Abstract
Oxidative stress is a principal cause of DNA damage, and mechanisms to repair this damage are among the most highly conserved of biological processes. Oxidative stress is also used by phagocytes to attack bacterial pathogens in defence of the host. We have identified and characterised two apurinic/apyrimidinic (AP) endonuclease paralogues in the human pathogen Neisseria meningitidis. The presence of multiple versions of DNA repair enzymes in a single organism is usually thought to reflect redundancy in activities that are essential for cellular viability. We demonstrate here that these two AP endonuclease paralogues have distinct activities in DNA repair: one is a typical Neisserial AP endonuclease (NApe), whereas the other is a specialised 3′-phosphodiesterase Neisserial exonuclease (NExo). The lack of AP endonuclease activity of NExo is shown to be attributable to the presence of a histidine side chain, blocking the abasic ribose-binding site. Both enzymes are necessary for survival of N. meningitidis under oxidative stress and during bloodstream infection. The novel functional pairing of NExo and NApe is widespread among bacteria and appears to have evolved independently on several occasions.
Keywords: DNA repair, exonuclease, Neisseria meningitidis, repair phosphodiesterase, X-ray structure
Introduction
DNA repair is one of the most highly conserved biological processes and is fundamental to the survival of all free-living organisms. One of the principal causes of DNA damage is oxidative stress (Lindahl, 1993), and base excision repair (BER) is the primary mechanism used to counter the genotoxic effects of oxidation. Apurinic/apyrimidinic (AP) endonucleases are central to BER (Lindahl, 1993; Bjelland and Seeberg, 2003). These enzymes excise abasic residues from the DNA backbone that arise spontaneously (through depurination or depyrimidination) or by the action of DNA glycosylases (which recognise and remove bases damaged by oxidation). AP endonucleases bind abasic residues and cleave the DNA 5′ to the abasic site, leaving a 3′-hydroxyl, which is then repaired by DNA polymerases (Demple and Harrison, 1994; Dianov et al, 2003).
Many organisms possess two functional AP endonucleases. For example, Escherichia coli expresses ExoIII and EndoIV, representatives of the structurally distinct Xth and Nfo AP endonuclease families, respectively (Ljungquist et al, 1976). Humans and Saccharomyces cerevisiae also have two functional AP endonucleases. S. cerevisiae has the same complement of enzymes as E. coli, but the major enzyme is from the Nfo family (Boiteux and Guillet, 2004), whereas the human AP endonucleases, HAP1 and HAP2, are both members of the Xth family (Hadi and Wilson, 2000); HAP1 is the major human AP endonuclease with an essential function in higher eukaryotes (Xanthoudakis et al, 1996; Ludwig et al, 1998; Meira et al, 2001; Demple and Sung, 2005; Izumi et al, 2005). The duplication of AP endonuclease activity is thought to reflect the fundamental importance of their role in cellular viability.
DNA repair is also vital for the survival and propagation of bacterial pathogens. Bacteria undergo frequent replication, providing the opportunity for rapid accumulation of potentially cytotoxic mutations. Bacteria are also subject to oxidative stress during pathogenesis and must overcome the devastating damage caused by the oxidative burst within phagocytic cells of the immune system. To date, there have been few studies that have examined the role of BER during pathogenesis despite its importance in maintenance of DNA integrity in the face of the innate host response to infection. Salmonella typhimurium requires its full complement of DNA glycosylases to survive effectively within host macrophages, and the putative AP endonuclease in Brucella abortus is necessary for the pathogen to resist oxidative stress (Suvarnapunya and Stein, 2005; Hornback and Roop, 2006). However, in neither instance was the biochemical activity of the gene product determined; the studies relied upon the characterisation of strains lacking genes identified by sequence comparison alone.
Neisseria meningitidis is a Gram-negative bacterium, which is a causative agent of meningitis and septicaemia in humans. This pathogen is closely associated with the human host and undergoes a wide range of interactions with host cells. During progression of disease, N. meningitidis is internalised by phagocytic cells of the immune system and is subjected to the oxidative burst. The importance of meningococcal genome dynamics has been recognised, and several genes encoding putative BER enzymes have been identified by bioinformatic analysis (Davidsen and Tonjum, 2006). However, only MutY, a DNA glycosylase, has been characterised at a biochemical level, although its role during pathogenesis has not been examined (Davidsen et al, 2005).
In this work, we identify two AP endonuclease paralogues within the meningococcal genome. Structural and biochemical characterisation of the products of these genes reveals these BER enzymes, Neisserial exonuclease (NExo) and Neisserial AP endonuclease (NApe), have distinct activities, and are both necessary for resistance to oxidative stress and for meningococcal virulence.
Results
Characterisation of NExo, an AP endonuclease paralogue
A search for homologues of E. coli ExoIII in the trEMBL database identified a N. meningitidis gene, NMB0399, whose predicted product shares 34% amino-acid identity with ExoIII (Supplementary Table S1). We examined the activity of recombinant NMB0399 against two 25 bp double-stranded DNA substrates. The first (25U) contains a uracil 11 bases from the 5′ end, whereas the second (25AP) is 25U after treatment with uracil DNA glycosylase (UDG) to generate an abasic site at the position previously occupied by the uracil. With 25U, NMB0399 exhibited 3′-5′ exonuclease activity, similar to known AP endonucleases, although at a significantly lower rate than ExoIII (Figure 1A); the reaction product had a 3′-OH, which can be extended by DNA polymerases (data not shown). Treatment of the abasic substrate 25AP with ExoIII yielded the expected 10 bp product, resulting from AP endonuclease cleavage (Figure 1A). Surprisingly, recombinant NMB0399 did not exhibit AP endonuclease activity (Figure 1A) even under different conditions of pH, varied concentrations of NaCl and Mg2+ and in the presence of other divalent cations (data not shown). The enzyme did not show any nonspecific DNAse activity observed in other XthA family members (Weston et al, 1992; Miertzschke and Greiner-Stoffele, 2003). We therefore designated NMB0399 as Nexo, because, distinct from any other related enzyme, it has no measurable AP endonuclease activity but still displays 3′-5′ exonuclease activity.
Figure 1.
