Abstract
DNA replication of eukaryotic chromosomes initiates at a number of discrete loci, called replication origins. Distribution and regulation of origins are important for complete duplication of the genome. Here, we determined locations of Orc1 and Mcm6, components of pre-replicative complex (pre-RC), on the whole genome of Schizosaccharomyces pombe using a high-resolution tiling array. Pre-RC sites were identified in 460 intergenic regions, where Orc1 and Mcm6 colocalized. By mapping of 5-bromo-2′-deoxyuridine (BrdU)-incorporated DNA in the presence of hydroxyurea (HU), 307 pre-RC sites were identified as early-firing origins. In contrast, 153 pre-RC sites without BrdU incorporation were considered to be late and/or inefficient origins. Inactivation of replication checkpoint by Cds1 deletion resulted in BrdU incorporation with HU specifically at the late origins. Early and late origins tend to distribute separately in large chromosome regions. Interestingly, pericentromeric heterochromatin and the silent mating-type locus replicated in the presence of HU, whereas the inner centromere or subtelomeric heterochromatin did not. Notably, MCM did not bind to inner centromeres where origin recognition complex was located. Thus, replication is differentially regulated in chromosome domains.
Keywords: centromere, DNA microarray, fission yeast, replication origin, subtelomere
Introduction
To ensure that a complete set of the eukaryotic genome is precisely duplicated during the limited period of S phase in every cell cycle, DNA replication initiates at a number of replication origins on chromosomes (Gilbert, 2001; Bell and Dutta, 2002). As each chromosome region replicates in a specific period within S phase, timing of origin activation must be regulated. Although we have a growing understanding of protein factors involved in initiation and elongation of replication, the mechanisms of origin activation at the chromosome level are yet to be clarified in detail. Thus, it is important to determine locations of all replication origins on chromosomes. However, only small numbers of replication origins have so far been identified in most organisms other than budding yeast Saccharomyces cerevisiae (MacAlpine and Bell, 2005).
The process of initiation of replication at individual replication origins is composed of two major steps, licensing of replication origins in G1 phase and activation of the origins in S phase. In G1 phase, pre-replicative complexes (pre-RCs) are formed at replication origins (Bell and Dutta, 2002; Kearsey and Cotterill, 2003). This requires binding of the origin recognition complex (ORC) to a replication origin, followed by assembly of the minichromosome maintenance (MCM) complex, depending on the loading factors, Cdc6/Cdc18 and Cdt1 (Diffley et al, 1994; Bell and Dutta, 2002). Although pre-RC formation is essential for initiation of replication, it is not in itself sufficient. Origin activation in S phase is regulated by two conserved protein kinases, cyclin-dependent protein kinase (CDK) and Cdc7–Dbf4 protein kinase (Dbf4-dependent kinase, DDK). These kinases are required for assembly of several other protein factors, including Cdc45 and GINS onto pre-RCs. This may lead to activation of MCM helicase and origin DNA unwinding, and the replication machinery is established through assembly of RPA and DNA polymerases onto the single-stranded DNA (Bell and Dutta, 2002).
Although proteins involved in initiation of replication are conserved among eukaryotes, the nucleotide sequences of replication origins are very diverse among organisms (Gilbert, 2001), mainly because of differences in DNA-binding properties of ORCs. In budding yeast, ORC recognizes the specific sequence called the ARS consensus sequence (ACS). In contrast, no clear consensus sequence has been found in origins in fission yeast, Schizosaccharomyces pombe (Clyne and Kelly, 1995; Dubey et al, 1996; Okuno et al, 1999), although AT-rich sequences to which ORC preferentially binds are required (Chuang and Kelly, 1999). Requirements for specific sequences become less clear in multicellular organisms such as metazoans, and ORC exhibits little sequence specificity in DNA binding in vitro (Vashee et al, 2003; Remus et al, 2004). Therefore, it is important to determine the locations of ORC binding and DNA synthesis experimentally.
Genome-wide analyses of replication kinetics and distribution of ORC and MCM proteins using DNA microarrays have been performed in budding yeast (Raghuraman et al, 2001; Wyrick et al, 2001). The majority of proposed ARS (pro-ARS) sites identified by ChIP-based analysis exhibit ARS activity, and the correlation with actual initiation sites has been demonstrated (Feng et al, 2006). On the other hand, replication timing analyses using microarrays with human, mouse and Drosophila chromosomes have suggested links between early replication timing and active transcription in large chromatin domains (MacAlpine and Bell, 2005). However, because of difficulties in genome-wide analysis of replication factor binding sites in metazoans, it has not been possible to clarify the relationship between pre-RC sites and selection of active origins. Fission yeast is a suitable model organism to study genome-wide regulation of chromosome replication, because both the structures of replication origins and chromatin configuration have similarities with those in metazoan organisms.
In fission yeast, owing to preferred binding of ORC to AT-rich sequences (Chuang and Kelly, 1999), and the requirement of multiple ORC binding sites for origin activity (Takahashi et al, 2003), replication origins have been predicted to be ‘A+T-rich islands' located preferentially in intergenic regions (Segurado et al, 2003). Locations of single-stranded DNA regions under nucleotide-depleting conditions are consistent with this prediction (Feng et al, 2006). Because the ARS activity of intergenic regions correlates with the AT content and the length, it has been proposed that replication origins fire stochastically in fission yeast (Dai et al, 2005). Single-molecule analyses using DNA combing also support the stochastic model (Patel et al, 2006). However, owing to the lack of information on genome-wide distribution of pre-RC sites, it has remained an open question whether all the pre-RCs are activated or only a subset is selected to fire.
In this study, we conducted high-resolution mapping of Orc1 and Mcm6 binding sites in G1 phase, using a tiling array covering almost the entire genome of fission yeast, resulting in identification of pre-RC sites precisely on the whole genome. Mapping of nascent DNA in the presence of HU in wild-type and checkpoint-deficient cds1Δ cells allowed us to identify early-firing replication origins and late and/or inefficient origins. Replication timing is not intrinsic to origin function but dependent on its surrounding regions. The centromeric and subtelomeric heterochromatin regions behaved differently, suggesting distinct regulation in chromatin domains.
Results
Mapping of pre-RCs on fission yeast chromosomes
Initiation of DNA replication in eukaryotic cells requires the ordered assembly of replication factors at specific sites on the chromosome. To determine the site of pre-RC formation on fission yeast chromosomes, DNA immunoprecipitated with Orc1 and Mcm6 in G1-arrested cells (Ogawa et al, 1999; Takahashi et al, 2003) was analyzed with a tiling array that covers almost the entire genome of fission yeast at 250 base pair (bp) resolution, except for telomeres and rDNA repeats. Both Orc1 and Mcm6 were located exclusively at intergenic regions, and 84% of Orc1-binding sites colocalized with Mcm6 (Figure 1 and Supplementary Figures S1, S2 and Supplementary Table S1). A total of 460 pre-RC sites, where peaks of Orc1 and Mcm6 shown in Supplementary Figures S1 and S2 colocalized, were identified (black triangles in Figure 1, Supplementary Table S1). The pre-RC sites were distributed throughout the chromosomes, with an average separation of 26.7 kb and enriched at the centromeres and the subtelomeric regions. Enrichment at the subtelomeric regions was not observed on chromosome III, where both ends of the array were flanked with rDNA repeats. Orc4, another ORC complex component, was highly colocalized with Orc1 (>90%; Supplementary Figure S3 and Supplementary Table S1), suggesting that ORC complex was localized at pre-RC sites.
