Abstract
The motile behavior of the bacterium Escherichia coli depends on the direction of rotation of its flagellar motors. Binding of the phosphorylated signaling molecule CheY to a motor component FliM is known to enhance clockwise rotation. It is difficult to study this interaction in vivo, because the dynamics of phosphorylation of CheY by its kinase CheA and the hydrolysis of CheY (accelerated by CheZ) are not under direct experimental control. Here, we examine instead the interaction with the flagellar motor of a double mutant CheY13DK106YW that is active without phosphorylation. The behavioral assays were carried out on tethered cells lacking CheA and CheZ. The effects of variation in intracellular concentration of the mutant protein were highly nonlinear. However, they can be explained by a thermal isomerization model in which the free energies of clockwise and counterclockwise states depend linearly on the amount of CheY bound.
Keywords: Escherichia coli, signal transduction, CheY, molecular motors
Motile bacteria such as Escherichia coli actively respond to a variety of stimuli by modulating the direction of rotation of their flagella. Addition of a chemical attractant (e.g., aspartate) or removal of a chemical repellent (e.g., leucine) enhances counterclockwise (CCW) rotation, causing cells to extend runs that carry them in a favorable direction. This occurs through regulation of a kinase, CheA, which phosphorylates an effector molecule, CheY. Phospho CheY (CheY-P) when bound to a component at the base of the flagellar motor, FliM, promotes clockwise (CW) rotation (for reviews, see refs. 1–4). This interaction has been studied in vivo (5), but interpretation of the results is complicated by the dynamics of phosphorylation and hydrolysis: the intracellular concentration of CheY-P was not measured.
One solution to this problem is to use CheY mutants that are active without phosphorylation. Wild-type CheY (CheYwt) is activated by phosphorylation at Asp-57 (6). Replacement of Asp-13 by Lys (or Arg) results in a CW phenotype (7). CheY13DK can be phosphorylated to some extent, but phosphorylation is not required for activity. Replacement of Tyr-106 by Trp results in an even stronger CW phenotype (8), but only when CheY106YW is phosphorylated. The double mutant CheY13DK106YW—we will call this protein CheY**—is active without phosphorylation (X. Zhu and P. Matsumura, private communication). We chose this mutant to study the dynamics of the interaction of CheY with the flagellar motor.
There are two kinds of mechanisms that might explain how CheY controls the direction of the flagellar motor. In one, the switch is thrown when a certain number of CheY molecules are bound. In the other, the number bound only determines the probability of CW or CCW rotation, and the switch is thrown by thermal fluctuations. Our results argue for the latter, stochastic mechanism.
MATERIALS AND METHODS
Bacteria, Phage, and Plasmids.
(See Table 1.) The following were gifts: pXYZ202 from Xiangyang Zhu and Phil Matsumura, EC0 and phage f1R408 from Jon Beckwith, pBIP from Steven Slater, MM5008 from Mike Manson, pACYC184-Iq from Karen McGovern, pBR322/hag93 and pFD313 from Goro Kuwajima, and RP9535 and RP4979 from Sandy Parkinson.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genes | Reference or source | Remarks |
|---|---|---|---|
| Strains | |||
| DH5α | recA1 endA1 gyrA96 | GIBCO/BRL | Host for plasmid propagation |
| EC0 | F′ts 114 lac+/Δ(lac-proB)XIII | 9 | Source of F′ts |
| HCB5* | fliC726 | P. Conley | Source of fliC null mutation |
| HCB758* | wild type for chemotaxis | 10 | Source of MluI genomic fragment |
| HCB899† | F′ts 114 lac+/Δ(cheA)1643 | This work | F′ts derivative of RP9535 |
| HCB900 | F− Δ(cheA)1643 cat Ptrc420 cheY13DK106YW ΔcheZ | This work | ΔcheZ derivative of HCB899 with IPTG-inducible cheY13DK106YW |
| HCB901 | F− Δ(cheA)1643 cat Ptrc420 cheY13DK106YW ΔcheZ fliC726 uvrC-279∷Tn10 | This work | Nonmotile derivative of HCB900 |
| HCB902 | F− Δ(cheA)1643 cat Ptrc420 cheY13DK106YW ΔcheZ fliC726 uvrC-279∷Tn10/pBES38 | This work | lacIq, fliCst derivative of HCB901 |
| HCB1264* | fliC726 uvrC-279∷Tn10 | This work | Tn10-linked fliC726 derivative of HCB5 |
| JM109 | F′ traD36/recA1 | 11 | Host for f1R408 |