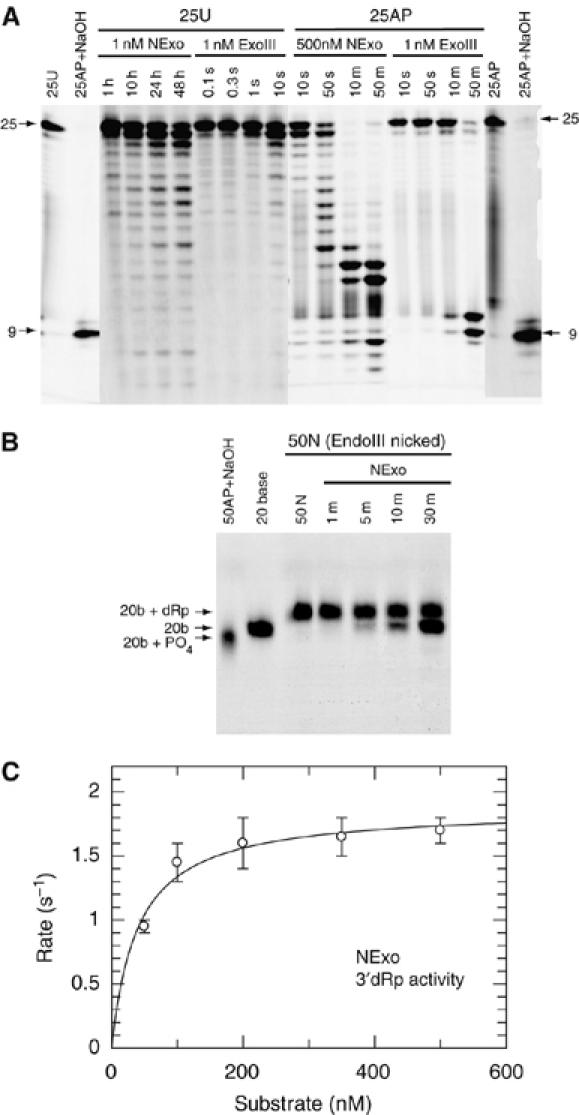
Biochemical activity of the N. meningitidis protein NExo (NMB0399). (A) Substrates 25U (100 nM) and 25AP (100 nM) were incubated with the indicated concentrations of NExo (NMB0399) or ExoIII and the products separated by denaturing PAGE. Control lanes show the unreacted 25U and 25AP substrates and the 25AP substrate after treatment with NaOH. (B) A nicked substrate with a 3′-dRp (50N) was generated by reacting a 50 bp DNA duplex with a U at position 21 with UDG (to give 50AP) and then Endonuclease III (50N). The 50N substrate (100 nM) was then reacted with 0.1 nM NExo (NMB0399) for the time periods shown before separation by denaturing PAGE. NExo is able to efficiently remove the 3′-dRp moiety from the 50N substrate before the further removal of nucleotides by exonuclease activity. Control lanes show the 50N substrate, 50AP after treatment with NaOH and a 20-base oligonucleotide. (C) The rate of 3′-dRp activity was measured for NExo by the appearance of products from the end processing of the 50N substrate. Reactions were performed as described in Materials and methods with substrate concentrations as shown and NExo at 1000-fold less. Data are shown with the best fit to the Michaelis–Menten equation: NExo has a kcat of 1.9±0.09 s−1 and KM of 40±9 nM.
We next examined whether NExo had 3′-deoxyribose phosphodiesterase (3′-dRpase) activity as this can also be catalysed by AP endonucleases. 3′-dRpases process 3′-blocked ends that are produced by single-strand breaks following oxidative damage (Takemoto et al, 1998; Dahm-Daphi et al, 2000) or by DNA glycosylases with lyase activity, which cleave abasic sites leaving a 3′-unsaturated aldehyde or phosphate (Michaels and Miller, 1992; Demple and Harrison, 1994; Agnez et al, 1996). Failure to remove these blocking residues is detrimental to cellular survival (Izumi et al, 1992; Takemoto et al, 1998; Boiteux and Guillet, 2004). Interestingly, NExo is a highly efficient 3′-dRpase. This was demonstrated experimentally using a nicked substrate with a 3′-dRp (50N), which was generated by reacting a 50 bp DNA duplex with a U at position 21 (50U) with UDG and the E. coli AP lyase endonuclease III. Incubating NExo with the 50N substrate resulted in the efficient removal of the 3′-dRp, leaving a 3′-OH (Figure 1B). A steady-state analysis of this reaction exhibited a kcat of 1.9±0.09 s−1 and KM of 40±9 nM (Figure 1C). Substitution of NExo Asp146 (the equivalent residue to the catalytically essential Asp210 in HAP1; Rothwell et al, 2000) with Asn resulted in an inactive enzyme (data not shown), demonstrating functional conservation of this key residue, even though NExo lacks AP endonuclease activity. These data indicate that NExo is a unique member of the Xth family with significant 3′-dRpase activity, but no detectable AP endonuclease activity.
A single amino-acid substitution can restore AP endonuclease activity on NExo
To investigate the molecular basis of these unique properties of NExo, we solved the NExo crystal structure at a 1.9 Å resolution, and found that it displays the characteristic four-layered α/β sandwich structure common to ExoIII and HAP1 (Mol et al, 1995; Gorman et al, 1997) (Figure 2A). The structures of NExo, ExoIII and HAP1 are closely related, having r.m.s.d. values of less than 1.5 Å over more than 230 Cα atoms (Supplementary Table S1). All the active site-residues necessary for AP endonuclease activity are present in NExo (i.e., Asp146, Asn148, Asp217 and His247; Figure 2B). More detailed examination of the NExo structure revealed a significant difference between this enzyme and the functional AP endonucleases, ExoIII and HAP1. The structure of HAP1 bound to an abasic DNA substrate shows that the DNA backbone is bent by 35°, with the abasic ribose extruded from the helix and bound in a conserved hydrophobic pocket formed by residues Asn226, Gly231, Phe266, Trp280 and Leu282 (Mol et al, 2000). Sequence and structural comparisons of Xth AP endonucleases indicate that a small amino acid (Gly231 in HAP1 and Ser178 in ExoIII) forms one side of the binding pocket. The equivalent residue in NExo is His167 (Figure 3A), which has a side chain that could sterically impair binding of the abasic ribose, preventing subsequent cleavage (Figure 3B).
Figure 2.
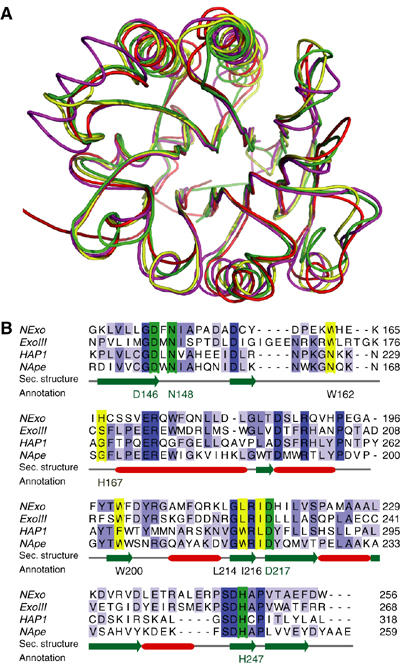
Crystal structure of the N. meningitidis protein, NExo. (A) Comparison of the AP endonuclease folds, HAP1 (pdb: 1dew; red) and ExoIII (pdb: 1ako; magenta) with NExo (NMB0399; pdb: 2jc4: yellow). Also included is the structure of NApe (NMB2082; pdb: 2jc5; green), the second AP endonuclease paralogue described in this paper. (B) Alignment of the C-terminal 117 residues of NExo, ExoIII and HAP1 with residues involved in abasic ribose binding (yellow) and catalysis (green) indicated. The sequence of NApe, the second AP endonuclease described in this paper, is included. Absolutely conserved residues in the alignment are coloured in dark blue and light blue for partial identity. Secondary structure elements are shown in red for α-helices and green for β-strands. A complete alignment of the proteins is provided in supporting online material (Supplementary Figure S2).
Figure 3.
