled oligomerization could lead to more elaborate information transfer and coding. Thus, analyzing compositional assemblies may help define a rational pathway for the spontaneous passage from the “random chemistry” of prebiotic organosynthesis to the highly constrained monomer repertoires and intricate polymer chemistry as seen in living cells.
Acknowledgments
We thank Ora Kedem, Avshalom Elitzur, Luca Peliti, Shmeior Lifson, Yitzhak Pilpel, and Eytan Domany for helpful discussions. This research was supported by the Israel Ministry of Science, the Krupp Foundation, and the Crown Human Genome Center. D.L. is the Ralph and Lois Silver Chair in Neuro-genomics.
Abbreviation
- QSS
quasi-stationary state
References
- 1.Miller S L. Science. 1953;117:528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- 2.Hargreaves W R, Mulvihill S, Deamer D W. Nature (London) 1977;266:78–80. doi: 10.1038/266078a0. [DOI] [PubMed] [Google Scholar]
- 3.Rao M, Eichenberg J, Oró J. J Mol Evol. 1982;18:196–202. doi: 10.1007/BF01733046. [DOI] [PubMed] [Google Scholar]
- 4.Dyson F. Origins of Life. Cambridge: Cambridge Univ. Press; 1999. [Google Scholar]
- 5.Lifson S, Lifson H. J Theor Biol. 1999;199:425–433. doi: 10.1006/jtbi.1999.0969. [DOI] [PubMed] [Google Scholar]
- 6.Ballester P, Rebek J. J Am Chem Soc. 1990;112:1249–1250. [Google Scholar]
- 7.Li T, Nicolaou K C. Nature (London) 1994;369:218–221. doi: 10.1038/369218a0. [DOI] [PubMed] [Google Scholar]
- 8.Sievers D, Von-Kiedrowski G. Nature (London) 1994;369:221–224. doi: 10.1038/369221a0. [DOI] [PubMed] [Google Scholar]
- 9.Lee D H, Granja J R, Martinez J A, Severin K, Ghadiri M R. Nature (London) 1996;382:525–528. doi: 10.1038/382525a0. [DOI] [PubMed] [Google Scholar]
- 10.Eigen M, Schuster P. J Mol Evol. 1982;19:47–61. doi: 10.1007/BF02100223. [DOI] [PubMed] [Google Scholar]
- 11.Küppers B. Molecular Theory of Evolution. Berlin: Springer; 1983. [Google Scholar]
- 12.Stein D L, Anderson P W. Proc Natl Acad Sci USA. 1984;81:1751–1753. doi: 10.1073/pnas.81.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orgel L E. Nature (London) 1992;358:203–209. doi: 10.1038/358203a0. [DOI] [PubMed] [Google Scholar]
- 14.Cech T R. Gene. 1993;135:33–36. doi: 10.1016/0378-1119(93)90046-6. [DOI] [PubMed] [Google Scholar]
- 15.Szostak J W. Trends Biochem Sci. 1992;17:89–93. doi: 10.1016/0968-0004(92)90242-2. [DOI] [PubMed] [Google Scholar]
- 16.Wright M C, Joyce G F. Science. 1997;276:614–617. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- 17.Shapiro R. Origins Life Evol Biosphere. 1984;14:565–570. doi: 10.1007/BF00933705. [DOI] [PubMed] [Google Scholar]
- 18.Oparin A I. The Origin of Life. New York: Dover; 1953. [Google Scholar]
- 19.Oparin A I, Gladilin K L. BioSystems. 1980;12:133–145. doi: 10.1016/0303-2647(80)90011-8. [DOI] [PubMed] [Google Scholar]
- 20.Dyson F J. J Mol Evol. 1982;18:344–350. doi: 10.1007/BF01733901. [DOI] [PubMed] [Google Scholar]
- 21.Kauffman S A. J Theor Biol. 1986;119:1–24. doi: 10.1016/s0022-5193(86)80047-9. [DOI] [PubMed] [Google Scholar]
- 22.Farmer J D, Kauffman S A, Packard N H. Physica D. 1986;22:50–67. [Google Scholar]
- 23.Morowitz H J, Heinz B, Deamer D W. Origins Life Evol Biosphere. 1988;18:281–287. doi: 10.1007/BF01804674. [DOI] [PubMed] [Google Scholar]
- 24.Bagley R J, Farmer J D, Fontana W. In: Artificial Life II. Langton C G, Taylor C, Farmer J D, Rasmussen S, editors. X. Reading, MA: Addison–Wesley; 1991. pp. 141–158. [Google Scholar]
- 25.Stadler P F, Fontana W, Miller J H. Physica D. 1993;63:378–392. [Google Scholar]
- 26.Fontana W, Buss L W. Proc Natl Acad Sci USA. 1994;91:757–761. doi: 10.1073/pnas.91.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanford C. Science. 1978;200:1012–1018. doi: 10.1126/science.653353. [DOI] [PubMed] [Google Scholar]
- 28.Luisi P L, Walde P, Oberholzer T. Ber Bunsenges Phys Chem. 1994;98:1160–1165. [Google Scholar]
- 29.Deamer D W. Microbiol Mol Biol Rev. 1997;61:239–261. doi: 10.1128/mmbr.61.2.239-261.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walde P, Goto A, Monnard P A, Wessicken M, Luisi P L. J Am Chem Soc. 1994;116:7541–7547. [Google Scholar]