Figure 1.

Locations of Orc1- and Mcm6-binding sites and BrdU incorporation sites on fission yeast chromosomes. For mapping of Orc1 and Mcm6 localization sites, HM568 (h− nda3-KM311 cdc10-129 ura4-D18 leu1-32 orp1-5flag/pREP82-cdc18 pREP81-cdt1) cells expressing Cdc18 and Cdt1 were arrested at the cdc10 arrest point in G1 phase and used for ChIP. The orange and blue vertical bars represent the binding ratios of loci showing enrichment of ChIP fractions with anti-Flag-Orc1 (orange bars in top panels) and anti-Mcm6 (blue bars in middle panels) antibodies, respectively, for regions 1000–1100 kb on chromosome I, 1500–1600 kb on chromosme II and 1800–1900 kb on chromosome III. For mapping of nascent DNA synthesis, HM668 (h− cdc25-22 nmt1-TK) cells arrested at the G2/M boundary were released at 25°C for 90 min in the presence of 10 mM HU and 200 μM BrdU. Cellular DNA was digested with HaeIII and centrifuged in a CsCl gradient. The experimental scheme is shown in Figure 2A. Relative enrichment of BrdU-labeled DNA compared with the control whole cell DNA is presented (green bars in bottom panels). Black triangles indicate pre-RC sites identified as colocalization sites of Orc1 and Mcm6, which were programmatically picked up (Supplementary Figure S1, S2 and Supplementary Table S1). Names of known replication origins colocalized with pre-RCs are shown. Horizontal bars show open reading frames. The scale of the vertical axis is log2.
Identification of early-firing replication origins that incorporate BrdU in the presence of HU
For identification of active replication origins, it is crucial to label newly synthesized DNA around replication origins. We labeled newly synthesized DNA by incorporation of 5-bromo-2′-deoxyuridine (BrdU), a heavy-density nucleotide analogue. Because fission yeast cells do not normally intake BrdU, owing to lack of thymidine kinase activity, the herpes simplex virus thymidine kinase gene was expressed from the inducible promoter. The experimental scheme is shown in Figure 2A. Fission yeast cells expressing thymidine kinase were synchronously released from the G2/M boundary in the presence of BrdU and hydroxyurea (HU) that depletes dNTPs. BrdU-labeled DNA was separated in an equilibrium gradient of CsCl by centrifugation. To examine whether BrdU was selectively incorporated around replication origins, the amounts of BrdU–DNA for the ars2004 locus, an early-firing replication origin (Okuno et al, 1999), and a non-ARS (non-origin) region were analyzed by real-time PCR. After 90 min of BrdU labeling in the presence of HU, about 50% of the ars2004 region was recovered in the heavy–light (HL) density fractions, whereas non-ARS fragment, about 30 kb distant from the origin, remained in the light–light density fractions (Figure 2B), indicating selective incorporation of BrdU around the origin. We also confirmed that both ars2004 and non-ARS regions were fully substituted with BrdU under HU-free conditions (Supplementary Figure S4), excluding the possibility that selective incorporation of BrdU around the ars2004 was due to a shortage of BrdU. As recovery of the nascent DNA by the method was verified, the HL DNA fractions were pooled and subjected to the whole-genome analysis.
Figure 2.
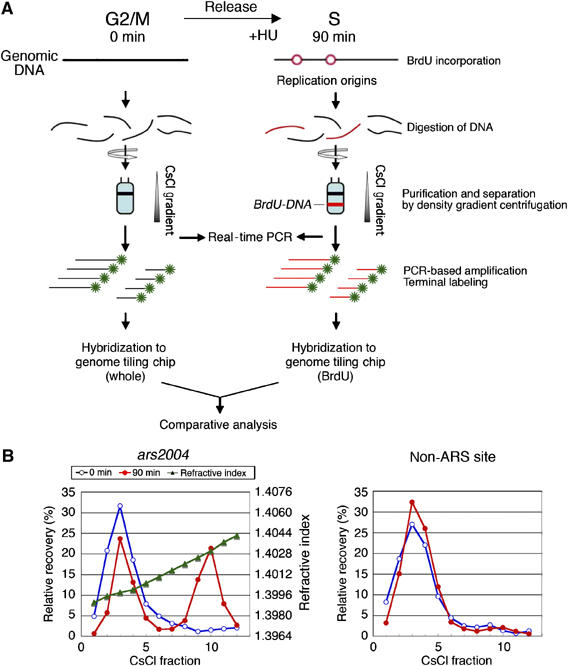
Incorporation of BrdU preferentially into origin proximal regions. (A) A scheme of the experiment is shown. HM668 (h− cdc25-22 nmt1-TK) cells were synchronized and labeled with BrdU in the presence of HU as described in Figure 1. Cellular DNA at 0 and 90 min after release from G2/M block was digested with HaeIII and centrifuged in a CsCl gradient. (B) DNA in each fraction at 0 (blue open circles) and 90 min (red filled circles) was analyzed by real-time PCR using primers for ars2004 (left) and non-ARS (right) regions. Relative recovery (%) among total DNA recovered is presented together with the refractive index (green triangles). For DNA microarray analysis, the heavy–light density fractions 8–12 of 90 min (BrdU–DNA) and light fractions 1–6 of 0 min (whole DNA) were pooled and used for comparative analysis with tiling array.
The results of microarray analysis showed that BrdU-labeled DNA was colocalized with Orc1 and Mcm6 at a very high frequency (Figure 1, green bars in bottom panels; Supplementary Figure S5 and Supplementary Table S1). At the ars2004 origin locus, BrdU-labeled DNA spanned about 10 kb around the intergenic region, where Orc1 and Mcm6 were confined (Figure 1, middle set of panels). This is consistent with bidirectional DNA synthesis initiated from the origin in early S phase (Okuno et al, 1997; Takahashi et al, 2003). The colocalization sites of Orc1, Mcm6 and BrdU, a total of 307 loci, 119, 107 and 81, on chromosomes I, II and III, respectively, were defined as early-firing replication origins (called as early origins) that initiated replication in the presence of HU (red diamonds in Figure 3 and Supplementary Table S1). In contrast, 153 Orc1-Mcm6 colocalization sites, 88, 62 and 3, on chromosomes I, II and III, respectively, did not incorporate BrdU in the presence of HU. Because these BrdU-negative origins are composed of late-firing origins and inefficient origins, they are collectively designated as late origins below (blue diamonds in Figure 3 and Supplementary Table S1). We show that some late origins are repressed by replication checkpoint in the later section.
Figure 3.
