Abstract
Cationic antimicrobial peptides (AMPs), a component of the innate immune system, play a major role in defense of mucosal surfaces against a wide spectrum of microorganisms such as viral and bacterial co-pathogens of the polymicrobial disease otitis media (OM). To further understand the role of AMPs in OM, we cloned a cDNA encoding a cathelicidin homolog (cCRAMP) from upper respiratory tract (URT) mucosae of the chinchilla, the predominant host used to model experimental OM. Recombinant cCRAMP exhibited alpha-helical secondary structure and killed the three main bacterial pathogens of OM. In situ hybridization showed cCRAMP mRNA production in epithelium of the chinchilla Eustachian tube and RT-PCR was used to amplify cCRAMP mRNA from several other tissues of the chinchilla URT. Quantitative RT-PCR analysis of chinchilla middle ear epithelial cells (CMEEs) incubated with either viral (influenza A virus, adenovirus, or RSV) or bacterial (nontypeable H. influenzae, M. catarrhalis, or S. pneumoniae) pathogens associated with OM demonstrated distinct microbe-specific patterns of altered expression. Collectively, these data showed that viruses and bacteria modulate AMP messages in the URT, which likely contributes to the disease course of OM.
Keywords: NTHI, cathelicidin, antimicrobial peptide, RSV, S. pneumoniae, M. catarrhalis
1. Introduction
Cathelicidins are one of two major families of antimicrobial peptides (AMPs) (Lehrer and Ganz, 2002a; Lehrer and Ganz, 2002b; Zanetti et al., 1995), distinguished by their conserved homology to a N-terminal pro-region in the pig leukocyte protein, cathelin (Ritonja et al., 1989). Full-length cathelicidins are formed as holoproteins that include a signal peptide, the cathelin domain, and a functional C-terminal peptide that has antimicrobial activity (Zanetti, 2004). The region between the cathelin domain and the C-terminus is cleaved by one of several serine proteinases, depending on both the host and the site of expression of the cathelicidin, to release the functional peptide (Sorensen et al., 2001; Sorensen et al., 2003b; Tjabringa et al., 2005). Cathelicidins are important components of the innate immune system on mucosal surfaces (Bals et al., 1999; Nizet et al., 2001) and, in addition to being potent antimicrobials, are also chemotactic for dendritic cells, monocytes, and neutrophils (Agerberth et al., 2000; Davidson et al., 2004; De et al., 2000). They also modulate the expression of numerous genes in macrophages (Hancock and Scott, 2000), promote histamine release from mast cells (Gudmundsson and Agerberth, 1999), neutralize LPS (Gough et al., 1996) and have been implicated in wound repair (Dorschner et al., 2001; Sorensen et al., 2003a).
Due to their antimicrobial activity and importance in protecting mucosal surfaces, it is likely that AMPs play a significant role in preventing the pediatric disease, OM. In addition, a recent report (Mygind et al., 2005) details the promising future of AMPs in human therapeutics which is especially relevant to OM since it is estimated that 83% of all children will experience at least one episode of acute OM (AOM) by 3 years of age, and more than 40% of children will experience three or more episodes of AOM by this age (Teele et al., 1989). Moreover, OM is the most frequently diagnosed illness in children under 15 yrs. and is the primary cause for emergency room visits (Cassell et al., 1994) leading to an economic cost in the U.S. that exceeds $5 billion annually (Alsarraf et al., 1999; Cassell, 1997; Kaplan et al., 1997). Clearly a need exists to develop novel strategies for both treatment and prevention of OM. Gaining an understanding of effectors of innate immunity operational in the uppermost airway is the first step towards this end.
OM is a disease of opportunity, not caused by highly pathogenic organisms but rather is a polymicrobial disease due to the synergistic activity of both upper respiratory tract (URT) viruses (predominately influenza A virus, respiratory syncytial virus [RSV], adenovirus, and rhinovirus) (Heikkinen and Chonmaitree, 2003; Monto and Ullman, 1974) and one or more of three bacteria [S. pneumoniae, M. catarrhalis, and nontypeable Haemophilus influenzae (NTHI)] (Faden, 2001). Typically, viral URT infection either precedes, or is concomitant with, ascension of the Eustachian tube by specific members of the normal flora from the colonized nasopharynx into the middle ear. We hypothesize that periodic increases in the bacterial load in the nasopharynx (Bakaletz, 2002) that is coincident with URT viral infection is potentiated by viral dysregulation of innate immune system components, specifically AMPs.
To better understand innate immune system factors that influence the outcome of OM, we have begun to investigate the role of AMPs in the uppermost respiratory tract of the chinchilla host, a model for investigating OM. Chinchillas do not naturally develop OM, but can be colonized and/or infected with many of the organisms (bacteria and viruses) that predominate in OM (Giebink et al., 1980; Suzuki and Bakaletz, 1994). The experimental disease course in the chinchilla superinfection model (Bakaletz, 2002) begins with viral compromise of the URT followed by bacterial invasion of the middle ear, with resolution of infection occurring within 5–6 weeks, similar to the human OM disease course (Kennedy et al., 2000). Thus, the chinchilla provides an excellent model in which to study OM pathogenesis and can likely be used to assess the role that AMPs play in altering the disease process. To date, we have shown that a β-defensin family member, cBD-1, is expressed in the upper airway of the chinchilla and further, that recombinant cBD-1 kills the three main bacterial pathogens of OM (Harris et al., 2004). As an additional step in assessing the relevance of AMPs in OM, we now describe the identification and initial characterization of the first cathelicidin homolog (cCRAMP) identified in the chinchilla. Herein, we show that this cathelicidin is produced by several tissues of the upper airway of this host, that recombinant cCRAMP can kill the predominant bacterial pathogens of OM, and that, in vitro, the expression of both cCRAMP and cBD-1, is modulated by both viral and bacterial co-pathogens of OM.
2. Materials and methods
2.1 Animals
Healthy adult chinchillas (Chinchilla lanigera), ~500–700 g, were purchased from Rauscher’s Chinchilla Ranch (LaRue, OH) and fed chinchilla chow (Cincinnati Lab and Pet Supply, Cincinnati, OH) and water ad libitum. The animals were free of middle ear disease as evidenced by otoscopy and tympanometry. Animals were deeply anesthetized with xylazine (2 mg/kg, Fort Dodge Animal Health, Fort Dodge, IA) and ketamine (10 mg/kg, Phoenix Scientific Inc., St. Joseph, MO) and then were sacrificed. Tissues of interest were dissected, snap frozen in liquid nitrogen, and stored at −80°C until needed. All studies involving chinchillas were performed under an Institutional Animal Care and Use Committee-approved protocol.
2.2 Cloning of cCRAMP cDNA
To clone cCRAMP cDNA, a modified 3′ RACE strategy was used as described previously (Harris et al., 2004; Tarver et al., 1998). Total RNA was isolated from the Eustachian tube and nasopharyngeal mucosae of a chinchilla and a first-strand cDNA product was amplified using a modified oligo(dT) primer (Table I) (Marathon cDNA synthesis primer, Clontech, Palo Alto, CA), according to the manufacturer’s suggestions. The resulting cDNA was amplified to make a double-stranded PCR product using 2X premixed HotStart Taq polymerase (Qiagen, Valencia, CA), the Clontech anchor primer AP1 and primer CATH2 (Table I). The sequence of CATH2 was based on conserved regions in the cathelin domain of human CAP-18 and rat CRAMP. The cDNA product was size-fractionated in an agarose gel and a band of approximately 450 bp was extracted using the Qiaquick PCR purification kit (Qiagen). This amplicon was cloned into pGEM-T (Table I; Promega, Madison, WI) via annealing of terminal 3′-thymidine overhangs on the vector to non-template deoxyadenosine bases on the amplified product and was transformed into E. coli. Plasmids from selected recombinants, which produced white colonies on LB agar containing 0.5 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG), 80 μg 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) /ml, and 100 μg ampicillin/ml, were sequenced using BigDye terminator chemistry (Applied Biosystems, Foster City, CA) with M13 forward and reverse primers (Table I) (Yanisch-Perron et al., 1985) on an Applied Biosystems Model 3100 Genetic Analyzer at the DNA Sequencing Core Facility of Columbus Children’s Research Institute. Clones with similarity to known cathelicidins were identified by BLASTX analysis (http://www.ncbi.nlm.nih.gov) and a single plasmid containing the cCRAMP insert was saved as pcCRAMP1 (Table I).