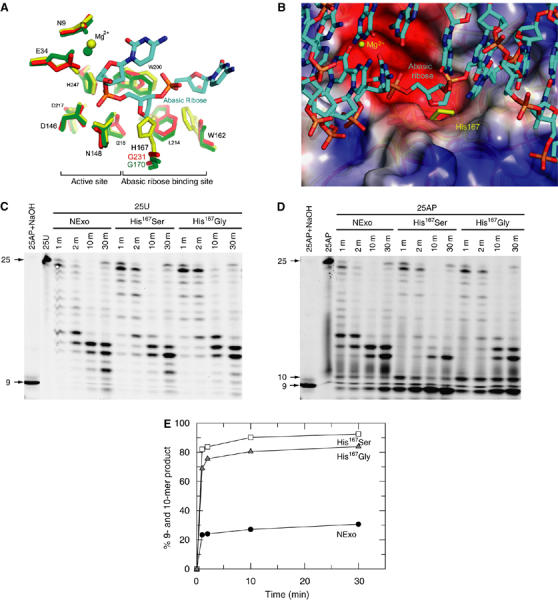
His167 is a structural determinant for the lack of AP endonuclease activity in NExo. (A) Comparison of the active site and the abasic ribose-binding pockets in NExo (yellow), NApe (green) and HAP1 (red), with the DNA from the HAP1/15mer DNA complex (pdb: 1dew). The numbering is for NExo, the equivalent residue numbers for other structures are shown in Figure 2B and Supplementary Figure S2. (B) The abasic ribose-binding site of HAP1 with a 15-mer DNA complex is shown with the electrostatic potential surface of the HAP1 protein (blue and red representing positive and negative charge, respectively) and the superimposed NExo protein (yellow), showing the presence of His167 of NExo in the proposed abasic ribose-binding site. Atoms in the DNA structure are coloured cyan for carbons, red for oxygen, blue for nitrogen and orange for phosphate. Residues Arg177 and Met270 were removed from the HAP1 structure to reveal the binding site beneath. (C) The 25U substrate (100 nM) was incubated with NExo, NExo His167Ser or NExo His167Gly (500 nM) for the time periods shown before separation by denaturing PAGE. Control lanes show the unreacted 25U and 25AP substrate after treatment with NaOH. (D) The 25AP substrate (100 nM) was incubated with NExo, NExo His167Ser or NExo His167Gly (500 nM) for the time periods shown before separation by denaturing PAGE. Control lanes show the unreacted 25AP substrate and the 25AP substrate after treatment with NaOH. Bands corresponding to 9 and 10 base products may arise through progressive exonuclease activity or through direct AP endonuclease activity to give a 10 base product followed by subsequent exonuclease activity to the 9 base product. (E) Quantification of the combined intensity in the 9 and 10 base products in panel D demonstrates that these accumulate only slowly with wild-type NExo (filled circles) but with NExo His167Gly (grey triangles) and His167Ser (open squares), these products accumulate rapidly and to near completion.
To test this possibility, we mutated NExo His167 to a Gly or a Ser, the corresponding residues in HAP1 and ExoIII, respectively. The modified enzymes retained exonuclease activity at similar levels to wild-type NExo (Figure 3C). Remarkably, modification of His167 to Gly or Ser was sufficient to confer AP endonuclease activity on NExo. Both enzymes cleaved the 25AP substrate at the abasic site, generating the expected 10 bp product, which was then fragmented by exonuclease activity (Figure 3D and E). Analysis of the crystal structures of the modified NExo enzymes confirmed that neither mutation caused any local or global structural perturbation in the structure (data not shown), and further kinetic analysis with a longer substrate allowed determination of the kcat and KM for AP endonuclease activity of the two mutants: for His167Ser kcat=0.41±0.02 s−1, KM=24±8 nM; for His167Gly kcat=0.15±0.01 s−1, KM=22±7 nM (Supplementary Figure S1). Remarkably, for a point mutation these values of kcat are only two- to five-fold lower than those determined for the wild-type NApe (see Identification of the functional N. meningitidis AP endonuclease). Therefore, His167 or its equivalent residue is a structural determinant for the activity of AP endonuclease paralogues.
Identification of the functional N. meningitidis AP endonuclease
As NExo had no apparent AP endonuclease activity, we searched the N. meningitidis genome for further AP endonuclease paralogues using the HAP1 sequence and identified NMB2082 (Supplementary Figure S2). The crystal structure of NMB2082 was solved at a 1.5 Å resolution and showed a high degree of structural similarity with NExo, HAP1 and ExoIII (rmsd <1.7 Å over 210 Cαs; Figure 2A; Supplementary Table S1), including the conservation of key catalytic residues (Figure 2B). Notably, NMB2082 has a glycine (Gly170) at the equivalent position to NExo His167 (Figure 3A), suggesting that NMB2082 could be a functional AP endonuclease. Using the 25AP substrate, NMB2082 indeed displayed significant specific AP endonuclease activity, with a kcat of 0.77±0.06 s−1 and KM of 33±12 nM (Figure 4A and C). Like other bona fide AP endonucleases, NMB2082 also has 3′-5′exonuclease and 3′-dRpase activities (Figure 4A and B); the latter has a kcat of 2.3±0.17 s−1 and KM of 73±19 nM (Figure 4D). We therefore designated N. meningitidis NMB2082 as NApe. Remarkably for a bacterial enzyme, NApe is more closely related to the equivalent human enzyme (HAP1, 33% sequence identity) than the one in E. coli (ExoIII, 23% sequence identity; Supplementary Table S1).
Figure 4.
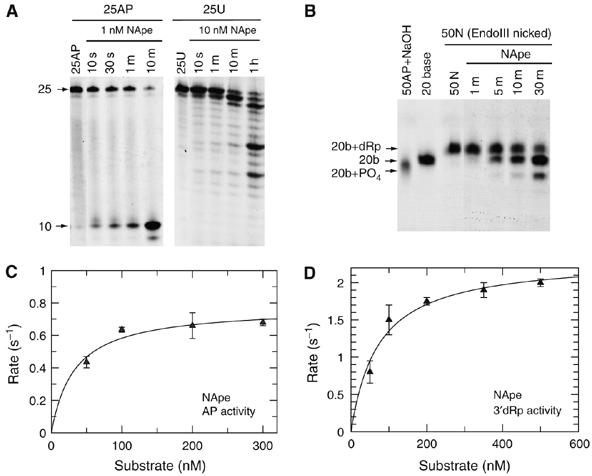
NApe protein catalytic activities. (A) Substrates 25AP (100 nM) and 25U (100 nM) were incubated with the indicated concentrations of NApe (NMB2082) for the time periods shown and separated by denaturing PAGE. Control lanes show the unreacted 25AP and 25U substrates. (B) Nicked substrate 50N (see Figure 1B) (100 nM) was reacted with 0.1 nM NApe for the time periods shown before separation by denaturing PAGE. NApe is capable of efficiently removing the 3′-dRp moiety from the 50N substrate before the further removal of nucleotides by exonuclease activity. Control lanes show the 50N substrate, 50AP after treatment with NaOH and a 20 base oligonucleotide. (C) AP endonuclease activity for NApe was measured by the appearance of product band on cleavage of substrate 25AP. Reactions were performed as described in Materials and methods with substrate concentrations as shown and NApe at 100-fold less than substrate. Data are shown with the best fit to the Michaelis–Menten equation with a kcat of 0.77±0.06 s−1 and KM of 33±12 nM. (D) 3′-dRp activity was measured for NApe by the appearance of products from the end processing of the 50N substrate. Reactions were performed as described in Materials and methods with substrate concentrations as shown and NApe at 1000-fold less than substrate. Data are shown with the best fit to the Michaelis–Menten equation with a kcat of 2.3±0.17 s−1 and KM of 73±19 nM.
NExo and NApe are necessary for resistance to oxidative stress
Oxidative stress is an important aspect of innate immunity against bacterial pathogens, and AP endonucleases are critical for repairing DNA lesions induced by oxidative stress. Therefore, we examined the role of NApe and NExo in bacterial survival under oxidative stress. We first confirmed by Western blot that NExo and NApe are expressed by N. meningitidis (Figure 5A). Strains were constructed lacking NExo or NApe, or lacking both enzymes. The mutants lacking NExo or NApe were significantly more sensitive to H2O2 and paraquat (which generate free radicals in the cytoplasm and periplasm, respectively) compared with the wild-type strain (Figure 5B). Interestingly, survival of the double mutant under conditions of oxidative stress is significantly impaired compared with the single mutants, such that the overall response is additive when compared to the individual mutants. Therefore, both NExo and NApe protect N. meningitidis from the harmful effects of oxidative stress; the additive response in the double mutant indicates that these enzymes have distinct roles in DNA repair.
Figure 5.
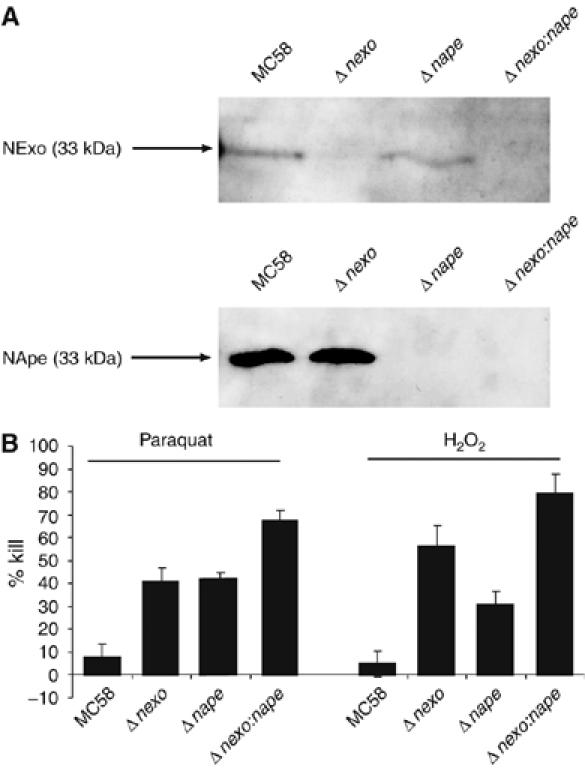
Western blots with anti-NExo and NApe antibodies showed protein expression in N. meningitidis. (A) Whole-cell lysates of N. meningitidis serotype B were separated by 12% SDS–PAGE and transferred to membranes, which were incubated with anti-NExo (panel A, 1:5000 dilution) or anti-NApe (Panel B, 1:10 000 dilution) sera (generated by immunisation of adult female BALB/c mice on three occasions with 25 mg of recombinant NExo or NApe with an equal volume of Freund's incomplete adjuvant). Binding was detected with the ECL bioluminescence kit (Amersham). Strains are indicated above each lane, and the predicted molecular mass of each protein is indicated. (B) Δnexo, Δnape and Δnexo:nape strains were plated to medium with or without the oxidising agents paraquat (0.075 mM) or H2O2 (13 mM). The percentage killing was calculated by comparing viable colony counts in the presence and the absence of the oxidising agent. Error bars show the s.e. The strains are indicated under each lane. Both Δnexo and Δnape are significantly susceptible to both agents, and the double mutant was more susceptible than either single mutant, exhibiting an additive effect.
NExo and NApe are necessary for virulence
The generation of oxidative stress is an important aspect of defence against microbial infection in vivo. Therefore, we evaluated the physiological relevance of the sensitivity to oxidative stress of the NExo and NApe mutant strains by assessing their ability to cause bacteraemia, an essential step during the pathogenesis of meningococcal infection. We initially determined that the mutations do not attenuate normal growth of the bacterium by performing growth curves (Supplementary Figure S3). The mutants were recovered from the bloodstream of infected infant rats at significantly lower levels than the wild-type strain, demonstrating that both NApe and NExo are required for full virulence (Figure 6). The marked additive attenuation of the double mutant indicates that NApe and NExo contribute to pathogenesis through distinct pathways.
Figure 6.
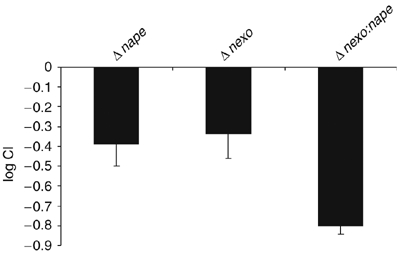
CI of Δnexo, Δnape and Δnexo:nape. Animals received a 1:1 mixture of wild-type and mutant bacteria, and the C.I. was determined from the ratio of wild-type to mutant in the bloodstream 8 h after inoculation. Both Δnexo and Δnape were significantly attenuated, and Δnexo:nape is more markedly attenuated than either of the single mutant strains, exhibiting an additive effect from the combined mutations. Error bars show the s.e.
The NExo/NApe pairing occurs frequently among bacterial species
Duplication of AP endonuclease enzymes as seen in numerous organisms may result from the critical nature of the BER pathway for cellular viability. However, our structure–function characterisation of AP endonuclease paralogues in the meningococcus demonstrates that there is specialisation in the activity of these enzymes. We considered whether having enzymes with distinct functions (such as NExo and NApe) could be unique to N. meningitidis, because, unlike most bacteria, it lacks an SOS response (Morelle et al, 2005) and thus might employ atypical mechanisms for dealing with DNA damage. Therefore, we analysed complete bacterial genome sequences for NExo and NApe homologues, paying particular attention to the nature of the amino acid at the residue equivalent to NExo His167. We identified a wide range of bacteria that contain a NExo/NApe pairing (Figure 7 and Supplementary Table S2), including human and plant pathogens and soil-living organisms. The alternative EndoIV family of AP endonucleases are found in some, but not all, of the organisms that have a NExo/NApe pairing. EndoIV enzymes may thus provide an additional AP activity in some organisms, but there is no direct correlation to the presence or absence of NExo and/or NApe.
Figure 7.
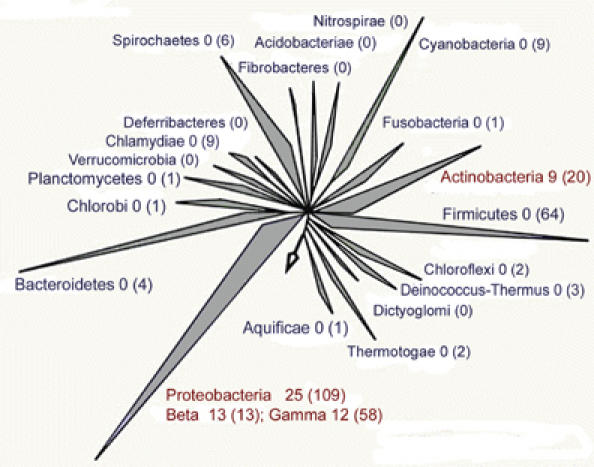
Phylogenetic distribution of the NExo/NApe pairing. The NExo/NApe pairing is an ancient occurrence, being found in three bacterial phyla, the actinobacteria, gammaproteobacteria and betaproteobacteria. Two hundred and ninety-seven completely sequenced genomes were analysed for the presence of both NApe and NExo paralogues. The difference between NExo and ExoIII paralogues was defined by the presence of a small or large side chain in the position equivalent to NExo His167; ExoIII paralogues were excluded from this analysis. The number of species with both NExo and NApe paralogues in each phylum is given after the phylum name and the number of genomes searched in each phylum is given in brackets. Phyla with pairs of these enzymes are shown in red.
Discussion
Functional variation within AP endonucleases
The Xth family of AP endonucleases are well conserved at the structural level, with a four-layered αβ-sandwich structure. The enzymes in this family are able to perform a wide variety of excision and incision reactions on DNA substrates, including AP endonuclease, 3′-5′ exonuclease, 3′-dRpase and 3′-phosphatase, and occasionally nonspecific DNase activities (Weston et al, 1992; Miertzschke and Greiner-Stoffele, 2003). The class II AP endonucleases typically perform the first three or four of these reactions. In contrast to DNaseI, these enzymes have additional surface loops that interact with the DNA and are required for abasic site recognition (Gorman et al, 1997; Cal et al, 1998; Mol et al, 2000). Both NExo and NApe possess the surface loops expected in an AP endonuclease enzyme, but with variations in sequence.
Structural analysis has revealed that a determinant of NExo activity is a His residue, which blocks the position where the flipped-out abasic ribose would bind in an AP endonuclease. For DNA to bind to an AP endonuclease, the abasic ribose must first be everted from the DNA helix and the DNA bent by approximately 35° (Gorman et al, 1997; Mol et al, 2000). His167 would prevent NExo from flipping out the abasic ribose. Mutation of this residue to a smaller residue, opening up the abasic ribose-binding site, conferred AP endonuclease activity on NExo and clearly demonstrates the importance of this residue in defining enzyme activity.
The region immediately before His167 in NExo forms a loop, which may interact with DNA. The equivalent of His167 in NExo in the nonspecific endonuclease DNAse1 is Tyr175. It has been shown that insertion of the helical sequence from ExoIII immediately before Tyr175 confers AP endonuclease activity onto DNase1 (Cal et al, 1998). As there is no structure for DNase1 with this insertion, it is unclear whether the recovery of AP endonuclease activity is because of the presence of the helix, or movement of the Tyr175 side chain, resulting in creation of an abasic ribose-binding site. We predict that both the abasic ribose-binding site blocking residues and the DNA-binding loops will determine the balance of AP endonuclease, 3′-dRpase and exonuclease activities found in individual enzymes.
It is now becoming apparent that different AP endonuclease enzymes have different efficiencies by which they catalyse the different reactions, and this most likely reflects their in vivo function. For instance, E. coli ExoIII is an active AP endonuclease and also a strong 3′-5′ exonuclease and 3′-dRpase, whereas the human AP endonuclease HAP1 is a very efficient AP endonuclease, but a poor 3′-5′ exonuclease and 3′-dRpase (Wilson et al, 1995; Suh et al, 1997). In contrast, the second human AP endonuclease HAP2 has low AP endonuclease activity, but strong 3′-5′ exonuclease and 3′-dRpase activities (Burkovics et al, 2006). NExo is a very active 3′-dRpase, but a weak 3′-5′ exonuclease and lacks detectable AP activity. Our characterisation of NExo thus represents the first description of an enzyme, which by both sequence and structure closely resembles other class II AP endonucleases of the Xth family, yet has no discernable AP activity. NExo appears to be a highly specialised 3'-dRpase. We consider this specialisation to be indicative of the importance of removing 3'-blocking lesions, which probably contribute more to cell lethality than abasic DNA sites (Izumi et al, 1992; Takemoto et al, 1998; Boiteux and Guillet, 2004). Although abasic sites are mutagenic, they may be bypassed by DNA polymerases, whereas blocks to replication are more detrimental.
In vivo function of NExo and NApe
The distinct enzyme activities of NExo and NApe suggest that they have evolved to perform different functions within the cell. In vivo assays demonstrate this to be the case, as the effects of the individual mutations are additive in the double mutant. This is true both for survival assays in the face of oxidative stress, and in infectivity assays. NExo and NApe therefore operate in discrete pathways, although they possess overlapping activity. This may be owing to differences in substrate specificity for the two enzymes: a number of different 3′-blocking lesions can arise through oxidative or other damage processes in addition to the 3′-dRp that we have tested, including 3′-glycolate and 3′-phosphate. Alternatively, AP endonucleases can interact with some DNA glycosylases to facilitate DNA repair through a hand-over process that protects mutagenic intermediates in the repair pathway (Waters et al, 1999; Hosfield et al, 2001). The functionality of NExo and NApe may therefore be due to the separation of discrete DNA repair pathways through interactions of protein networks.
Implications for bacterial pathogenesis
To survive periods of sustained high levels of DNA damage, many bacterial pathogens possess an SOS response. This damage-inducible system is characterised by a general upregulation of a range of components of DNA repair pathways following detection of DNA damage. The SOS response has been well studied in a number of pathogenic and non-pathogenic bacterial species, including E. coli, Staphylococcus aureus, Vibrio cholerae and Bacillus subtilis (Wojciechowski et al, 1991; Michel, 2005; Goerke et al, 2006; Quinones et al, 2006). Interestingly, N. meningitidis does not possess an SOS response, and is thus thought to be unable to upregulate expression of components of the repair pathway. Comparison of the expression of NExo and NApe in the presence and absence of oxidative agents confirms that there is no change in the production of the two enzymes when the bacterium is exposed to oxidative stress (data not shown). Therefore, the lack of upregulation of NExo and NApe production is entirely consistent with the absence of an SOS response.
The mechanisms by which pathogenic Neisseria neutralise exposure to oxidising agents have been studied extensively. In the gonococcus, these include the production of superoxide dismutases (Sods), catalase and peroxidase (Seib et al, 2004). In N. meningitidis, Sod is required to evade internalisation by phagocytic cells and provides protection against the action of reactive oxygen species (Dunn et al, 2003; Seib et al, 2004). Production of both glutathione peroxidase and catalase aid resistance to paraquat and hydrogen peroxide, respectively (Moore and Sparling, 1996; Seib et al, 2004). Despite the presence of these protective systems, our results clearly show that the ability to repair oxidative DNA damage still contributes significantly to the survival of the bacterium under oxidative stress. Furthermore, these repair enzymes are necessary for wild-type levels of virulence. The attenuation of the mutant strains lacking NExo and NApe is likely to be because of their inability to prevent accumulation of deleterious mutations arising as a result of stress induced by products of the host cellular metabolism or phagocyte oxidative burst.
Diversity and evolution
Diverse bacteria have evolved to contain a NExo/NApe pairing, defining a novel combination of structurally related, but functionally distinct enzymes in BER. Characterisation of the NApe and NExo knockout strains demonstrates that their functions appear to be independent. The widespread distribution of NApe paralogues among bacteria (including those with a saprophytic lifestyle) indicates that they were unlikely to have been acquired recently by horizontal transfer from a eukaryotic host. Instead, the pairing of NApe and NExo appears to have arisen on several occasions in distinct prokaryotic lineages, indicating that it must provide a significant evolutionary advantage to certain microbes.
Conclusion
We have characterised two Xth family members from N. meningitidis, which have distinct activities in DNA repair. This specialisation of enzyme function within the BER pathway, which is distinct from the redundancy observed in other organisms, is a novel paradigm present in a wide variety of bacteria that is relevant to the infectivity of human pathogens such as N. meningitidis.
Materials and methods
Bacterial strains and growth
N. meningitidis was grown on BHI media with 5% Levanthal's supplement, and incubated at 37°C in the presence of 5% CO2. E. coli XL1-blue cells (Invitrogen, for recombinant work) and ER2566 (New England Biolabs), for protein expression, were propagated in LB media. Antibiotics were used at the following final concentrations: ampicillin, 100 μg/ml; kanamycin, 75 μg/ml; and tetracycline, 2 μg/ml.
Identification and site-directed mutagenesis of NExo and NApe
NMB0399 was identified as a possible AP endonuclease paralogue by searching the trEMBL database (Boeckmann et al, 2003) using the programme BLAST (Altschul et al, 1997), implemented at the European Bioinformatics Institute (www.ebi.ac.uk). NMB2082 was identified by searching the serogroup B N. meningitidis genome, with the HAP1 gene, using the Sanger Institute's version of the programme BLAST. NMB0399 and NMB2082 were amplified by PCR from N. meningitidis with the primers NMB0399A and NMB0399B for NMB0399, and primers NMB2082A and NMB2082B for NMB2082 (Supplementary Table S3).
Relevant restriction sites are underlined. PCR products were digested with BamHI and HindIII, cloned into pProEX HTb (Invitrogen) and transformed into E. coli XL1-Blue (Invitrogen). Subsequently, plasmids were transformed into E. coli ER2566 for protein expression.
The mutants, NExoD146N, NExoH167S and NExoH167G, were obtained using the Stratagene QuikChange Site-Directed Mutagenesis kit with primers D146Nf and D146Nr for NExoD146N; and H167Sf and H167Sr for NExoH167S; H167Gf and H167Gr for NExoH167G (Supplementary Table S3). All mutations were verified by DNA sequencing.
Protein expression and purification
E. coli cultures of 3–6 l were grown to an OD A600 of 0.4–0.6 in LB medium and protein expression was induced by the addition of 1 mM IPTG, followed by continued growth for 3 h at 37°C. Cells were harvested by centrifugation. Pellets were resuspended in buffer containing 25 mM Tris (pH 8.0), 25 mM imidazole, and 0.5 M NaCl (buffer A), and lysed by pulse sonication. The lysate was cleared by centrifugation. His-tagged proteins were purified using a metal-affinity HisTrap chelating HP column (GE Healthcare) pre-equilibrated with CoCl2. Proteins were eluted with an imidazole gradient and dialysed into buffer containing 50 mM Tris (pH 8), 0.5 mM EDTA, and 1.0 mM DTT. The His tag was cleaved by the addition of TEV protease (Invitrogen) and the sample was then dialysed against buffer A. The cleaved proteins were applied to a cobalt-loaded HisTrap column and the protein of interest was found in the flow-through. NApe was further purified by size-exclusion chromatography using HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare) pre-equilibrated in 10 mM Tris (pH 7.0), 100 mM NaCl. NExo samples were crystallised without prior size exclusion chromatography.
Biochemical assays
Double-stranded DNA substrates 25U, 50U and 71AP were made by mixing equimolar amounts of complementary strands, heating to 90°C and cooling slowly to room temperature. One strand contained a uracil residue and a 5′-hexachlorofluorescein residue (HEX) for visualisation; the complementary strand was unmodified. The sequence of the double-stranded 25U oligonucleotide was 5′-HEXGGATCACTATUATAGGTAGTTTAT–3′; the sequence of the double-stranded 50U oligonucleotide was 5′-HEXCGTGTATGACATCTAACTATUATAGCGCT CATCGTCATCGTCATCCTCGGCACCGT-3′ (U was paired with A in both cases). The double-stranded 71AP substrate contained an abasic furanose analogue at position 36, the sequence was 5′-(HEX)-GAG CAG AGT CAG TGC TGC TAC GAG CGG ATC ACT ATF ATA GGT AGT TTA TCC TAC GAA CTC CGT CCG TAC CG-3′, where F is the abasic furanose analogue. Oligonucleotides were synthesised and HPLC-purified by MWG Biotech (Ebersberg, Germany) or Eurogentec (71AP). The 25U and 50U substrates were reacted with 100 nM Herpes simplex virus type-1 uracil DNA glycosylase (UDG) for 2 h at 25°C to create the abasic 25AP and 50AP DNAs (Bellamy and Baldwin, 2001). Nicked DNA was produced by reacting 200 nM of 50AP DNA with 25 U of endonuclease III (New England Biolabs) in standard reaction buffer (20 mM Tris pH 7.5, 1 mM EDTA, 20 mM NaCl, and 0.1 mg/ml BSA) at 37°C for 90 min.
Nuclease assays were performed in a standard reaction buffer supplemented with 15 mM MgCl2 at 25°C. Aliquots were removed at specified time points as detailed in the figure legends, and quenched in formamide loading buffer (0.01% xylene cyanol, 0.01% bromophenol blue, 30 mM EDTA in formamide) before separation via denaturing polyacrylamide gel electrophoresis. All assays used 100 nM of the respective DNA substrate and an equivalent of 500 fmol of DNA substrate per lane was loaded onto acrylamide gels. Enzyme concentrations vary as described in the figure legends. Exonuclease assays were analysed using 15% PAGE, whereas end-processing assays used 20% PAGE. Data were fitted using Grafit 5 (Erithacus software).
Crystallization and X-ray crystallography
NExo was dialysed into 10 mM Tris, pH 7.0, 100 mM NaCl and concentrated to 9 mg/ml before crystallisation. Crystals were obtained in 96-well Corning plates using 1 μl of protein and 1 μl of well solution. The well solution consisted of 0.1 M sodium cacodylate, pH 6.5, 0.2 M sodium acetate and 15% PEG 8000. Crystals were transferred into a cryobuffer containing all the well-solution components and 25% PEG400. Data were collected to 1.9 Å on beamline 14.1 at the SRS synchrotron in Daresbury, UK (see Table I).
Table 1.
Crystallographic statistics for the NExo and NApe wild-type crystals
| NExo | NApe | |
|---|---|---|
| Beamline | SRS, 14.1 | ESRF, id14.2 |
| Space group | P212121 | P21 |
| Unit-cell parameters a, b, c (Å) | 46.19, 58.62, 88.63 | 38.16, 79.5, 41.0 |
| β (deg) | 90.0 | 107.2 |
| Resolution (Å) | 20.0–1.9 (1.97-1.90) | 30.0–1.50 (1.55–1.50) |
| Completeness (%)* | 97.6 (91.4) | 94.6 (86.3) |
| Redundancy* | 6.8 | 3.5 (3.1) |
| 〈I〉/〈σI〉* | 24.5 (5.1) | 18.3 (4.5) |
| Unique reflections | 19164 | 35560 |
| Rsym (%)*† | 9.4 (32.1) | 5.6 (26.9) |
| Refinement resolution range (Å) | 20.0–1.9 | 30.0–1.5 |
| No reflections working/test | 18130/982 | 33629/1774 |
| Rcryst§/Rfree¶ (%) | 15.2/20.2 | 12.1/15.9 |
| Rmsd bonds (Å) | 0.016 | 0.010 |
| Rmsd on angles (deg) | 1.501 | 1.330 |
| Both the structures and the structure factors have been deposited with the protein databank. NExo: 2jc4.pdb; NApe: 2jc5.pdb. | ||
| aValues in parentheses correspond to the highest resolution shell. | ||
| †Rsym=∑h∑i∣I i (h)−〈I(h)〉∣/∑h ∑i (h), where Ii(h) is the ith measurement. | ||
| §R factor=∑∣∣F(h)obs∣−∣F(h)calc∣∣/∑∣F(h)obs. | ||
| ¶Rfree is calculated in the same way as the R factor, using only 5% of reflection randomly selected to be excluded from the refinement. | ||
The NApe crystals were obtained from hanging-drop experiments using 24-well Linbro plates with drops of 0.5 μl protein (20 mg/ml) in 10 mM Tris, pH 7.0 and 100 mM NaCl, and 0.5 μl well solution at 20°C. The well solution contained 20% PEG 20000, 0.1 M bicine, pH 9.0 and 2% dioxane. NApe crystals were placed in a cryobuffer with 25% glycerol and all the components of the well solution briefly before cryocooling. X-ray crystallographic data were collected on beamline id 14.2 at ESRF.
All data were processed with DENZO and reduced with SCALEPACK (Otwinowski and Minor, 1997). Both structures were solved by molecular replacement (Vagin and Teplyakov, 1997) using the structure of the uncomplexed human enzyme (Gorman et al, 1997) (pdb code 1bix). ARP/WARP (Perrakis et al, 2001) was used for automatic model building and O50 for manual rebuilding. The structure was refined using both the CNS (Brunger et al, 1998) and REFMAC5 (Murshudov et al, 1997) packages with anisotropic B factors applied to the NApe structure only. The quality of all structures was assessed using both PROCHECK (Laskowski et al, 1993) and WHATCHECK (Hooft et al, 1996). Structures were superimposed using the lsq_exp and lsq_imp functions in O (Jones et al, 1991). Structural alignments were initially generated with CLUSTALW (Thompson et al, 1994), then manually adjusted using the programme JalView (Clamp et al, 2004) to match the structural alignment. Structure figures were generated using PyMol (DeLano Scientific).
Bioinformatic analysis
A database containing over 297 complete bacterial and eukaryotic genomes was searched to identify genes with similarity to NApe and NExo using NCBI BLAST (Altschul et al, 1997). Each gene identified as a possible paralogue was then used to search the N. meningitidis genome and matches were only accepted if the gene used for the original search was found in the N. meningitidis genome. All searches were performed with protein sequences. Organisms were identified which had both NApe and NExo paralogues and these genes were further analysed to identify which residue aligned with the His167 in NExo. Alignments were produced using ClustalW. The alignments were assisted by forcing matches to catalytically important residues. A total of 35 organisms were identified that clearly had both enzymes (Supplementary Table S2). These organisms were mapped onto a version of the tree of life and found to come from many but not all evolutionary families. Sequence processing and analysis were carried out using custom Perl scripts.
Construction of bacterial strains and analysis of growth curves
A 1771 bp fragment containing the open reading frame (orf) of NMB0399 was amplified by PCR from MC58 genomic DNA with primers NG489 and NG490 (Supplementary Table S3). The product was ligated into pCR TOPO2.1 (Invitrogen). The resulting plasmid was digested with SphI to remove a portion of the cassette encoding resistance to kanamycin, and then self-ligated to yield in pMW34. This plasmid was subjected to in vitro mutagenesis with Tn5 (Epicentre) and the sites of transposon insertion mapped by restriction enzyme analysis and sequencing. pMW34 contains Tn5 at nt 900 of the NMB0399 ORF. A 700 bp upstream fragment of NMB2082 was amplified with primers NG491 and NG492 and a 700 bp downstream region with primers NG493 and NG494 (Supplementary Table S3). The PCR products were ligated into pCR2.1TOPO. The upstream fragment was excised and ligated into the vector containing the downstream fragment. The gene encoding tetracycline resistance was removed from pCMT18 and inserted into between the up- and downstream fragments at the unique EcoRI site. N. meningitidis MC58 was transformed to kanamycin resistance by standard methods, and transformants were examined by PCR and Southern analysis to confirm that allelic replacement had occurred. For the analysis of bacterial growth, 10 ml of BHI broth was inoculated with bacteria to an of 0.15 and incubated at 37°C with shaking at 150 r.p.m. The optical density at 600 nm was measured at hourly intervals for up to 8 h.
Assays for survival under oxidative stress
Strains were grown overnight, harvested into PBS and the bacterial suspensions were adjusted to 1 × 108 colony-forming units (CFU)/ml and serial dilutions made. Each strain (10 μl, 103 CFU/ml) was then spread onto BHI agar containing either hydrogen peroxide (13 mM; Biochemika) or paraquat (0.075 mM; Sigma) or onto BHI alone. The number of CFU were counted and the survival of each strain on media containing paraquat or hydrogen peroxide was expressed as the proportion of survival on BHI alone. The assays were performed in triplicate on at least three independent occasions.
Virulence assays
To examine the effect loss of NExo and NApe on virulence, mixed litters of 5-day-old rats (Wistar) were inoculated with 1 × 107 CFU/ml N. meningitidis in PBS. Bacteria were grown overnight on solid media, resuspended in PBS and then enumerated by measuring the A260 of an aliquot of the suspension in lysis buffer (1% SDS/0.1 M NaOH). The virulence of the mutants was compared directly with MC58 in individual animals given a 1:1 ratio of wild-type to mutant bacteria. The number of mutant and wild-type bacteria recovered from the blood of animals 8 h later was established by plating to media with or without antibiotics. The competitive index (CI) was calculated as the (number of mutant/wild-type bacteria recovered from animals)/(number of mutant/wild-type bacteria in the inoculum).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Acknowledgments
We thank the European Synchrotron Facility in Grenoble, France and the Synchrotron radiation facility, Daresbury, UK, for data collection and the MRC for financial support. Work in CMT's laboratory is supported by the Meningitis Research Foundation. We would also like to thank Mathieu Rappas, Valerie Pye and Ingrid Dreveny for assistance with X-ray crystallographic data collection and Gayle Bartlett and Derek Huntley of the ICL Bioinformatics service for assistance with the bioinformatics analysis.