| MM5008† | uvrC-279∷Tn10 | 12 | Source of Tn10 marker near fliC |
| RBB1041† | Δ(cheA-cheZ)∷ZeoR | W. N. Abouhamad and R. B. Bourret | Host for testing cross-in constructs |
| RP3098 | Δ(flhA-flhD) | 13 | cheY, cheZ deleted strain |
| RP4979† | Δ(cheY)m43-10 | J. S. Parkinson | Host for selection of pKAF118 |
| RP9535† | Δ(cheA)1643 (eda+) lacY1 | 14 | Background ΔcheA strain |
| Plasmids | |||
| pACYC184-Iq | tet lacIq | 15 | Source of lacIq (1.1 kb EcoRI insert) |
| pBES32 | bla ′cheR cheB cat cheY13DK106YW ′cheZ | This work | Cross-in construct in pBSIISK(+) |
| pBES36 | npt sacB ′cheR cat cheY13DK106YW ′cheZ | This work | Cross-in construct in pBIP |
| pBES38 | bla fliCst lacIq | This work | Provides LacI and FliCst in HCB902 |
| pBIP | npt sacB | 16 | Phagemid-based integrating plasmid |
| pBSIISK(+)‡ | bla | Stratagene | Cloning vector |
| pBR322/hag93 | bla fliCwt | 17 | Source of FliCwt in pBR322 |
| pFD313 | bla fliCst | 18 | Source of FliCst in pBR322 |
| pKAF118 | bla ′cheR cheB cheY cheZ′ | This work | Genomic MluI insert in pSE280 |
| pKAF119 | bla ′cheR cheB cheY cheZ′ | This work | Reversed MluI insert of pKAF118 |
| pMAK705 | cat | 19 | Source of cat gene |
| pRL22 | bla ′cheB cheY cheZ flhB′ | 20 | Source for construction of ΔcheZ |
| pSE280 | bla | Invitrogen | Cloning/expression vector |
| pSE420 | bla | Invitrogen | Cloning/expression vector |
| pSE420ΔStyI | bla | This work | pSE420 without transcriptional start site |
| pXYZ202 | bla cheY13DK106YW | X. Zhu and P. Matsumura | Source of cheY13DK106YW |
pSE420 contains an initiating ATG at its unique NcoI site (base pair 412) at the start of the 353-bp SuperLinker (SL2). This translational start site was eliminated by restriction with StyI, which recognizes both the NcoI site and a downstream StyI site at base pair 945, and religation of the larger 4380-bp fragment. The resulting plasmid, pSE420ΔStyI, was checked for the loss of the NcoI site.
A genomic region containing part of cheR, all of cheB and cheY, and part of cheZ was obtained from a pool of 1.6- to 2.0-kb MluI-restricted DNA from HCB758. These fragments were cloned into the MluI site of pSE280, and the resulting plasmids were transformed into the cheY deletion strain RP4979. Transformants were tested for restoration of swarming on soft agar plates (0.3% agar/1% tryptone/0.5% NaCl). One complementing plasmid, pKAF118, was obtained. The MluI genomic insert was recloned in the reverse orientation, giving pKAF119.
Cross-In Construction.
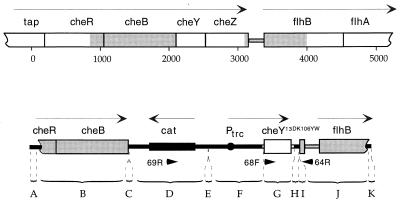
A construct was assembled in pBluescript IISK(+) (pBES32) that contains the following: cheY13DK106YW under control of the isopropyl β-d-thiogalactoside (IPTG)-inducible promoter, Ptrc; a deletion in the adjacent gene, cheZ; the cat (chloramphenicol transacetylase) gene; and DNA flanking the cheY locus to provide homology for crossing in. This construct, pBES32, is shown in Fig. 1. Its constituent parts, sources, and the restriction sites used in assembly are listed in Table 2. When necessary for ligation, restricted DNA fragments with overhangs were blunt-ended by treatment with T4 DNA polymerase and/or E. coli DNA Polymerase I Large Fragment (New England Biolabs). In addition to Ptrc, the construct contains the following elements derived from pSE420: the lac operator (lacO), a sequence from rrnB that inhibits premature termination, and the bacteriophage T7 gene 10 mini-cistron with an internal ribosome-binding site and translation termination codon. The ability of pBES32 to induce CW rotation with added IPTG was tested in RBB1041, a strain deleted for chemotaxis genes cheA through cheZ. The insert of pBES32 contains flanking NotI sites and was inserted into the NotI site of the bacterial integrating plasmid, pBIP, to give pBES36.
Figure 1.
The segment of DNA (Lower) crossed into the chromosome of a cheA deletion strain (Upper) to yield strain HCB900, drawn to the same scale. The shaded regions are regions of homology used in the cross-in. cheY was replaced by cheY13DK106YW and most of cheZ was deleted. Base pairs are numbered and fragments are labeled as in Table 2. Arrows, transcription units; arrowheads, hybridization sites for primers; open boxes and lines, genomic DNA; solid boxes, lines, and circle, plasmid DNA.
Table 2.
