Abstract
Chromatin clusters containing CENP-A, a histone H3 variant, are found in centromeres of multicellular eukaryotes. This study examines the ability of alpha-satellite (alphoid) DNA arrays in different lengths to nucleate CENP-A chromatin and form functional kinetochores de novo. Kinetochore assembly was followed by measuring human artificial chromosome formation in cultured human cells and by chromatin immunoprecipitation analysis. The results showed that both the length of alphoid DNA arrays and the density of CENP-B boxes had a strong impact on nucleation, spreading and/or maintenance of CENP-A chromatin, and formation of functional kinetochores. These effects are attributed to a change in the dynamic balance between assembly of chromatin containing trimethyl histone H3-K9 and chromatin containing CENP-A/C. The data presented here suggest that a functional minimum core stably maintained on 30–70 kb alphoid DNA arrays represents an epigenetic memory of centromeric chromatin.
Keywords: alphoid DNA, artificial chromosome, centromere chromatin, epigenetics, heterochromatin
Introduction
The centromere is a conserved chromosomal structural element that is essential for segregation of duplicated sister chromatids to daughter cells during eukaryotic cell division. Many of the structural components of centromeres have been identified, including protein components such as CENP-A–C, -E, -F, -H, -I, -K–U and hMis12 (Foltz et al, 2006; Izuta et al, 2006; Okada et al, 2006). CENP-A, a centromere-specific histone H3 variant, is an essential centromeric protein found in human, mouse, chicken, nematode and yeast. CENP-A has been used as a marker of centromeric DNA, because centromere assembly appears to be blocked in CENP-A-deficient cells (Howman et al, 2000; Goshima et al, 2003; Amor et al, 2004). Although CENP-A is required to nucleate and maintain centromeric chromatin, the mechanism by which it promotes and stabilizes the centromere in eukaryotic cells is not yet known.
In normal human chromosomes, CENP-A colocalizes with alpha-satellite DNA (alphoid DNA). Alphoid DNA regions are 0.2–5 Mb in length and consist of tandem copies of an AT-rich ∼171-bp sequence (Lee et al, 1997). Type-I alphoid DNA forms homogeneous higher-order repeats and includes multiple CENP-B boxes, a 17-bp motif that binds CENP-B (Earnshaw et al, 1987; Masumoto et al, 1989a; Muro et al, 1992). Type-I alphoid DNA colocalizes and co-immunoprecipitates with CENP-A, CENP-B and CENP-C (Ando et al, 2002; Spence et al, 2002; Nakano et al, 2003; Lam et al, 2006).
Previous studies showed that type-I alphoid DNA from human chromosomes 5, 17, 21 and X support efficient formation of human artificial chromosomes (HACs) in human HT1080 cells. In contrast, type-II alphoid DNA from chromosome 21, neocentromere-derived sequences (see below) and Y alphoid DNA, which lack CENP-B boxes, do not form HACs in cultured human cells (Harrington et al, 1997; Ikeno et al, 1998; Ebersole et al, 2000, 2005; Saffery et al, 2001; Grimes et al, 2002; Mejia et al, 2002). Other substrates that lack the ability to form HACs include synthetic type-I alphoid DNA with mutant CENP-B boxes and GC-rich synthetic repetitive DNA containing CENP-B boxes (Ohzeki et al, 2002). These results indicate that both alphoid DNA and CENP-B boxes may be essential for nucleation of a functional CENP-A-containing centromeric structure. However, under rare circumstances, chromosome regions lacking alphoid DNA form a functional centromere-like structure known as a neocentromere (Amor and Choo, 2002; Warburton, 2004), and one of canonical centromeres on dicentric fusion chromosomes is frequently inactivated (Earnshaw et al, 1989; Sullivan and Schwartz, 1995; Warburton et al, 1997). These observations suggest that epigenetic mechanisms regulate assembly and/or disassembly of functional human centromeres.
Recent studies reported that human and Drosophila melanogaster centromeres carry interspersed dimethylated histone H3-K4 clusters and CENP-A clusters (Blower et al, 2002; Sullivan and Karpen, 2004). CENP-A chromatin clusters have also been found in a human neocentromere and in the centromere of rice chromosome 8, which contains relatively little repetitive DNA (Nagaki et al, 2004; Chueh et al, 2005). Our previous studies showed that HACs form efficiently on multimers of a 60–70 kb alphoid DNA containing YAC/BAC, and that CENP-A chromatin clusters assemble preferentially on the alphoid DNA insert interspersed with histone H3 clusters on the vector sequences with telomeres (Ikeno et al, 1998; Nakano et al, 2003; Nakashima et al, 2005). Thus, striking structural similarities as chromatins were observed for centromeres assembled on naked DNA and natural chromosomes. Moreover, HACs in living mitotic cells can align at the metaphase plate, and the chromatids of the HACs are resolved with the same timing as natural sister chromatids (Tsuduki et al, 2006). On the basis of these observations, it appears that HAC formation and stability in cultured cells may provide a model for investigating the assembly of eukaryotic centromeres. We suppose that if each CENP-A chromatin cluster required for the centromere function is smaller enough than the insert alphoid DNA in size or if CENP-A chromatin cluster easily spreads out irrespective of the underlying vector DNA sequences, the efficiency of the HAC formation is expected not to change by the size of the insert alphoid DNA.
This study investigates whether the size of alphoid DNA arrays influences assembly and maintenance of centromeres and/or their function. YACs were constructed containing 10, 30, 50 or 70 kb α21-I alphoid arrays and BACs were constructed containing 60, 120 or 240 kb α21-I alphoid DNA with wild-type or mutant CENP-B boxes. Chromatin assembly was followed by measuring HAC formation and by chromatin immunoprecipitation (ChIP) analysis. The results demonstrated that the length of alphoid DNA arrays and the density of wild-type CENP-B boxes had a strong impact on HAC formation and assembly of CENP-A chromatin.
Results
Construction of YACs and BACs with alphoid DNA inserts of different lengths
Previous studies reported construction of YAC clone α7C5, which contains 70 kb type-I alphoid DNA from the centromere of human chromosome 21 (α21-I). YAC clone α7C5 forms HACs with high efficiency in human cells (Ikeno et al, 1998). Three deletion variants of α7C5 containing 10, 30 or 50 kb alphoid DNA (del.22, del.20 and del.24, respectively) were generated by homologous recombination in yeast (Figure 1A and B). A BAC was also constructed previously with an insert of 60 kb synthetic alphoid DNA (pWTR11.32; Ohzeki et al, 2002). Both YAC clone α7C5 and this BAC are based on the same repeating 11-mer motif (α21-I) and both readily form HACs. Because a long alphoid DNA is very unstable in YAC vector (Neil et al, 1990; Ikeno et al, 1998), BACs containing 120 and 240 kb alphoid DNA inserts (pWTR11.64 and pWTR11.128, respectively) were constructed from the 60 kb BAC by in vitro tandem ligation (Figure 1D and E).
Figure 1.
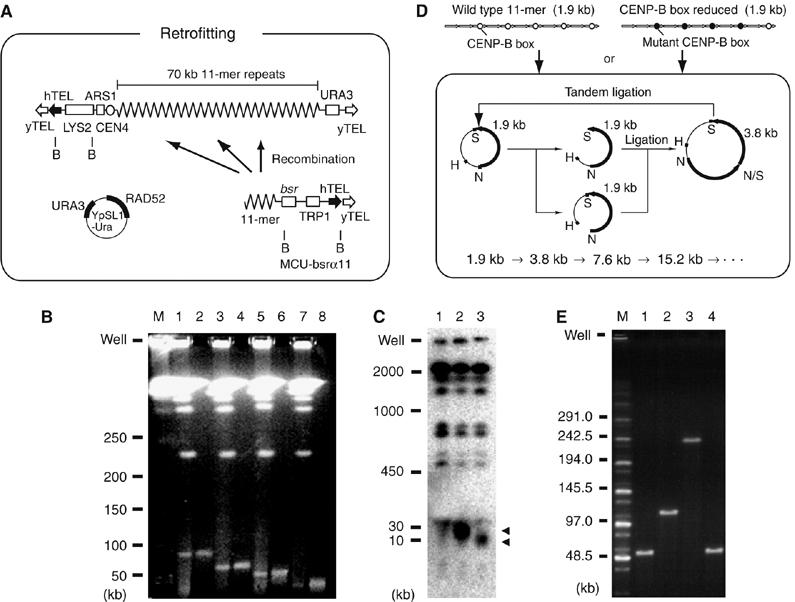
Construction of YACs and BACs with shorter or longer alphoid DNA inserts. (A) The method for construction of deleted YACs using homologous recombination in yeast. B indicates the BamHI site. (B) Constructs were analyzed by PFGE, followed by ethidium bromide staining. Gel-purified YAC DNA was analyzed in lanes 2, 4, 6 and 8. Lanes 1 and 2, α7C5htel; lanes 3 and 4, del.24; lanes 5 and 6, del.20; lanes 7 and 8, del.22; M indicates DNA size marker. (C) HT1080 genomic DNA transformed with the YAC constructs were digested with BamHI and analyzed by PFGE, followed by Southern blot using a 32P-labeled 11-mer probe. The arrowheads indicate input alphoid DNAs. Lane 1, HT1080; lane 2, del.20HT1–5; lane 3, del.22HT1–3. (D) BACs with alphoid DNA with wild-type or mutant CENP-B boxes were constructed as shown. H, N and S indicate HindIII, NheI and SpeI sites, respectively. (E) BACs were digested with NheI and SpeI and analyzed by PFGE, followed by ethidium bromide staining. Lane 1, pWTR11.32; lane 2, pWTR11.64; lane 3, pWTR11.128; lane 4, pCBB11.1.32. M indicates DNA size marker.