- 31.Bachmann P, Luisi P, Lang J. Nature (London) 1992;357:57–59. [Google Scholar]
- 32.Mayer B, Rasmussen S. Int J Mod Phys C. 1998;9:157–177. [Google Scholar]
- 33.Varela F J, Maturana H R, Uribe R. BioSystems. 1974;5:187–196. doi: 10.1016/0303-2647(74)90031-8. [DOI] [PubMed] [Google Scholar]
- 34.Segré D, Lancet D. In: Mutually Catalytic Amphiphiles: Simulated Chemical Evolution and Implications to Exobiology. Chela-Flores J, Raulin F, editors. Trieste, Italy: Kluwer; 1998. pp. 123–131. [Google Scholar]
- 35.Segré, D., Ben-Eli, D., Deamer, D. & Lancet, D. (2000) Origins Life Evol. Biosphere, in press. [DOI] [PubMed]
- 36.Wächtershauser G. Proc Natl Acad Sci USA. 1990;87:200–204. doi: 10.1073/pnas.87.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oparin A I. The Origin of Life on the Earth. London: Oliver and Boyd; 1957. [Google Scholar]
- 38.Segré D, Pilpel Y, Lancet D. Physica A. 1998;249:558–564. [Google Scholar]
- 39.Segré D, Lancet D, Kedem O, Pilpel Y. Origins Life Evol Biosphere. 1998;28:501–514. [PubMed] [Google Scholar]
- 40.Kuppers B-O. Information and the Origin of Life. Cambridge, MA: MIT Press; 1990. [Google Scholar]
- 41.Eigen M, Schuster P. The Hypercycle. Berlin: Springer; 1979. [Google Scholar]
- 42.Morowitz H J. Beginnings of Cellular Life. New Haven: Yale Univ. Press; 1992. [Google Scholar]
- 43.Bolli M, Micura R, Eschenmoser A. Chem Biol. 1997;4:309–320. doi: 10.1016/s1074-5521(97)90074-0. [DOI] [PubMed] [Google Scholar]
- 44.Ourisson G, Nakatani Y. Chem Biol. 1994;1:11–23. doi: 10.1016/1074-5521(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 45.Deamer D W. Origins Life Evol Biosphere. 1989;19:21–38. doi: 10.1007/BF01808285. [DOI] [PubMed] [Google Scholar]
- 46.Bagley R J, Farmer J D. In: Artificial Life II. Langton C G, Taylor C, Farmer J D, Rasmussen S, editors. X. Reading, MA: Addison–Wesley; 1991. pp. 93–140. [Google Scholar]
- 47.Jain S, Krishna S. Phys Rev Lett. 1998;81:5684–5687. [Google Scholar]
- 48.Devaux P F. Annu Rev Biophys Biomol Struct. 1992;21:417–439. doi: 10.1146/annurev.bb.21.060192.002221. [DOI] [PubMed] [Google Scholar]
- 49.Kust P R, Rathman J F. Langmuir. 1995;11:3007–3012. [Google Scholar]
- 50.Cuccovia I M, Quina F H, Chaimovich H. Tetrahedron. 1982;38:917–920. [Google Scholar]
- 51.Talhout R, Engberts B F N. Langmuir. 1997;13:5001–5006. [Google Scholar]
- 52.Fendler J H. Membrane Mimetic Chemistry. New York: Wiley; 1982. [Google Scholar]
- 53.von-Gottberg F K, Smith K A, Hatton T A. J Chem Phys. 1997;106:9850–9857. [Google Scholar]
- 54.Bolhuis P G, Frenkel D. Physica A. 1997;244:45–58. [Google Scholar]
- 55.Pohorille A, Wilson M A. Origins Life Evol Biosphere. 1995;25:21–46. doi: 10.1007/BF01581571. [DOI] [PubMed] [Google Scholar]
- 56.Segré D, Lancet D. Chemtracts Biochem Mol Biol. 1999;12:382–397. [Google Scholar]
- 57.Lancet D, Sadovsky E, Seidemann E. Proc Natl Acad Sci USA. 1993;90:3715–3719. doi: 10.1073/pnas.90.8.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lancet D, Kedem O, Pilpel Y. Ber Bunsenges Phys Chem. 1994;98:1166–1169. [Google Scholar]
- 59.Safran S A. Statistical Thermodynamics of Surfaces, Interfaces, and Membranes. Reading, MA: Addison–Wesley; 1994. [Google Scholar]
- 60.Nygren H. Adv Colloid Interface Sci. 1995;62:137–159. doi: 10.1016/0001-8686(95)00277-w. [DOI] [PubMed] [Google Scholar]
- 61.Rusanen M, Koponen I, Heinonen J, Sillanpaa J. Nuclear Instru Methods Phys Res B. 1999;148:116–120. [Google Scholar]
- 62.Hamano K, Ushiki H, Tsunomori F, Sengers J V. Int J Thermophys. 1997;18:379–386. [Google Scholar]
- 63.Gillespie D T. Physica A. 1979;95:69–103. [Google Scholar]
- 64.Buhse T, Pimienta V, Lavabre D, Micheau J-C. J Chem Phys. 1997;101:5215–5217. [Google Scholar]
- 65.Morowitz H J. Energy Flow in Biology. New York: Academic; 1979. [Google Scholar]
- 66.Nicolis G, Prigogine I. Self-Organization in Nonequilibrium Systems: From Dissipative Structures to Order Through Fluctuations. Toronto: Wiley; 1977. [Google Scholar]
- 67.Lahav N. Biogenesis: Theories of Life's Origin. Oxford: Oxford Univ. Press; 1999. [Google Scholar]
- 68.Gillespie D T. J Phys Chem. 1977;81:2340–2361. [Google Scholar]