Segments comprising NotI insert of pBES32 (and pBES36)
| Segment* | Nucleotide number† | Source‡ | Restriction site§ |
|---|---|---|---|
| A | 1–95 | pSE280 (456–550) | KpnI-MluI |
| B | 96–1414 | pKAF119 | MluI-(DraI) |
| C | 1415–1431 | pBSIISK(+) | (HincII)-(HindIII) |
| D | 1432–2582 | pMAK705 | (Bsu36I)-(AatII) |
| E | 2583–2592 | pBSIISK(+) | (HindIII)-(EcoRV) |
| F | 2593–3368 | pSE420ΔStyI (4536–699) | (PvuII)-(NruI) |
| G | 3369–3817 | pXYZ202 | (DraI)-(BsmI) |
| H | 3818–3882 | pSE420ΔStyI (702–759) | (NruI)-(HindIII) |
| I | 3883–3905 | pSE280 (331–352) | (SalI)-(SmaI) |
| J | 3906–4916 | pRL22 | (AvaII)-BamHI |
| K | 4917–4941 | pBSIISK(+) | BamHI-NotI |
Segments, as labeled in Fig. 1. A, C, E, H, I, and K are fragments of polylinkers. B, G, and J are che coding regions containing 3′(cheR)-cheB, cheY13DK106YW, and 3′(cheZ)-5′(flhB), respectively. D is a fragment containing cat, and F comprises regulatory elements derived from pSE420 (see Section titled Cross-In Construction).
Deduced from published sequences and expected results of ligations.
Numbers in parentheses are nucleotide number of source.
Restriction sites used to generate fragment from source. Parentheses indicate that the original site was not regenerated after ligation.
Strain Construction.
The cheY region of RP9535, a strain with a preexisting cheA deletion, was replaced with that constructed in pBES36 by recombination by using the phagemid-based scheme of Slater and Maurer (16). In this method, transfer is accomplished by infection with recombinant filamentous phage; therefore, an F factor was introduced into RP9535 (cheA lacY1) by mating with EC0, a Pro− strain that contains a temperature-sensitive F factor that carries the lac operon, and selecting for the ability to use lactose as a carbon source in the absence of proline, at 30°C. The resulting strain, HCB899, was infected with the helper phage strain, f1R408, which had been grown on JM109 transformed with pBES36. f1R408 preferentially packages ss plasmid DNA (23). Recombinants were selected for resistance to kanamycin (50 μg/ml), followed by a second selection for resolved products by resistance to both chloramphenicol (10 μg/ml) and sucrose (5%), as described (16). The resulting strain, now kanamycin-sensitive, was passaged at 42°C to promote loss of the episome. This strain, HCB900, proved negative for infection by M13 (no plaque formation).
To confirm the deletion of the cheZ gene, PCR amplification of HCB900 DNA by using flanking primers 64R (5′-ACACCGGCTTTGCTGGTATC-3′) and 68F (5′-GTTATGGATTTGTTATCTCCGAC-3′) (Fig. 1) generated a fragment of the expected 315-bp size, compared with a fragment of 844 bp generated by parental strain DNA. Furthermore, no CheZ could be detected in immunoblots by using anti-CheZ mAb (see below). To confirm the transfer of cheY13DK106YW, PCR amplification by using the primer pair 69R (5′-GTGGTATTCACTCCAGAGCG-3′) and 64R generated a single fragment of the expected ≈1.3 kb, compared with the absence of any fragment generated by the parental strain DNA.
To facilitate analysis of the switching behavior of tethered cells, we utilized the self-tethering property of flagellar filaments composed of an internally truncated flagellin (24). First, a nonreverting null mutation, fliC726 (25), was introduced into HCB900 by P1 transduction, giving HCB901. Then, pFD313, which encodes a mutant flagellin in which 57 centrally located residues are replaced by 6 other residues (18), referred to as FliCst, was transformed into HCB901.
Preliminary swimming assays suggested that expression of CheY** in uninduced cells of HCB901 transformed with pBR322/hag93 was high enough to cause significant switching. To increase the range of inducibility, an EcoRI fragment encoding LacI from pACYC184-Iq was inserted into the unique EcoRI site of pFD313, and the resulting plasmid, pBES38, was transformed into HCB901, giving the final strain, HCB902. This was effective in reducing the uninduced concentration of CheY**, as confirmed by comparative immunoblots.
Purification of CheYwt, CheY**, and CheZ.
Proteins were isolated essentially as described (26) by using the host strain RP3098 (ΔflhA-flhD). CheYwt and CheZ were produced from plasmid pRL22, and CheY** was produced from plasmid pXYZ202. Concentrations of purified proteins were determined from the Ala and Phe contents by amino acid analysis performed by the Microchemistry Facility, Bio Labs, Harvard University.
Purification of mAb.
Clones of mouse mAb against CheYwt and CheZ were generated at the Max-Planck-Institut für molekulare Physiologie, Dortmund, Germany, and propagated in DMEM/10% fetal bovine serum/10% FetalClone I (HyClone) with 50 μg/ml gentamycin sulfate (GIBCO/BRL). Antibodies were purified from tissue culture supernatants by ammonium sulfate precipitation, followed by chromatography on Protein G Sepharose 4 (Pharmacia) according to the manufacturer’s instructions. Protein concentrations were determined with Bio-Rad Protein Assay.
Cell Cultures.