Dependence of HAC formation on the length of alphoid DNA
Alphoid DNA-containing YACs were purified and transfected into human HT1080 cells. Stable transformants were identified based on resistance to blasticidin S (BS) and HACs were identified in transfected cells using fluorescence in situ hybridization (FISH) with probes for alphoid DNA (the 11-mer) and the YAC vector. The efficiency of HAC formation was similar for YACs with 50 or 70 kb alphoid DNA (33 or 35% HAC-positive cell lines, respectively); however, the efficiency of HAC formation was significantly lower for YACs with a 30 kb alphoid DNA insert (9%) (Figure 2A and B; Table IA, P<0.001), and YACs with a 10 kb alphoid DNA insert did not form at all (Figure 2C; Table IA). FISH analysis suggested that HACs with 70, 50 and 30 kb alphoid DNA arrays contained no host chromosomal DNA signals (Supplementary Figure 2). YAC copy number per cell was estimated using pulse-field gel electrophoresis (PFGE), Southern blot and real-time PCR (Tables IIA and B; Figure 1C). These data indicated that multiple YAC copies concatenated and integrated as an extra artificial chromosome or into a host chromosomal insertion site. HACs were mitotically stable in HT1080 cells for 60 days in the absence of drug selection (R=0.00010–0.0086; chromosomal loss rate per cell division, Table IIA). The efficiency of HAC formation decreased from approximately 26 to 17 to 5%, for BACs with 60, 120 and 240 kb alphoid DNA inserts, respectively (Table IB; Figure 2D). The transformation efficiency for all BACs was similar. These results indicate that the ability to form HACs is strongly influenced by the length of the alphoid DNA region in YACs and BACs.
Figure 2.
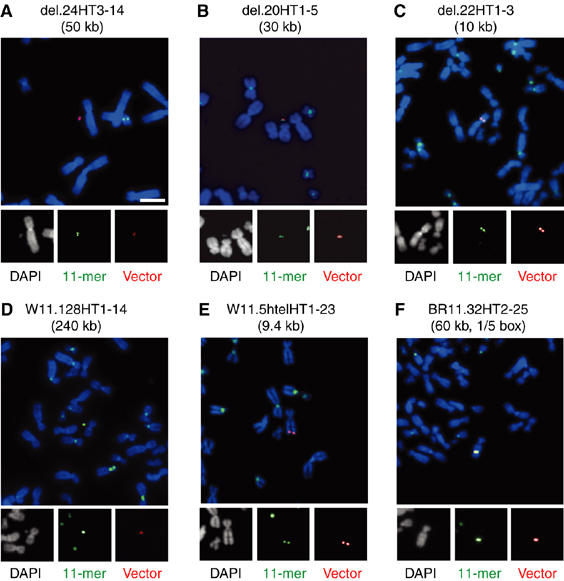
FISH analysis of HAC formation and integration. Metaphase cells were prepared from cell lines with non-integrated (A: del.24HT3–14, B: del.20HT1–5 and D: W11.128HT1–14) and integrated YACs/BACs (C: del.22HT1–3, E: W11.5htelHT1–23 and F: BR11.1.32HT2–25). FISH analysis was carried out using probes for the α21-I alphoid 11-mer (green) and YAC vector sequences (red). Chromosomes were counterstained with DAPI (white and blue in merged panels). Scale bar, 10 μm.
Table 1a.
HAC formation assay: DNA constructs containing short series of alphoid DNA (linear constructs)
| Size of input DNA (kb) | ||||||
|---|---|---|---|---|---|---|
| Input DNA | Alphoid | Vector left arm | Vector right arm | Total | Analyzed cell lines | HAC formed cell linesa |
| α7C5htel | 70 | 15 | 11 | 96 | 34 | 12 (35%) |
| del.24 | 50 | 15 | 11 | 76 | 9 | 3 (33%) |
| del.20 | 30 | 15 | 11 | 56 | 34 | 3 (9%) |
| del.22 | 10 | 15 | 11 | 36 | 41 | 0 |
| pNeo11.5 | 9.5 | 2.5 | 3.5 | 15.5 | 23 | 0 |
| pNeo11.5htel | 9.5 | 4 | 5 | 18.5 | 23 | 0 |
| aCell lines in which >50% of spreads (n>20) contained HACs were categorized into HAC formed cell lines. A χ2-test of the predominant pattern for HAC formation efficiency between α7C5htel cell line and other cell lines except for del.24 showed highly significant differences (P<0.001), but that of del.24 did not (P>0.90). | ||||||
Table 2a.
CENPs assembly and centromere function on introduced alphoid DNAs: HAC formation cell lines
| Input DNA size (vector) | Cell lines | Multiplicity of input DNA | CENPs assembly (%)a | de novo HACb | Loss ratec | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Left arm (copies) | Right arm (copies) | Total size of alphoid (kb)d | A | B | C | E | ||||
| Linear constructs | ||||||||||
| 70 kb (α7C5htel) | HT1–2e | 34 | 35 | 2500 | 100 | 100 | 100 | 100 | Yes | 0.0086 |
| 50 kb (del.24) | del.24HT3–14 | 35 | 42 | 1900 | 100 | 100 | 100 | 100 | Yes | 0.0049 |
| 30 kb (del.20) | del.20HT1–5 | 74 | 68 | 2100 | 100 | 100 | 100 | 100 | Yes | 0.00010 |
| del.20HT5–2 | 160 | 160 | 4800 | 100 | 100 | nt | 100 | Yes | nt | |
| Circular constructs | ||||||||||
| 60 kb (pWTR11.32) | W0210R-8f | − | − | 1400 | 100 | 100 | 100 | 100 | Yes | 0.0011 |
| 240 kb (pWTR11.128) | W11.128HT1–14 | − | − | 1500 | + | + | nt | nt | Yes | nt |
| aPercentage of centromere protein assembly was assessed based on colocalized signals from indirect immunostaining of CENP and FISH analysis of vector probes (n>20). | ||||||||||
| bNon-acquisition of host chromosomal DNA fragments was examined by FISH analysis using inter-/intra-Alu, other alphoid probes. | ||||||||||
| cThe number of HACs in 30–50 metaphase cells was scored by FISH using YAC probes and 11-mer probes at day 0 (N0) or at day 60 (N60) without selection. Values were calculated using the following formula: N60=N0 × (1–R)60. | ||||||||||
| dTotal size of alphoid DNA in multimer of input alphoid YAC was calculated by multiplying alphoid DNA size of input alphoid YAC by copy number of average of left arm and right arm. | ||||||||||
| eThese cell lines were produced by Ikeno et al (1998). | ||||||||||
| fThese cell lines were produced by Ohzeki et al (2002) | ||||||||||
| nt, not tested; +, assembly detected with ChIP assay. | ||||||||||
Table 2b.