Table I.
Primers and plasmids used in this study.
| Name | Description | Source |
|---|---|---|
| Primer | ||
| Oligo(dT) | 5′-CTAATACGACTCACTATAGGGCTCGAGCGGC(T18V N-3′ | Clontech |
| AP1 | 5′-CCATCCTAATACGACTCACTATAGGGC-3′ | Clontech |
| CATH2 | 5′-CGGAGCAGTGTGACTTCAAG-3′ | This study |
| CATH5+ | 5′-CGCGGATCCTCTAAAACTGGTACCGATACT-3′ | This study |
| M13F | 5′-GTAAAACGACGGCCAG T-3′ | Promega |
| M13R | 5′-CAGGAAACAGCTATGAC-3′ | Promega |
| T7 promoter | 5′-TAATACGACTCACTATAGGG-3′ | Novagen |
| CATH3 | 5′-CTAAAACTGGTACCGATACTG-3 | This study |
| ST CCW | 5′-ATCAGCCATGGCCTTGTCGTCGTCGTCGGT-3′ | This study |
| ST CW# | 5′-CCCGGGCTGGTGCCACGCGGTTCTGCAAAAGGGGCGGGTTCTGGCGCAAAGTC-3′ | This study |
| cBD-1 RT 5′ | 5′-TATTTCCTTGTTCGGCAGCATTTC-3′ | This study |
| cBD-1 RT 3′ | 5′-GCTCTTCTTGTTCTTGATGCCAGT-3′ | This study |
| GAPDH RT 5′ | 5′-AGCCAAAAGGGTCATCATCTCT-3′ | This study |
| GAPDH RT 3′ | 5′-CTTGGCCAGGGGTGCTAA-3′ | This study |
| cCRAMP RT 5′ | 5′-GCAAAGAGGGGCGGGTTCTGG-3′ | This study |
| cCRAMP RT 3′ | 5′-TTCCGGATTCCTTTGCCCAGCTT-3′ | This study |
| Plasmid | ||
| pcCRAMP1 | Partial cDNA of cCRAMP cloned into pGEM-T easy | This study |
| pcCRAMP2 | cDNA of the predicted mature cCRAMP cloned in-frame into pET-30(a) | This study |
| pET-30(a) | Expression vector encoding ampicillin resistance | Novagen |
| pET-32 (a) | Expression vector encoding kanamycin resistance | Novagen |
| pGM-4 | Expression vector containing thioredoxin in-frame with cCRAMP | This study |
| pGM-5 | Expression vector containing thioredoxin in-frame with a 6xHis tag and cCRAMP with an intervening thrombin digestion site | This study |
| pGEM-T easy | Cloning vector encoding ampicillin resistance | Promega |
| pGEM-T | Cloning vector encoding ampicillin resistance | Promega |
| pcCRAMP3 | cDNA of cCRAMP cloned into pGEM-T for in situ hybridization | This study |
The underlined sequence denotes a BamHI digestion site.
The underlined sequence denotes a SmaI digestion site and the bold nucleotides show a thrombin recognition sequence.
2.3 Construction of recombinant cCRAMP expression vectors
The sequence for the cDNA encoding the predicted active peptide of cCRAMP was submitted to the GenBank database (http://www.ncbi.nlm.nih.gov/Genbank/index.html) under accession number DQ097506. A 129-bp fragment encoding the predicted mature cCRAMP was amplified via PCR from pcCRAMP1 and cloned into pET-30(a) giving the pcCRAMP2 plasmid (Table I; Novagen, La Jolla, CA). This plasmid failed to produce recombinant protein as evaluated by SDS-PAGE analysis of E. coli total cell lysates, therefore we next constructed a series of vectors to achieve expression of soluble cCRAMP.
The cCRAMP cDNA was amplified from pcCRAMP2 using primers CATH3 and CATH5 (Table I) with cloned Pfu DNA polymerase (Stratagene) and the resulting amplicon was gel extracted (Qiagen) and blunt-ligated into the single EcoRV site of pET32-(a) (Table I; Novagen). This plasmid allowed for fusion of cCRAMP to an upstream thioredoxin (trxA) peptide, a common protein fusion partner used to increase solubility of proteins, and a 6-residue His tag (Figure 2A). Ligation products were transformed into E. coli Top10 and recombinants were selected for growth on LB agar containing 50 μg ampicillin per ml. A colony blot hybridization (Sambrook and Russell, 2001) was then done to screen for cCRAMP-positive clones using a cCRAMP-specific probe (the same amplicon used in cloning cCRAMP into pET-30(a)). One plasmid, pGM-4 (Table I and Figure 2A), contained the cCRAMP cDNA in the correct orientation to create a fusion with the thioredoxin protein encoded in pET-32(a) and was also found to contain a single nucleotide deletion in the stop codon when compared to the cCRAMP cDNA sequence in pcCRAMP2. This mutation changed the UAG amber codon to a UAA ochre codon which allowed us to use this construct.
Fig. 2.
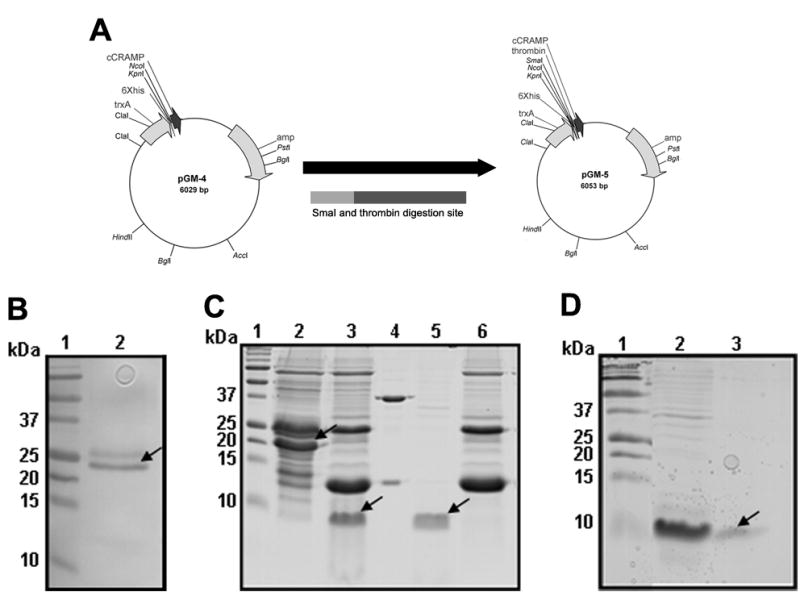
The pGM-5 plasmid in E. coli BL21 STAR (DE3) expresses recombinant cCRAMP. (A) Maps of vectors constructed to express (r)cCRAMP as detailed in experimental procedures. (B) E. coli-derived total cell lysates separated in a SDS-PAGE gel containing cCRAMP expressed from pGM-5 (arrow indicates recombinant cCRAMP fusion protein containing a His-tag). (C) A SDS-PAGE gel stained with Coomassie brilliant blue showing purification of cCRAMP from E. coli BL21 STAR (DE3) harboring pGM-5, as detailed in experimental procedures. Protein samples were loaded as follows: Protein bound to the first Ni2+-NTA column (lane 2), Ni2+-NTA column bound protein digested with thrombin (lane 3), a control protein (Novagen) digested with thrombin (lane 4), thrombin digested protein that did not bind to a second Ni2+-NTA column (lane 5), and bound protein from the same second column (lane 6). Arrows show (r)cCRAMP. (D) Further purification of (r)cCRAMP. Thrombin digested protein retained on a 5-kDa size-exclusion column (Lane 2) and (r)CRAMP isolated as a single band after retention on a 3-kDa size-exclusion column (Lane 3). Lane 1 in all gels contains a molecular weight standard.