Cells were prepared in the same way for quantitative immunoblots and behavioral assays. Strain HCB902 was grown overnight from frozen stocks in a 125-ml culture flask containing 10 ml tryptone broth (TB) (1% tryptone/0.5% NaCl/0.1% yeast extract/100 μg/ml ampicillin). Saturated cultures were diluted 1:200 in a 250-ml flask containing 40 ml TB and 100 μg/ml ampicillin. After 4 h, IPTG from a freshly thawed 0.01 or 0.1 M stock solution was added and incubation was continued for 2 h, to OD610 ≈ 0.8. All incubations were at 33°C and 200 rpm. Strain RP3098 was grown as HCB902, omitting ampicillin and IPTG. Actual cell densities were determined by viable cell counts of serially diluted cultures plated on Luria–Bertani (LB) agar plates. Because cells of HCB902 stick to glass and yield an artificially low count, cells of HCB901 transformed with pBR322/hag93 were used for this determination. At OD610 = 0.8 (measured with Hitachi spectrophotometer U-3000), cell density was (6.2 ± 0.7) × 108 cells per ml.
Dry weights of cells were determined as follows. Cells were grown as above. Four 30-ml samples were harvested by centrifugation, resuspended in 77 mM ammonium acetate, pH 7.0, transferred to tared Eppendorf tubes, centrifuged, washed in the same buffer, and lyophilized for 2–3 days. Medium and buffer were prefiltered (0.2 μm). A value of 0.26 ± 0.01 mg/ml of culture (mean ± SD for five determinations) was obtained.
Quantitative Immunoblots.
Aliquots of strain HCB902 (0.2–1.0 ml) were used with aliquots of strain RP3098 to bring the final volume to 1.0 ml. Cells were harvested by centrifugation, washed once in 50 mM Tris⋅HCl/0.5 mM EDTA/2 mM DTT/10% (vol/vol) glycerol, pH 7.5, resuspended in 40 μl of SDS–sample buffer, and heated to 100°C for 6 min. Samples of this mixture (called cell extracts) were stored at −20°C. Control samples were prepared by adding 10–60 ng of purified CheY** to an extract prepared from a 1-ml aliquot of RP3098. Four samples and nine standards (at three different concentrations) were applied to each gel. Electrophoresis was performed by the procedure of Laemmli (27) in a 1-mm-thick gel in a linear gradient from 12.5 to 20% acrylamide (National Diagnostics). Electrophoretic transfer of proteins from gels to 0.45 μm nitrocellulose (Hybond ECL, Amersham) was done in a tank blot device (Bio Labs, Harvard University) for 1.5 h at 500 mA by using Bjerrum and Schafer–Nielsen buffer (28). Nitrocellulose blots (4 cm × 14 cm) were blocked overnight at room temperature in 80 mM Na2HPO4/20 mM NaH2PO4/100 mM NaCl/0.1% (vol/vol) Tween 20 (Bio-Rad), pH 7.5/5% instant nonfat dry milk (Carnation) on a rocking platform. The remaining steps were carried out in the same medium in a similar manner. Blots were probed with 0.01 mg/ml anti-CheYwt mAb for 3 h, washed three times (10 min each), incubated for 2 h with sheep anti-mouse horseradish peroxidase-linked whole Ig antibody (Amersham) diluted 1:1000, and washed four times (10 min each). The detection reaction, by using enhanced chemiluminescence (ECL, Amersham), was performed according to the manufacturer’s protocol. Preflashed Hyperfilm ECL (Amersham) was used for detection with a series of exposures. Films were scanned in the presence of a calibrated Step Tablet (Eastman Kodak) by using a Scan Jet IIcx/T (Hewlett–Packard) and deskscan ii 2.3 software. Analysis of scans was performed by using nih image 1.59 and kaleidagraph 3.8.
Tethering.
A glass coverslip silanized with Rain-X (Unelco, Scottsdale, AZ) was supported over a glass slide at its edges by two other coverslips, which were greased together with Apiezon L (Fisher). Cells were sheared 45 times (29) and added to the space between the coverslip and the slide. The slide was inverted and, after 3–5 min, inverted again and rinsed with several volumes of 10 mM potassium phosphate (pH 7.0)/67 mM NaCl/0.1 mM EDTA. Data were collected for up to 2 h.
Data Acquisition and Analysis.