CENPs assembly and centromere function on introduced alphoid DNAs: ectopic alphoid DNA integration sites
| Input DNA size (vector) | Cell lines | Multiplicity of input DNA | CENPs assembly (%)a | TSA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Left arm (copies) | Right arm (copies) | Total size of alphoid (kb)b | A | B | C | E | CENP-A assemblya (%) | Extra chromosome formationc (%) | ||
| Linear constructs | ||||||||||
| 70 kb (α7C5htel) | HT1-19d | 34 | 35 | 2400 | 17 | 20 | 17 | nt | 65 | 30 |
| HT2-1d | nt | nt | − | 13 | 39 | 23 | nt | 50 | 16 | |
| 50 kb (del.24) | del.24HT5–11 | 26 | 24 | 2500 | 13 | 100 | nt | nt | 45 | 20 |
| del.24HT5–20 | 27 | 32 | 1500 | 13 | 100 | nt | nt | 40 | 15 | |
| 30 kb (del.20) | del.20HT5–6 | 58 | 53 | 1700 | 25 | 100 | nt | 23 | nt | nt |
| del.20HT5–24 | 18 | 14 | 480 | 20 | 100 | nt | 25 | 40 | 20 | |
| 10 kb (del.22) | del.22HT1–3 | 94 | 101 | 980 | 0 | 1008 | 0 | nt | 23 | 0 |
| del.22HT5–7 | 53 | 38 | 480 | 0 | 100 | nt | nt | nt | nt | |
| del.22HT5–18 | 27 | 23 | 250 | 0 | 100 | nt | nt | 15 | 0 | |
| 9.4 kb (pNeo11.5) | W11.5HT1–16 | nt | 173 | 1600 | 0 | 100 | nt | nt | 10 | 0 |
| 9.4 kb (pNeo11.5htel) | W11.5htelHT1–23 | nt | 120 | 1100 | 0 | 100 | nt | nt | 10 | 0 |
| Circular constructs | ||||||||||
| 60 kb (pWTR11.32) | W0210R-1e | − | 14f | 1000 | 23 | 100 | nt | nt | nt | nt |
| 240 kb (pWTR11.128) | W11.128HT1–5 | − | 5f | 1200 | + | + | nt | nt | nt | nt |
| 240 kb (pWTR11.128) | W11.128HT4–3 | − | 6f | 1100 | + | + | nt | nt | nt | nt |
| 60 kb (pCBB11.1.32) | BR11.32HT2–25 | − | 42f | 4000 | + | + | nt | nt | nt | nt |
| 60 kb (pMTR11.32) | M1319e | − | 12f | 1100 | 0 | 0 | 0 | 0 | nt | nt |
| aPercentage of centromere protein assembly was assessed based on colocalized signals from indirect immunostaining of CENP and FISH analysis of vector probes (n>20). | ||||||||||
| bTotal size of alphoid DNA in multimer of input DNA was calculated by multiplying alphoid DNA size of input DNA by copy number of average of left arm and right arm. | ||||||||||
| cFormation of extra-chromosomes with active centromere formed on ectopic integration of input DNA were examined by immunostaining of CENP and FISH analysis of vector probes. | ||||||||||
| dThese cell lines were produced by Ikeno et al (1998). | ||||||||||
| eThese cell lines were produced by Ohzeki et al (2002). | ||||||||||
| fThese constructs are circular, therefore Neo gene in BAC vector sequence was quantified by real time-PCR. | ||||||||||
| gAmong them, 17% could be detected by the mixture of antibodies against CENP-B. | ||||||||||
| nt, not tested; +, assembly detected only with ChIP assay. | ||||||||||
Table 1b.
HAC formation assay: DNA constructs containing long series of alphoid DNA or CENP-B box-reduced alphoid DNA (circular constructs)
| Input DNA | Size of input DNA (kb) | Experiments | Analyzed cell lines | HAC formed cell lines | ||
|---|---|---|---|---|---|---|
| Alphoid | Vector | Total | ||||
| pWTR11.32 | 60 | 7 | 67 | 1st | 11 | 3 (27%) |
| 2nd | 12 | 3 (25%) | ||||
| pWTR11.64 | 120 | 7 | 127 | 1st | 24 | 4 (17%) |
| 2nd | 18 | 3 (17%) | ||||
| pWTR11.128 | 240 | 7 | 247 | 1st | 19 | 1 (5%) |
| 2nd | 23 | 1 (4%) | ||||
| pCBB11.1.32 | 60 | 7 | 67 | 1st | 23 | 0 |
| 2nd | 18 | 0 | ||||
| pMTR11.32a | 60 | 7 | 67 | 1st | 20 | 0 |
| 2nd | 18 | 0 | ||||
| A χ2-test of the predominant pattern of HAC formation efficiency between pWTR11.32 cell line and other cell lines except for pWTR11. 64 showed highly significant differences (P<0.002), but that of pWTR11.64 did not (P>0.15). | ||||||
| This construct contains 60 kb alphoid DNA with mutant-type CENP-B boxes, and is previously analyzed by Ohzeki et al (2002). | ||||||
Assembly and spreading of CENP-A chromatin in HACs
The protein and DNA components of HACs were analyzed using immunofluorescence and FISH analysis of metaphase chromosomes (Figure 3A; Table IIA). The results confirmed the presence of CENP-A, CENP-B and CENP-E in the HACs (CENP-E is a kinesin-like kinetochore motor protein, which is also involved in mitotic checkpoints; Thrower et al, 1995). ChIP was used to examine the assembly of CENP-A chromatin in six HAC regions. HACs with 30 or 70 kb alphoid DNA arrays were used for this experiment. The HAC sites examined included three regions (regions 1–3) on the left arm of the YAC located at 8, 4 or 2 kb from alphoid DNA, respectively; two regions (regions 4 and 5) bridging the left and right ends of alphoid DNA, and the bsr gene in the right arm of the YAC at 3 kb from alphoid DNA (region 6). 5S ribosomal DNA (5SrDNA) was used as a negative control and the 11-mer alphoid DNA motif was used as a positive control. The presence of CENP-A and CENP-B in these regions was quantified by real-time PCR. Relative enrichment of CENP-A in HAC regions was calculated (normalized to 5SrDNA negative control) and is presented in Figure 3B and C. The results indicated that for HACs with 70 kb alphoid DNA, CENP-A nucleosomes are only enriched in regions 4 and 5 (boundaries of alphoid DNA at the left and the right vector arm junctions, P<0.01) but not in YAC vector DNA (regions 1–3 or 6, P>0.10). These observations are consistent with previous results (Nakano et al, 2003; Nakashima et al, 2005).
Figure 3.
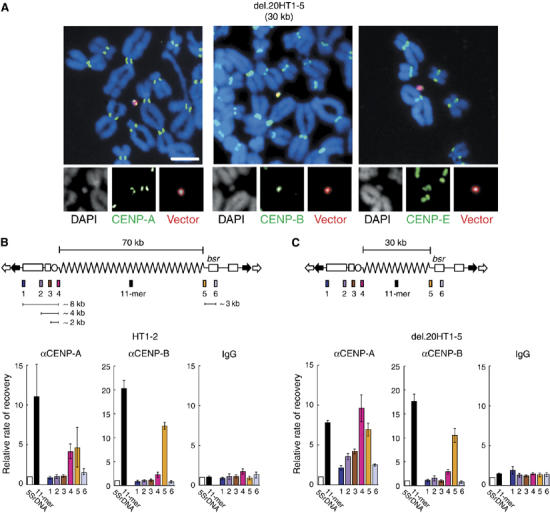
Analysis of CENP-A chromatin in HACs. (A) Indirect immunostaining and FISH analysis of del.20HT1–5. Green fluorescent secondary antibodies were used to detect CENP-A, -B or -E. Red fluorescent signal indicates hybridization to YAC vector probes. Scale bar, 10 μm. (B, C) ChIP and real-time PCR analysis of HAC cell lines 7C5HT1–2 and del.20HT1–5. The diagrams of YAC structures indicate one unit of the concatenated multimers found in HACs. Relative enrichment of YAC regions 1–6 was quantified as described. Results were normalized to 5S ribosomal DNA and are presented as a bar graph. Normal IgG was used as a negative control. Error bars indicate s.e.m. (n=3). Enrichment of CENP-A in regions 4 and 5 in (B) and in regions 2–5 in (C) was statistically significant (P<0.01) by χ2 test, relative to values with normal IgG; values for regions 1, 2, 3 and 6 in (B), and regions 1 and 6 in (C) were not statistically significant (P>0.10) by χ2 test.
