Abstract
Many organisms use polar localization of signalling proteins to control developmental events in response to completion of asymmetric cell division. Asymmetric division was recently reported for Brucella abortus, a class III facultative intracellular pathogen generating two sibling cells of slightly different size. Here we characterize PdhS, a cytoplasmic histidine kinase essential for B. abortus viability and homologous to the asymmetrically distributed PleC and DivJ histidine kinases from Caulobacter crescentus. PdhS is localized at the old pole of the large cell, and after division and growth, the small cell acquires PdhS at its old pole. PdhS may therefore be considered as a differentiation marker as it labels the old pole of the large cell. Moreover, PdhS colocalizes with its paired response regulator DivK. Finally, PdhS is able to localize at one pole in other α-proteobacteria, suggesting that a polar structure associating PdhS with one pole is conserved in these bacteria. We propose that a differentiation event takes place after the completion of cytokinesis in asymmetrically dividing α-proteobacteria. Altogether, these data suggest that prokaryotic differentiation may be much more widespread than expected.
Keywords: asymmetry, Brucella, Caulobacter, differentiation, polar localization
Introduction
Asymmetry and cellular differentiation are intrinsic features important for cell diversity in both eukaryotic and prokaryotic organisms. Bacillus subtilis and Caulobacter crescentus are two well-documented bacterial models of differentiation. Indeed, sporulation of B. subtilis is initiated by an asymmetric division immediately followed by maturation of the spore (Errington, 2003). C. crescentus is an α-proteobacterium displaying reproductive dimorphism, which is maintained by invariable asymmetric division at each cell cycle, giving rise to two genetically identical but morphologically different daughter cells, a sessile stalked cell and a motile swarmer cell (Ausmees and Jacobs-Wagner, 2003). The swarmer cell must differentiate into a stalked cell before initiating a new round of DNA replication. The specific developmental program associated with the differentiation of the swarmer progeny (Swarmer progeny-specific program, i.e. SwaPS program) is directly linked to the completion of the previous cell cycle and cytokinesis (Huguenel and Newton, 1982). The fact that the SwaPS program is initiated only after the completion of cytokinesis ensures the generation of different cell types by asymmetric cell division. A complex regulatory network composed of several two-component signal transduction systems, involving histidine kinases (HK) and response regulators (RR), is implicated in the control of differentiation in C. crescentus (Ausmees and Jacobs-Wagner, 2003). At the core of this regulatory network, PleC and DivJ, two asymmetrically distributed HK (Jacobs-Wagner, 2004), couple SwaPS development with cell division, through the spatial regulation of a conserved single-domain RR named DivK. Indeed, DivJ is localized at the stalked pole and PleC at the flagellated pole of the predivisional cell (Wheeler and Shapiro, 1999). During the cell cycle, DivK is first localized at the stalked pole, where it interacts with and is phosphorylated by DivJ. This phosphorylation allows DivK to localize at the opposite pole, that is, the flagellated pole (Lam et al, 2003). When cytokinesis is completed, the release of DivK from the flagellated pole through the phosphatase activity of PleC induces the initiation of the SwaPS program (Matroule et al, 2004). Accordingly, DivK is completely delocalized in a ΔdivJ strain by remaining dispersed in the cytoplasm of all cell types, whereas in ΔpleC cells, DivK is found at both poles because of its inability to be released from the flagellated pole. Thus, dynamic localization of DivK, controlled by the opposite activity and position of DivJ and PleC, provides a simple molecular mechanism to coordinate cytokinesis with the developmental program in C. crescentus (Matroule et al, 2004). Little is known about the mechanisms by which DivK activates or represses the SwaPS program, but it was reported that DivK controls the activity of CtrA, a major transcriptional regulator controlling both cell cycle and morphogenesis (Quon et al, 1996; Laub et al, 2000; Hung and Shapiro, 2002).
Interestingly, other α-proteobacteria such as Agrobacterium tumefaciens, Sinorhizobium meliloti and B. abortus also display an asymmetric division (Hallez et al, 2004) despite the fact that they have very different lifestyles, but we do not know if all these bacterial species display a differentiation program. Moreover, components of the DivK regulatory network are well conserved among these α-proteobacteria, suggesting that species other than C. crescentus may go through a developmental program (Hallez et al, 2004). B. abortus, a class III pathogen, is a facultative intracellular bacterium causing brucellosis in domestic mammals like cows and Malta fever in humans (Boschiroli et al, 2001). In B. abortus, the role of CtrA is partially conserved compared with C. crescentus, which indicates plasticity of this regulatory network along evolution (Bellefontaine et al, 2002). An additional HK homologous to PleC and DivJ, called PdhS (PleC-DivJ homolog sensor), was also identified. PdhS is a predicted large HK without transmembrane segments, conserved in several α-proteobacteria (Hallez et al, 2004).
Here, we show in B. abortus that PdhS is cytoplasmic and belongs to the DivK regulatory network. PdhS is located at the old pole of the large B. abortus cell and, after division, the small cell maturates to acquire PdhS at its old pole, whereas PdhS remains at the old pole of the large cell. As the pdhS gene is essential for viability in B. abortus, this maturation of a small cell into a large one is proposed to be obligatory. The cell-cycle pattern of PdhS suggests that the small cell maturates into the large one in a manner reminiscent of the swarmer-to-stalked cell differentiation in Caulobacter. PdhS being also able to localize at one pole of C. crescentus and S. meliloti, we suggest not only that a functional asymmetry occurs at a molecular level in at least three different α-proteobacterial species, but also that a differentiation event could take place after the completion of asymmetric division.
Results
The functional DivK homolog displays a phosphorylation-dependent polar localization in B. abortus
We first characterized the B. abortus DivK homolog (AAF14690). The divK gene is predicted to encode a 123-amino-acid protein with a molecular mass of 14 kDa that shares 79% identity with DivK from C. crescentus (ccDivK) (Table I). If B. abortus DivK is able to play a role similar to ccDivK, it is expected to complement a divK mutation in C. crescentus. As ccdivK is an essential gene, by transducing a ΔdivK∷SpecR lysate we demonstrated that B. abortus divK expressed from the medium-copy plasmid pRH223 was able to support viability in C. crescentus as the only copy of divK in the cells (data not shown). Immunoblot analysis of this strain confirmed that the only copy of divK expressed in this strain was B. abortus divK (data not shown). All attempts to delete divK in B. abortus failed, suggesting that this gene is also essential in B. abortus. Moreover, as indicated below, DivK is able to interact with its predicted kinases in a yeast two-hybrid (Y2H) test. These results argue that B. abortus DivK is the functional homolog of ccDivK.
Table 1.
Identification of B. abortus DivK, PleC and DivJ orthologs
| BaDivK (123)a | BaPleC (783)a | BaDivJ (609)a | PdhS (1035)a | |
|---|---|---|---|---|
| CcDivK (125)a | 79%b (123/123)c | — | — | — |
| CcPleC (842)a | — | 37%b (229/604)c | 38%b (100/257)c | 36%b (142/391)c |
| CcDivJ (597)a | — | 45%b (112/245)c | 45%b (112/246)c | 39%b (104/261)c |
| Numbers in parentheses represent the length of the corresponding protein-coding ORF. | ||||
| Represents the percentage identity. | ||||
| Numbers in parentheses represent the portion of the protein aligned. | ||||
Then we postulated that the subcellular distribution of DivK could be polar, at least in a fraction of B. abortus cells, as it is the case in C. crescentus and S. meliloti. To test this possibility, we first localized DivK-YFP with a divK-yfp fusion expressed from the pRH336 in the B. abortus 544 wild-type (WT) strain and found that DivK-YFP was localized at one pole of the cells in approximately one half of the population (Figure 1). Similar results were obtained with the strain XDB1106, in which a divK-yfp fusion was integrated at the divK locus (see Supplementary Figure S9). When the conserved phosphorylatable Asp53 of DivK was substituted with an alanine, the DivKD53A-YFP fusion expressed from pRH409 was never found polarly localized (Supplementary Figure S1), suggesting that the polar localization of DivK is dependent upon its phosphorylation state, as it was already demonstrated in C. crescentus and S. meliloti (Lam et al, 2003).