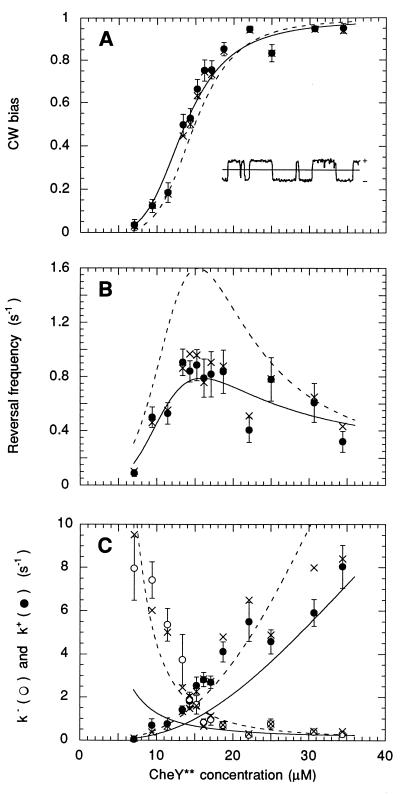
The angular positions of tethered cells, rotating under the coverslip, were measured individually with a linear-graded filter apparatus (30), which generates two output voltages, x and y, proportional to the cosine and sine of the angle of the cell image. These output voltages were low-pass filtered (two-pole filter, time constant, 4.7 ms) and sampled 256 times per second, in blocks of 128 s, by using labview 3.1 (National Instruments, Austin, TX). The number of blocks per cell varied from 2 to 10. The raw data were smoothed by averaging successive data points, and an envelope-detection routine was used to eliminate DC offset and correct for drift. This routine normalized the peak values of the cosine and sine functions to 1. The angle θ was calculated as arctan(sin/cos). Rotation speeds, ν = (1/2π)dθ/dt, were calculated as ν(i) = 256 (1/4π) [θ(i+1) − θ(i − 1)] and then smoothed by applying a median filter of rank 2. Speed records resembled square waves, with positive values for CCW rotation intervals and negative values for CW rotation intervals (Fig. 3A Inset). Speeds ranged from 4 to 20 Hz. The CW bias (the fraction of time a cell spun CW) was calculated as the number of data points with negative values divided by the total number of data points, for all blocks. The reversal frequency was calculated from the number of zero crossings divided by the duration of the blocks. A two-state model was assumed, and the forward (CCW-to-CW) and reverse (CW-to-CCW) rate constants were calculated as k+ = reversal frequency/[2(1 − bias)] and k− = reversal frequency/[2(bias)], respectively (29, 31). For cells with a sufficient number of reversals (80 or more), these rate constants also were estimated from exponential fits to distributions of CCW and CW interval lengths. Intervals were not counted at the beginning or the end of a block, where initial and final reversal times were not known. Bias CW was computed from k+/(k+ + k−), and reversal frequency was computed from 2 k+ k−/(k+ + k−).
Figure 3.
Behavioral effects of induction of CheY**. The circle values were obtained by measuring the bias and reversal rate for each cell, computing k+ and k− for each cell, and taking the means (and standard errors) over the cell population (at least 30 cells at each level of induction). The × values were obtained from exponential fits to interval distributions (where possible), computing the bias and reversal frequency for each cell, and taking the means over the cell population (with fewer than 30 cells at low and high bias, where reversal frequencies were infrequent). The dashed line values were obtained from fits of the model mechanism to k+ and k− (Fig. 4). The solid line values were obtained from fits of the model mechanism to CW bias and reversal frequency. See the text. (Inset) A segment of an angular speed record spanning 10 sec; CCW is +.
RESULTS
Strain HCB902.
This strain carries in single copy cheY13DK106YW under control of the promoter Ptrc (inducible by IPTG) as a replacement for wild-type cheY, on a chromosome deleted for cheA and cheZ (Fig. 1). CheY13DK106YW (CheY**) promotes clockwise flagellar rotation in the absence of phosphorylation (X. Zhu and P. Matsumura, private communication). Its expression is enhanced by inclusion of genetic elements that improve the efficiency of transcription and translation and tightened by expression, from the plasmid pBES38, of repressor LacI. The plasmid also carries the gene for a flagellin that forms sticky filaments, obviating the need for antifilament antibody in tethering.
CheY** Induction.
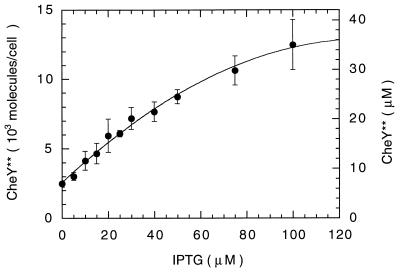
The amount of CheY** produced by HCB902 at various levels of IPTG induction was determined by immunoblotting by using anti-CheYwt mAb 1E7B11 and ECL detection. The response of this system was nonlinear to both purified CheY** and CheYwt. Consequently, all samples were chosen to contain an optimum range of CheY** (between 10 and 60 ng), amounts that gave the steepest linear responses. The assay also was sensitive to the total amount of protein in each sample, so this was held constant by addition of extracts from a congenic strain deleted for most of flagellar region II. The induction profile is shown in Fig. 2. About 2,500 molecules per cell are expressed in uninduced cultures, and this number is doubled at about 17 μM IPTG.
Figure 2.
Induction of CheY**. At least six immunoblots (24 samples) were analyzed for each induction level. The error bars are standard deviations of the means of each immunoblot. The line is a polynomial fit. Ng of CheY** were determined at each IPTG level and the molecules per cell were computed from the molecular mass (14.3 kDa) and the measured cell density (6.2 × 108 cells per ml). Corresponding concentrations were computed from the measured dry weight/ml cell culture (0.26 mg) and the cytoplasmic volume/mg dry weight (1.4 μl, ref. 32).
Behavioral Response.
The CW bias and reversal frequency of tethered cells observed at different levels of CheY** induction are shown in Fig. 3, together with the forward and reverse rate constants k+ and k−. The scatter is a result of variations from cell to cell and was largest for reversal frequency. The Inset in Fig. 3A is a short segment of a speed record, ν(t), of the type from which directions of rotation were determined. The values of the rate constants deduced from measurements of bias and reversal frequency agreed closely with those made from exponential fits to interval distributions. This agreement, together with the fact that the distributions were exponential (data not shown), support the view that the motor is essentially a two-state system, with switching probabilities per unit time (k+ and k−) that depend on the amount of CheY** bound. To a first approximation, the impact of larger levels of CheY** is symmetric: k+ rises and k− falls; their values are the same at a concentration of about 14 μM, near where the reversal frequency peaks and the CW bias approaches 0.5.