Unexpectedly, for HACs with 30 kb alphoid DNA arrays, regions 2 and 3 were enriched for CENP-A nucleosomes (P<0.01; Figure 3C), whereas regions 1 and 6 were not (P>0.10; Figure 3C). Similar results were obtained using an independent HAC cell line carrying this YAC (del.20HT5–2) and at the integration site (del.20HT5–6) of this YAC (Supplementary Figure 3A and B; Tables IIA and B). CENP-B was enriched exclusively at region 5 (right boundary of alphoid DNA, which includes a CENP-B box), but not in the YAC arms (regions 1, 2, 3 and 6), and only slightly at region 4 (the left boundary of alphoid DNA, which does not include a CENP-B box; Figure 3B and C). CENP-B distribution was similar for HACs with 30 and 70 kb alphoid DNA arrays. These results suggest that the assays used here are highly reliable and have a high enough resolution to detect one or two alphoid DNA units of 171 bp each.
HACs with 30 kb alphoid DNA formed less efficiently than HACs with 70 kb alphoid DNA, but CENP-A nucleosomes assembled more efficiently at the alphoid–vector junctions and at the left vector arm sites in HACs with 30 kb alphoid DNA. It should be noted that the latter sites do not nucleate CENP-A nucleosomes by themselves. However, the chromatin structure noted above was observed in two independent HACs with 30 kb alphoid DNA arrays. One possible interpretation of these results is that CENP-A chromatin initiates in the 30 kb alphoid DNA array, but the size of this region is not sufficient to support HAC formation. Therefore, CENP-A chromatin may propagate beyond the boundaries of the insert to include flanking vector sequence. This suggests that a region greater than 30 kb in size is necessary to achieve a stable CENP-A chromatin domain and to establish normal centromere function in the context of a YAC-derived HAC. This structure may also be essential for centromere function in HACs containing 30 kb alphoid DNA. Using FISH and immunostaining, we estimated that 9–33% of the total 70 copies of the 30 kb alphoid YAC units on the contiguously extended HAC chromatin are possibly involved in the maintenance of the CENP-A chromatin areas (Supplementary Figure 4).
CENP-A chromatin assembly and the length of alphoid DNA at chromosomal insertion sites
The presence of centromere proteins was also assessed at the chromosomal insertion sites of YACs with 10, 30 or 70 kb alphoid DNA. For cells transfected with the YAC with 70 kb alphoid DNA or with the BAC with 60 kb alphoid DNA, 10–30% of the cells had variegated assembly of CENP-A, -B, -C and -E at the insertion site. This was true regardless of YAC copy number or the site of integration (Figure 4A; Table IIB; Ohzeki et al, 2002; Nakashima et al, 2005). The presence of CENP-A and -E always correlated with a phenotype of sister chromatid cohesion at the insertion sites in 20–25% of cells with 30 kb alphoid DNA arrays (del.20HT5–6 and 5–24 cell lines, respectively, Table IIB; Figure 4A). However, centromere protein components and sister chromatid cohesion were not associated with the remaining 75–80% of cells with 30 kb alphoid DNA arrays or at any insertion sites of 10 kb alphoid DNA arrays (Table IIB). By ChIP analysis, CENP-A nucleosomes were slightly enriched at the insertion sites of 30, 60 or 70 kb alphoid DNA arrays (Supplementary Figure 3B; Table IIB; see also Figure 5C), but not at insertion sites for 10 kb alphoid arrays (Figure 4C). This is consistent with the cytological observations noted above. These data confirm the conclusion that the length of an alphoid DNA array (up to 70 kb) correlates positively with the capacity to form and maintain functional HACs, to assemble CENP-A and other centromeric proteins, and to promote sister chromatid cohesion at ectopic insertion sites in HT1080 cells.
Figure 4.
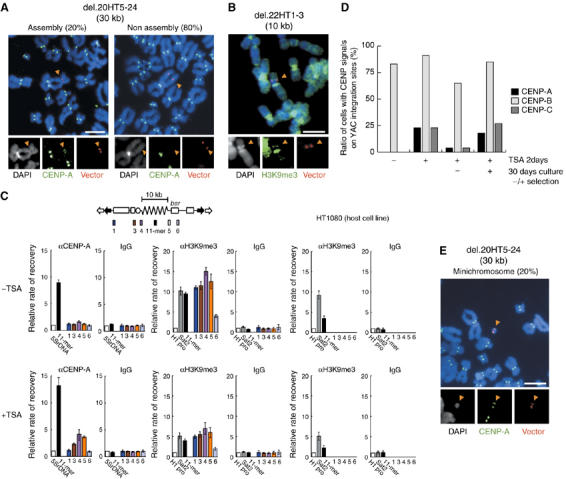
Analysis of CENP-A and H3K9me3 chromatin in alphoid DNA arrays. (A) Detection of CENP-A in 30 kb alphoid DNA arrays in del.20HT5–24 cell line. (B) Detection of H3K9me3 in 10 kb alphoid DNA array in del.22HT1–3 cell line. (A, B) Indirect immunostaining (green) and FISH (red) were carried out. Arrowheads indicate YAC integration site. Scale bar, 10 μm. (C) ChIP and real-time PCR analysis of CENP-A and H3K9me3 in 10 kb alphoid DNA array in del.22HT1–3 cells and parental HT1080 cells before or after treatment with TSA. The diagram of the YAC structure indicates one unit of the concatenated multimers found at ectopic chromosomal insertion sites. Enrichment of CENP-A or H3K9me3 was calculated as described. Results were normalized to 5S ribosomal DNA in CENP-A panels and H1 promoter in H3K9me3 panels, and are presented as a bar graph. Normal IgG was used as a negative control. DNA probes were for 11-mer, Sat2, regions 1–6 on the YAC and the H1 promoter. Error bars indicate s.e.m (n=3). Enrichment of CENP-A in regions 3, 4 and 5 was statistically significant in TSA treated cells (P<0.01) by χ2 test, relative to values with normal IgG; values in regions 1 and 6 were not statistically significant (P>0.10) by χ2 test. (D) Cells were treated with TSA for 2 days, incubated for 30 days in the presence or absence of blastocidin S and analyzed for CENPs assembly by immunofluorescence and FISH. The fraction of del.22HT1–3 cells positive for CENPs assembly are indicated. (E) Stable reformed minichromosomes (20% of cells) were detected by activated centromere signal (CENP-A, green) in the 30 kb alphoid DNA array in TSA-treated del.20HT5–24 cells. YAC probe is shown in red. Scale bars, 10 μm.
Figure 5.
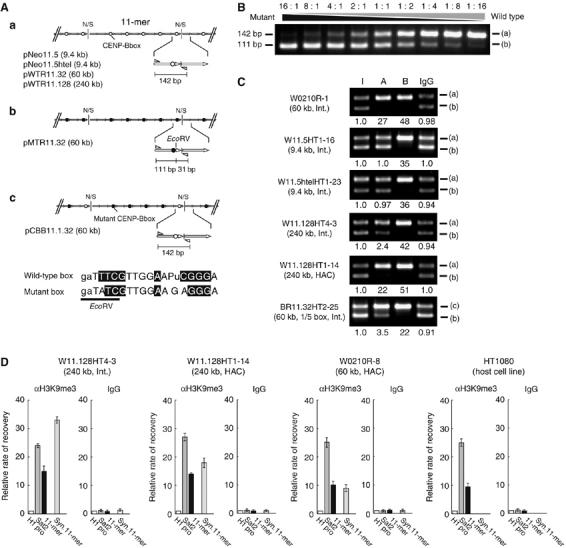
ChIP analysis of CENP-A, CENP-B and H3K9me3 in alphoid DNA arrays with wild-type or mutant CENP-B boxes. (A) Schematic diagram of synthetic alphoid DNA 11-mers. (a) Synthetic wild-type 11-mer constructs with different lengths. (b) Synthetic 11-mer with mutant CENP-B boxes. (c) Synthetic 11-mer with four mutant CENP-B boxes. The primer sites (arrow heads) used for the competitive PCR are shown. N/S indicates cohesive end of the ligated NheI and SpeI sites. The 17-bp motif of CENP-B box is shown as wild-type or mutant. The EcoRV site exists only on the mutant CENP-B box. (B) Control for competitive PCR. W0210R-1 cells carrying wild-type CENP-B boxes and M1319 cells carrying mutant CENP-B boxes were mixed in the indicated ratios (16:1–1:16). Competitive PCR was carried out with genomic DNA, and EcoRV-digested PCR products were analyzed by electrophoresis. (C) ChIP and competitive PCR analyses assays were carried out as described. I indicates input DNA. Antibodies to CENP-A, CENP-B or normal mouse IgG were used for immunoprecipitation. Fold enrichment was calculated by the ratio of the upper and lower bands and normalized to input DNA. (a), (b) and (c) are the DNA fragments corresponding to the alphoid constructs (a), (b) and (c), respectively, in panel A. (D) ChIP and real-time PCR analysis of H3K9me3 assembly on the synthetic alphoid DNA array in W11.128HT4–3, W11.128HT1–14 and W0210R-8 cells. Enrichment of H3K9me3 was calculated as described. Results were normalized to the H1 promoter DNA and are presented as a bar graph. Normal IgG was used as a negative control. DNA probes were for Sat2, endogenous 11-mer, synthetic 11-mer and the H1 promoter. Error bars indicate s.e.m. (n=3).