Figure 1.
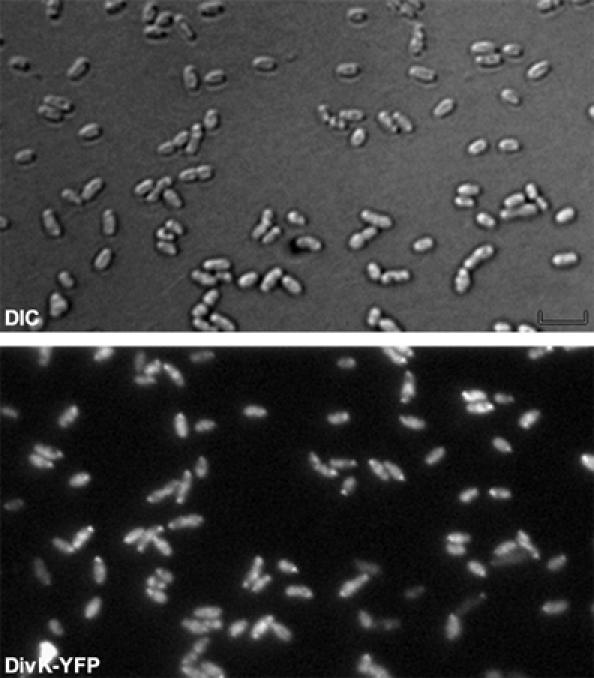
DivK is polarly localized in B. abortus. DIC and corresponding fluorescence images were taken of B. abortus WT cells producing DivK-YFP from the low-copy plasmid pRH336.
B. abortus DivK is able to directly interact with histidine kinases similar to PleC and DivJ
To identify paired histidine kinases of DivK in B. abortus, we performed a Y2H matrix in which the interactions of all HK and RR predicted from the Brucella genomic sequences (DelVecchio et al, 2002; Paulsen et al, 2002; Halling et al, 2005) (20 HK including two hybrid HK, CckA and PrlS, and 21 RR) were assayed (Supplementary Figures S2 and S3). We used the available entry clones (Dricot et al, 2004) for the RR and HK predicted to be cytoplasmic. For HK containing predicted transmembrane segments, truncated coding sequences (CDS) were amplified by PCR in order to construct new entry clones carrying CDS for C-terminal domains only, that is, the protein parts containing the HK domains (Supplementary Figure S2). The CDS from entry clones were inserted into destination vectors by recombinational cloning, in fusion with either activating domain (AD) or DNA binding domain (BD) of GAL4, as detailed in the Materials and methods section. Positive interactions were detected using the lacZ and HIS3 reporter genes.
As a positive control, we found six positive interactions in the Y2H test among the 10 pairs of HK and RR predicted to be organized in operons in the genome (TcbRS, TccRS, TcdRS, TceRS, TcfRS, NtrBC, NtrYX, FeuPQ, BvrRS and NodVW) (Supplementary Figure S4). No interaction between these pairs (HK with RR, HK with HK or RR with RR) was found, arguing in favor of the specificity of these Y2H interactions. We detected physical interactions between the single-domain RR DivK and the four HK DivL, PleC, DivJ and PdhS (see below) (Figure 2A). These were the only interactions made by these four HK in the Y2H tests (Supplementary Figures S2–S4), which again suggests that positive interactions are specific. We also observed that the C-terminal part of PdhS (aa 611–1035) was sufficient for interaction with DivK, and that this is the only interaction we detected with PdhS611−1035 in fusion with either AD or BD of Gal4. Moreover, PdhS is able to interact with itself in the Y2H experiments (Figure 2B). This later interaction is probably, at least partially, made by the N-terminal part of the protein, as the first 613 aa alone are able to interact with the complete PdhS protein (Figure 2B). This suggests that PdhS contains at least two functional parts: an N-terminal multimerization part and a C-terminal histidine kinase part interacting with DivK.
Figure 2.
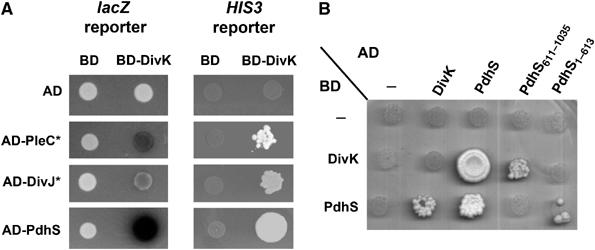
B. abortus PleC, DivJ and PdhS interact with DivK through their C-terminal domain. (A) Yeast two-hybrid assay showing physical interactions between the B. abortus response regulator DivK and the histidine kinases PleC, DivJ and PdhS. Activation of lacZ reporter gene was demonstrated as formation of blue colonies on plates containing X-Gal. Activation of HIS3 reporter gene was measured as growth on plates without histidine but with 3-AT. ‘*' Represents truncated forms of PleC and DivJ fused to AD, which comprised the HATPase_C domain and excluded the predicted transmembrane domains. (B) Yeast two-hybrid assay demonstrating that the N-terminal domain of PdhS is required for its dimerization, and its C-terminal domain, comprising the HATPase_C and PAS domains, is involved in the interaction with DivK. As in (A), activation of HIS3 reporter gene was measured as growth on plates without histidine but with 3-AT. AD and BD represent the activation and binding domains of Gal4, respectively. At least three independent experiments were performed for each yeast two-hybrid assay.
Among the four histidine kinases interacting with DivK, three are highly similar to C. crescentus PleC and DivJ, the kinases of DivK (Table I). These CDS are conserved with >99% identity at the amino-acid level between B. abortus 9–941, B. melitensis 16M and B. suis 1330. The interaction between DivK and DivL was also demonstrated in C. crescentus (Ohta and Newton, 2003), but the in vivo implications of this interaction are not understood as it was reported that DivL could control the phosphorylation state of CtrA but not DivK (Wu et al, 1999). C. crescentus PleC and DivJ share 46% identity, and therefore belong to the same subfamily of HK. B. abortus PleC (YP_414082) is aligned with ccPleC on an extended length (Supplementary Figure S5) and presents a similar predicted topology, that is, an N-terminal periplasmic domain flanked by transmembrane segments (Supplementary Figure S6). In C. crescentus, the deletion of pleC leads to lack of motility and complete resistance to φCbK and CR30 phages (Ohta et al, 1992; Wang et al, 1993; Wheeler and Shapiro, 1999), both phenotypes directly associated with a misregulation of the SwaPS program (Matroule et al, 2004). Expression of B. abortus pleC on the low-copy plasmid pRH246 in a C. crescentus ΔpleC strain restored motility and phage sensitivity to the WT level (data not shown). Taken together, these results strongly suggest that PleC is the functional homolog of ccPleC in B. abortus. The proposed B. abortus DivJ homolog (AAX73976) is slightly more identical to the ccDivJ sequence compared with ccPleC (Table I) and this deduced protein has a similar size and predicted topology compared with ccDivJ, that is, several N-terminal transmembrane segments without extensive regions in the periplasm (Supplementary Figure S6). Finally, the third HK (AAX74918) is called PdhS for PleC-DivJ homolog sensor, because it presents a similar degree of conservation with ccPleC and ccDivJ. The domain's composition of B. abortus PleC, DivJ and PdhS is represented in Supplementary Figure S6. The most striking feature is that PdhS presents a large N-terminal portion of more than 700 aa without predicted functions. Moreover, the absence of predicted transmembrane domains suggests that PdhS is a cytoplasmic histidine kinase, which is not the case with B. abortus or C. crescentus PleC and DivJ proteins. An immunoblot with anti-PdhS antibodies performed on fractionated B. abortus WT cells demonstrated that PdhS could be detected in the cytoplasmic soluble fraction. CtrA, a predicted cytoplasmic protein, was also detected in the same fraction, whereas VirB10, a protein predicted to be anchored to the inner membrane of B. abortus, was not detected in this soluble fraction (Supplementary Figure S7). These results indicate that PdhS is a large cytoplasmic histidine kinase produced by B. abortus at least during culture in bacteriological medium.