A Model Mechanism.
We assume that the behavior of the motor can be described by a thermal isomerization model, specified by the free-energy diagram shown in the Inset of Fig. 4, where GCCW, GCW, and GT are the free energies of the CCW, CW, and transition states, respectively [similar to the model proposed by Khan and Macnab (33)]. The free-energy difference ΔG = GCCW − GCW determines the equilibrium rotational bias, whereas the activation energies ΔG+‡ = GT − GCCW and ΔG−‡ = GT − GCW set the transition rates k+ and k−, respectively. Binding of CheY** shifts GCCW upwards and GCW downwards, altering bias and transition rates. We assume that this binding occurs by mass action to several, independent sites (presumably on different molecules of FliM; see refs. 34 and 35) and that the occupancy of any site changes the free energies by a fixed amount. More specifically, let the binding of n molecules of CheY** raise GCCW by np, lower GCW by nq, and decrease their energy difference by nr, where r = p + q are constants. If the value for ΔG in the absence of binding is ΔGo, then
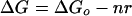 |
1 |
A value for ΔGo of 14.4 kT is obtained from a linear extrapolation (to 23°C) of the free-energy vs. temperature plot of Turner et al. (figure 5C of ref. 31), where we express ΔGo in units of energy per motor rather than energy per mole, and kT is Boltzmann’s constant times absolute temperature, evaluated (by convention) at 289°K.
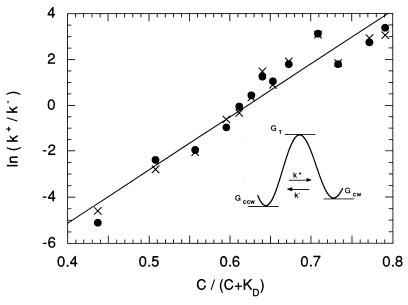
Figure 4.
Fit to Eq. 3 for ΔGo = 14.4 kT, yielding Mr = 23.1 kT, and KD = 9.1 μM. (Inset) The free-energy diagram.
If M is the number of binding sites per motor and C is the concentration of free CheY**, then by mass action the number of bound sites, n, is
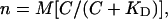 |
2 |
where KD is the dissociation constant for the CheY** binding site. The quantity in square brackets is the fraction of binding sites occupied (or the probability that a given site is occupied). The ratio of the probabilities of being in the CW or CCW state, (CW bias)/(1 − CW bias) = k+/k−, is equal to the Boltzmann factor exp(−ΔG/kT). Therefore,
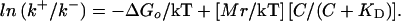 |
3 |
If this equation is fit to the data points of Fig. 3C, given ΔGo = 14.4 kT, we obtain Mr = 23.1 kT, and KD = 9.1 μM (Fig. 4). We have assumed that C, the concentration of free CheY**, is the same as the total amount of CheY**, i.e., that the total number of binding sites in the cell is small compared with the total number of molecules of CheY**. The abscissa in Fig. 4 is the fraction of binding sites occupied, C/(C + KD).
From transition-rate theory (see ref. 31) it follows that ln k+ is equal to a constant plus [Mp/kT] [C/(C + KD)], and ln k− is equal to a different constant minus [Mq/kT] [C/(C + KD)]. For the value of KD determined above, these fits yield Mp = 12.5 kT and Mq = 10.6 kT. As expected, Mp + Mq = Mr. Because p is not very different from q, binding of CheY** destabilizes the CCW state by roughly the same degree that it stabilizes the CW state. If there are, say, 26 CheY** binding sites (36, 37), then the shift in activation energy per CheY** bound, p or q, is about 0.4 kT, a value substantially smaller than the change in activation energy effected by a typical enzyme. Given these fits, the values for k+ and k− can be plotted again as a function of the concentration of CheY**, as shown by the dashed lines in Fig. 3C. The corresponding values for CW bias and reversal frequency are shown by the dashed lines in Fig. 3 A and B. The fit for the reversal frequency is not very good, but the reason for this lies with cell-to-cell scatter. The points shown in Fig. 3 are averages over the cell population of each parameter measured or computed for each cell separately. One gets different values for CW bias and reversal frequency when these values are computed from the average values for k+ and k−, or alternatively, one gets different values for k+ and k− when these values are computed from the average values of CW bias and reversal frequency. The latter fits are shown by the solid lines in Fig. 3 (for Mr = 21.1 kT and KD = 6.4 μM). The fit shown by the solid line in Fig. 3A was obtained from the bias data alone, because bias is an equilibrium property, and k+/k−, Eq. 3, is equal to (CW bias)/(1 − CW bias). However, the fits shown by the solid lines in Fig. 3 B and C require, in addition, measurements of rates (the data in Fig. 3B). Our estimates of Mr and KD are probably no better than about ±10% and ±40%, respectively.