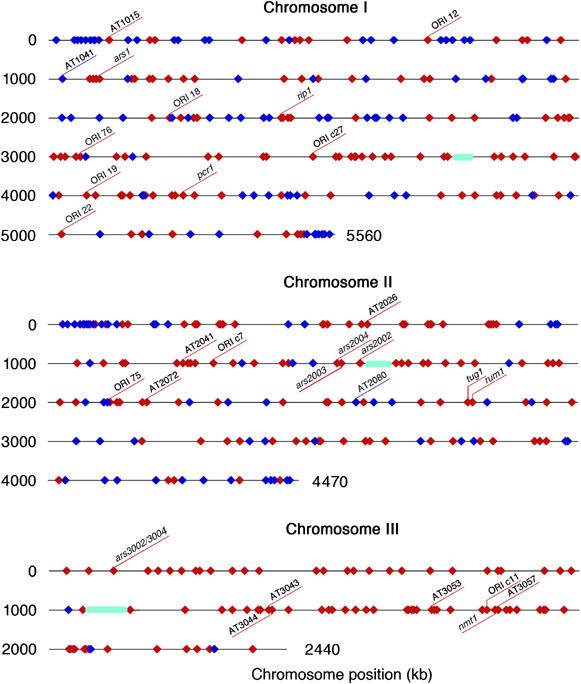
Distributions of early and late origins. Locations of the early origins (red diamonds) and the late and/or inefficient origins (blue diamonds) are shown on chromosomes I, II and III. Positions of known replication origins are shown. Positions of centromeres are shown by green ellipses.
Among 36 origins previously identified by two-dimensional (2D) gel electrophoresis, 28 coincide with the early origins identified here, whereas two match to the late origins (Figure 3 and Supplementary Table S1) (Dubey et al, 1994; Wohlgemuth et al, 1994; Smith et al, 1995; Okuno et al, 1997; Sanchez et al, 1998; Gomez and Antequera, 1999; Segurado et al, 2002, 2003). At the remaining six known origin loci, Orc1 signals were below the standard, although at least either BrdU or Mcm6 signals were detected. When the replication origins identified in this study were compared with those obtained in the previous genome-wide analyses, 189 out of 307 early origins (62%) and 69 out of 153 late origins (45%) coincide precisely with the origins (A+T-rich islands) predicted from AT content calculation (Segurado et al, 2003) (Supplementary Table S1). On the other hand, the early origins are colocalized at a high frequency (239 origins, 78%) with the origins identified as peaks of DNA content increase, whereas the late origins coincide at a lower frequency (34 origins, 22%) (Heichinger et al, 2006). In comparison with the origins identified as the center of single-stranded DNA formed in the presence of HU (Feng et al, 2006), 50% (154 loci) of the early origins and 16% (24 loci) of the late origins match with the previously identified origins (Supplementary Table S1).
Early origins clustered in narrow regions
It should be noted that Orc1-Mcm6 colocalization sites are frequently clustered within a broad BrdU-labeled region extending 20–30 kb, such as at positions 510–530 of chromosome II (Figure 4A). The presence of multiple peaks of BrdU-labeled DNA corresponding to Orc1-Mcm6-binding sites is consistent with the initiation from closely located several origins, although the possibility that DNA synthesis extended from a unique origin remains. To distinguish these possibilities, we first examined whether each of Orc1-Mcm6 colocalization sites exhibited the ARS activity. Among 11 fragments derived from the region, five generated transformants at a high frequency (Figure 4B), indicating that the ARS fragments are clustered.
Figure 4.

ARS activity and two-dimensional gel electrophoresis analysis of clustered early-firing origins. (A) Locations of Orc1 (orange, top panel), Mcm6 (blue, middle panel) and BrdU-labeled DNA (green, bottom panel) in the 500–540 kb region of chromosome II are presented. (B) Eleven fragments containing intergenic regions, shown by horizontal lines in (A), were cloned into the pYC11 vector and used for transformation of HM123 (h− leu1-32). Transformants formed on minimal media plates after 4 days at 30°C are presented. Vector alone and the ars2004 plasmid were used as controls. Plus signs, (+++, ++ and +) below panels represent large, middle and small colony size, respectively, whereas a minus sign shows the absence of any visible colony. (C) HM668 (h− cdc25-22 nmt1-TK) cells released from the G2/M block were cultured at 25°C for 90 min in the presence of 10 mM HU, and replication intermediates were analyzed by 2D gel electrophoresis. Locations of the restriction fragments analyzed by 2D gel methods are shown above the map of open reading frames and the relevant restriction enzyme sites: B, BamHI; Xb, XbaI; N, NdeI; H, HindIII; C, ClaI; S, SpeI; E, EcoRI. Positions of the hybridization probes, which correspond to the fragments used for the ARS assay in (B), are shown as gray bars.
Next, we examined whether replication initiated from these ARSs on the chromosome. Chromosomal DNA of cells synchronously released from G2/M in the presence of HU was analyzed by 2D gel electrophoresis. The results presented in Figure 4C show that bubble arcs, which are indicative of initiation of replication, were detected for fragments 3, 7, 8 and 11 (black triangles in Figure 4C). Fragments 7, 8 and 11 correspond to ori2031E, 2032E and 2033E, respectively (Supplementary Table S1). Another early origin exists in fragment 3, although this site was not identified as the origin by genome-wide analysis owing to weak Orc1 signal (Supplementary Figure S1). These results demonstrate that clustered pre-RC sites act as early origins. Initiation of replication from closely located origins has been reported for the ura4+ locus on chromosome III by 2D gel and DNA combing analyses (Dubey et al, 1994; Patel et al, 2006), and it seems to be common on fission yeast chromosomes.
Repression of late origins by checkpoint kinase Cds1
Late-firing origins in budding yeast are repressed by checkpoint pathway under replication stress such as depletion of nucleotides by HU (Santocanale and Diffley, 1998; Feng et al, 2006). Mapping of single-stranded DNA in the HU-arrested fission yeast cells has suggested that similar regulation exists in fission yeast (Feng et al, 2006). On the other hand, deletion of Rad3, the ATR homologue in fission yeast, affects initiation from a small number of origins (Heichinger et al, 2006). We tested whether the late origins identified in this study might be activated in the absence of replication checkpoint kinase Cds1/Chk2. Wild-type and cds1Δ cells were synchronously released from G2/M block and labeled with BrdU in the presence of HU for 150 min. The BrdU DNA purified by CsCl centrifugation was analyzed by DNA microarray. The results of wild type at 150 min were very similar to those at 90 min, except that BrdU DNA extended further than those at 90 min, which is consistent with slow DNA synthesis in the presence of HU (top panels in Figure 5; Supplementary Figure S6). In contrast, small but significant BrdU incorporation was detected in the subtelomeric regions of chromosomes I and II specifically in cds1Δ cells, although BrdU DNA did not form peaks at most of late origins (middle panels in Figure 5; Supplementary Figure S7). These results suggested that the majority of late origins did not fire at a comparable efficiency to the early origins even in cds1Δ mutant. However, when the ratio of BrdU DNA in cds1Δ to that in wild type was calculated, cds1Δ-specific BrdU incorporation was observed at subtelomeric regions and at the late-origin loci, but not at the early-origin loci (brown bars in bottom panels of Figure 5; Supplementary Figure S8). BrdU incorporation was increased at 90 late-origin loci (59% of the late origins) and at 10 early-origin loci (3% of the early origins), showing specific firing of late origins in cds1Δ cells. We also examined locations of BrdU-labeled DNA prepared at 210 min after release in the presence of HU, and the results were very similar to those at 150 min (data not shown). Furthermore, the results of real-time PCR analysis showed that BrdU incorporation increased in cds1Δ cells compared with wild type at the late origin AT2080 and at the subtelomere locus but not at the early origin or non-ARS locus (Supplementary Figure S9). These results suggest that subsets of late origins in the arm and the subtelomeric regions are repressed in part by replication checkpoint regulation.