H3K9me3 chromatin is antagonistic to CENP-A chromatin in a 10 kb alphoid DNA array
Previous studies indicated that marker gene expressions are repressed in chromatin with a low level of acetylated histone H3 and/or a high level of lysine 9 tri-methyl histone H3 (H3K9me3), and that CENP-A nucleosomes were also depleted from the ectopic insertion sites of the alphoid YAC/BAC DNA (Nakano et al, 2003; Nakashima et al, 2005). Here, the distribution of H3K9me3 was examined as a possible marker of heterochromatin and associated gene repression (Lachner et al, 2003; Peters et al, 2003). Indirect immunofluorescence and FISH studies revealed dense regions of H3K9me3 nucleosomes overlapping with the insertion site of YACs with 10 kb alphoid DNA arrays (del.22HT1–3 cell line) (Figure 4B). Stronger staining was observed in pericentromeric heterochromatin than on chromosome arms.
ChIP analysis was also used to correlate density of H3K9me3 with specific loci. Pericentromeric satellite 2 (sat2), used as a positive control, correlated with high levels of H3K9me3 (Espada et al, 2004), as expected, and low levels of H3K9me3 were found in the promoter of the housekeeping H1 gene (Baer et al, 1990), a negative control (Figure 4C). To assess the amount of H3K9me3 at sites of insertion of YACs with 10 kb alphoid DNA, data were normalized to the negative control (the H1 promoter). In the del.22 HT1–3 cell line, H3K9me3 was >10-fold enriched in regions 1, 3, 4 and 5, whereas it was only four-fold enriched in region 6 (the bsr gene) (Figure 4C). H3K9me3 was nine-fold enriched in the 11-mer alphoid DNA repeat using a probe that detects ectopic alphoid DNA as well as endogenous alphoid DNA on chromosomes 13 and 21, whereas it was only three-fold-enriched in control HT1080 cells (Figure 4C). These data indicate that H3K9me3 was enriched in most of the integrated YAC with 10 kb alphoid DNA, suggesting that H3K9me3 chromatin may prevent assembly of CENP-A chromatin on ectopic alphoid DNA. To examine this possibility, del.22 HT1–3 cells were treated with the histone deacetylase inhibitor Trichostatin A (TSA), which causes histone hyperacetylation and disrupts heterochromatin assembly (Ekwall et al, 1997; Taddei et al, 2001). For cells with insertions of YACs with 10 kb alphoid DNA, H3K9me3 was on the whole lower, but CENP-A was enriched 4.1-, 3.6- and 2.3-fold at regions 4, 5 and 3 (P<0.01), respectively (Figure 4C), and unchanged at regions 1 and 6 by ChIP analysis. CENP-A and CENP-C assemblies were also detected at the ectopic insertion sites in 23% of the TSA-treated cells, as indicated by immunofluorescence and FISH analysis (Figure 4D; Table IIB; Supplementary Figure 5). When TSA-treated cells were grown for 30 days without selecting for the bsr marker, the fraction of cells with CENP-A/CENP-C signals decreased from 23 to 4%. However, in cells grown for 30 days with selection for the bsr marker gene, CENP-A/CENP-C chromatin was stable. These data suggest that 10 kb alphoid DNA arrays can form CENP-A chromatin when histone deacetylase is inhibited and, therefore, heterochromatin is disrupted, but this CENP-A chromatin conformation is unstable under non-selective conditions.
Stable reformation of minichromosomes with an activated centromere via chromosome breakage events were frequently observed when cells with 70 or 30 kb ectopic alphoid DNA arrays were treated with TSA or were maintained for long periods under selection for bsr (Figure 4E; Table IIB). However, this phenomenon was not observed in cells with ectopic 10 kb alphoid DNA arrays (Table IIB). This result confirms that a minimum of 30 kb alphoid DNA is required to form a functional centromere in transfected HT1080 cells.
Effect of alphoid array length and density of CENP-B boxes on stability of CENP-A nucleosomes
It is possible that YAC arms (total 26 kb) prevent spreading of CENP-A chromatin throughout ectopic alphoid DNA arrays in chromosomes with integrated YACs. This idea was tested with two constructs, pNeo11.5 or pNeo11.5htel, in which a 9.4 kb alphoid array (11-mer × 5) was cloned into a 6 kb plasmid vector with or without 1.2 kb human telomeric sequences, respectively. Neither HACs nor CENP-A chromatin was detected in cells carrying chromosomal insertions of these constructs (Figure 2E; Tables IA and IIB). Moreover, this result was confirmed using a highly sensitive ChIP assay. This assay is based on competitive PCR with an integrated copy of a BAC containing alphoid DNA with a mutant CENP-B box as a reference (Figure 5A and B). The assay showed that CENP-A was not enriched in the 9.4 kb alphoid DNA (1.0–0.97 relative to control), whereas CENP-B was enriched approximately 36-fold (Figure 5A and C). In contrast, heterochromatin readily spreads from flanking regions via vector DNA or via alphoid DNA itself, displacing or preventing stable formation of CENP-A chromatin on short segments of alphoid DNA (Figure 4B and D; Table IIB).
ChIP and competitive PCR analysis showed that CENP-A nucleosomes were 22-fold enriched in HACs with 240 kb alphoid DNA; however, CENP-A nucleosomes were only enriched approximately 2.4-fold at the integration site of the same BAC and CENP-B was enriched 42- to 51-fold (Figure 5A and C). In addition, H3K9me3 was abundant in the 240 kb alphoid DNA array (Figure 5D). Thus, CENP-A chromatin was much less stable in ectopic 10 or 240 kb alphoid DNA arrays than in 30–70 kb alphoid DNA arrays.
HACs formed more efficiently from the 60 kb BAC construct with five CENP-B boxes per 11-mer repeating unit than from the same construct with one CENP-B box (pCBB11.1.32; Figures 1D, E, 2F and 5A; Table IB). Fewer CENP-B boxes also reduced assembly of CENP-A chromatin eight-fold (Figure 5C; Table IIB; 3.5-fold enrichment in BR11.32HT2–25 versus W0210R-1). This result indicates that CENP-A can assemble on the alphoid unit with one CENP-B box. However, CENP-A chromatin does not spread over the whole 11-mer with one CENP-B box and does not form a functional centromere core in a 60 kb alphoid DNA array with reduced numbers of CENP-boxes. Therefore, a functional centromere requires a minimum of 30 kb alphoid DNA and a minimum density of CENP-B boxes.
Discussion
The minimal length of alphoid DNA for a functional human centromere
One of the goals of this study was to determine the minimum requirements to form a functional human centromere. The minimum required amount of centromere-specific repeated sequence DNA, such as type-I alphoid DNA, was examined by measuring the ability of linear YACs or BACs with different lengths of alphoid DNA to form HACs in human HT1080 cells. Previous studies indicated that HACs composed of a concatenated structure of the input alphoid YAC DNA with human telomere sequences are a reasonable model of natural chromosomes, so this system was considered a reasonable indicator of normal centromere function in human cells (Ikeno et al, 1998: Tsuduki et al, 2006). The minimum length of alphoid DNA that supported formation of stable HACs in HT1080 cells was 30 kb, with improved efficiency if the length of alphoid DNA increased to 50 or 70 kb. The result agrees with a previous study based on a different HAC formation efficiency of two isogenic alphoid DNA clones from chromosome 17 (35 and 80 kb) (Grimes et al, 2002). In our study, HACs with 30 kb alphoid DNA were as stable as or more stable than HACs with longer alphoid DNA regions (i.e. R=0.00010 for HACs with 30 kb alphoid DNA versus R=0.0049–0.0086 for HACs with 50 or 70 kb alphoid DNA). HACs did not form when the YACs carried 10 kb alphoid DNA. This result was not because of interference from long YAC vector arms, as the same result was observed for a construct with 6 kb vector arms and 10 kb alphoid DNA. Cells (10–30%) carrying YACs with 30–70 kb alphoid DNA arrays established and maintained variegated CENP-A chromatin (containing CENP-A, -B, and -C; Table IIB) irrespective of the integration site or YAC copy number. Moreover, normal centromere function was associated with ectopic CENP-A chromatin, which formed at sites of integrated 30–70 kb alphoid DNA arrays in TSA-treated cells (Table IIB; Figure 4E). These observations strongly indicate that a wild-type 30 kb alphoid DNA array is sufficient to support the assembly of functional centromeric chromatin in human HT1080 cells. It is worth noting that this result is consistent with the reported size of CENP-A chromatin clusters in naturally occurring chromosomes in human, mouse and Drosophila cells (Blower et al, 2002; Chueh et al, 2005).