DISCUSSION
There are two kinds of mechanisms by which the binding of molecules of CheY (CheY-P or CheY**) might affect the direction of rotation of a flagellar motor: one deterministic and the other stochastic. In a deterministic mechanism, the direction of rotation depends, at any moment in time, on the amount of CheY bound. Thus, in a two-state model considered (and rejected) by Kuo and Koshland (5), the motor spins CCW when no CheY is bound, or CW when one CheY is bound. In this special case, binding and rotational states are equivalent. More generally, in a deterministic mechanism, state refers to a binding state, not a rotational state. Thus, for example, in the five-state model considered by Bray et al. (38), the motor spins CCW when zero or one CheY is bound, or CW when three or four CheYs are bound. It is asserted that the sigmoidal nature of the bias plot reflects cooperative binding, and this is built into the model to achieve a fit (5, 38). A Hill plot of our data of Fig. 3A has a slope of 4.2 (not shown).
In a stochastic mechanism, on the other hand, the binding of CheY determines only the probability of CW or CCW rotation. For any given fraction of sites bound, switching can still occur; it is driven by thermal fluctuations. Here, state refers to direction of rotation, not to CheY binding. The deterministic model asserts that CheY throws the switch. The stochastic model asserts, merely, that CheY changes the stabilities of the two rotational states.
One argument in favor of a stochastic mechanism that also provides justification for a theory involving hopping between states is that switching events are rare (occur on a time scale that is long compared with periods of molecular vibration; ref. 33) and distributions of waiting times (of CCW or CW rotation intervals) are exponential (39). A multistate deterministic model does not generally have the latter property, particularly if the rate constants coupling different states have similar orders of magnitude. This is evident, for example, in figure 4 of Bray et al. (38), which shows a run-length distribution with a long tail. Another argument in favor of a stochastic model is that transitions can be induced without any CheY binding at all: rotation in a cell devoid of CheY can be shifted from exclusively CCW to predominantly CW simply by lowering the temperature (31).
As evident in Fig. 4, our behavioral data can be fit by a stochastic model remarkably well, given two simple assumptions: (i) that CheY binds to a set of identical, independent sites, and (ii) that this binding shifts the energy level of the CCW state up and the energy level of the CW state down by amounts directly proportional to the number of molecules bound. In such a model, motor switching rates do not depend on CheY-binding off-rates. For a KD in the micromolar range, one expects binding-site dwell times to be relatively short. If the binding site on FliM is approximated as a sticky patch of radius s = 1 nm and the on-rate for binding is assumed to be diffusion-limited, then the mean dwell time of CheY on the binding site will be (4DsKD)−1—see ref. 40—where D is the diffusion coefficient for CheY (about 10−7 cm2/s), and KD = 5.4 × 1015 molecules/cm3 (9.1 μM). This yields a dwell time of 4.6 ms. If this is so, CheY will visit each binding site about 220 times a second or any one of 26 independent sites 5,700 times a second, i.e., at frequencies much higher than the reversal frequency. As a consequence, the free-energy difference between CW and CCW states will fluctuate, but the averages over rotation intervals and thus, the probabilities of hopping from one state to the other, will remain well defined.
Kuo and Koshland (5), working with CheYwt weakly activated by a CheA/Z fusion protein, obtained CW bias and k+, k− curves of approximately the same shape as those shown in Fig. 3. This equality is expected if CheY-P and CheY** have similar effects on the motor and if the same fraction of CheYwt is phosphorylated at different CheYwt concentrations. In addition, the values of k+ and k− obtained in the two sets of experiments for a given bias, say 0.5, are approximately the same, even though the KD for binding of CheY-P must be substantially smaller than that for CheY**. This implies that the motor-switching rates are the same in the two sets of experiments, even though the dwell times of CheY on the binding sites are different. As noted above, this is expected for a stochastic model. It is not expected for a deterministic one. Finally, our model is similar to one proposed by Macnab (41) but without the assumption that the binding of CheY is highly cooperative. We have shown that one can obtain a sigmoid bias curve that displays sizable gain over a narrow concentration range (10–20 μM for CheY**, Fig. 3A) without that condition.
Acknowledgments
We thank Fricke Pietruschka for preparing hybridoma cell lines, Richard Berry for the tethered-cell analysis program, Aravi Samuel for theoretical insights, and David DeRosier and Karel Svoboda for comments on the manuscript. B.E.S. was a recipient of an Otto Hahn Prize Fellowship from the Max Planck Society. This work was supported by National Institutes of Health Grant AI16478 and by the Rowland Institute for Science.