Figure 5.

Incorporation of BrdU at late origins in subtelomere and chromosome arm in cds1Δ cells. HM668 (h− cdc25-22 nmt1-TK) and HM1405 (h− cdc25-22 nmt1-TK cds1Δ∷kanMX6) were released from G2/M block and labeled with BrdU for 150 min at 25°C in the presence of HU and the genomic DNA was analyzed with DNA microarray, as described in Figure 1. Green vertical bars represent relative enrichment of BrdU-incorporated DNA in wild type (top panels) and in cds1Δ (middle panels) in 300 kb region from the left end of chromosome I. The bottom panels show ratios of enrichment of BrdU DNA in cds1Δ to that in wild type (brown vertical bars). Red and blue triangles above panels show locations of the early and late origins identified in this work, respectively. Horizontal bars show open reading frames. The scale of the vertical axis is log2. BrdU signals were detected in spite of the absence of replication origins in gray-shaded regions, where localization of Orc1 or Mcm6 was not analyzed (in Supplementary Figures S1 and S2) because of the presence of homologous sequences.
Replication timing of early and late origins is affected by their surrounding regions
To examine whether replication timing of the early and late origins is intrinsic to replication origins or affected by their surrounding regions, we constructed a strain in which an early origin, ars2004, and a late origin, AT2080, were mutually replaced (Figure 6A). These origins were chosen because initiation of replication from the loci were confirmed by 2D gel analysis (Okuno et al, 1997; Segurado et al, 2003) and because two flanking genes are not essential for the viability, allowing manipulation of the loci. Fragments retaining ARS activity in plasmid transformation assay were used for replacement (data not shown). Wild type and ars2004/AT2080 cells were synchronized and labeled with BrdU in the presence of HU, and the HL (replicated) and light–light (unreplicated) DNA was separated in a CsCl gradient. The replication efficiencies of ars2004 and AT2080 as well as the early origin and non-ARS controls were measured by real-time PCR analyses. In the case of wild type, replication efficiencies of AT2024 and ars2004 were about 20–35% in the presence of HU, whereas the efficiency of AT2080 was about 1%, similar to the level of non-ARS locus, consistent with early firing at ars2004 but not at AT2080 (Figure 6B). In the origin-replaced strain ars2004/AT2080, however, the efficiency of ars2004 fragment integrated at AT2080 locus was about 3%, which was similar to the non-ARS locus, whereas the efficiency of AT2080 placed at the ars2004 locus was 17%, similar to the value of the early origin AT2024 (24%) (Figure 6B). These results show that replication timing of the origin is affected by its surrounding regions.
Figure 6.

Replication of the early and late origin fragments at the mutually exchanged loci. (A) Schematic drawing of chromosome II in wild type and the origin-exchange strains. Positions of AT2024 (early origin), non-ARS, ars2004 (early origin), cen2 and AT2080 (late origin) are shown. (B) Replication efficiency in the presence of HU. HM668 (h− cdc25-22 nmt1-TK) and HM1347 (h− cdc25-22 nmt1-TK ars2004∷AT2080 AT2080∷ars2004) synchronously released from G2/M block were labeled with BrdU in the presence of HU for 120 min and BrdU DNA was separated by CsCl centrifugation as described in Figure 1. The amounts of DNA in the light–light (LL) and heavy–light (HL) density fractions were measured by real-time PCR, and the replication efficiency (Re %) was obtained by the equation Re=[(HL/2)/(LL+HL/2)] × 100.
Coordinated distribution of early and late origins in large chromatin regions
Because the above results suggest that timing of origin firing is not intrinsic to origin function but dependent on its location, distribution of origins was statistically analyzed. The distribution of inter-pre-RC distances fitted well to an exponential curve (Supplementary Figure S10A), suggesting that pre-RCs are randomly distributed along chromosomes (Patel et al, 2006). Then, we asked whether a pre-RC is chosen to be an early origin independently of the neighboring origins or a subset of pre-RCs are coordinately chosen. In Figure 3, the early and late origins tend to cluster separately, spanning several hundred kb on chromosome I and on the left arm of chromosome II. This distribution could result from either random or coordinated choice of pre-RCs. To examine these possibilities, a statistical analysis, Wald–Wolfowitz run test (Chang, 2000), was carried out against a null hypothesis that a pre-RC is randomly chosen to be an early origin (see Materials and methods and Supplementary Figure S10B). The actual distribution of early origins does not likely result from randomness on chromosome I and on the left arm of chromosome II (P<0.01), whereas randomness was not rejected on the right arm of chromosome II (P=0.0198; α=0.01). The analysis was not applied on chromosome III, where only a few late origins exist. These results suggest that activation of pre-RCs tends to occur coordinately in large chromosome regions.
Initiation of replication is differently regulated in centromeric and subtelomeric heterochromatins
It is of interest how initiation of replication is regulated in heterochromatins, which have been reported to replicate in late S phase in higher eukaryotes (Woodfine et al, 2004; Jeon et al, 2005). In fission yeast, three regions, centromeres, telomeres and the mating-type controlling locus, are known to form constitutive heterochromatin structures. In 100–250 kb subtelomeric regions from the ends of chromosomes I and II, BrdU incorporation was rare despite the existence of highly clustered pre-RCs (Figures 3 and 7A), suggesting that they were not activated at least in early S phase. Checkpoint regulation is involved in the inactivation of the subtelomeric origins in late S phase under replication stress, as described in Figure 5.
Figure 7.