Since the cCRAMP peptide encoded by pGM-4 could not be separated from the thioredoxin fusion protein, another vector was constructed using the Failsafe PCR system with (Epicenter, Madison, WI) primers directed outward which allowed amplification of the entire pGM-4 plasmid with the addition of a unique SmaI restriction site (Table I; primer ST CCW) and a thrombin protease site (Table I; ST CW) immediately upstream of the cCRAMP cDNA sequence. The amplicon from this Failsafe PCR was treated with polynucleotide kinase (Invitrogen, Carlsbad, CA) for 30 minutes at 37°C in T4 DNA ligase buffer (New England Biolabs, Beverly, MA), and phosphorylated products were ligated at room temperature for 2 hours before transformation into E. coli Top10. The SmaI site (which was not present in pGM-4) allowed for identification of plasmid molecules that were successfully phosphorylated and ligated. After DNA sequencing one plasmid, pGM-5 (Table I and figure 2A), was found to have the thioredoxin coding region in-frame with the thrombin digestion site and the open reading frame for cCRAMP (Table I and figure 2A). DNA sequencing of both strands verified that no mutations had occurred throughout the cloning process and that a full-length fusion protein could be made in an appropriate bacterial host.
2.4 Expression and purification of recombinant cCRAMP
To express the recombinant cCRAMP [(r)cCRAMP] peptide, we electroporated pGM-5 into several E. coli strains, and looked for a combination of strain and vector that produced the greatest amount of non-truncated (r)cCRAMP. This DNA construct was transformed into E. coli BL21 trxB (DE3) (Novagen), E. coli Rosetta (DE3) (Novagen) and E. coli BL21 STAR (DE3) (Invitrogen), and the appearance of an IPTG-inducible protein band of appropriate molecular mass was evaluated in Coomassie brilliant blue-stained SDS-PAGE gels of total cell lysates. Plasmid pGM-5 in E. coli BL21 STAR (DE3) produced the greatest amount of full-length protein and was thereby used in the subsequent purification of (r)cCRAMP.
For isolation of recombinant cCRAMP, three liters of cells in LB broth grown to mid-log growth phase were induced overnight with 1 mM IPTG to express the fusion protein. Bacterial cells were pelleted by centrifugation at 8000 X g for 10 minutes and the cell pellet was suspended in a mixture of Bugbuster reagent (Novagen) (5 ml per gram of wet weight), 20 μg lysozyme per ml, and 1/1000 volume Benzonase nuclease (Novagen). Cell lysis was allowed to occur for 15 minutes at room temperature and insoluble debris was separated from soluble protein by centrifugation at 20,000 X g for 20 minutes. The supernatant containing soluble protein was applied to a prepared Ni2+-NTA column according to the manufacturers suggestions for soluble proteins (Invitrogen). The His-rich proteins were allowed to bind to the Ni2+-agarose slurry for 1 hour at room temperature and loosely bound protein was stripped off the column using 4 separate washes of 1 X Native Wash buffer (10 mM sodium phosphate buffer, 500 mM NaCl, and 20 mM imidazole pH 8.0). Proteins bound to the Ni2+-agarose slurry were then eluted with Elution buffer (10 mM sodium phosphate buffer, 500 mM NaCl, and 200 mM imidazole pH 8.0). One milliliter fractions were collected and pooled fractions were dialyzed overnight at 4°C against 1 X Native purification buffer (Native wash buffer without imidazole). The dialyzed protein sample was then digested with recombinant thrombin (Novagen) for 2 hours at 20°C using 1/25 unit of enzyme per 20 μg of total protein. After digestion, the protein sample was applied to a newly prepared Ni2+-NTA column and protein was again allowed to bind to the Ni2+-agarose slurry for 1 hour. The column was centrifuged at 800 X g for 1 minute and the supernatant was aspirated and kept for further purification. The supernatant from the column was placed in a 5,000 molecular weight cutoff (MWCO) column (Millipore, Billerica, MA) and centrifuged at 3000 x g until approximately 100 μl of liquid was retained by the column. The flow-through from the column was then applied to a 3,000 MWCO column (Millipore) and centrifuged as described above except that approximately 500 μl of sample remained. This retentate from the 3,000 MWCO column contained purified (r)cCRAMP and was dialyzed against 10 mM sodium phosphate buffer pH 7.2.
2.5 Western blotting
Western blots were performed essentially as described (Ausubel et al., 1993) except that the running and stacking gels were polymerized in the absence of SDS to enhance resolution of the low molecular weight and cationic AMPs. Proteins were transferred to nitrocellulose overnight at 4°C using 7 volts. A mouse anti-Penta•His antibody (Qiagen) (1:2000 dilution) was used as the primary antibody to detect His-tagged proteins and protein A conjugated to horseradish peroxidase (1:2000) allowed visualization of immune complexes after development with the chromogenic CN/DAB substrate (Pierce, Rockford, IL).
2.6 Antimicrobial assays
The liquid bactericidal assay used here with modification has been previously described (Harris et al., 2004). Briefly, E. coli ML35, NTHI strain 86-028NP, and S. pneumoniae (serotype 14) were cultured statically to mid-log phase in brain heart infusion (BHI) broth supplemented with 2 μg of NAD /ml (Sigma, St. Louis, MO) and 2 μg hemin /ml (Sigma). M. catarrhalis 1857 was grown in similar conditions but with aeration. E. coli ML35 is a standard strain used to assay microbicidal activity since it expresses a mutated form of lipopolysaccharide that renders it highly sensitive to the action of AMPs (Zasloff, 1987). NTHI strain 86-028NP is a minimally passaged clinical isolate obtained from a pediatric patient who underwent typanostomy and tube insertion (Harrison, 2005) and M. catarrhalis strain 1857 in a minimally passaged pediatric middle ear effusion isolate (Bakaletz, 1995). S. pneumoniae serotype 14 strains are frequently isolated from pediatric middle ear fluids (Hausdorff, 2002). The assay was performed as described, except 1x108 cells were incubated with increasing concentrations of (r)cCRAMP. Colony forming units were counted and percent killing was calculated relative to identical cultures incubated with sodium phosphate buffer (pH 7.2) in the absence of (r)cCRAMP. Data from duplicate assays per strain are presented as mean percentage killed ± standard deviation relative to concentration of recombinant cCRAMP.
2.7 Isolation of total RNA from chinchilla tissues and performance of RT-PCR
After removing chinchilla tissues from storage at −80°C, one milliliter of Trizol reagent (Invitrogen) was added and flasks were incubated for 5 minutes, with rocking, at room temperature. Total nucleic acid was purified by organic extraction with chloroform, followed by precipitation with first isopropanol, then 75% ethanol (Sambrook and Russell, 2001). The pellet remaining after the final ethanol precipitation was then air-dried for 5 minutes before suspending in 30 μl of diethylpyrocarbonate (DEPC)-treated water. Contaminating DNA was degraded by treatment with 2 units of rDNase I (Ambion, Austin, TX) for 30 minutes at 37°C and this enzyme was then inactivated according to the manufacturers directions (Ambion). The RNA sample volume was 35 μl. Finally, RNA samples were purified by passage through a RNeasy column following the manufacturers protocol for RNA clean-up (Qiagen). RNA bound to the column was eluted with DEPC-treated water in a volume of 30 μl. Integrity of purified RNA was evaluated using the Agilent 2100 Bioanalyzer (Agilent, Foster City, CA). For RT-PCR, cCRAMP RT 5′ and cCRAMP RT 3′ primers (Table 1) at 0.5 μM each with 2 nanograms of total RNA were used in 25 μl amplification reactions using the QuantiTect SYBR green RT-PCR system (Qiagen). Reaction conditions for the one-step procedure were 30 minutes of reverse transcription at 50°C, followed by heating to 95°C for 15 minutes. A thirty-five cycle 3-step procedure was then used which consisted of repeated denaturation at 94°C for 15 seconds, annealing at 60°C for 30 seconds, then extension at 72°C for 30 seconds. Amplicons generated from reactions with or without reverse transcriptase, to confirm the absence of contaminating DNA, were separated using the FlashGel™ system (Cambrex, East Rutherford, NJ).