ABBREVIATIONS
- CheY**
CheY13DK106YW
- CheYwt
wild-type CheY
- CheY-P
phospho CheYwt
- CW
clockwise
- CCW
counterclockwise
- ECL
enhanced chemiluminescence
- FliCst
flagellin yielding sticky filaments
- IPTG
isopropyl β-d-thiogalactoside
References
- 1.Amsler C D, Matsumura P. In: Two-Component Signal Transduction. Hoch J H, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 89–103. [Google Scholar]
- 2.Eisenbach M. Mol Microbiol. 1996;20:903–910. doi: 10.1111/j.1365-2958.1996.tb02531.x. [DOI] [PubMed] [Google Scholar]
- 3.Macnab R M. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 123–145. [Google Scholar]
- 4.Stock J B, Surette M G. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1103–1129. [Google Scholar]
- 5.Kuo S C, Koshland D E., Jr J Bacteriol. 1989;171:6279–6287. doi: 10.1128/jb.171.11.6279-6287.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders D A, Gillece-Castro B L, Stock A M, Burlingame A L, Koshland D E., Jr J Biol Chem. 1989;264:21770–21778. [PubMed] [Google Scholar]
- 7.Bourret R B, Hess J F, Simon M I. Proc Natl Acad Sci USA. 1990;87:41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu X, Amsler C D, Volz K, Matsumura P. J Bacteriol. 1996;178:4208–4215. doi: 10.1128/jb.178.14.4208-4215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beckwith J R, Signer E R. J Mol Biol. 1966;19:254–265. doi: 10.1016/s0022-2836(66)80003-7. [DOI] [PubMed] [Google Scholar]
- 10.Fung D C Y K. Ph.D. Thesis. Cambridge, MA: Harvard University; 1994. [Google Scholar]
- 11.Yanisch-Perron C, Vieira J, Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 12.Garza A G, Bronstein P A, Valdez P A, Harris-Haller L W, Manson M D. J Bacteriol. 1996;178:6116–6122. doi: 10.1128/jb.178.21.6116-6122.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith R A, Parkinson J S. Proc Natl Acad Sci USA. 1980;77:5370–5374. doi: 10.1073/pnas.77.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanatinia H, Kofoid E C, Morrison T B, Parkinson J S. J Bacteriol. 1995;177:2713–2720. doi: 10.1128/jb.177.10.2713-2720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J C, Peck L J, Becherer K. Cold Spring Harbor Symp Quant Biol. 1982;47:85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- 16.Slater S, Maurer R. J Bacteriol. 1993;175:4260–4262. doi: 10.1128/jb.175.13.4260-4262.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuwajima G, Asaka J-I, Fujiwara T, Fujiwara T, Node K, Kondoh E. J Bacteriol. 1986;168:1479–1483. doi: 10.1128/jb.168.3.1479-1483.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuwajima G. J Bacteriol. 1988;170:3305–3309. doi: 10.1128/jb.170.7.3305-3309.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumura P, Rydel J J, Linzmeier R, Vacante D. J Bacteriol. 1984;160:36–41. doi: 10.1128/jb.160.1.36-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Armstrong J B, Adler J, Dahl M M. J Bacteriol. 1967;93:390–398. doi: 10.1128/jb.93.1.390-398.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkinson J S. J Bacteriol. 1978;135:45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russel M, Kidd S, Kelley M R. Gene. 1986;45:333–338. doi: 10.1016/0378-1119(86)90032-6. [DOI] [PubMed] [Google Scholar]
- 24.Fahrner K A. Ph.D. Thesis. Cambridge, MA: Harvard University; 1995. [Google Scholar]
- 25.Silverman M, Simon M. J Bacteriol. 1973;113:105–113. doi: 10.1128/jb.113.1.105-113.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hess J F, Bourret R B, Simon M I. Method Enzymol. 1991;200:188–204. doi: 10.1016/0076-6879(91)00139-n. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Bjerrum O J, Schafer-Nielsen C. In: Electrophoresis ’86. Dunn M J, editor. New York: VCH; 1986. pp. 315–327. [Google Scholar]
- 29.Block S M, Segall J E, Berg H C. Cell. 1982;31:215–226. doi: 10.1016/0092-8674(82)90421-4. [DOI] [PubMed] [Google Scholar]
- 30.Berg H C, Turner L. Biophys J. 1993;65:2201–2216. doi: 10.1016/S0006-3495(93)81278-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner L, Caplan S R, Berg H C. Biophys J. 1996;71:2227–2233. doi: 10.1016/S0006-3495(96)79425-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stock J B, Rauch B, Roseman S. J Biol Chem. 1977;252:7850–7861. [PubMed] [Google Scholar]
- 33.Khan S, Macnab R M. J Mol Biol. 1980;138:563–597. doi: 10.1016/s0022-2836(80)80018-0. [DOI] [PubMed] [Google Scholar]
- 34.Welch M, Oosawa K, Aizawa S-I, Eisenbach M. Proc Natl Acad Sci USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welch M, Oosawa K, Aizawa S-I, Eisenbach M. Biochemistry. 1994;33:10470–10476. doi: 10.1021/bi00200a031. [DOI] [PubMed] [Google Scholar]
- 36.Jones C J, Macnab R M, Okino H, Aizawa S-I. J Mol Biol. 1990;212:377–387. doi: 10.1016/0022-2836(90)90132-6. [DOI] [PubMed] [Google Scholar]
- 37.Sosinsky G E, Francis N R, DeRosier D J, Wall J S, Simon M N, Hainfeld J. Proc Natl Acad Sci USA. 1992;89:4801–4805. doi: 10.1073/pnas.89.11.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bray D, Bourret R B, Simon M I. Mol Biol Cell. 1993;4:469–482. doi: 10.1091/mbc.4.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Block S M, Segall J E, Berg H C. J Bacteriol. 1983;154:312–323. doi: 10.1128/jb.154.1.312-323.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berg H C, Purcell E M. Biophys J. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macnab R M. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 181–199. [Google Scholar]