Spreading of CENP-A chromatin into vector DNA
This study demonstrated that CENP-A chromatin can spread into flanking YAC vector DNA. This was an unexpected result, because YAC vectors with no inserted or with type-II alphoid DNA do not form HACs or CENP-A chromatin in human cells (Ikeno et al, 1998; Nakano et al, 2003). In addition, the spreading is not efficient, does not encompass the entire 26 kb vector arm and only occurred in constructs with 30 kb alphoid DNA arrays. Considering these factors, it seems possible that once formed in alphoid DNA, a CENP-A cluster propagates to a size greater than 30 kb. In the context of the HAC with a 30 kb alphoid DNA insert, this includes non-alphoid vector DNA. Thus, a region greater than 30 kb may be necessary for achieving a stable CENP-A chromatin domain and for establishing normal centromere function in the context of a YAC-derived HAC (Figures 3C and 6, model A; Supplementary Figure 4). This result clearly shows that CENP-A chromatin can spread into flanking vector DNA if the length of alphoid DNA is insufficient and if spreading into vector DNA is required to support centromere function. Such extended CENP-A chromatin domains are presumably maintained by epigenetic mechanisms that support the assembly of functional centromeres.
Figure 6.
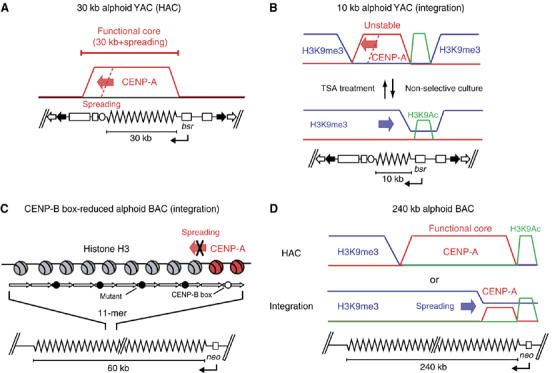
Hypothetical models for assembly and spreading of CENP-A chromatin. (A) Chromatin assembly on the 30 kb alphoid HAC. The CENP-A chromatin (red line) spread into the YAC vector arm regions (red arrow) following nucleation of the assembly on the insert 30 kb alphoid DNA array, and was maintained. Therefore, the functional centromere core of CENP-A chromatin (at least 30 kb plus spreading) may exist as an epigenetic memory. (B) Chromatin assembly on the short 10 kb alphoid integration site. The multimer of the 10 kb alphoid YACs acquired strong heterochromatic H3K9me3 state (blue line) at its integration site. CENP-A chromatin could nucleate the assembly if the heterochromatin structure was compulsorily broken by TSA treatment. However, this kind of CENP-A assembly without fulfilling the functional length was unstable, and thus, dynamically encroached by the spreading of H3K9me3 (blue arrow) under the non-selective culture. (C) Chromatin assembly on the CENP-B box-reduced alphoid BAC at the integration site. CENP-A nucleosomes can assemble on an alphoid unit containing only one CENP-B box in the 11-mer unit. However, such a CENP-A nucleosome cannot spread and generate a functional centromere core unit on the 60 kb CENP-B box-reduced alphoid DNA. (D) Chromatin assembly on the long 240 kb alphoid HAC or the integration site. The 240 kb alphoid BAC also has an ability to form a HAC, but a high level of H3K9me3 also covered the integration site of the 240 kb alphoid BAC.
Similar observations were made in a study of Drosophila minichromosomes formed by γ-irradiation (Maggert and Karpen, 2001). The authors of this study suggested that centromeric chromatin spreads into flanking chromosomal sequences when there is an inversion in a centromere domain. The other group also proposed a model for centromere expansion/evolution based on the insertion of multiple transposons in or near centromeric domains, followed by spreading of centromeric chromatin (Henikoff et al, 2001). The results presented here support these hypotheses especially in the context of artificial chromosomes containing insufficient alphoid DNA to form a functional centromere. It should be noted that excess CENP-A binds nonspecifically to entire chromosome arms, but does not form functional centromeric chromatin in cells engineered to overexpress this protein (Sullivan et al, 1994; Van Hooser et al, 2001). Thus, functional spreading of CENP-A chromatin is a specialized mechanism that supports centromere assembly. It is likely that this process involves recruitment of other essential kinetochore proteins.
H3K9me3 chromatin antagonizes CENP-A chromatin on ectopic alphoid DNA arrays
This study uses cytological analysis and ChIP to show that CENP-A chromatin does not form at ectopic 10 kb alphoid DNA arrays, but forms at 30–70 kb alphoid DNA arrays. One explanation for this result is that 10 kb alphoid DNA regions may preferentially bind H3K9me3, which promotes formation of H3K9me3 heterochromatin and precludes formation of CENP-A chromatin (Figures 4B, C and 6, model B). Assembly and maintenance of H3K9me3 heterochromatin are partly dependent on HP1, histone metyltransferase Suv39 and RNA interference (RNAi) (Grewal and Moazed, 2003; Lachner et al, 2003). Treatment of cells with TSA leads to histone hyperacetylation, breakdown of heterochromatin structure in mouse and Schizosaccharomyces pombe (Ekwall et al, 1997; Taddei et al, 2001), and centromere reactivation in human isodicentromic chromosomes (Higgins et al, 2005).
In this study, treatment with TSA increased binding of CENP-A and CENP-C in 10 kb alphoid DNA arrays and decreased the amount of H3K9me3 chromatin. However, CENP-A chromatin was unstable unless cells were maintained under selection for the expression of bsr. This suggests that chromatin remodeling associated with transcriptional activation of bsr may have synergistic effect on the assembly of CENP-A chromatin, and in contrast, assembly of H3K9me3 heterochromatin may be antagonistic to assembly or maintenance of CENP-A chromatin (Figure 6, model B). Interestingly, transcriptional activities were reported in the natural complex centromeres of rice and human (Saffery et al, 2003; Nagaki et al, 2004). Even when TSA-treated cells carrying 10 kb alphoid DNA arrays were maintained under selection for bsr, reformed minichromosomes were not observed. In contrast, stable reformed minichromosomes with functional centromeres were easily observed in cells with 30 or 70 kb alphoid DNA arrays (Figure 4E; Table IIB).
In summary, these studies indicate that a minimum of 30 kb alphoid DNA is required for de novo HAC assembly or for centromere reactivation in human cultured cells. It is reasonable to propose that molecular interactions between DNA and chromatin proteins and specific structural constraints contribute to this size threshold, because the DNA component is otherwise identical in YACs with 10, 30 and 70 kb alphoid DNA arrays (Supplementary Figure 1). Alphoid DNA arrays (10 kb) may fail to form functional centromeric chromatin because of a preferential tendency of these arrays to form H3K9me3 heterochromatin.
CENP-B box density and alphoid length affect the CENP-A assembly
Large alphoid DNA arrays and CENP-B boxes are two essential components of a functional human centromere (Ohzeki et al, 2002). Recent studies indicate that HACs form more efficiently in synthetic alphoid DNA containing CENP-B boxes in every alphoid monomer (Basu et al, 2005), and that CENP-B boxes promote CENP-A binding in native human centromeres (Irvine et al, 2004). The present study demonstrated that alphoid DNA with a very low density of CENP-B boxes (one CENP-B box in the 11-mer; 32 CENP-B boxes in 60 kb) only supports inefficient formation of centromeric chromatin and does not support formation of HACs. The CENP-A nucleosomes that do form fail to spread through alphoid DNA with mutant CENP-B boxes, and thus do not form a functional centromere (Figures 5C and 6, model C). These results indicate that a minimum density of CENP-B boxes is required to form stable CENP-A chromatin core in alphoid DNA. Nevertheless, 10 kb alphoid DNA arrays including 25–27 CENP-B boxes showed no capacity to maintain CENP-A nucleosomes. This result suggests that both the length of alphoid DNA and the density of CENP-B boxes are critical in supporting the assembly of functional centromeric chromatin.