Altogether, these results strongly suggest that PdhS, PleC and DivJ interact with DivK and thus form a regulatory network comparable to the DivK regulatory network reported in C. crescentus, except for the presence of an additional identified HK, PdhS.
Neither PleC nor DivJ controls the localization of DivK in B. abortus
The pleC and divJ genes are both non-essential in C. crescentus, and deletion of these genes gave very different phenotypes. Whereas a divJ-null mutant displays a cell division defect with a weak filamentation, the deletion of pleC generates a characteristic differentiation default (Ohta et al, 1992; Wang et al, 1993; Wheeler and Shapiro, 1999). To investigate the biological role of pleC, divJ and pdhS in B. abortus, we tried to delete these genes. Deleting pleC and divJ was possible, but we were not able to delete pdhS without a plasmid-borne copy of pdhS, suggesting that this gene is essential (see below). As illustrated by differential interference contrast (DIC) microscopy, the ΔdivJ strain (XDB1102) generated Y-shaped bacteria, a phenotype associated with a cell division defect in several α2-proteobacteria such as S. meliloti and A. tumefaciens (Latch and Margolin, 1997) (Figure 3A). The ΔpleC strain (XDB1101) did not display any obvious and reproducible morphological defect (data not shown). These results suggest that divJ could play a role in cell-cycle control in B. abortus, as it was reported for C. crescentus.
Figure 3.
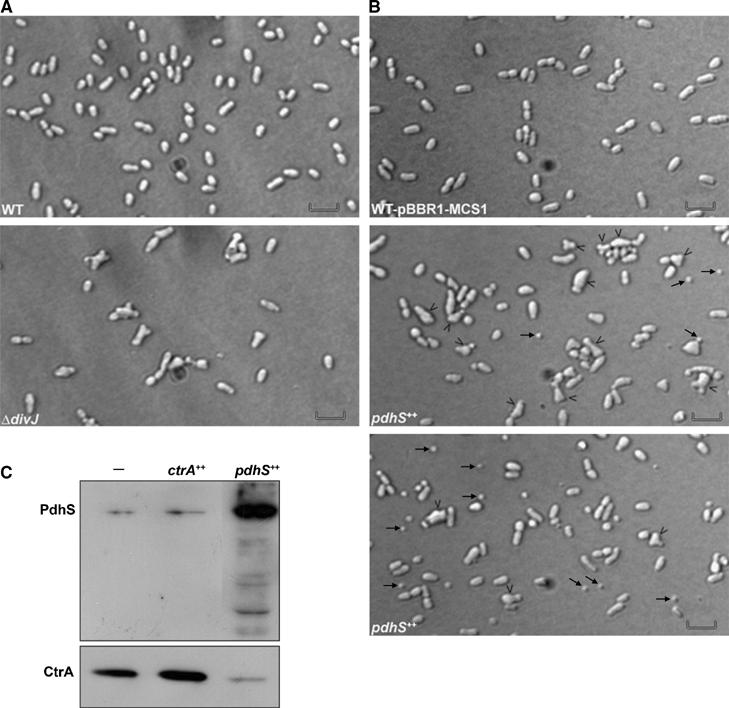
Morphotypes of the ΔdivJ and pdhS overexpressing strains. (A) DIC micrographs of the WT B. abortus 544 (WT) and ΔdivJ (XDB1102) strains from mid-exponential cultures. Scale bar, 2 μm. (B) DIC micrographs of the WT B. abortus 544 strain bearing the empty plasmid pBBR1-MCS1 (WT pBBR1-MCS1, upper panel) and overexpressing pdhS (pdhS++, middle and lower panels). Black arrows indicate minicells. Black arrowheads point to cells with abnormal morphology. Scale bar, 2 μm. (C) Western blot analysis of relative CtrA and PdhS protein levels in strains overexpressing ctrA (ctrA++) and pdhS (pdhS++) in comparison to the wild-type strain (−). ‘−' Represents the empty vector pBBR1-MCS1 (Kovach et al, 1995) in the WT strain.
As polar localization of DivK in B. abortus seems to be dependent on its phosphorylation state, we checked if the absence of divJ or pleC gene could mislocalize a DivK-YFP fusion. Surprisingly, the polar DivK-YFP distribution was not modified in B. abortus ΔpleC or ΔdivJ strains when compared with the WT control (data not shown), suggesting that PleC or DivJ could not control phosphorylation of DivK in vivo, in the conditions tested here. PdhS could therefore constitute a good candidate for such functions.
PdhS is essential for viability in B. abortus and its overexpression leads to a cell division defect
To test the hypothesis of pdhS essentiality, we first deleted pdhS in the presence of a rescue copy expressed from pRH232. After checking for the deletion of the pdhS genomic copy by PCR, we tried to swap the rescue copy by introducing another vector from the same incompatibility group expressing pdhS (pRH404) or not (pBBR1-MCS4). As shown in Table II, pRH232 was replaced in the ΔpdhS strain only in the presence of another WT copy of pdhS (pRH404). In contrast, pRH232 could be easily cured from the B. abortus 544 WT strain in the presence of either the empty plasmid pBBR1-MCS4 or pRH404. The requirement for a plasmid-borne copy of pdhS in a ΔpdhS strain demonstrates that pdhS is essential for viability in B. abortus.
Table 2.
pdhS is an essential gene in B. abortus
| pRH003 | ||||
|---|---|---|---|---|
| Strainsa | — | pdhS | pdhS611–1035 | pdhS1–613 |
| Wild type | 100 | 96 | 100 | 94 |
| ΔpdhS | 0 | 98 | 0 | 0 |
| The wild-type and ΔpdhS strains contained plasmid pRH232 (pRH002-pdhS) before swap experiments. | ||||
| Values are expressed as percentages of swapping plasmid pRH232 (clones AmpR/CmS, n=100) in WT and ΔpdhS backgrounds. | ||||
To test whether a fragment of PdhS is sufficient to carry out the essential functions of PdhS, two parts of the CDS were inserted in pRH003, coding for either the first 613 amino acids (pRH405) or the last 424 amino acids (pRH406). These constructs were used in our plasmid swap experiment. The ΔpdhS strain was unable to replace the plasmid pRH232 by either pRH405 or pRH406 (Table II). On the other hand, pRH232 could be easily cured from WT strain in the presence of either of these plasmids. Therefore, these two parts of PdhS cannot carry out separately the essential functions missing in the ΔpdhS strain, even if these parts of PdhS are functional in other tests, such as Y2H and subcellular localization (see below). Because of our inability to obtain a null mutant for pdhS and the current absence of molecular tools allowing the construction of depleted strains in Brucella spp, we decided to study the effects of its overexpression. Indeed, overexpressing an essential gene may give important information about its functions, as it was successfully performed with the ctrA gene of B. abortus (Bellefontaine et al, 2002). Overexpression of pdhS generated the characteristic Y-shaped phenotype, but also a high proportion of minicells in comparison to the WT strain (Figure 3B). This minicell phenotype was very well described in Escherichia coli (de Boer et al, 1989), where it is due to the alteration of the MinCDE system, encoded by the minB operon. Given that (i) the minB operon is a putative target of CtrA in B. abortus (Bellefontaine et al, 2002), (ii) CtrA proteolysis could be under the control of the DivK regulatory network, as it is the case in C. crescentus (Hung and Shapiro, 2002), and (iii) PdhS interacts with DivK and could therefore control its activity, we evaluated the abundance of CtrA in a pdhS++ strain using immunoblot analysis with anti-CtrA antibodies. The CtrA level was lower in pdhS++ strain compared with WT control (Figure 3C), suggesting that CtrA abundance is indeed under the control of PdhS.