2.7 Cell culture and quantitative real-time RT-PCR
Chinchilla middle ear epithelial cells (CMEEs) are a cell strain generated in our laboratory from outgrowth of primary culture of inferior bullar mucosa. CMEEs were cultured as previously described (Nakamura et al., 1991; Nakamura et al., 1992) and were passaged no more than seven times prior to conduct of quantitative real-time RT-PCR (qRT-PCR) analysis. To prepare cells for qRT-PCR assays, approximately 2.75 x 105 cells were seeded into 12.5 cm2 flasks and were incubated for 48 hours until confluent. CMEE monolayers were then incubated with either RSV (strain A2), adenovirus (serotype 1), or influenza A virus (Alaska 6/77 H3/N3) diluted in prewarmed growth medium or were sham inoculated with medium alone. For adenovirus and influenza A virus, CMEEs were inoculated with ~5 x 106 50% tissue culture infective doses and for RSV the multiplicity of infection was 100 PFU per CMEE cell. Cells plus virus were incubated for 1 hour at 37°C with 5% CO2 and then the medium containing virus was replaced with fresh CMEE growth medium (Nakamura et al., 1991; Nakamura et al., 1992). Medium was also applied to the uninfected control cultures. The cells inoculated with influenza A virus were allowed to incubate for 12 hours, whereas the adenovirus- and RSV-infected cells were incubated for 48 hours so as to allow for efficient viral replication but to minimize destruction of the cell monolayer.
To assay the effect of bacterial interactions with CMEE cells on relative expression of AMPs, Moraxella catarrhalis strain 1857 and NTHI strain 86-028NP were grown in supplemented BHI broth and these cultures were used to inoculate CMEEs (MOI of 100:1) prior to incubation for 6 hours. CMEEs were incubated with S. pneumoniae (serotype 14) at an MOI of 10:1 to minimize cell death.
After the specified incubation period had elapsed, total RNA was isolated as stated above, except the cell culture medium was removed and CMEEs were quickly washed twice with sterile PBS warmed to 37°C before the addition of one milliliter of Trizol reagent. For qRT-PCR, combinations of primers (Table I; cBD-1 RT 5′ and cBD-1 RT 3′; GAPDH RT 5′ and GAPDH RT3′; and cCRAMP RT 5′ and cCRAMP RT 3′) were used in reactions under the conditions used in RT-PCR. Real-time fluorescence measurements were taken with an Applied Biosystems 7700 or 7000 sequence detector. CMEE cells were inoculated with virus and qRT-PCR reactions were run in triplicate. Incubation of CMEE cells with bacteria was done in duplicate and each RT-PCR reaction was run twice. Values reported are the geometric mean ratios of transcript abundance of infected to uninfected cultures normalized against transcripts from the glyceraldelyde-3-phosphate dehydrogenase (GAPDH) gene, with standard error of the mean and statistical significance calculated using a Student’s T-test. Significance was accepted at a p-value of ≤ 0.05.
2.8 In situ hybridization
Preparation of tissues and hybridization with sense and antisense cCRAMP probes was done essentially as described (Harris et al., 2004). To generate a plasmid for preparation of the riboprobes, pcCRAMP1 was used as template in a PCR reaction using primers CATH2 and CATH5 and the amplicon was purified (Qiagen). This amplicon was ligated into pGEM-T, the ligation mixture was transformed into E. coli, and plasmids from selected recombinants were sequenced. One plasmid contained the cCRAMP cDNA sequence and was saved as pcCRAMP3. The plasmid clone was linearized which allowed for production of a digoxigenin-labeled sense and antisense RNA probe (Roche Applied Science) when transcribed using the T7 and Sp6 RNA polymerases. Dissected, paraformaldehydefixed, and paraffin-embedded Eustachian tube was prepared and cCRAMP-specific mRNA was detected as reported (Harris et al., 2004).
2.9 Computer modeling
A computer-simulated structural model of cCRAMP was derived based on known solution structures of other cathelicidin molecules (Johansson et al., 1998; Tack et al., 2002). The central α-helical region was modeled on the rabbit CAP-18 structure (Chen, 1995) due to sequence similarity and apparent amphipathic character of the resultant helix. Residues prior to the helix-breaking glycine pair were left unstructured and the alternating polar aromatic residues near the carboxy-terminus were manually assigned to a β-sheet conformation. Quanta software (MSI) was used to subject the resulting structure to 2000 CHARMm minimization steps to a gradient tolerance of 0.01 Kcal/mole, and 1000 iterations of CHARMm molecular dynamics at ~300 Kelvin using a 0.001 picosecond time step in a 10 Å water shell.
2.10 Circular dichroism (CD)
CD measurements were performed on a 62A DS spectropolarimeter (AVIV Associates, Lakewood, NJ) with temperature controlled at 20°C using a VWR 1155 constant temperature circulator with 40% ethylene glycol. Spectra were recorded in a 1-mm pathlength cuvette at 1 nm spacing with a 5 second accumulation interval and a total of 4 scans per datum point, over a range from 190 to 250 nm. Peptide concentration was 25 μM in a buffer of 10 mM sodium phosphate (pH 7.2). Helical content was estimated by K2d (http://www.emblheidelberg.de/~andrade/k2d.html) analysis and mean residue ellipticity was expressed as [Θ]MRE (deg•cm2dmol−1)x10−3. CD measurements were also obtained for cCRAMP in phosphate buffer at pH 8.6 and pH 9.9 in the presence or absence of 20 mM sodium dodecyl sulfate (Fisher Scientific, Pittsburg, PA).
3. Results
3.1 Cloning of the cDNA for cCRAMP
To clone the cDNA product containing cCRAMP, a modified 3′ RACE strategy was employed using total RNA isolated from chinchilla Eustachian tube and nasopharyngeal mucosae. The RACE procedure generated several amplicons from which a ~450-bp fragment was cloned, sequenced, and the resulting plasmid was saved as pcCRAMP1. BLASTX analysis of the ~450-bp clone showed that the derived amino acid sequence of the insert was similar to several known cathelicidin molecules, including guinea pig CAP-11 (66% identity) (Nagaoka et al., 1997), rhesus monkey RL-37 (53% identity) (Zhao et al., 2001), human CAP18 (46% identity) (Larrick et al., 1995), mouse CRAMP (45% identity) (Gallo et al., 1997), and rabbit CAP-18 (44% identity) (Larrick et al., 1991). The term chinchilla cathelin-related antimicrobial peptide or ‘cCRAMP’ was thus coined to be in keeping with nomenclature of other rodent-derived cathelicidins including rat CRAMP (Termen et al., 2003).
Since the antimicrobial peptide of the cathelicidin precursors are located adjacent to the C-terminus of the conserved cathelin domain, the corresponding 129-bp fragment of the pcCRAMP1 insert, encoding the predicted mature cCRAMP, was conceptually translated and analyzed using DNASTAR software. The mature cCRAMP peptide was predicted to have 42 amino acids, a molecular mass of 5008 Da, and an isoelectric point of 12.24. A DNASTAR CLUSTAL W peptide alignment of the antimicrobial domains from selected cathelicidin molecules showed that the translated cCRAMP peptide had 43.2 % similarity to rhesus monkey RL-37 and also displayed sequence conservation to human LL-37 (35.1 %), mouse CRAMP (33.3 %), rat CRAMP (33.3%), and guinea pig CAP-11 (28.6%) (Fig. 1A). Interestingly, this modest similarity to human LL-37 is in contrast to the high homology (77%) we showed between human β-defensin-3 and the recently described chinchilla β-defensin-1 (Harris et al., 2004).
Fig. 1.