References
- Agnez LF, Costa de Oliveira RL, Di Mascio P, Menck CF (1996) Involvement of Escherichia coli exonuclease III and endonuclease IV in the repair of singlet oxygen-induced DNA damage. Carcinogenesis 17: 1183–1185 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellamy SR, Baldwin GS (2001) A kinetic analysis of substrate recognition by uracil-DNA glycosylase from herpes simplex virus type 1. Nucleic Acids Res 29: 3857–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelland S, Seeberg E (2003) Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res 531: 37–80 [DOI] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, Pilbout S, Schneider M (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S, Guillet M (2004) Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair 3: 1–12 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Burkovics P, Szukacsov V, Unk I, Haracska L (2006) Human Ape2 protein has a 3′-5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res 34: 2508–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cal S, Tan KL, McGregor A, Connolly BA (1998) Conversion of bovine pancreatic DNase I to a repair endonuclease with a high selectivity for abasic sites. EMBO J 17: 7128–7138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20: 426–427 [DOI] [PubMed] [Google Scholar]
- Dahm-Daphi J, Sass C, Alberti W (2000) Comparison of biological effects of DNA damage induced by ionizing radiation and hydrogen peroxide in CHO cells. Int J Radiat Biol 76: 67–75 [DOI] [PubMed] [Google Scholar]
- Davidsen T, Tonjum T (2006) Meningococcal genome dynamics. Nat Rev Microbiol 4: 11–22 [DOI] [PubMed] [Google Scholar]
- Davidsen T, Bjoras M, Seeberg EC, Tonjum T (2005) Antimutator role of DNA glycosylase MutY in pathogenic Neisseria species. J Bacteriol 187: 2801–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B, Harrison L (1994) Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem 63: 915–948 [DOI] [PubMed] [Google Scholar]
- Demple B, Sung JS (2005) Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair 4: 1442–1449 [DOI] [PubMed] [Google Scholar]
- Dianov GL, Sleeth KM, Dianova II, Allinson SL (2003) Repair of abasic sites in DNA. Mutat Res 531: 157–163 [DOI] [PubMed] [Google Scholar]
- Dunn KL, Farrant JL, Langford PR, Kroll JS (2003) Bacterial [Cu,Zn]-cofactored superoxide dismutase protects opsonized, encapsulated Neisseria meningitidis from phagocytosis by human monocytes/macrophages. Infect Immun 71: 1604–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerke C, Koller J, Wolz C (2006) Ciprofloxacin and trimethoprim cause phage induction and virulence modulation in Staphylococcus aureus. Antimicrob Agents Chemother 50: 171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MA, Morera S, Rothwell DG, de La Fortelle E, Mol CD, Tainer JA, Hickson ID, Freemont PS (1997) The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J 16: 6548–6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi MZ, Wilson DM III (2000) Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ Mol Mutag 36: 312–324 [PubMed] [Google Scholar]
- Hooft RWW, Vriend G, Sander C, Abola EE (1996) Errors in protein structures. Nature 381: 272. [DOI] [PubMed] [Google Scholar]
- Hornback ML, Roop RM II (2006) The Brucella abortus xthA-1 gene product participates in base excision repair and resistance to oxidative killing but is not required for wild-type virulence in the mouse model. J Bacteriol 188: 1295–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosfield DJ, Daniels DS, Mol CD, Putnam CD, Parikh SS, Tainer JA (2001) DNA damage recognition and repair pathway coordination revealed by the structural biochemistry of DNA repair enzymes. Prog Nucleic Acid Res Mol Biol 68: 315–347 [DOI] [PubMed] [Google Scholar]
- Izumi T, Brown DB, Naidu CV, Bhakat KK, Macinnes MA, Saito H, Chen DJ, Mitra S (2005) Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci USA 102: 5739–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Ishizaki K, Ikenaga M, Yonei S (1992) A mutant endonuclease IV of Escherichia coli loses the ability to repair lethal DNA damage induced by hydrogen peroxide but not that induced by methyl methanesulfonate. J Bacteriol 174: 7711–7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM (1993) Procheck—a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362: 709–715 [DOI] [PubMed] [Google Scholar]
- Ljungquist S, Lindahl T, Howard-Flanders P (1976) Methyl methane sulfonate-sensitive mutant of Escherichia coli deficient in an endonuclease specific for apurinic sites in deoxyribonucleic acid. J Bacteriol 126: 646–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig DL, MacInnes MA, Takiguchi Y, Purtymun PE, Henrie M, Flannery M, Meneses J, Pedersen RA, Chen DJ (1998) A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat Res 409: 17–29 [DOI] [PubMed] [Google Scholar]
- Meira LB, Devaraj S, Kisby GE, Burns DK, Daniel RL, Hammer RE, Grundy S, Jialal I, Friedberg EC (2001) Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Research 61: 5552–5557 [PubMed] [Google Scholar]
- Michaels ML, Miller JH (1992) The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J Bacteriol 174: 6321–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B (2005) After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol 3: e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miertzschke M, Greiner-Stoffele T (2003) The XthA gene product of Archaeoglobus fulgidus is an unspecific DNase. Eur J Biochem 270: 1838–1849 [DOI] [PubMed] [Google Scholar]
- Mol CD, Kuo CF, Thayer MM, Cunningham RP, Tainer JA (1995) Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature 374: 381–386 [DOI] [PubMed] [Google Scholar]
- Mol CD, Izumi T, Mitra S, Tainer JA (2000) DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature 403: 451–456 [DOI] [PubMed] [Google Scholar]
- Moore TD, Sparling PF (1996) Interruption of the gpxA gene increases the sensitivity of Neisseria meningitidis to paraquat. J Bacteriol 178: 4301–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelle S, Carbonnelle E, Matic I, Nassif X (2005) Contact with host cells induces a DNA repair system in pathogenic Neisseriae. Mol Microbiol 55: 853–861 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS (2001) ARP/wARP and molecular replacement. Acta Crystallogr D 57: 1445–1450 [DOI] [PubMed] [Google Scholar]
- Quinones M, Davis BM, Waldor MK (2006) Activation of the Vibrio cholerae SOS response is not required for intestinal cholera toxin production or colonization. Infect Immun 74: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell DG, Hang B, Gorman MA, Freemont PS, Singer B, Hickson ID (2000) Substitution of Asp-210 in HAP1 (APE/Ref-1) eliminates endonuclease activity but stabilises substrate binding. Nucleic Acids Res 28: 2207–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seib KL, Tseng HJ, McEwan AG, Apicella MA, Jennings MP (2004) Defenses against oxidative stress in Neisseria gonorrhoeae and Neisseria meningitidis: distinctive systems for different lifestyles. J Infect Dis 190: 136–147 [DOI] [PubMed] [Google Scholar]
- Suh D, Wilson DM, Povirk LF (1997) 3′-phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res 25: 2495–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvarnapunya AE, Stein MA (2005) DNA base excision repair potentiates the protective effect of Salmonella Pathogenicity Island 2 within macrophages. Microbiology 151: 557–567 [DOI] [PubMed] [Google Scholar]
- Takemoto T, Zhang QM, Matsumoto Y, Mito S, Izumi T, Ikehata H, Yonei S (1998) 3′-blocking damage of DNA as a mutagenic lesion caused by hydrogen peroxide in Escherichia coli. J Radiat Res 39: 137–144 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30: 1022–1025 [Google Scholar]
- Waters TR, Gallinari P, Jiricny J, Swann PF (1999) Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J Biol Chem 274: 67–74 [DOI] [PubMed] [Google Scholar]
- Weston SA, Lahm A, Suck D (1992) X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution. J Mol Biol 226: 1237–1256 [DOI] [PubMed] [Google Scholar]
- Wilson DM, Takeshita M, Grollman AP, Demple B (1995) Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J Biol Chem 270: 16002–16007 [DOI] [PubMed] [Google Scholar]
- Wojciechowski MF, Peterson KR, Love PE (1991) Regulation of the SOS response in Bacillus subtilis: evidence for a LexA repressor homolog. J Bacteriol 173: 6489–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T (1996) The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA 93: 8919–8923 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3