It should be pointed out that very long (240 kb) alphoid DNA arrays may not be optimal for de novo centromere assembly. For example, CENP-A chromatin was maintained with extremely low efficiency in 240 kb alphoid DNA arrays at the ectopic integration sites, and these long alphoid DNA arrays form H3K9me3 heterochromatin with relatively high efficiency. This process may be related to the inactivation of centromeres in dicentromeric fusion chromosomes, which is thought to be an epigenetic phenomenon (Sullivan and Schwartz, 1995; Warburton et al, 1997), and may involve spreading of heterochromatin in alphoid DNA (Figure 6, model D). These results suggest that the assembly of centromeric chromatin, influenced by the length of repetitive alphoid DNA and the density of CENP-B boxes, is envisioned as a dynamic balance between the nucleation and spreading of CENP-A chromatin, and the disassembly and/or prevention of heterochromatin spreading. TSA treatment and selective pressure for transcriptional activity also alter this dynamic balance and disfavor heterochromatization, thereby promoting CENP-A nucleation and spreading of centromeric chromatin at ectopic sites of alphoid DNA (Figure 6, model B). Nevertheless, it is still unclear why 120 or 240 kb alphoid DNA arrays do not support HAC formation and CENP-A assembly more efficiently than 50–70 kb alphoid DNA arrays. Our recent results suggest a possible role for CENP-B in satellite DNAs, as CENP-B/CENP-B box interactions play an important role in de novo assembly of mouse CENP-A chromatin and in the assembly of heterochromatin in ectopic satellite DNA insertion sites (Okada et al, submitted). In addition to these dynamic chromatin assemblies, vector sequences can also nucleate heterochromatic structures (H3K9me3 and HP1α) on HACs or ectopic integration sites (Nakashima et al, 2005). In any case, more precise analyses of these interactions are required in both human and mouse cells.
In summary, the present study demonstrated that the length of alphoid DNA arrays and the density of CENP-B boxes influence the assembly and maintenance of CENP-A chromatin in human HT1080 cells. These factors may also influence spreading of H3K9me3 heterochromatin, which appears to be antagonistic to CENP-A chromatin. It is hoped that these and future studies will contribute to a more complete understanding of the mechanisms that regulate assembly and maintenance of unique chromatin domains, including native human centromeres.
Materials and methods
Construction of deleted variants of the 70 kb α21-I alphoid YAC
The YAC right arm replacement vector (MCU-bsr α11) was constructed by insertion of the α21-I alphoid 11-mer fragment from p11-4 (Ikeno et al, 1994) by partial digestion with EcoRI and end-filling with the Klenow large fragment, into the end-filled ClaI site of the MCU-bsr (Ikeno et al, 1998). Retrofitting was carried out by method described previously (Ikeno et al, 1998; Figure 1A).
Construction of alphoid BACs and alphoid plasmids
BACs containing 120 or 240 kb synthetic α21-I (pWTR11.64 and pWTR11.128, respectively) were constructed from pWTR11.32 by the tandem concatenation method described previously (Ohzeki et al, 2002; Figure 1D). The alphoid plasmid, pNeo11.5, was generated by insertion of a neomycin resistance gene and a 9.4 kb synthetic α21-I alphoid (the 11-mer × 5 times) fragment into the XbaI–DraI-digested and end-filling site of the plasmid PET30a (Novagen) with linker DNAs. To generate pNeo11.5htel, two fragments of 1.1 kb hTEL sequence inverted on each other with ampicilin resistance gene between them, were inserted at the HindIII site at the linker site of pNeo11.5.
Cell culture and DNA transfection
Human fibrosarcoma cell line HT1080 (ATCC CCL121) was grown in DMEM medium, supplemented with 10% FBS (Gibco BRL), penicillin, streptomycin and glutamine. The alphoid YAC DNAs and BAC DNAs were purified as previously described (Ikeno et al, 1998; Ohzeki et al, 2002). The alphoid plasmids, pNeo11.5 and pNeo11.5htel were purified with a Qiagen column and linearized at HindIII or AatII site at the end of hTELs, respectively. PFGE gel-purified YAC DNA (35–95 ng), alphoid BAC DNA (400 ng) or plasmid DNA were transfected into 6 × 105 HT1080 cells using lipofectamine reagent (Gibco BRL). Stable transformants generated were selected with 4 μg/ml BS (Kaken Seiyaku) or 400 μg/ml G418 (Wako), as appropriate. For treatment with TSA (TSA), aliquots of 2.44 × 106 cells were seeded in 10 ml medium with 1 μg/ml TSA (Wako).
Cytological assays
Standard FISH assays were carried out as described previously (Masumoto et al, 1989b). Probes used were p11–4 for the alphoid 11-mer alphoid DNA, pYAC4 for YAC arm DNA, PCR products amplified from pBAC108L using primers BACX and BACS for BAC vector DNA and pET30a for alphoid plasmid DNA (Ohzeki et al, 2002). Plasmid DNA or PCR products were labeled using digoxigenin-11dUTP or biotin16-dUTP (Roche Diagnostics; Nick Translation kit). Indirect immunofluorescence and simultaneous staining by FISH were carried out as described previously (Masumoto et al, 1989b). Antibodies used were anti-CENP-A (mAN1, Masumoto et al, 1998), anti-CENP-B (2D8D8; Ohzeki et al, 2002), anti-CENP-E (mAb177; Yen et al, 1991) and anti-trimethyl H3 lys9 (Upstate).
DNA multiplicity by real-time PCR
Genomic DNA was prepared by standard techniques. Real-time PCR was carried out using the iCyclerIQTM MultiColor Real-Time PCR System (Bio-Rad) according to the manufacturer's instruction. The copy number of lys2 in YAC left arm and bsr in YAC right arm were quantified using real-time PCR with primer sets, LYS2–1 and LYS2–3, bsr-F and bsr-R (Nakano et al, 2003). HT1–2 cells were used as a reference for determining lys2 and bsr copy numbers (HT1–2 copy numbers: lys2, 34; bsr, 35; Ikeno et al, 1998). W0210R-1 cells were used as a reference for determining Neo copy number (Neo copy number 14). Neo copy number was determined with primer sets N1 and N2 (Ohzeki et al, 2002).
ChIP and real-time PCR
ChIP was carried out as described previously (Nakano et al, 2003) using the following antibodies: anti-CENP-A (mAN1), anti-CENP-B (2D8D8 and 5E6C1), anti-trimethyl H3 lys9 (Upstate), normal mouse IgG (Santa Cruz) and normal rabbit IgG (Santa Cruz). Relative recovery of immunoprecipitated DNA was quantified by real-time PCR using primer sets indicated in Supplementary Figure 6.
ChIP and competitive PCR assay for the cell lines generated from synthetic alphoid constructs
Cells with ectopic synthetic alphoid DNA containing wild-type CENP-B boxes were compared with reference M1319 cells with 60 kb synthetic alphoid DNA with mutant CENP-B boxes. Cells were mixed, fixed and sonicated. The ratio of input cells was calculated to have the same amount of alphoid DNA per cell line. Previous studies showed that CENP-A and CENP-B do not bind to alphoid DNA with mutant CENP-B boxes in M1319 cells (Ohzeki et al, 2002). ChIP assays were carried out as described above. Target DNA was quantified by competitive PCR as described previously (Ohzeki et al, 2002).
Supplementary Material
Supplementary Figures
Acknowledgments
We thank M Hirano and T Higuchi for experimental support, T Okada and H Nakashima for discussion and N Kouprina for critical reading of the manuscript. We also thank T Yen (Fox Chase Cancer Center, Philadelphia, PA) for anti-CENP-E antibody, K Yoda (Nagoya University) and N Nozaki (Kanagawa Dental College) for producing anti-CENP-A and -B antibodies. This work was supported by a grant-in-aid for Scientific Research on Priority Areas (B) and Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Science, Sports and Culture of Japan to HM. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, USA.