Altogether, these results suggest that PdhS has essential functions in controlling some aspects of the B. abortus division process, maybe through the control of CtrA activity.
PdhS is localized at one pole in B. abortus
The homology of B. abortus PdhS, PleC and DivJ with the C. crescentus HK PleC and DivJ prompted the question of whether these B. abortus proteins also display spatial regulation in B. abortus. To address this question, we fused the CDS of pleC, divJ and pdhS to yfp on the pRH011 suicide vector and introduced these constructions into the B. abortus 544 WT strain. After a single recombination event at the corresponding loci, the translational fusions were expressed from the native promoters (XDB1105, XDB1107 and XDB1104 strains, respectively). Using fluorescence microscopy, we observed that PdhS-YFP accumulated at one pole in more than 90% of the cells (Figure 4A). PleC-YFP was not polar in B. abortus. However, surprisingly we observed that PleC-YFP was able to localize either at mid-cell or at new poles (Figure 4B). This ambiguity is due to the resolution of fluorescence microscopy, as we cannot determine whether PleC-YFP appears before or during the separation of daughter cells. The same localization patterns were observed when PdhS or PleC was fused to other fluorescent proteins (CFP and eGFP), or when these translational fusions were expressed from low-copy plasmids (data not shown). The PdhS localization pattern was also the same in the XDB1109 strain, in which the only copy of pdhS expressed was the pdhS–yfp fusion (data not shown). The fact that this strain is viable and does not present any apparent morphological defects or growth delay demonstrates that YFP fusion at the C terminus of PdhS is not affecting PdhS essential functions. Control immunoblots with anti-GFP and anti-PdhS antibodies performed on the XDB1109 strain revealed that the only protein detected was PdhS-YFP, and that this fusion protein was stable (data not shown). Finally, DivJ-YFP did not accumulate at any particular subcellular location and appeared diffused when examined by fluorescence microscopy (data not shown).
Figure 4.
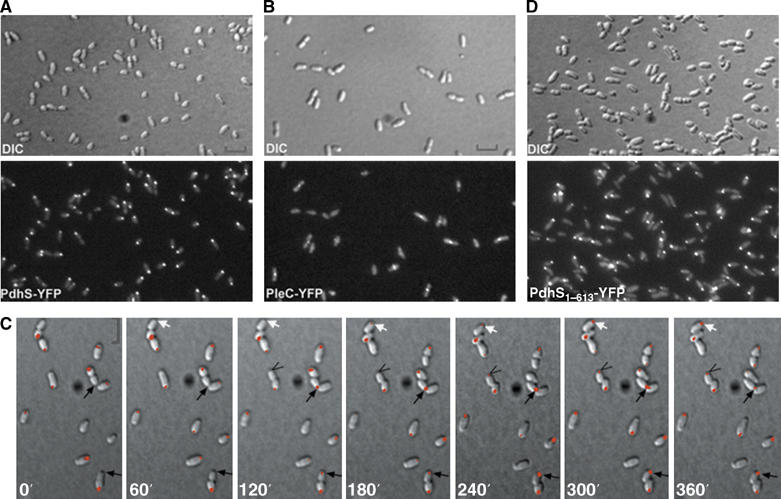
PdhS is dynamically localized at one pole of B. abortus through its N-terminal domain, whereas PleC localizes at mid-cell position. (A) PdhS localizes at one pole of B. abortus. DIC and corresponding fluorescence images were taken of B. abortus cells producing PdhS-YFP (XDB1104 strain). (B) BaPleC localizes at mid-cell in B. abortus. DIC and corresponding fluorescence images were taken of B. abortus cells producing PleC-YFP (XDB1105 strain). (C) PdhS dynamically localizes at the old pole of B. abortus cells. A time-lapse microscopy experiment was performed on the XDB1104 strain expressing pdhS-yfp by taking DIC and corresponding fluorescence images each 60 min. Arrows show that polar fluorescence signal appears 60–120 min following the physical separation of the daughter cells. (D) The N-terminal domain of PdhS is important for its polar localization. DIC and corresponding fluorescence images were taken of B. abortus cells producing PdhS1–613-YFP from the low-copy plasmid pRH407. Scale bar, 2 μm.
From pictures presented in Figure 4A, we hypothesized that PdhS-YFP dynamically changes its spatial distribution during the B. abortus cell cycle. To investigate this possibility, we performed time-lapse fluorescence microscopy on XDB1104 strain as illustrated in Figure 4C. The PdhS-YFP fluorescent signal was concentrated at one pole. Following several cells in the time-lapse experiment allowed us to determine that PdhS-YFP was accumulating at the old pole of the large cells, as opposed to the new pole originating from the last cell division. As the cells grew, PdhS-YFP remained localized at the old pole until cell division was initiated. It is only after a short time (about 60–120 min) following cell separation that PdhS-YFP accumulated at the old pole of the small sibling cells (arrows in Figure 4C). The same time-lapse sequence with separated DIC and fluorescence fields is presented in Supplementary Figure S8. Because of the very low growth rate of B. abortus on an agarose pad at room temperature, we could not follow more than one cell-cycle equivalent per time-lapse experiment. These results strongly suggest that PdhS-YFP accumulated at the old pole of B. abortus only when the small progeny has differentiated into a stage corresponding to the large cell.
The N-terminal portion of PdhS is sufficient for its polar localization
As it was demonstrated for several polarly localized HK that their N-terminal part was involved in their subcellular localization (Jacobs et al, 1999; Sciochetti et al, 2002), we investigated the possibility that the large N-terminal part of PdhS is sufficient for its polar localization. For this purpose, we expressed the N-terminal (aa 1–613) part of PdhS in fusion to YFP from the low-copy plasmid pRH407. As illustrated in Figure 4D, PdhS1–613-YFP was able to localize at one pole of B. abortus despite the fact that the PAS, HisKA and HATPase_c predicted domains were absent. The same results were obtained with the XDB1108 strain, in which pdhS1–613-yfp was integrated at the pdhS locus (data not shown). These results suggest that the N-terminal domain of PdhS is sufficient for polar localization in B. abortus. As the N-terminal domain of PdhS is able to interact with the full-length PdhS (Figure 2B) and a full-length copy of PdhS is always present in all our strains because of the essentiality of pdhS (Table II and data not shown), it is possible that the PdhS1–613-YFP fusion is polarly localized through its interaction with the polar PdhS WT copy. However, the same N-terminal domain of PdhS fused to CFP (PdhS1–613-CFP) is polarly localized in C. crescentus (data not shown), which does not carry a close ortholog of pdhS. Taken together, our data indicate that the N-terminal domain of PdhS is sufficient for polar localization in C. crescentus and B. abortus.
PdhS colocalizes with DivK at one pole in B. abortus
As PdhS can physically interact with DivK and both proteins localize at one pole in B. abortus, we postulated that DivK and PdhS colocalize at the same pole, at least in a fraction of B. abortus cells. To test this possibility, we introduced the low-copy plasmid pRH324 producing PdhS-CFP into the XDB1106 strain expressing a divK-yfp fusion at the divK locus. We observed that DivK-YFP is located at the same pole as PdhS-CFP (Figure 5), which is expected if PdhS is a kinase or a phosphatase of DivK. Moreover, if PdhS controls the phosphorylation state of DivK, we can predict that the overexpression of pdhS could increase or decrease the polar localization of DivK-YFP. Analysis of the subcellular pattern of DivK localization confirmed this prediction (Supplementary Figure S9). Indeed, compared to the WT control, the pdhS overexpression strain keeps the polar localization of a PdhS-YFP fusion and displays an increase of the polar fluorescence signal of a DivK-YFP fusion (Supplementary Figure S9). These results suggest that PdhS could enhance the phosphorylation of DivK and its subsequent polar localization.
Figure 5.