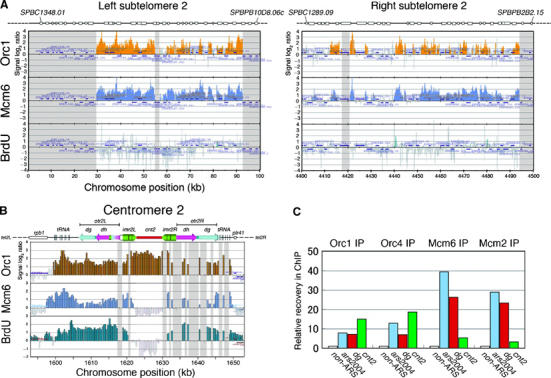
Locations of Orc1, Mcm6 and BrdU DNA in subtelomeric and centromeric heterochoromain regions. (A) Locations of Orc1 (orange bars, top), Mcm6 (blue bars, middle) and BrdU incorporation (green bars, bottom) in left and right subtelomeric regions on chromosome II in wild-type HM668 (h− cdc25-22 nmt1-TK) are presented. White boxes above the panels denote open reading frames. Telomere repeats and telomere-associated sequence (TAS) are not included in the analysis. Signals are absent in gray-shaded regions because of the presence of homologous sequences. (B) Locations of Orc1, Mcm6 and BrdU incorporation at centromere on chromosome II are shown. The physical map of fission yeast cen2 above the panels denotes otr comprised of dg and dh elements, imr and cnt. Vertical lines indicate tRNA genes. Signals are not present in gray-shaded regions at imr2R and otr2R because of the presence of identical sequences with imr2L and otr2L, respectively. (C) Localization of Orc1 and Orc4 but not of Mcm6 and Mcm2 at cnt2. ChIP with Orc1-5Flag, Orc4, Mcm6 and Mcm2 was carried out from G1-arrested cells as described in Figure 1. DNA recovered in ChIP was analyzed by real-time PCR using primers amplifying non-ARS, ars2004, dg and cnt2 regions, and relative recovery normalized to non-ARS region is presented.
Fission yeast centromeres are composed of two functional domains, the unique central core region (cnt), where the kinetochore complex is formed, and the inverted repeats (imr) and the outer centromeric repeats (otr), where heterochromatin and cohesion are formed (Pidoux and Allshire, 2004). It is remarkable that all BrdU, Orc1 and Mcm6 signals were detected at the otr, indicative of efficient initiation from the otr in early S phase (Figure 7B). In addition, we confirmed that BrdU-Orc1-Mcm6 signals were colocalized at the mating-type K locus in h90 strain, where the heterochromatin structure is formed depending on the otr-like sequence (data not shown). In contrast, the inner centromere cnt2 region did not replicate in the presence of HU (Figure 7B). Surprisingly, Mcm6 was not localized at the cnt2 region despite enriched binding of Orc1 (Figure 7B). Localization of Orc1 without Mcm6 or BrdU incorporation at the cnt was a common feature on all the chromosomes (Supplementary Figure S1, S2, S5 and data not shown). As we anticipated that epitopes in Mcm6 could be sequestered from antibody in cnt-specific protein complex, chromatin immunoprecipitation (ChIP) in G1-arrested cells (Takahashi et al, 2003) was carried out with polyclonal antibody against Mcm2 and Orc4 in addition to Orc1 and Mcm6, and immunoprecipitated DNA was analyzed by real-time PCR. The cnt2 DNA was recovered by Orc1-IP and Orc4-IP at a higher efficiency than the ars2004 and the otr DNA (Figure 7C). In contrast, recoveries of the cnt2 DNA by Mcm2-IP and Mcm6-IP were about one-tenth of those of the ars2004 and the otr DNA (Figure 7C). These results show that neither Mcm2 nor Mcm6 is efficiently located at cnt2, which is consistent with the results of DNA microarray. Thus, initiation of replication is differently regulated in the inner and outer centromere domains.
Discussion
In this study, high-resolution whole-genome mapping of Orc1 and Mcm6 binding sites allowed us to identify precise locations of 460 pre-RC sites on S. pombe chromosomes. We found that 307 pre-RC sites acted as early origins that initiated DNA synthesis in the presence of HU, whereas the rest of pre-RC sites were considered as late and/or inefficient origins (called collectively as late origins). Interestingly, pericentromeric heterochromatin and the silent mating-type locus replicated in the presence of HU, whereas the inner centromeres or subtelomeric regions did not, suggesting specific regulation of replication in heterochromatin regions.
Because more than 80% of Orc1-binding sites were colocalized with Mcm6, the majority of ORC binding sites serve for pre-RC assembly. The pre-RCs are formed exclusively in long and AT-rich intergenic regions as described previously (Gomez and Antequera, 1999; Dai et al, 2005). Pre-RCs are distributed randomly along the entire chromosomes except for enriched localization at pericentromeric and subtelomeric regions. However, only subsets of pre-RCs are activated in early S phase. An origin exchange experiment suggests that timing of firing is not intrinsic to ARS fragment but dependent on the context of the locus. Furthermore, the results of statistical analysis suggest that distribution of the early and late origins does not result from random choice of pre-RCs at least in chromosome I and in the left arm of chromosome II. These results are consistent with the idea that replication origins are coordinately regulated in broad chromosome regions, such as chromatin domains, which has been documented in higher eukaryotes (Schwaiger and Schubeler, 2006).
In metazoan organisms, different chromatin structures representing chromatin banding patterns replicate with different timing in S phase (Takebayashi et al, 2005). Growing evidence that actively-transcribed chromatin domains replicate early in S phase suggests coordinated regulation of replication timing in chromatin domains (Schubeler et al, 2002; MacAlpine et al, 2004; Woodfine et al, 2004; Schwaiger and Schubeler, 2006). We have been unsuccessful in finding correlation between transcription and origin activities in fission yeast (Jianhua Liu, personal communication). Swi6/HP1-dependent heterochromatin is not likely to be involved in replication timing regulation in arm regions, because neither Swi6 nor methylation of histone H3 lysine 9 is distributed on chromosome arms in fission yeast (Cam et al, 2005). However, various histone modifications could be involved in regulation of chromatin activities. Histone deacetylase inhibitors alter replication timing in mammalian cells (Bickmore and Carothers, 1995). In budding yeast, the Rpd3-Sin3 histone deacetylase affects replication timing of origins, which is independent of its role in transcriptional repression (Aparicio et al, 2004). Thus, it is of interest whether histone modifications are involved in regulation of replication timing on fission yeast chromosomes.
We showed that the late origins in subtelomeric and arm regions incorporated BrdU in HU-arrested late S phase in cds1Δ mutant (Figure 5). These results are consistent with the finding that single-stranded DNA accumulates at subtelomeric regions and other chromosome loci in HU-arrested cds1Δ cells (Feng et al, 2006). It should be noted, however, that cds1Δ-specific BrdU incorporation at the late origins was much less efficient than those at the early origins (Figure 5), suggesting that replication initiation occurs only in small population of cells. This is consistent with the report that deletion of Rad3 does not significantly change the number of origins identified by DNA content increase (Heichinger et al, 2006). Therefore, both checkpoint-dependent and checkpoint-independent regulations may account for suppression of initiation from the late origins.
Using a tiling array covering almost the entire genome of fission yeast, overall replication profiles of heterochromatin regions were shown in this study. Replication origins in constitutive heterochromatic regions, subtelomeres, centromeres and the silent mating-type locus behave differently from those in other chromosome regions. Pre-RCs are highly abundant in subtelomeric regions as far as 100 kb from the chromosome ends, but they are not activated in early S phase. In budding yeast, mutations in Sir3 result in both increased gene expression (Wyrick et al, 1999) and firing of silent replication origins in subtelomeric regions (Stevenson and Gottschling, 1999). In fission yeast, the heterochromatin protein Swi6 is required for repression of transcription by heterochromatin structures extending from the telomere ends (Kanoh et al, 2005), although it is not known whether Swi6 is involved in repression of the subtelomeric late origins. On the other hand, we showed that, in the absence of Cds1, BrdU incorporation increased at subtelomeric regions after prolonged incubation with HU (Supplementary Figure S9), suggesting that Cds1 has an important role in repression of telomere replication under replication stress. Progression of replication fork at telomeric region may be particularly sensitive to replication stress, and this may be avoided by strict suppression of initiation. Efficient telomeric fork progression is dependent on the telomere-specific protein Taz1 (Miller et al, 2006), suggesting that telomere is hard to replicate even in unperturbed S phase. Highly clustered pre-RCs in the subtelomeric regions might be required for efficient replication of heterochromatin DNA in very late S phase. Alternatively, they might play some role in telomeric heterochromatin maintenance.
The results of DNA microarray analysis in the present study clearly demonstrated distinct replication profiles at centromere; replication initiates at the pericentromeric repeats (otr) but not at the core regions (cnt) in all three centromeres. These results are consistent with the previous observation of centromeric ARSs using 2D gel analysis (Smith et al, 1995; Kim et al, 2003). Although the cnt1 and cnt2 fragments cloned on plasmids exhibit ARS activity, these replicators do not fire in the centromere (Takahashi et al, 1992; Smith et al, 1995). Remarkably, neither Mcm6 nor Mcm2 was localized at the core regions despite Orc1 and Orc4 enrichment (Figure 7B and C). It should be noted that suppression of pre-RC formation at ORC-bound origins is specifically observed in the core centromere, suggesting that interactions of ORC with pre-RC components might be interfered by those with centromere core specific proteins. As ORC but not MCM is allowed to bind to the inner centromere, it would be possible that ORC is needed for centromere/kinetochore functions. Possible involvement of human Orc2 in mitosis and cytokinesis has been suggested (Prasanth et al, 2004).
Early replication at the pericentromeric repeats and the absence of initiation at the cores may ensure establishment of cohesion at pericentromeric heterochromatin before kinetochore reassembly on duplicated cnt. Thus, distinct replication patterns of centromeric subdomains might be important for centromere functions. Earlier replication of pericentromeric heterochromatin (major satellite) than kinetochore-forming minor satellite on mouse chromosomes has been reported (Hollo et al, 1996; Guenatri et al, 2004).
High-resolution mapping of pre-RC sites and active replication origins on fission yeast chromosomes have provided genome-wide information of assembly sites for initiation factors and allowed prediction of replication fork movement. From the present study, we envision several advances in understanding of chromatin functions. First, as replication is likely to be coupled with various chromatin functions such as sister chromatid cohesion, condensation, DNA repair, checkpoint and chromatin structures, the data on replication machinery assembly sites and direction of replication fork progression will help to identify locations and movement of relevant proteins. Second, the combination of ChIP-based mapping and BrdU incorporation provide a powerful tool to investigate chromosome replication under various genetic conditions with different chromatin structures and histone modifications, or in replication mutants. This approach should be useful for identification of replication origins in more complex organisms, if locations of ORC and other initiation factors can be mapped using non-repetitive sequence tiling arrays.
Materials and methods
Yeast strains and plasmids
All strains used in this study are listed in Supplementary Table S2 and are available on request. A fission yeast strain expressing the herpes simplex virus thymidine kinase gene (TK) was obtained as below. For integration of TK-ura4+ fragment into ura4 locus, PCR-amplified upstream (−1078 to −579 bp) and downstream (+1244 to +1594 bp) locus of ura4+ gene was cloned into BamHI site of pBluescript II SK(+) by using primers 5′-AAAGGATCCTGCAGGCATGAAGAATTGGTTATCC-3′ and 5′-AAAGGATCCAAGCTTCTGTCAAAGTTTAAC-3′, and 5′-AAAGGATCCGCGGCCGCTAGTATACTTTTTCTCGGAG-3′ and 5′-AAAGGATCCTCGAGCCTGCAGGAGACGGTTCA-3′, respectively, resulting in pBS-ura4up and pBS-ura4dw. To obtain pMH2, a HindIII and SseI8387 fragment of the pBS-ura4up was cloned into modified pBluescript II, where XhoI–NotI fragment was replaced with a fragment carrying XhoI–NotI–ClaI–HindIII-SseI8387 recognition sites. A 1.8 kb ura4+ fragment was inserted at the HindIII site of the pMH2 generating pMH3, and then both the XhoI–NotI fragment of pBS-ura4dw and NotI fragment of pUC-nmt-Not were cloned into the XhoI–NotI site of pMH3, resulting in pMH5. The TK gene was re-cloned from the pGAD-TK plasmid (Katou et al, 2003) into the BamHI site between the nmt1 promoter and terminator sequences (Maundrell, 1993) on pMH5 to create pMH6. pMH6 was digested by SseI8387 and used for transformation of MH52 (h− ura4-D18 leu1-32), resulting in HM654.
The origin-exchanged strain was constructed as below. To construct a strain carrying ars2004 fragment at AT2080 locus, two segments near the ends of intergenic region containing AT2080 origin were PCR-amplified using primers 5′-AAAGAATTCGGATCCAACTCCGTAATCTTTTC-3′ and 5′-AAAAAGCTTGCGGCCGCAAACAGAGGCTTTGCATC-3′, and 5′-AAAAAGCTTCTAGAATTGTATACGTTCGC-3′ and 5′-AAAGGATCCGCAATGATGGTTGAACAG-3′, and cloned into modified pBluescript II, where HindIII–NotI fragment was replaced with EcoRI–HindIII–BamHI recognition sequence without XbaI or NotI, resulting in pAT2080-d1d2. The 1.8 kb ura4+ fragment was inserted at the HindIII site of pAT2080-d1d2 and the BamHI fragment carrying ura4+ gene flanked with segments homologous to AT2080 locus was used for transformation of HM83 (h+ ura4-D18). HM1170, an integrant of ura4+ at the locus was selected by PCR among Ura+ transformants. A 3.2 kb NotI–XbaI fragment of pARS2004 lacking an internal BamHI site was inserted into the NotI–XbaI sites of pAT2080-d1d2 and the BamHI fragment carrying ars2004 flanked with segments homologous to AT2080 locus was used for transformation of HM1176, and Ura− transformants were selected on 5-fluoro-orotic acid (5-FOA)-containing plates. Integration of ars2004 fragment at AT2080 locus was confirmed by PCR, resulting in HM1280. For construction of a strain carrying AT2080 fragment at the ars2004 locus, two fragments were amplified from the ars2004 locus using primers 5′-AAAGAATTCGGATCCGAAGATTCGCGAGGCACC and 5′-AAAAAGCTTCTGGCGAGCTATCTGTG, and 5′-AAAAAGCTTCCAAATCAACACACCCTAAC and 5′-AAAGGATCCGAATTCGTTAGATGTCTGTACAGG, respectively, and cloned into modified pBluescript II. A 2.3 kb AT2080 fragment amplified by PCR using 5′-AAAAAGCTTCAACTCCGTAATCTTTTC and 5′-AAAAAGCTTGCAATGATGGTTGAACAG was inserted at the HindIII site and the BamHI fragment carrying AT2080 flanked with homologous segments with ars2004 was used for transformation of HM198 (h+ ura4-D18 leu1-32 ars2004∷ura4+). HM1345 (h+ ura4-D18 leu1-32 ars2004∷AT2080) was selected by PCR among ura− transformants formed on 5-FOA-containing plates. HM1347 (h− cdc25-22 ars2004∷AT2080 AT2080∷ars2004 ura4-D18∷ura4+-nmt1-TK) was made by a standard genetic crosses.
ChIP on chip analysis
S. pombe chromosome II–III high-density oligonucleotide tiling arrays and whole chromosome tiling arrays were produced by Affymetrix Custom Express Service (S_pombea520106F, P/N 550106; pombeAlla520099, P/N 520099). Sequences and positions of oligonucleotides on the array are available from Affymetrix. For immunoprecipitation, HM568 (h− nda3-KM311 cdc10-129 orp1∷orp1-5flag ura4-D18 leu1-32) cells harboring pREP82-cdc18 and pREP81-cdt1 were cultured at 28°C for 16 h to express Cdc18 and Cdt1 from inducible nmt1 promoters, and then at 20°C for 4 h for M-phase arrest. Cells synchronously reentering the cell cycle were incubated at 36°C, the restrictive temperature for the cdc10-129, for 3 h to arrest them in G1 phase. Cell extracts were used for ChIP assays with anti-Flag mouse IgG (Sigma-Aldrich) or anti-Mcm6 rabbit IgG, as described earlier (Takahashi et al, 2003). Amplification of ChIP DNA, labeling with biotin-N6-ddATP, hybridization and primary data analyses were performed as described (Katou et al, 2003). For discrimination of positive and negative signals for binding, we compared the ChIP fraction with total cellular DNA without immunoprecipitation using three criteria. First, reliability of the strength of signals was judged by the detection P-value for each locus (P⩽0.025). Second, reliability of binding ratios was judged by change in P-values (P⩽0.025). Third, clusters consisting of at least three contiguous loci that fulfilled the above two criteria were selected. Microarray data presented in this paper can be obtained from GEO (http://www.ncbi.nlm.nih.gov/geo) with the accession number GSE6523.
BrdU incorporation
HM668 (h− cdc25-22 ura4-D18∷ura4+nmt1-TK+) or HM1405 (h− cdc25-22 cds1Δ∷kanMX6 ura4-D18∷ura4+nmt1-TK+) cells grown in EMM medium lacking thiamine to induce transcription of the TK gene at 25°C for 18 h to 1 × 107 cells/ml were arrested at the G2/M boundary for 3 h at 36°C, and then released for the indicated time at 25°C in the presence of 200 μM BrdU and 10 mM HU. Cells (1 × 108) were fixed with cold water containing 0.1% sodium azide and total cellular DNA was purified as described (Raghuraman et al, 2001). DNA was digested with HaeIII and centrifuged in 1.7 ml of the CsCl solution containing 10 mM Tris–HCl (pH 7.4), 1 mM EDTA and 150 mM NaCl (the refractive index at 25°C adjusted to 1.4030) in a Hitachi RP120VT rotor at 80 000 r.p.m. for 14 h at 20°C. Fractions (120 μl each) collected from the top were dialyzed using the Micro Dialysis System (GIBCO BRL), and recovery of DNA in each fraction was analyzed after PCR amplification with ars2004 and non-ARS primers. DNA in the HL density fractions was pooled and used for tiling array analysis as described above. BrdU DNA in HL fractions and total genomic DNA as control were analyzed with chromosome II and III tiling array and whole genome tiling array (Affymetrix). Amplification of BrdU DNA, labeling with biotin-N6-ddATP, hybridization and primary data analyses were performed as described (Katou et al, 2003). For quantitative analysis, DNA in each fraction was analyzed by real-time PCR using SYBR green I in 7300 Real-Time PCR System (Applied Biosystems). Primers used for real-time PCR are listed in Supplementary Table S3.
ARS assay
Intergenic regions were PCR-amplified and cloned into pYC11, a derivative of pBluescript II SK(−) carrying the LEU2 gene (Okuno et al, 1997), and used for transformation of HM123 (h− leu1-32). After 4 days incubation on EMM plates at 30°C, ARS plasmids yielded visible transformants at a high frequency.
2D gel electrophoresis
HM668 cells arrested at G2/M block were released at 25°C for 90 min in the presence of 10 mM HU. Genomic DNA was prepared and digested with the indicated restriction enzymes in low melting temperature agarose plugs (Arcangioli, 1998). Fractions enriched for replication intermediates were obtained by melting the agarose plug as described below (Furuya K, personal communication). Following 2 h digestion of genomic DNA with restriction enzymes (40 U/plug), the plugs were incubated at 70°C for 10 min, and then incubated at 37°C for 1 h with the restriction enzymes (40 U/plug), B-Agarase I (2 U/plug, New England BioLabs Inc.) and RNaseA (1 μg/plug, Sigma-Aldrich Inc.). Supernatant was collected after centrifugation at 13 000 r.p.m. for 10 min and DNA was precipitated with isopropanol. The replication intermediates were analyzed by neutral/neutral 2D gel electrophoresis as described previously (Brewer and Fangman, 1987).
Statistical analysis
To test the randomness of distribution of the early and late origins, we used Wald–Wolfowitz run test (Chang, 2000). A ‘run' means a sequence of adjacent equal symbols. For example, the sequence ‘EEELLLELLLLLEELE' is divided in seven runs: four of them are made of ‘E' and the others are made of ‘L'. If the two symbols are generated randomly, an expected number of runs, μ, and the variance, σ2, are given as μ=1+(2NENL)/N and σ2=2NENL(2NENL−N)/N2(N−1)=(μ−1)(μ−2)/(N−1), respectively, where NE is the number of E, NL that of L, and N=NE+NL. The z statistic is given by  , where Ra is the actual number of runs in the pattern. If the z statistic is negative, the actual number of runs is smaller than the expected number of runs, which means that each symbol has a tendency to be clustered. We set the significance level, α, at 0.01 in the analysis and calculated the z statistic for the distribution of the early and late origins on the left and right arms of each chromosome except for chromosome III, where the number of late origins is too small for valid statistics. All the values of z were negative and we could successfully reject null hypothesis that the distribution pattern of the early and late origins is random on the whole chromosome I and on the left arm of chromosome II (P<0.01), whereas the randomness was not rejected on the right arm of chromosome II (P=0.0198).
, where Ra is the actual number of runs in the pattern. If the z statistic is negative, the actual number of runs is smaller than the expected number of runs, which means that each symbol has a tendency to be clustered. We set the significance level, α, at 0.01 in the analysis and calculated the z statistic for the distribution of the early and late origins on the left and right arms of each chromosome except for chromosome III, where the number of late origins is too small for valid statistics. All the values of z were negative and we could successfully reject null hypothesis that the distribution pattern of the early and late origins is random on the whole chromosome I and on the left arm of chromosome II (P<0.01), whereas the randomness was not rejected on the right arm of chromosome II (P=0.0198).
Supplementary Material
Supplementary Legends
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Figure S9
Supplementary Figure S10
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Acknowledgments
We thank Drs Toshiki Tsurimoto and Hiroyuki Araki for critical reading, Jianhua Liu and Jun-ichi Nakayama for providing unpublished information, Kanji Furuya for valuable advice on two-dimensional gel electrophoresis and Shin-ichi Kotani for discussion on statistical analysis. This work was supported in part by a Grant-in-aid from the Ministry of Education, Science, Technology, Sports, and Culture, Japan, to HM and to KS.
References
- Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM (2004) The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol 24: 4769–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli B (1998) A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J 17: 4503–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Bickmore WA, Carothers AD (1995) Factors affecting the timing and imprinting of replication on a mammalian chromosome. J Cell Sci 108: 2801–2809 [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471 [DOI] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI (2005) Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Chang YC (2000) Residuals analysis of the generalized linear models for longitudinal data. Stat Med 19: 1277–1293 [DOI] [PubMed] [Google Scholar]
- Chuang RY, Kelly TJ (1999) The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci USA 96: 2656–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne RK, Kelly TJ (1995) Genetic analysis of an ARS element from the fission yeast Schizosaccharomyces pombe. EMBO J 14: 6348–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Chuang RY, Kelly TJ (2005) DNA replication origins in the Schizosaccharomyces pombe genome. Proc Natl Acad Sci USA 102: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A (1994) Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78: 303–316 [DOI] [PubMed] [Google Scholar]
- Dubey DD, Kim SM, Todorov IT, Huberman JA (1996) Large, complex modular structure of a fission yeast DNA replication origin. Curr Biol 6: 467–473 [DOI] [PubMed] [Google Scholar]
- Dubey DD, Zhu J, Carlson DL, Sharma K, Huberman JA (1994) Three ARS elements contribute to the ura4 replication origin region in the fission yeast, Schizosaccharomyces pombe. EMBO J 13: 3638–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Collingwood D, Boeck ME, Fox LA, Alvino GM, Fangman WL, Raghuraman MK, Brewer BJ (2006) Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat Cell Biol 8: 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM (2001) Making sense of eukaryotic DNA replication origins. Science 294: 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez M, Antequera F (1999) Organization of DNA replication origins in the fission yeast genome. EMBO J 18: 5683–5690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenatri M, Bailly D, Maison C, Almouzni G (2004) Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol 166: 493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bahler J, Nurse P (2006) Genome-wide characterization of fission yeast DNA replication origins. EMBO J 25: 5171–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollo G, Kereso J, Praznovszky T, Cserpan I, Fodor K, Katona R, Csonka E, Fatyol K, Szeles A, Szalay AA, Hadlaczky G (1996) Evidence for a megareplicon covering megabases of centromeric chromosome segments. Chromosome Res 4: 240–247 [DOI] [PubMed] [Google Scholar]
- Jeon Y, Bekiranov S, Karnani N, Kapranov P, Ghosh S, MacAlpine D, Lee C, Hwang DS, Gingeras TR, Dutta A (2005) Temporal profile of replication of human chromosomes. Proc Natl Acad Sci USA 102: 6419–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Sadaie M, Urano T, Ishikawa F (2005) Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol 15: 1808–1819 [DOI] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Kearsey SE, Cotterill S (2003) Enigmatic variations: divergent modes of regulating eukaryotic DNA replication. Mol Cell 12: 1067–1075 [DOI] [PubMed] [Google Scholar]
- Kim SM, Dubey DD, Huberman JA (2003) Early-replicating heterochromatin. Genes Dev 17: 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine DM, Bell SP (2005) A genomic view of eukaryotic DNA replication. Chromosome Res 13: 309–326 [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Rodriguez HK, Bell SP (2004) Coordination of replication and transcription along a Drosophila chromosome. Genes Dev 18: 3094–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123: 127–130 [DOI] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP (2006) Semi-conservative DNA replication through telomeres requires Taz1. Nature 440: 824–828 [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Takahashi T, Masukata H (1999) Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol Cell Biol 19: 7228–7236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, Okazaki T, Masukata H (1997) Identification of a predominant replication origin in fission yeast. Nucleic Acids Res 25: 530–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, Satoh H, Sekiguchi M, Masukata H (1999) Clustered adenine/thymine stretches are essential for function of a fission yeast replication origin. Mol Cell Biol 19: 6699–6709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N (2006) DNA replication origins fire stochastically in fission yeast. Mol Biol Cell 17: 308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Allshire RC (2004) Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosome Res 12: 521–534 [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B (2004) Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J 23: 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL (2001) Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Remus D, Beall EL, Botchan MR (2004) DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC–DNA binding. EMBO J 23: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JA, Kim SM, Huberman JA (1998) Ribosomal DNA replication in the fission yeast, Schizosaccharomyces pombe. Exp Cell Res 238: 220–230 [DOI] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF (1998) A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618 [DOI] [PubMed] [Google Scholar]
- Schubeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M (2002) Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet 32: 438–442 [DOI] [PubMed] [Google Scholar]
- Schwaiger M, Schubeler D (2006) A question of timing: emerging links between transcription and replication. Curr Opin Genet Dev 16: 177–183 [DOI] [PubMed] [Google Scholar]
- Segurado M, de Luis A, Antequera F (2003) Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep 4: 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segurado M, Gomez M, Antequera F (2002) Increased recombination intermediates and homologous integration hot spots at DNA replication origins. Mol Cell 10: 907–916 [DOI] [PubMed] [Google Scholar]
- Smith JG, Caddle MS, Bulboaca GH, Wohlgemuth JG, Baum M, Clarke L, Calos MP (1995) Replication of centromere II of Schizosaccharomyces pombe. Mol Cell Biol 15: 5165–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JB, Gottschling DE (1999) Telomeric chromatin modulates replication timing near chromosome ends. Genes Dev 13: 146–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M (1992) A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell 3: 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Ohara E, Nishitani H, Masukata H (2003) Multiple ORC-binding sites are required for efficient MCM loading and origin firing in fission yeast. EMBO J 22: 964–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebayashi S, Sugimura K, Saito T, Sato C, Fukushima Y, Taguchi H, Okumura K (2005) Regulation of replication at the R/G chromosomal band boundary and pericentromeric heterochromatin of mammalian cells. Exp Cell Res 304: 162–174 [DOI] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC (2003) Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev 17: 1894–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth JG, Bulboaca GH, Moghadam M, Caddle MS, Calos MP (1994) Physical mapping of origins of replication in the fission yeast Schizosaccharomyces pombe. Mol Biol Cell 5: 839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodfine K, Fiegler H, Beare DM, Collins JE, McCann OT, Young BD, Debernardi S, Mott R, Dunham I, Carter NP (2004) Replication timing of the human genome. Hum Mol Genet 13: 191–202 [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM (2001) Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294: 2357–2360 [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA (1999) Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418–421 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Legends
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Figure S9
Supplementary Figure S10
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3