References
- Amor DJ, Choo KH (2002) Neocentromeres: role in human disease, evolution, and centromere study. Am J Hum Genet 71: 695–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor DJ, Kalitsis P, Sumer H, Choo KH (2004) Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol 14: 359–368 [DOI] [PubMed] [Google Scholar]
- Ando S, Yang H, Nozaki N, Okazaki T, Yoda K (2002) CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol 22: 2229–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer M, Nilsen TW, Costigan C, Altman S (1990) Structure and transcription of a human gene for H1 RNA, the RNA component of human RNase P. Nucleic Acids Res 18: 97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu J, Stromberg G, Compitello G, Willard HF, Van Bokkelen G (2005) Rapid creation of BAC-based human artificial chromosome vectors by transposition with synthetic alpha-satellite arrays. Nucleic Acids Res 33: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH (2002) Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2: 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chueh AC, Wong LH, Wong N, Choo KH (2005) Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere. Hum Mol Genet 14: 85–93 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Ratrie H III, Stetten G (1989) Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma 98: 1–12 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Sullivan KF, Machlin PS, Cooke CA, Kaiser DA, Pollard TD, Rothfield NF, Cleveland DW (1987) Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J Cell Biol 104: 817–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole T, Okamoto Y, Noskov VN, Kouprina N, Kim JH, Leem SH, Barrett JC, Masumoto H, Larionov V (2005) Rapid generation of long synthetic tandem repeats and its application for analysis in human artificial chromosome formation. Nucleic Acids Res 33: e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersole TA, Ross A, Clark E, McGill N, Schindelhauer D, Cooke H, Grimes B (2000) Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum Mol Genet 9: 1623–1631 [DOI] [PubMed] [Google Scholar]
- Ekwall K, Olsson T, Turner BM, Cranston G, Allshire RC (1997) Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91: 1021–1032 [DOI] [PubMed] [Google Scholar]
- Espada J, Ballestar E, Fraga MF, Villar-Garea A, Juarranz A, Stockert JC, Robertson KD, Fuks F, Esteller M (2004) Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J Biol Chem 279: 37175–37184 [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR III, Cleveland DW (2006) The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 8: 458–469 [DOI] [PubMed] [Google Scholar]
- Goshima G, Kiyomitsu T, Yoda K, Yanagida M (2003) Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J Cell Biol 160: 25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Moazed D (2003) Heterochromatin and epigenetic control of gene expression. Science 301: 798–802 [DOI] [PubMed] [Google Scholar]
- Grimes BR, Rhoades AA, Willard HF (2002) Alpha-satellite DNA and vector composition influence rates of human artificial chromosome formation. Mol Ther 5: 798–805 [DOI] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF (1997) Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 15: 345–355 [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102 [DOI] [PubMed] [Google Scholar]
- Higgins AW, Gustashaw KM, Willard HF (2005) Engineered human dicentric chromosomes show centromere plasticity. Chromosome Res 13: 745–762 [DOI] [PubMed] [Google Scholar]
- Howman EV, Fowler KJ, Newson AJ, Redward S, MacDonald AC, Kalitsis P, Choo KH (2000) Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc Natl Acad Sci USA 97: 1148–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H (1998) Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol 16: 431–439 [DOI] [PubMed] [Google Scholar]
- Ikeno M, Masumoto H, Okazaki T (1994) Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range alpha-satellite DNA arrays of human chromosome 21. Hum Mol Genet 3: 1245–1257 [DOI] [PubMed] [Google Scholar]
- Irvine DV, Amor DJ, Perry J, Sirvent N, Pedeutour F, Choo KH, Saffery R (2004) Chromosome size and origin as determinants of the level of CENP-A incorporation into human centromeres. Chromosome Res 12: 805–815 [DOI] [PubMed] [Google Scholar]
- Izuta H, Ikeno M, Suzuki N, Tomonaga T, Nozaki N, Obuse C, Kisu Y, Goshima N, Nomura F, Nomura N, Yoda K (2006) Comprehensive analysis of the ICEN (interphase centromere complex) components enriched in the CENP-A chromatin of human cells. Genes Cells 11: 673–684 [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Sullivan RJ, Jenuwein T (2003) An epigenetic road map for histone lysine methylation. J Cell Sci 116: 2117–2124 [DOI] [PubMed] [Google Scholar]
- Lam AL, Boivin CD, Bonney CF, Rudd MK, Sullivan BA (2006) Human centromeric chromatin is a dynamic chromosomal domain that can spread over noncentromeric DNA. Proc Natl Acad Sci USA 103: 4186–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Wevrick R, Fisher RB, Ferguson-Smith MA, Lin CC (1997) Human centromeric DNAs. Hum Genet 100: 291–304 [DOI] [PubMed] [Google Scholar]
- Maggert KA, Karpen GH (2001) The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics 158: 1615–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Ikeno M, Nakano M, Okazaki T, Grimes B, Cooke H, Suzuki N (1998) Assay of centromere function using a human artificial chromosome. Chromosoma 107: 406–416 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T (1989a) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 109: 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Sugimoto K, Okazaki T (1989b) Alphoid satellite DNA is tightly associated with centromere antigens in human chromosomes throughout the cell cycle. Exp Cell Res 181: 181–196 [DOI] [PubMed] [Google Scholar]
- Mejia JE, Alazami A, Willmott A, Marschall P, Levy E, Earnshaw WC, Larin Z (2002) Efficiency of de novo centromere formation in human artificial chromosomes. Genomics 79: 297–304 [DOI] [PubMed] [Google Scholar]
- Muro Y, Masumoto H, Yoda K, Nozaki N, Ohashi M, Okazaki T (1992) Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. J Cell Biol 116: 585–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36: 138–145 [DOI] [PubMed] [Google Scholar]
- Nakano M, Okamoto Y, Ohzeki J, Masumoto H (2003) Epigenetic assembly of centromeric chromatin at ectopic alpha-satellite sites on human chromosomes. J Cell Sci 116: 4021–4034 [DOI] [PubMed] [Google Scholar]
- Nakashima H, Nakano M, Ohnishi R, Hiraoka Y, Kaneda Y, Sugino A, Masumoto H (2005) Assembly of additional heterochromatin distinct from centromere–kinetochore chromatin is required for de novo formation of human artificial chromosome. J Cell Sci 118: 5885–5898 [DOI] [PubMed] [Google Scholar]
- Neil DL, Villasante A, Fisher RB, Vetrie D, Cox B, Tyler-Smith C (1990) Structural instability of human tandemly repeated DNA sequences cloned in yeast artificial chromosome vectors. Nucl Acids Res 18: 1421–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H (2002) CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159: 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR III, Desai A, Fukagawa T (2006) The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol 8: 446–457 [DOI] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Saffery R, Sumer H, Hassan S, Wong LH, Craig JM, Todokoro K, Anderson M, Stafford A, Choo KH (2003) Transcription within a functional human centromere. Mol Cell 12: 509–516 [DOI] [PubMed] [Google Scholar]
- Saffery R, Wong LH, Irvine DV, Bateman MA, Griffiths B, Cutts SM, Cancilla MR, Cendron AC, Stafford AJ, Choo KH (2001) Construction of neocentromere-based human minichromosomes by telomere-associated chromosomal truncation. Proc Natl Acad Sci USA 98: 5705–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JM, Critcher R, Ebersole TA, Valdivia MM, Earnshaw WC, Fukagawa T, Farr CJ (2002) Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha-satellite array. EMBO J 21: 5269–5280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Schwartz S (1995) Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum Mol Genet 4: 2189–2197 [DOI] [PubMed] [Google Scholar]
- Sullivan KF, Hechenberger M, Masri K (1994) Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol 127: 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Maison C, Roche D, Almouzni G (2001) Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol 3: 114–120 [DOI] [PubMed] [Google Scholar]
- Thrower DA, Jordan MA, Schaar BT, Yen TJ, Wilson L (1995) Mitotic HeLa cells contain a CENP-E-associated minus end-directed microtubule motor. EMBO J 14: 918–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuduki T, Nakano M, Yasuoka S, Yamazaki S, Okada T, Okamoto Y, Masumoto H (2006) An artificially constructed de novo human chromosome behaves almost identically to its natural counterpart during metaphase and anaphase in living cells. Mol Cell Biol 26: 7682–7695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, Goldberg ML, Yokomori K, Earnshaw WC, Sullivan KF, Brinkley BR (2001) Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci 114: 3529–3542 [DOI] [PubMed] [Google Scholar]
- Warburton PE (2004) Chromosomal dynamics of human neocentromere formation. Chromosome Res 12: 617–626 [DOI] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, Poirier GG, Earnshaw WC (1997) Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol 7: 901–904 [DOI] [PubMed] [Google Scholar]
- Yen TJ, Compton DA, Wise D, Zinkowski RP, Brinkley BR, Earnshaw WC, Cleveland DW (1991) CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J 10: 1245–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures