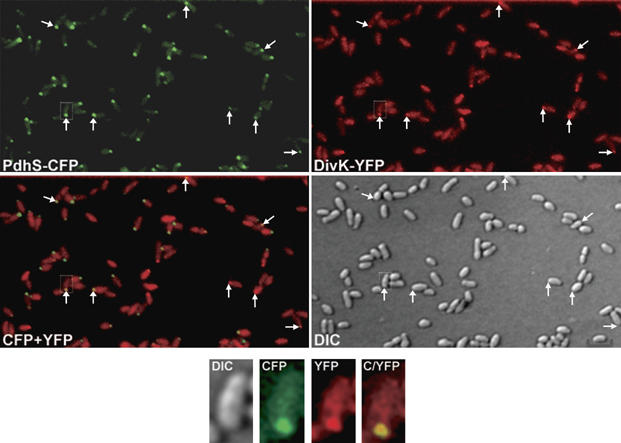
DivK colocalizes with PdhS at one pole of B. abortus. DIC and corresponding fluorescence images were taken with B. abortus cells from strain XDB1106/pRH324, coexpressing divK-yfp (red) and pdhS-cfp (green). An overlaid image shows the colocalization of both proteins within same cells (yellow). White arrows indicate polar colocalization of PdhS and DivK. Scale bar, 2 μm.
The polar localization of PdhS is maintained during a cellular infection
B. abortus is a facultative intracellular pathogen that encounters different environments during its life cycle. We therefore tested the possibility that polar localization of PdhS would be strictly dependent upon environmental stimuli absent in infected cells. To this end, the localization pattern of PdhS-YFP (XDB1104 strain) was monitored after infection of bovine macrophages. At 48 h post-infection, a polar fluorescence signal was observed in nearly all intracellular bacteria (Figure 6A). In contrast, a B. abortus strain expressing a transcriptional fusion between the pdhS promoter and gfp (PpdhS-gfp) gave a uniform and cytoplasmic fluorescence signal after 48 h of macrophage infection (Figure 6B), suggesting that the dots observed in Figure 6A represent the subcellular localization of PdhS-YFP. These observations indicate that at this stage of infection, which corresponds to a high rate of intracellular replication, PdhS-YFP is polarly localized, as in the case of B. abortus growth in bacteriological culture medium. These results suggest that the spatial distribution of PdhS is an intrinsic feature of PdhS rather than a physiological response to environmental stimuli encountered by B. abortus.
Figure 6.
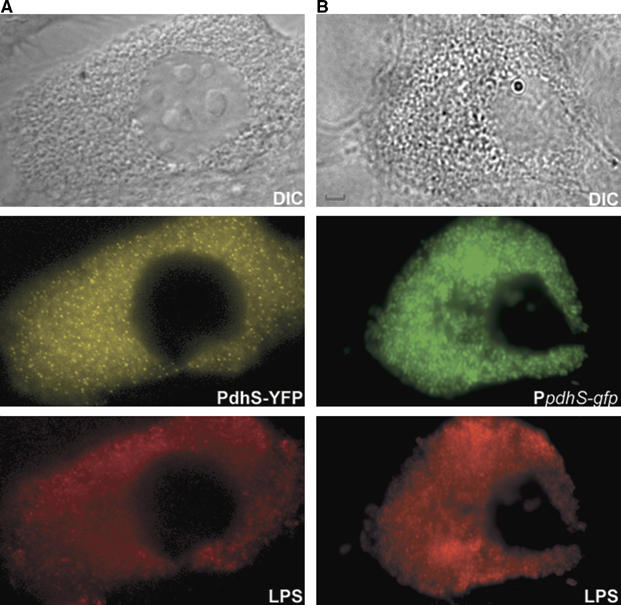
PdhS localizes at one pole of B. abortus cells during cellular infection. (A) DIC and corresponding fluorescence images were taken with bovine macrophages after 48 h of infection by the B. abortus strain expressing pdhS-yfp (XDB1104). (B) DIC and corresponding fluorescence images were taken with bovine macrophages after 48 h of infection by the B. abortus WT strain expressing the transcriptional fusion PpdhS-gfp from the plasmid pRH295. The LPS, representing the immunolabelling of the B. abortus lipopolysaccharide, the PdhS-YFP and the PpdhS-gfp fluorescence signals are shown in red, yellow and green, respectively. Scale bar, 2 μm.
PdhS is polarly localized in other α-proteobacteria
Recently we described that besides C. crescentus, at least three other α-proteobacteria (S. meliloti, A. tumefaciens and B. abortus) divide asymmetrically by generating two progeny cells of different size (Hallez et al, 2004). PdhS has close homologs in both S. meliloti and A. tumefaciens, that is, predicted large (>1000 aa) and cytoplasmic HK sharing 43% identity with PdhS over their whole length (Hallez et al, 2004). We investigated the possibility that B. abortus PdhS could be polarly localized in species other than B. abortus, for example, being associated with a polar structure conserved in α-proteobacteria. To test this hypothesis, we introduced the low-copy plasmid pRH324 expressing B. abortus pdhS fused to cfp into S. meliloti. Interestingly, we found that PdhS-CFP also accumulated at one pole in S. meliloti, probably the old pole of the large cells (Figure 7A). This pattern of localization is similar to the one described for B. abortus (Figure 4C). In addition, we also observed that the same pdhS-cfp fusion expressed in C. crescentus accumulated fluorescence at the stalked pole (Figure 7B). Using a timecourse fluorescence microscopy experiment performed on a synchronized population of C. crescentus, we observed that PdhS-CFP (i) remained associated with the stalked pole during all the cell-cycle stages and (ii) accumulated at the flagellated pole, but only after a short time following cytokinesis (data not shown). Altogether, these results suggest that PdhS recognizes a polar structure conserved in several α-proteobacteria, even those lacking a close PdhS homolog except PleC and DivJ, such as C. crescentus (Hallez et al, 2004). These data also suggest that the old pole of a large cell of B. abortus and the old pole of a C. crescentus stalked cell (i.e. stalked pole) share evolutionarily conserved characteristics.
Figure 7.
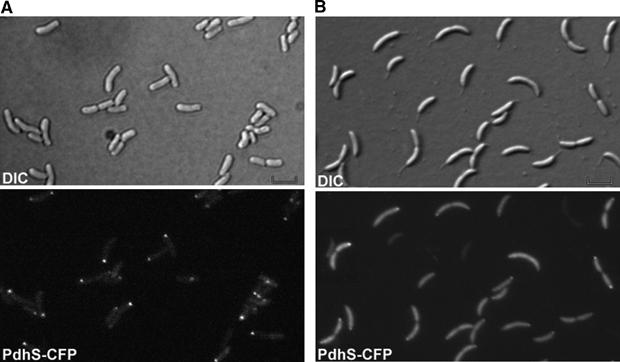
PdhS localizes at one pole of S. meliloti cells and at the stalked pole of C. crescentus cells. DIC and corresponding fluorescence images were taken with S. meliloti (A) or C. crescentus (B) cells producing PdhS-CFP from the low-copy plasmid pRH324. Scale bar, 2 μm.
Discussion
In this report, we describe the characterization of PdhS, a bacterial HK with unusual features. Indeed, it is much larger than other HK, cytoplasmic and essential. In bacteria, the majority of HK are integral membrane proteins. It is therefore not surprising that 18 out of 22 predicted HK of B. abortus comprise putative transmembrane segments. Besides PdhS, the CckA homolog, which in C. crescentus is anchored to the membrane and required for CtrA phosphorylation (Jacobs et al, 1999, 2003), seems to be also cytoplasmic in B. abortus (Supplementary Figure S3). Essential HK are rather rare, as examplified by the analysis of complete sets of knockout B. subtilis and E. coli strains, which revealed that there are, respectively, only one and none essential HK (Kobayashi et al, 2003; Baba et al, 2006). On the other hand, the systematic analysis of two-component signal transduction systems in C. crescentus identified four essential HK out of 62 (Skerker et al, 2005). Interestingly, two of these four essential HK, CckA and DivL are involved in the CtrA regulatory pathway. It is possible that the essential functions of PdhS in B. abortus is also dependent on CtrA. In S. meliloti, the closer pdhS homolog, called cbrA, is not essential (Gibson et al, 2006) and is involved in succinoglycan production. However, as previously suggested (Hallez et al, 2004), there are two PdhS homologs in S. meliloti, a long form (PdhS1) and a short one (PdhS2), and it is possible that these two proteins have redundant functions.
The functions of PdhS remain to be elucidated in detail, but it is likely that PdhS is involved in cell division control, as pdhS++ strain exhibits cell cycle defects, including minicells. In E. coli, the minB operon encodes MinC, MinD and MinE proteins, and alterations of the minB operon produce minicells (de Boer et al, 1989). This is particularly interesting as PdhS is expected to control DivK phosphorylation, itself probably involved in the regulation of CtrA, a transcriptional regulator of the minB operon. Indeed B. abortus His6-CtrA is able to bind to several sites in the minB promoter (Bellefontaine et al, 2002), and the level of a MinE-GFP-tagged protein is lower in a ctrA++ strain (R Hallez, unpublished data). In B. abortus, the products of the minB operon probably play a similar role than their counterparts in E. coli, as it was recently shown that a MinD-GFP fusion is able to oscillate from pole to pole (Hallez et al, 2007). PdhS probably controls the activity of CtrA indirectly. Indeed, it was recently demonstrated in C. crescentus that the DivJ/PleC/DivK pathway controls the activity of CtrA through the control of the CckA/ChpT phosphorelay, which itself regulates the phosphorylation and proteolytic turnover of CtrA (Biondi et al, 2006). The signal sensed by PdhS is unknown, it may be intracellular as PdhS is cytoplasmic and its subcellular distribution is maintained during both extra- and intracellular lifestyles of B. abortus.
Another particularity of PdhS is its polar localization in B. abortus. The asymmetric distribution of PdhS in the late predivisional cells implies that only the bigger cells inherit polar PdhS. The nature of the segregated functions of PdhS is presently unknown, but by homology to C. crescentus and S. meliloti, it is likely that DivK phosphorylation and/or distribution is affected by PdhS. Four observations are in favor of this hypothesis: (i) polar localization of DivK is dependent on its phosphorylation state, as the D53A mutation abolished the ability of DivK to localize at the pole(s), (ii) the phosphorylation-dependent localization of DivK is not under the control of PleC or DivJ, two other HK interacting with DivK in the Y2H test, as DivK-YFP remains associated with the pole(s) of B. abortus ΔpleC and ΔdivJ cells, (iii) DivK interacts with PdhS in a Y2H experiment (Figure 2) and (iv) in the cells where DivK-YFP is associated to one pole, PdhS-CFP colocalizes with it at the same pole (Figure 5). PdhS, therefore, constitutes the most likely candidate for the control of phosphorylation and/or distribution of DivK. However, we cannot exclude the possibility that DivJ and PleC act as phosphatases of DivK in B. abortus, which should lead to the accumulation of DivK at the pole(s) for example, or yet that both these HK control the phosphorylation state of DivK in other circumstances (e.g. in other culture media or during intracellular infection). Unexpectedly, after division, the small cells are mostly devoid of polar PdhS and as pdhS is essential, it is possible that the small cells are not able to enter into a new cell cycle as such, that is, without PdhS polarly localized. It would therefore be likely that the small cells must maturate, which would include the presence of PdhS at one pole, before being able to produce progeny cells. This model is analogous to the one described for C. crescentus (Figure 8). Indeed, the C. crescentus swarmer cells must differentiate into stalked cells to become competent for replication and division (Ausmees and Jacobs-Wagner, 2003; Quardokus and Brun, 2003). From the model proposed in Figure 8, it is also striking that B. abortus PdhS presents a subcellular localization very similar to the one described for DivJ in C. crescentus. However, ccDivJ and the N-terminal part of PdhS do not share detectable similarity. The presence of a supplemental HK potentially controlling the DivK activity suggests an evolutionary plasticity for this regulatory network. On the other hand, despite the fact that PleC is the most conserved HK of DivK (Hallez et al, 2004), it does present a completely different subcellular localization in B. abortus and C. crescentus (Figure 8), again illustrating the malleability of this network in the course of evolution.
Figure 8.
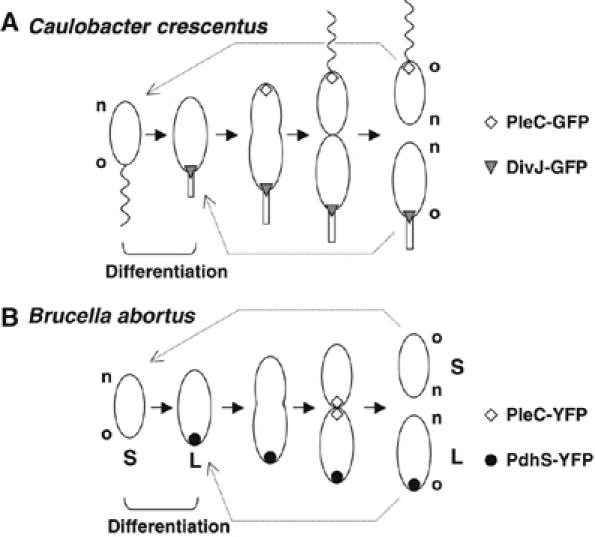
Hypothetical model for B. abortus cell cycle (A) compared with C. crescentus (B). The localization of PdhS and PleC is shown for B. abortus, whereas the localization of PleC and DivJ is shown for C. crescentus. The small (S) and large (L) cells are annotated for B. abortus. Old (o) and new (n) poles are indicated for B. abortus and C. crescentus.
As B. abortus PdhS is able to be polarly localized in S. meliloti and C. crescentus, it is likely that a conserved structure anchoring PdhS to the pole is present in these bacteria. PdhS-YFP or PdhS-CFP is not polarly localized in E. coli (J Mignolet, unpublished data). The nature of the polar anchoring structure remains to be discovered. The conservation of such a structure indicates that polar specialization, that is, the anchoring of particular functions to a bacterial cell pole, may be an evolutionarily ancient trait, maybe already present in the ancestor(s) of α-proteobacteria. This would mean that bacterial differentiation is probably not a recent acquisition in the course of evolution. Interestingly, conservation of polar localization of a protein between bacterial and eukaryotic species has been described for the B. subtilis DivIVA protein, which is targeted to the poles of both E. coli and notably Schizosaccharomyces pombe cells (Edwards et al, 2000).
The fact that PdhS localization is maintained during an intracellular infection indicates that polar specialization suggested here is not restricted to B. abortus grown in bacteriological culture medium. Furthermore, polar structures may be required for the appropriate function of virulence factors. Indeed, it was reported that all A. tumefaciens VirB proteins are polarly localized (Judd et al, 2005).
In several cell types, Brucellae invade and replicate in professional and non-professional phagocytes by inhibiting fusion between phagosome and lysosome, and subsequently creating an intracellular niche inside which they replicate (Gorvel and Moreno, 2002; Celli et al, 2003). When intracellular Brucellae avoid fusion with phago-lysosomes, they do not replicate. A possible reason would be that only the smaller progeny cells are able to successfully internalize phagocytes and deviate the intracellular traffic. Once the small cells reach the replication niche, they could differentiate into larger ones capable of initiating active replication. This would be in keeping with the fact that the small B. abortus cells correspond to the swarmer cells of C. crescentus, which are not competent for replication and must differentiate into stalked cells before initiating a new round of replication. It could also be proposed that several cell types have different functions regarding the control of the host immune response. The specific functions of each B. abortus daughter cell will have to be studied further. Identifying Brucella spp proteins specifically located in one cell type or at one cell pole could help in discovering functions segregated in the different cell types. The availability of the complete collection of all the B. melitensis predicted coding sequences (Dricot et al, 2004) constitutes a powerful post-genomic resource for such identifications.
Asymmetric division was also observed in other host-associated bacteria such as A. tumefaciens and S. meliloti (Hallez et al, 2004). The work performed by Lam et al (2003) to describe the subcellular localization of SMDivK in S. meliloti and the evidence that PdhS-CFP is present at one pole (probably the old pole of the large cells) in S. meliloti (Figure 7A) support the functionality of the asymmetric division of this symbiont. As Sinorhizobium spp is also an intracellular bacterium like Brucella spp, it is tempting to speculate that cell types generated by asymmetric division play different role(s) for establishing the complex host–bacteria relationship. Moreover, it is known that S. meliloti goes through a differentiation program during its intracellular lifestyle, that is, the formation of bacteroids indispensable for symbiosis establishment, suggesting that more than two cell types may be produced by this bacterium.
In conclusion, PdhS is the first polar differentiation marker identified in B. abortus, a bacterium that was not suspected to undergo asymmetric division until recently (Hallez et al, 2004). This suggests that cellular differentiation may be much more widespread than previously expected in the prokaryotic world.
Materials and methods
Strains, plasmids and media
All Brucella strains used in this study (Supplementary Table SII) were derived from B. abortus 544 NalR (spontaneous nalidixic acid-resistant mutant), and were routinely cultivated in 2YT. C. crescentus CB15N was grown in peptone–yeast extract (PYE complex media). S. meliloti was grown in Luria–Bertani (LB) broth supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2. E. coli strains DH10B (Invitrogen Life-Technologies), DB3.1 (Invitrogen Life-Technologies), S17-1 (Simon et al, 1983), DH5 (Bethesda Research Laboratories) and MT616 (Finan et al, 1986) were cultivated in LB broth. Antibiotics were used at the following concentrations when appropriate: nalidixic acid, 25 μg/ml; kanamycin, 20 μg/ml; chloramphenicol, 20 μg/ml; ampicillin, 100 μg/ml; gentamicin, 50 μg/ml. Plasmids (Supplementary Table SII) were mobilized from E. coli strain S17-1 into B. abortus and C. crescentus by bacterial conjugation (Ely, 1991), and from E. coli strain DH5 into S. meliloti by triparental mating, as described previously by Glazebrook and Walker (1991). Growth media and yeast genetic techniques have been described previously (Sherman, 1991). Full details about the Y2H assays are available in Supplementary data.
Molecular techniques
DNA manipulations were performed according to standard techniques (Ausubel, 1989). The mode of construction of strains and plasmids (Supplementary Table SII) as well as the sequences of all primers is available in Supplementary data and in Supplementary Table SI, respectively.
Plasmid swap experiment
To demonstrate the essentiality of pdhS, we developed a plasmid swap experiment (Table II) based on the instability of two vectors from the same incompatibility group in the same bacterial population. For this, we first introduced by conjugation the plasmid pBBR1-MCS4 and the pRH003 derivatives named pRH404, pRH405 and pRH406 (all derived from the pBBR1-MCS4 vector encoding resistance to ampicillin) into B. abortus 544 WT/pRH232 and B. abortus 544 ΔpdhS/pRH232 (XDB1103) strains. We selected only the presence of pRH003-derived vectors with ampicillin. Then three independent clones for each strain were cultivated in the presence of ampicillin without chloramphenicol (required for the selection of pRH232) during at least 10 generations and the cultures were on 2YT plates with ampicillin. Then, 100 colonies for each clone of each strain were replicated onto 2YT-Amp and 2YT-Cm. Values presented in Table II represent percentages of chasing plasmid pRH232 (clones AmpR/CmS, n=100).
Preparation of PdhS antibodies and immunoblotting experiments
N-terminally hexahistidine-tagged PdhS lacking the first 610 N-terminal residues was expressed from the pET-15b vector (Novagen) in E. coli BL21 (DE3) and purified as previously described (Bellefontaine et al, 2002). Purified His-tagged protein was used as an antigen to generate antibodies in rabbits as described for anti-CtrA antibodies production (Bellefontaine et al, 2002).
Western blot analysis was carried out as described previously (Dozot et al, 2006) with anti-PdhS and anti-CtrA sera at a dilution of 1/3000 and 1/2000, respectively. The monoclonal antibody anti-GFP (JL8, BD Biosciences) was used at a dilution of 1/1000 to check the stability of translational fusions to CFP, YFP and eGFP.
Microscopy
For fluorescence imaging, cell populations of B. abortus, C. crescentus or S. meliloti strains were placed on a microscope slide that was layered with a pad of 1% agarose containing PBS (Jacobs et al, 1999). Time-lapse microscopy was performed by placing strain XDB1104 on a microscope slide that was layered with a pad of 1% agarose containing 2YT-rich medium. These slides were placed on a microscope stage at room temperature (approximately 22°C). Samples were observed on a Nikon E1000 microscope through a differential interference contrast (DIC) × 100 objective with a Hamamatsu Orca-ER LCD camera. Images were taken and processed with Simple PCI (Hamamatsu).
Cellular infections and immunofluorescence labelling
Infections of bovine macrophages SV40 (Stabel and Stabel, 1995) by B. abortus XDB1104 strain were performed as described previously (Delrue et al, 2001). Anti-Brucella LPS O-chain monoclonal antibody 12G12 (Cloeckaert et al, 1993) was used. The secondary antibody used was Texas red-conjugated anti-rabbit IgG (Molecular Probes).
Supplementary Material
Supplementary data
Acknowledgments
We are very grateful to R-M Genicot for generous technical assistance during cloning procedures and to C Nijskens for performing cellular infections. We also thank L Van Melderen and B NKengfac for critical reading of the manuscript. We thank the anonymous reviewers for their interesting comments and suggestions. This work was supported by FRFC (Fonds de la Recherche Fondamentale Collective, convention 2.4521.04). R Hallez was supported by a short-term fellowship from EMBO (European Molecular Biology Organization) and FNRS (Fonds National de la Recherche Scientifique) and by a Wood-Whelan research fellowship. R Hallez was holding a PhD fellowship from the FRIA (Fonds pour la formation à la Recherche dans l'Industrie et dans l'Agriculture). J Mignolet is holding a PhD fellowship from the FNRS.
Contributions RH performed all the experiments except otherwise stated and JM contributed to the construction of divJ and pleC mutants, to the yeast two-hybrid presented in Figure 2 and to the construction of gfp fusions with pdhS and pleC. JM also tested PdhS-CFP localization in S. meliloti (Figure 7A). VVM and MW tested the interactions within and between two-component systems (Supplementary Figure S4). XDB, CJW, JJL and JV supervised the work.
References
- Ausmees N, Jacobs-Wagner C (2003) Spatial and temporal control of differentiation and cell cycle progression in Caulobacter crescentus. Annu Rev Microbiol 57: 225–247 [DOI] [PubMed] [Google Scholar]
- Ausubel FM (1989) Current Protocols in Molecular Biology. New York: John Wiley & Sons [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefontaine AF, Pierreux CE, Mertens P, Vandenhaute J, Letesson JJ, De Bolle X (2002) Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol Microbiol 43: 945–960 [DOI] [PubMed] [Google Scholar]
- Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT (2006) Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444: 899–904 [DOI] [PubMed] [Google Scholar]
- Boschiroli ML, Foulongne V, O'Callaghan D (2001) Brucellosis: a worldwide zoonosis. Curr Opin Microbiol 4: 58–64 [DOI] [PubMed] [Google Scholar]
- Celli J, de Chastellier C, Franchini DM, Pizarro-Cerda J, Moreno E, Gorvel JP (2003) Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J Exp Med 198: 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloeckaert A, Zygmunt MS, Dubray G, Limet JN (1993) Characterization of O-polysaccharide specific monoclonal antibodies derived from mice infected with the rough Brucella melitensis strain B115. J Gen Microbiol 139: 1551–1556 [DOI] [PubMed] [Google Scholar]
- de Boer PA, Crossley RE, Rothfield LI (1989) A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56: 641–649 [DOI] [PubMed] [Google Scholar]
- Delrue RM, Martinez-Lorenzo M, Lestrate P, Danese I, Bielarz V, Mertens P, De Bolle X, Tibor A, Gorvel JP, Letesson JJ (2001) Identification of Brucella spp. genes involved in intracellular trafficking. Cell Microbiol 3: 487–497 [DOI] [PubMed] [Google Scholar]
- DelVecchio VG, Kapatral V, Redkar RJ, Patra G, Mujer C, Los T, Ivanova N, Anderson I, Bhattacharyya A, Lykidis A, Reznik G, Jablonski L, Larsen N, D'Souza M, Bernal A, Mazur M, Goltsman E, Selkov E, Elzer PH, Hagius S, O'Callaghan D, Letesson JJ, Haselkorn R, Kyrpides N, Overbeek R (2002) The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc Natl Acad Sci USA 99: 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozot M, Boigegrain RA, Delrue RM, Hallez R, Ouahrani-Bettache S, Danese I, Letesson JJ, De Bolle X, Kohler S (2006) The stringent response mediator Rsh is required for Brucella melitensis and Brucella suis virulence, and for expression of the type IV secretion system virB. Cell Microbiol 8: 1791–1802 [DOI] [PubMed] [Google Scholar]
- Dricot A, Rual JF, Lamesch P, Bertin N, Dupuy D, Hao T, Lambert C, Hallez R, Delroisse JM, Vandenhaute J, Lopez-Goni I, Moriyon I, Garcia-Lobo JM, Sangari FJ, Macmillan AP, Cutler SJ, Whatmore AM, Bozak S, Sequerra R, Doucette-Stamm L, Vidal M, Hill DE, Letesson JJ, De Bolle X (2004) Generation of the Brucella melitensis ORFeome version 1.1. Genome Res 14: 2201–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DH, Thomaides HB, Errington J (2000) Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J 19: 2719–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B (1991) Genetics of Caulobacter crescentus. Methods Enzymol 204: 372–384 [DOI] [PubMed] [Google Scholar]
- Errington J (2003) Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol 1: 117–126 [DOI] [PubMed] [Google Scholar]
- Finan TM, Kunkel B, De Vos GF, Signer ER (1986) Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol 167: 66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson KE, Campbell GR, Lloret J, Walker GC (2006) CbrA is a stationary-phase regulator of cell surface physiology and legume symbiosis in Sinorhizobium meliloti. J Bacteriol 188: 4508–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Walker GC (1991) Genetic techniques in Rhizobium meliloti. Methods Enzymol 204: 398–418 [DOI] [PubMed] [Google Scholar]
- Gorvel JP, Moreno E (2002) Brucella intracellular life: from invasion to intracellular replication. Vet Microbiol 90: 281–297 [DOI] [PubMed] [Google Scholar]
- Hallez R, Bellefontaine AF, Letesson JJ, De Bolle X (2004) Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol 12: 361–365 [DOI] [PubMed] [Google Scholar]
- Hallez R, Letesson JJ, Vandenhaute J, De Bolle X (2007) Gateway-based destination vectors for functional analyses of bacterial ORFeomes: application to the Min system in Brucella abortus. Appl Environ Microbiol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halling SM, Peterson-Burch BD, Bricker BJ, Zuerner RL, Qing Z, Li LL, Kapur V, Alt DP, Olsen SC (2005) Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J Bacteriol 187: 2715–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguenel ED, Newton A (1982) Localization of surface structures during procaryotic differentiation: role of cell division in Caulobacter crescentus. Differentiation 21: 71–78 [DOI] [PubMed] [Google Scholar]
- Hung DY, Shapiro L (2002) A signal transduction protein cues proteolytic events critical to Caulobacter cell cycle progression. Proc Natl Acad Sci USA 99: 13160–13165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C, Ausmees N, Cordwell SJ, Shapiro L, Laub MT (2003) Functions of the CckA histidine kinase in Caulobacter cell cycle control. Mol Microbiol 47: 1279–1290 [DOI] [PubMed] [Google Scholar]
- Jacobs C, Domian IJ, Maddock JR, Shapiro L (1999) Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97: 111–120 [DOI] [PubMed] [Google Scholar]
- Jacobs-Wagner C (2004) Regulatory proteins with a sense of direction: cell cycle signalling network in Caulobacter. Mol Microbiol 51: 7–13 [DOI] [PubMed] [Google Scholar]
- Judd PK, Kumar RB, Das A (2005) Spatial location and requirements for the assembly of the Agrobacterium tumefaciens type IV secretion apparatus. Proc Natl Acad Sci USA 102: 11498–11503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O'Reilly M, O'Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JF, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N (2003) Essential Bacillus subtilis genes. Proc Natl Acad Sci USA 100: 4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM III, Peterson KM (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166: 175–176 [DOI] [PubMed] [Google Scholar]
- Lam H, Matroule JY, Jacobs-Wagner C (2003) The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev Cell 5: 149–159 [DOI] [PubMed] [Google Scholar]
- Latch JN, Margolin W (1997) Generation of buds, swellings, and branches instead of filaments after blocking the cell cycle of Rhizobium meliloti. J Bacteriol 179: 2373–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L (2000) Global analysis of the genetic network controlling a bacterial cell cycle. Science 290: 2144–2148 [DOI] [PubMed] [Google Scholar]
- Matroule JY, Lam H, Burnette DT, Jacobs-Wagner C (2004) Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell 118: 579–590 [DOI] [PubMed] [Google Scholar]
- Ohta N, Lane T, Ninfa EG, Sommer JM, Newton A (1992) A histidine protein kinase homologue required for regulation of bacterial cell division and differentiation. Proc Natl Acad Sci USA 89: 10297–10301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N, Newton A (2003) The core dimerization domains of histidine kinases contain recognition specificity for the cognate response regulator. J Bacteriol 185: 4424–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen IT, Seshadri R, Nelson KE, Eisen JA, Heidelberg JF, Read TD, Dodson RJ, Umayam L, Brinkac LM, Beanan MJ, Daugherty SC, Deboy RT, Durkin AS, Kolonay JF, Madupu R, Nelson WC, Ayodeji B, Kraul M, Shetty J, Malek J, Van Aken SE, Riedmuller S, Tettelin H, Gill SR, White O, Salzberg SL, Hoover DL, Lindler LE, Halling SM, Boyle SM, Fraser CM (2002) The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc Natl Acad Sci USA 99: 13148–13153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quardokus EM, Brun YV (2003) Cell cycle timing and developmental checkpoints in Caulobacter crescentus. Curr Opin Microbiol 6: 541–549 [DOI] [PubMed] [Google Scholar]
- Quon KC, Marczynski GT, Shapiro L (1996) Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84: 83–93 [DOI] [PubMed] [Google Scholar]
- Sciochetti SA, Lane T, Ohta N, Newton A (2002) Protein sequences and cellular factors required for polar localization of a histidine kinase in Caulobacter crescentus. J Bacteriol 184: 6037–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F (1991) Getting started with yeast. Methods Enzymol 194: 3–21 [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A (1983) A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 10: 783–791 [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT (2005) Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol 3: e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabel JR, Stabel TJ (1995) Immortalization and characterization of bovine peritoneal macrophages transfected with SV40 plasmid DNA. Vet Immunol Immunopathol 45: 211–220 [DOI] [PubMed] [Google Scholar]
- Wang SP, Sharma PL, Schoenlein PV, Ely B (1993) A histidine protein kinase is involved in polar organelle development in Caulobacter crescentus. Proc Natl Acad Sci USA 90: 630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L (1999) Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell 4: 683–694 [DOI] [PubMed] [Google Scholar]
- Wu J, Ohta N, Zhao JL, Newton A (1999) A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc Natl Acad Sci USA 96: 13068–13073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