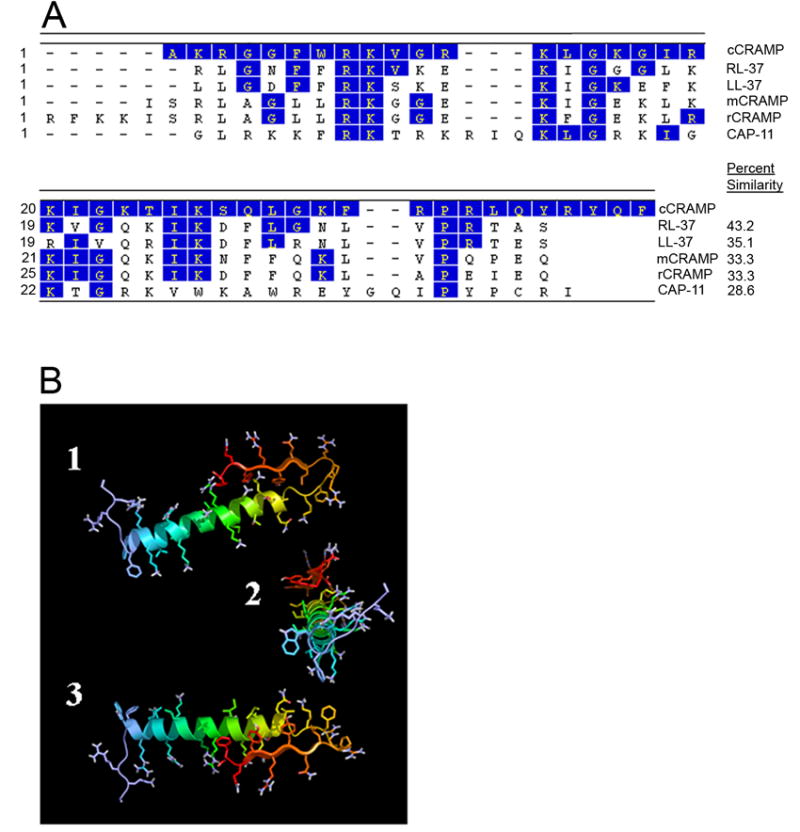
In silico analysis of cCRAMP. (A) DNASTAR Clustal W alignment of cCRAMP with rhesus monkey RL-37 (accession no. AF181954), human LL-37 (accession no. NM004345), mouse CRAMP (accession no. NM009921), rat CRAMP (accession no. AF484553), and guinea pig CAP-11 (accession no. D87405). Shaded residues indicate amino acids which are identical between cCRAMP and the respective cathelicidin. Numbers on the left represent residue numbers of the corresponding cathelicidin in the labeled row, and the percent similarity of an AMP with cCRAMP is shown on the right. (B) Three views of the predicted 3-D structural model of cCRAMP. In the top (1) and bottom (3) views of this computer-generated model, the N-terminus is to the left and in the middle (2) view, the N-terminus is facing the viewer. The orientation of the β-sheet, as folded against the α-helix, may be an artifact due to performing the molecular mechanics and dynamics simulations with the model isolated in aqueous solution rather than in a complex environment, such as in a membrane, in which it is naturally found. However, the predicted amphipathic nature of both the α-helix and β-sheet can be clearly seen in this model.
To contribute to our in silico analysis of mature cCRAMP, and further substantiate that cCRAMP was a cathelicidin homolog, we generated a theoretical model of the molecule using Quanta (MSI) molecular mechanics and molecular dynamics to predict its three-dimensional structure. Figure 1B shows a theoretical structure of cCRAMP, with the predominant α-helix typical of cathelicidin molecules clearly evident (Zanetti, 2004). The computer-generated model also suggested an amphipathic nature of the α-helix, with positively charged arginine and lysine residues on one helical face, and isoleucine on the other; an unordered N-terminus; and a C-terminus with several aromatic, hydrophobic amino acids on the same face of the predicted β sheet.
3.2 Purification of (r)cCRAMP
Two major obstacles typically hinder expression of an AMP by bacteria. The first is that these peptides are antimicrobial and thereby must be expressed in an inactive state, and second, the peptides are also highly charged and thus could be expressed poorly and/or as an insoluble product. We first attempted to overexpress recombinant cCRAMP [(r)cCRAMP] using methodology that has been used previously (Zaiou et al., 2003), however this resulted in a low yield of recombinant protein that made for inefficient purification. We therefore modified an approach described by Yang et al., that was used to isolate human LL-37 (Yang et al., 2004). Figure 2A illustrates the cloning strategy used to overexpress (r)cCRAMP. The plasmid pGM-5 encodes a fusion peptide containing soluble thioredoxin, a 6-residue His tag, and a thrombin digestion site engineered directly upstream of the cCRAMP sequence and allowed for production of recombinant cCRAMP in an inactive form and, when digested with thrombin, released the cCRAMP peptide from its His-containing fusion partner. When we transformed pGM-5 into E. coli BL21 STAR (DE3) (Invitrogen) and induced expression overnight with IPTG, recombinant protein containing a His tag was detected (Fig. 2B lane 2).
To purify (r)cCRAMP, three liters of cells were grown overnight after induction with IPTG, and soluble protein containing the His-tagged (r)cCRAMP was purified by Ni2+ affinity chromatography. Column-retained protein was dialyzed, digested with recombinant thrombin (Novagen), and the protein samples were further separated by additional affinity chromatography and size-exclusion chromatography. Figures 2C and D show Coomassie brilliant blue-stained SDS-PAGE gels, demonstrating isolation of His-tagged (r)cCRAMP and the subsequent release of (r)cCRAMP after digestion with thrombin as indicated by the presence of a single band at ~8 kDa (arrow in Fig. 2D, lane 3).
3.3 Antimicrobial activity of (r)cCRAMP
To assess activity of (r)cCRAMP, we tested the ability of the purified protein to kill an E. coli strain that is sensitive to the activity of AMPs, as well as each of the three bacteria commonly associated with OM: NTHI 86-028NP, S. pneumoniae serotype 14, and M. catarrhalis 1857. In a liquid bactericidal assay, ≥ 50% of a 1x108 colony forming unit inoculum of either NTHI or E. coli ML35 was killed by 20 μg (r)cCRAMP/ml in one hour (Fig. 3A). For comparison, 1.25 μg synthesized LL-37/ml (Phoenix pharmaceuticals) killed ~80% NTHI strain 86-028NP when assayed under the same conditions (data not shown). The concentration of (r)cCRAMP required to kill 50% of these organisms is comparable to that described for recombinantly expressed LL-37 (Yang et al., 2004). Interestingly, approximately 40% of M. catarrhalis was killed in a one-hour assay while only 10% of S. pneumoniae died when incubated in the presence of similar concentrations of (r)cCRAMP (Fig. 3B) suggesting either selectivity in AMP killing or the presence of active antimicrobial resistance mechanisms in these strains. Sodium phosphate buffer alone did not lead to a reduction in viable bacteria in these assays (data not shown).
Fig. 3.
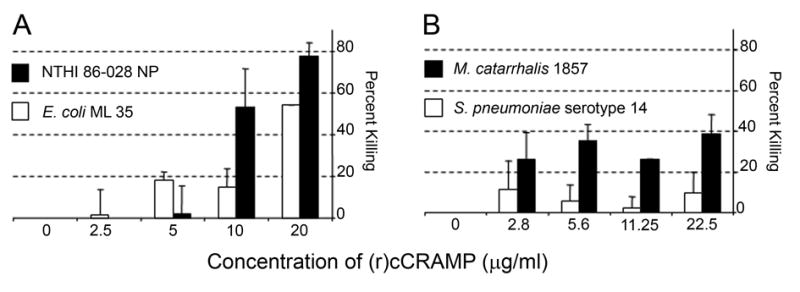
(r)cCRAMP is bactericidal at concentrations comparable to other antimicrobial peptides. (A) An E. coli strain sensitive to killing by antimicrobial peptides and NTHI were grown to mid-log phase, diluted to 1 x108 colony-forming units, incubated with increasing concentrations of (r)cCRAMP for 1 h at 37°C, and plated for determination of number of surviving colony forming units. (B) Same as in (A) except that M. catarrhalis, and S. pneumoniae were used as the test microorganisms.
3.4 Structural Properties of (r)cCRAMP
While some cathelicidins, including human LL-37, appear to be primarily comprised of amphipathic α-helices (Agerberth et al., 1995; Johansson et al., 1998), our computer-generated model of cCRAMP suggested that this AMP molecule had distinctly different N-terminal and C-terminal domains (Fig. 1B). To support our theoretical structure of cCRAMP, we used circular dichroism analyses to determine the solution structure of this AMP. As has previously been shown with the sheep leukocyte cathelicidin SMAP-29 (Tack et al., 2002), (r)cCRAMP in aqueous solution adopted a mostly disordered structure (Fig. 4). Raising the pH of the solution to either pH 8.6 or 9.9 resulted in an intensifying of the readable spectrum, as well as a shift of the negative peak from ~207 nanometers to ~198 nanometers. The spectra obtained under both of these conditions were similarly indicative of a less ordered molecule than was observed at pH 7.2 (Fig. 4). When SDS was added to the test solution at either pH 7.2 or 9.9, the resulting spectra of (r)cCRAMP were nearly indistinguishable. Both spectra had negative peaks at 222 nm and 207 nm, and a single positive peak at 196 nm. This spectral pattern was suggestive of a more strongly ordered, mixed α-helix, β-sheet structure. As a positive control, synthetic LL-37 was also subjected to CD analysis in the same phosphate buffer pH 7.2 (with and without SDS). The results were comparable to other published analyses for LL-37 and SMAP-29 (data not shown) (Johansson et al., 1998; Tack et al., 2002). K2d, an algorithm that estimates the percentage of secondary structure in a peptide based on CD spectra, analysis of 25 mM (r)cCRAMP (pH 7.2 or pH 9.9, with SDS) resulted in spectra suggesting 30% α-helix and 30% β-sheet character. The 1:1 α-helix to β-sheet ratio from the K2d analysis of (r)cCRAMP thus supports our helical-domain/sheet-domain 3-D computational model (Fig. 1B).
Fig. 4.
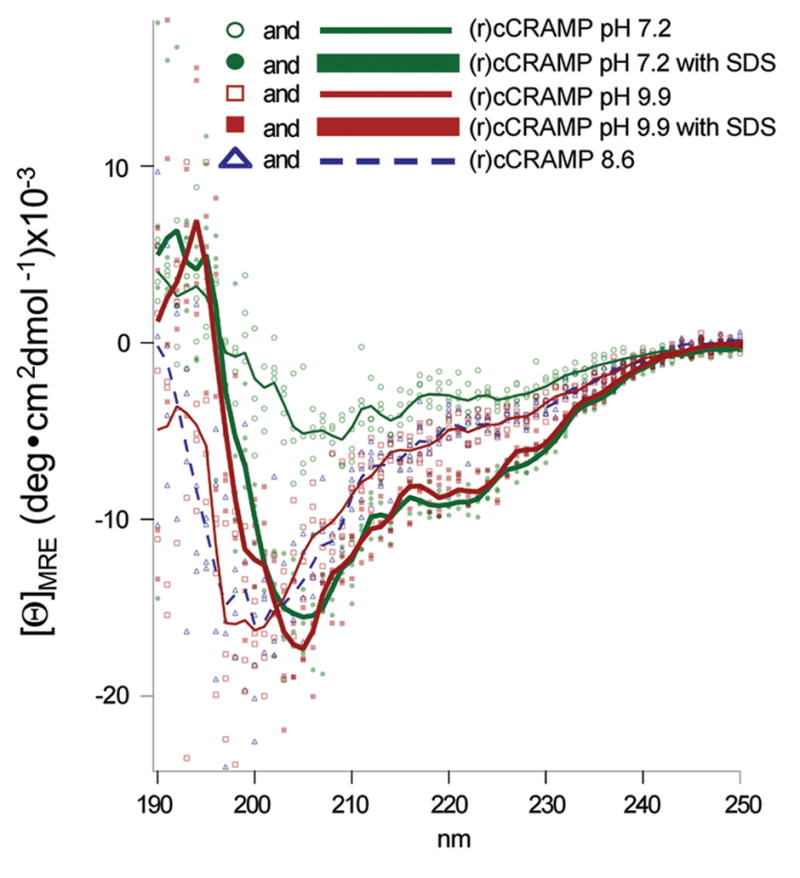
CD spectra of a 25 μM solution of (r)cCRAMP. Unprocessed data for all scans are shown as either filled or unfilled shapes whereas the corresponding averaged traces (with background subtracted) are depicted as lines. Increasing the pH to 8.6 or 9.9 resulted in a spectral shift to a more disordered conformation (see dashed blue and thin solid red lines). SDS rescued and intensified the structure seen in aqueous solution at pH 7.2 (see bold green and red lines).
3.5 Determination of relative expression of cCRAMP mRNA among tissues of the chinchilla URT
As a step to further characterize the AMP expression patterns in the URT of the chinchilla, we studied cCRAMP mRNA expression in selected tissue sites as we did previously for cBD-1 (Harris et al., 2004). Thereby, total RNA from tissues was isolated and analyzed by RT-PCR. In a naïve chinchilla, cCRAMP transcripts were found in the lower respiratory tract (bronchus and lung) and in several other tissues (testis, bladder, brain, and skin) similar to what has been reported for human LL-37 (Bals et al., 1998; Bergman et al. 2005; Dorschner et al., 2001). In addition, cCRAMP transcripts were detected in all tissues of the upper respiratory tract evaluated (Fig. 5A), including the Eustachian tube and middle ear mucosa. To date, cCRAMP and cBD-1(Harris et al., 2004) transcripts have been detected in all URT tissues tested suggesting an important role for both AMPs in protection of URT epithelial surfaces including the tubotympanum.
Fig. 5.
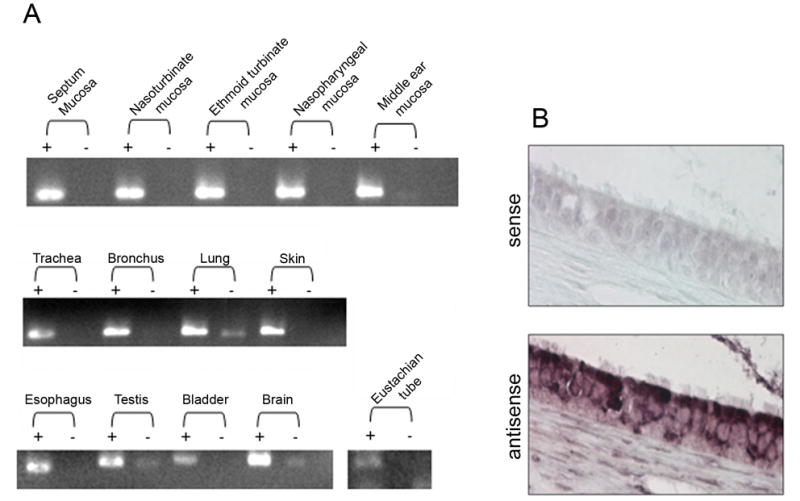
cCRAMP mRNA analysis. (A) RT-PCR analysis of cCRAMP transcripts in several chinchilla tissues. Lanes containing amplicons generated with reverse transcriptase (+) and without reverse transcriptase (−) are shown. cCRAMP transcripts were detected in every tissue evaluated. (B) in situ hybridization of chinchilla Eustachian tube using cCRAMP sense and antisense riboprobes. Images were obtained from the mid-portion of the Eustachian tube and cCRAMP mRNA was detected in the ciliated columnar epithelium of this tubal organ.
We then used in situ hybridization to further localize expression of cCRAMP mRNA to a specific cell type in Eustachian tube sections. We focused on this tissue, since Eustachian tube dysfunction, induced by URT viral infection, is a major predisposing factor in the development of bacterial OM (Bakaletz, 2002). Figure 5B shows that cCRAMP expression was detected in the ciliated columnar epithelium of the Eustachian tube (bottom panel), while there was no hybridization when the control (sense) probe was used (top panel).
3.6 Viral and bacterial modulation of cCRAMP mRNA expression
In keeping with our hypothesis that viral and bacterial co-pathogens of OM dysregulate effectors of innate immunity, we sought to determine the individual affect of several OM-relevant viral and bacterial microorganisms on the levels of expression of cCRAMP and cBD-1, a beta-defensin produced in the chinchilla airway, transcripts by URT mucosae. Toward this end, we inoculated a minimally passaged cell strain of chinchilla middle ear epithelial cells (CMEEs) with each of these microbes individually. Since the permissivity of CMEEs to infection with wild type influenza A virus (Alaska 6/77 H3/N3), adenovirus (serotype 1), or respiratory syncytial virus (RSV) (strain A2) was not known, we performed in vitro experiments to determine if CMEEs were permissive to infection with these human pathogens. Immunofluorescence imaging of CMEEs incubated with these viruses and stained with FITC-conjugated anti-influenza A virus, anti-adenovirus, or anti-RSV antibodies showed that CMEEs allowed for replication of all three viruses (data not shown).
To test how incubation with these predominant viral co-pathogens of OM as well as how incubation with M. catarrhalis, S. pneumoniae, and NTHI influenced expression of cCRAMP and cBD-1 transcripts, we used qRT-PCR. Incubation of CMEE cells with influenza A virus led to a ~50% reduction level in cCRAMP mRNA (p = 0.01), while incubation with RSV or adenovirus only minimally affected the cCRAMP message level (Fig. 6A, see white bars). In contrast, incubation of CMEEs with RSV reduced the message levels of cBD-1 approximately 40% (p = 0.05), whereas incubation with either adenovirus or influenza A virus did not (Fig. 6A, black bars). CMEEs responded differently to bacterial challenge. When CMEEs were incubated with either M. catarrhalis or S. pneumoniae, a 52% (p = 0.08) or 53% (p = 0.06) reduction in cBD-1 message was seen (Fig. 6B, black bars). No substantial change was observed when CMEEs were incubated with NTHI (Fig. 6B). When CMEEs were incubated with M. catarrhalis, the abundance of cCRAMP mRNA was reduced approximately 30%. Conversely, cCRAMP transcript levels were increased approximately 50% when NTHI was used as the pathogen, and increased 175% when S. pneumoniae was used. M. catarrhalis was the only organism tested that induced a downregulation of both cCRAMP and cBD-1 mRNA, suggesting that this causative agent of OM may be particularly adept at modulating expression of AMPs operational in the uppermost airway.
Fig. 6.
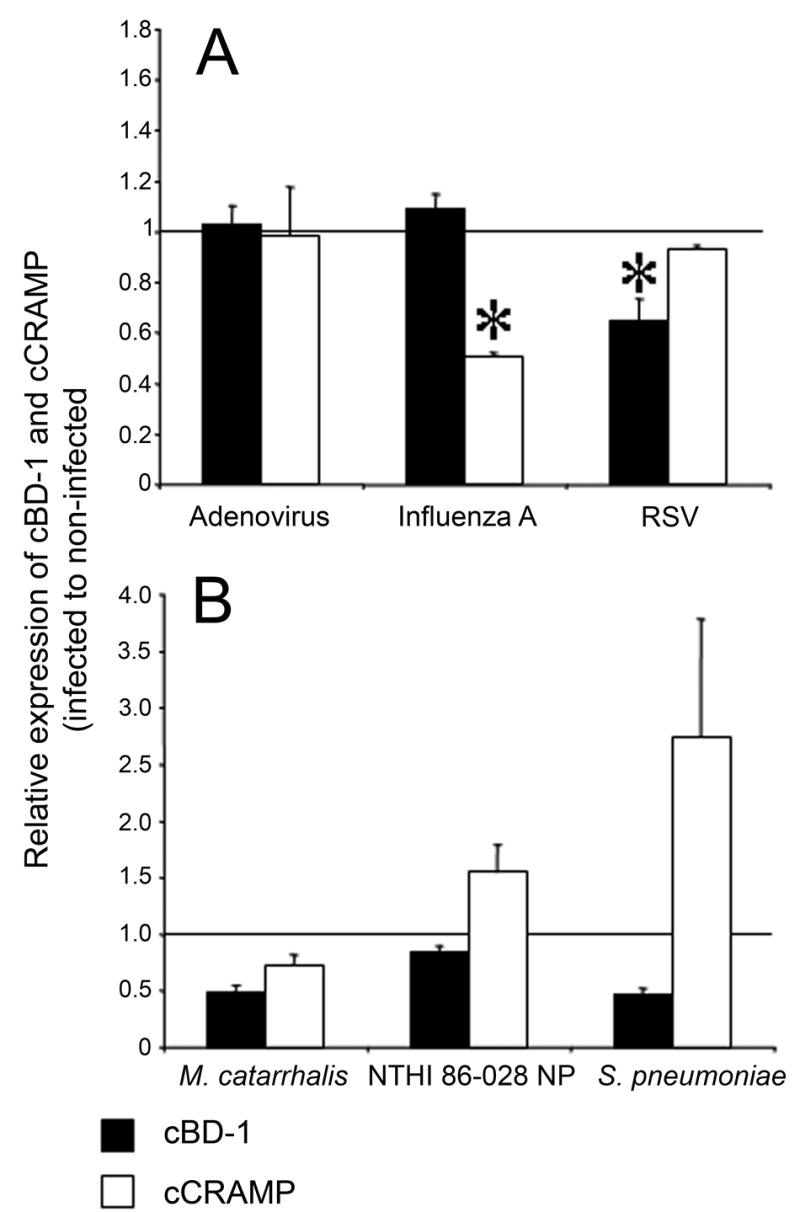
Quantitative real-time PCR analysis of cCRAMP and cBD-1 mRNA levels. Chinchilla middle ear epithelial cells were grown to confluency and were either untreated or inoculated with (A) adenovirus, influenza A virus, RSV or (B) M. catarrhalis, NTHI, or S. pneumoniae. The abundance of mRNA was normalized to GAPDH levels and values are reported as the ratio of normalized AMP mRNA levels from infected to uninfected cell cultures. Values greater than 1.0 represent samples in which mRNA levels increased after CMEEs were incubated with microorganisms, whereas values less than 1.0 represent samples in which the AMP mRNA abundance decreased after inoculated with a bacterium or virus. Data reported in (A) are the average of at least 3 experiments each performed in triplicate, whereas those in (B) are from two experiments each performed in duplicate. Asterisk denote statistically significant reduction (p ≤ 0.05) in either cBD-1 or cCRAMP mRNA levels obtained when CMEEs were infected with virus or bacteria relative to baseline, with standard error of the mean values shown.
4. Discussion
In order to determine the possible role of AMP expression and function in the establishment and/or resolution of OM, our laboratory has begun to characterize the AMP repertoire of the chinchilla URT, since it is the predominant animal model for OM research. We recently published the identification of a cDNA encoding a β-defensin family member, cBD-1 (Harris et al., 2004), in this host, and here we described the cloning, identification, and expression of a cDNA peptide with significant similarity to known cathelicidin molecules. Importantly, we demonstrated that pathogens relevant to OM modulate expression of these AMPs in a specific and discriminate manner.
We used an anchored PCR approach to amplify a partial cDNA from chinchilla Eustachian tube that encoded a cathelicidin peptide. This PCR reaction, which used a primer based on conserved sequence in the cathelin domain, did not amplify other cathelicidin products suggesting that only one cathelicidin was expressed in the chinchilla URT mucosae, as has been shown in humans and mice (Zanetti, 2004). Computer modeling and circular dichroism analysis of recombinant peptide indicated that cCRAMP possessed structural characteristics shared among cathelicidin molecules from many organisms, including the presence of a characteristic amphipathic α-helix. Of note was the observation that the secondary structure of this peptide was altered when diluted in buffer at a pH 8.6 or pH 9.9, which may have implications for AMP function in vivo considering that middle ear effusions are frequently alkaline (Wezyk and Makowski, 2000). Since our data demonstrate that this pH-dependent change in cCRAMP structure can be overcome by the addition of SDS, which mimics contact with a bacterial membrane, it is not likely that cCRAMP-mediated bacterial killing would be affected by a more basic environment. However, another cathelicidin-like activity, such as being chemotactic for neutrophils, monocytes, or dendritic cells (Agerberth et al., 2000; Davidson et al., 2004; De et al., 2000; Kurosaka et al., 2005), may be influenced by alkaline conditions which could lead to a compromise in host immunity.
When the mature antimicrobial fragments of other cathelicidin peptides were aligned with cCRAMP, via CLUSTAL W peptide alignment (Fig. 1A), the chinchilla derived cathelicidin showed greatest similarity with the rhesus monkey RL-37 peptide (43.2% similarity). We find it interesting that, in the active C-terminal domains, both cCRAMP and cBD-1 shared greatest similarity to AMPs isolated from higher primates (humans and monkeys), suggesting that the chinchilla can be effectively used as an animal model toward understanding the role of human AMPs in innate immunity, specifically as related to OM.
The (r)cCRAMP killed NTHI 86-028NP, E. coli ML-35, M. catarrhalis 1857, and, much less efficiently, S. pneumoniae serotype 14 at concentrations in the micromolar range. Although the amount of peptide required to kill these organisms was approximately double of what is typically reported (Turner et al., 1998), these assays were performed using a larger inoculum of bacterial cells (~1 x 108 cfu/ml) and were within a range observed with other cathelicidins (Brogden et al., 2001; Starner et al., 2005; Yang et al., 2004). As designed, cleavage of (r)cCRAMP with thrombin should leave a glycine and serine residue on the N-terminus of the peptide which are not predicted to be present on native mature cCRAMP. Thereby, it is possible that these amino acids could have ameliorated the antimicrobial activity of this recombinant peptide. Nevertheless, (r)cCRAMP effectively killed NTHI 86-028NP but not S. pneumoniae, suggesting that cCRAMP does not play a significant role in protecting mucosal surfaces from infection with this bacterium.
The observation that cCRAMP mRNA could be detected by RT-PCR in tissues of the chinchilla is consistent with cathelicidin expression in other species, including lung, trachea, bladder, and brain (Bals et al., 1998), providing support for the conclusion that cCRAMP is a member of the cathelicidin family. Our work has also expanded upon these data to show that cCRAMP was expressed in every tissue evaluated in the URT of the chinchilla including the mucosae of the nasal septum, nasoturbinates, ethmoid turbinates, nasopharynx, and the middle ear mucosa. We used in situ hybridization to demonstrate that cCRAMP mRNA was produced by the columnar epithelium of the Eustachian tube, a site-specific expression pattern which differs from that observed with respect to cBD-1 (Harris et al., 2004) since this latter AMP is not only expressed by the epithelium but also by submucosal glands (either mucus or serous). In humans and other mammals, the Eustachian tube acts as a conduit, bridging the heavily colonized nasopharynx to the sterile middle ear, therefore it is likely important that a member of both families of AMPs, a defensin and a cathelicidin, are expressed at a basal level in this site potentially providing a broader defense against invading pathogens.
Expression levels of both cathelicidins and some β-defensins are inducible and thereby change dramatically when exposed to microorganisms (Dorschner et al., 2001; Islam et al., 2001; Swanson et al., 2004; Wehkamp et al., 2003). Rhinovirus, a causative agent of OM, infected epithelial cells express more human β-defensin-2 than uninfected controls, and rhinovirus infection of cultured bronchial epithelial cells increases human β-defensin-2 and -3 transcripts; all in a manner that is dependent upon the presence of viral double-stranded RNA (Duits et al., 2003; Proud et al., 2004). In contrast, N. gonorrhoeae downregulates LL-37 expression when infecting a cervical cell line, and Shigella spp. similarly downregulate LL-37 and human β-defensin-1 mRNA as shown in analysis of gut biopsies recovered from infected patients (Bergman et al., 2005; Islam et al., 2001). Collectively, these analyses demonstrate that pathogens differentially alter levels of AMP mRNA and also provide evidence supporting the hypothesis that select microbes can decrease transcription of different AMPs, possibly providing a mechanism for immune evasion.
A similar scenario was presented in this work with regard to several co-pathogens of otitis media and their effect on AMP transcript levels in CMEEs, a cell which lines the middle ear cavity of the chinchilla. Incubation of CMEEs with influenza A virus resulted in decreased levels of cCRAMP transcripts, but had no effect on cBD-1. In contrast, RSV downregulated mRNA of cBD-1, but did not alter levels of cCRAMP transcripts. This microbe-specific reduction in AP message levels naturally leads to the question as to what specific influenza A or RSV gene product(s), or stage in the viral replication cycle, was responsible for the different affects on transcript levels of cCRAMP and cBD-1. We are currently conducting in vitro studies using RSV deletion mutants that lack specific gene products which should provide some details as to the mechanism behind the reduction in cBD-1 message levels.
When we incubated CMEEs with bacteria, we found that NTHI, the bacterium that predominates in chronic and recurrent OM, increased cCRAMP transcript levels by approximately 50%, however cBD-1 levels remained almost unchanged. In published work, an increase in human cathelicidin LL-37 mRNA was detected after incubation of pharyngeal carcinoma cells with H. influenzae and a similar increase was also observed in nasal mucosa recovered from individuals with rhinitis thus supporting our observations (Kim et al., 2003; Lysenko et al., 2000). When CMEEs were incubated with M. catarrhalis (MOI=100), a reduction of both cCRAMP and cBD-1 transcripts was demonstrated suggesting that this bacterium is proficient at dysregulating expression of AMPs. It is interesting that human cathelicidin LL-37 transcripts were reduced in a cervical cell line incubated with N. gonorrhoeae (MOI=70) and epithelial cells incubated with Shigella sp. (MOI=100) also showed reduction of LL-37 mRNA by 6 hours post infection whereas similar effects were not seen at earlier time points (Bergman et al., 2005; Islam et al., 2001). Although we did not evaluate AMP mRNA levels from CMEEs incubated with bacteria for less than six hours, the response to M. catarrhalis mimics that to the N. gonorrhoeae and Shigella dysenteriae type I-induced decrease in LL-37 transcripts and suggests that, in vitro, there is a delay between the co-culturing of mucosal pathogens with epithelial cells and the bacterial-mediated downregulation of AMP expression, likely because of the need for de novo synthesis of a bacterial product(s).
Although at a lower MOI, S. pneumoniae infection of CMEEs also produced a similar reduction of cBD-1 mRNA. This same affect was not seen with cCRAMP transcript levels which were upregulated 175%. It is of note that mRNA levels for this latter AMP were increased, even though (r)cCRAMP was least active against this pathogen in vitro, while mRNA levels of cBD-1, which is effective at killing S. pneumoniae (Harris et al., 2004), were reduced approximately 50%. These data provide evidence for the hypothesis that some microbes may downregulate AMP transcripts only for encoded proteins which are effective at killing the respective bacterium.
Similar to the situation in man, in the chinchilla, influenza A virus is an excellent copartner for S. pneumoniae-induced OM, whereas this viral pathogen does not increase the incidence of OM over single pathogen infection models when partnered with either NTHI or M. catarrhalis (Bakaletz, 2002). Although many mechanisms have been elucidated which contribute to the synergism between the pneumococcus and influenza A (Giebink et al., 1987; McCullers and Rehg, 2002; Plotkowski et al., 1986), our data suggested that downregulation of specific AMP messages may be an additional contributory component. It will be interesting to compare these data, obtained in vitro, with time course expression profiles for both cBD-1 and cCRAMP using chinchillas infected with the same pathogens used in this work (Gitiban et al., 2005). These in vivo studies are ongoing.
The identification and characterization of a cathelicidin that is expressed at all tested mucosal sites within the chinchilla URT, combined with our earlier characterization of a β-defensin (also expressed within the airway of this host), lays the foundation for increasing our understanding of the role of innate immunity in OM. Even more exciting and challenging is the potential for the chinchilla superinfection model to allow us to study the interaction among these organisms in a pathogen-, tissue-, and time-specific manner.
Acknowledgments
We thank Jennifer Neelans for manuscript preparation, Molly Bruggeman for conduct of microbicidal assays, and Joseph Jurcisek for performing histological techniques. We also thank John Hayes for statistical consultation and Kevin Mason for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agerberth B, Charo J, Werr J, Olsson B, Idali F, Lindbom L, Kiessling R, Jornvall H, Wigzell H, Gudmundsson GH. The human antimicrobial and chemotactic peptides LL-37 and alpha-defensins are expressed by specific lymphocyte and monocyte populations. Blood. 2000;96:3086–3093. [PubMed] [Google Scholar]
- Agerberth B, Gunne H, Odeberg J, Kogner P, Boman HG, Gudmundsson GH. FALL-39, a putative human peptide antibiotic, is cysteine-free and expressed in bone marrow and testis. Proc Natl Acad Sci U S A. 1995;92:195–199. doi: 10.1073/pnas.92.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsarraf R, Jung CJ, Perkins J, Crowley C, Alsarraf NW, Gates GA. Measuring the indirect and direct costs of acute otitis media. Arch Otolaryngol Head Neck Surg. 1999;125:12–18. doi: 10.1001/archotol.125.1.12. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. John Wiley & Sons, Inc; 1993. [Google Scholar]
- Bakaletz LO. Otitis Media. In: Brogden KA, Guthmiller JM, editors. Polymicrobial Diseases. ASM Press; Washington, D.C: 2002. pp. 259–298. [PubMed] [Google Scholar]
- Bakaletz LO. Adenovirus serotype 1 does not act synergistically with Moraxella (Branhamella) catarrhalis to induce otitis media in the chinchilla. Infect Immun. 1995;63:4188–4190. doi: 10.1128/iai.63.10.4188-4190.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bals R, Wang X, Zasloff M, Wilson JM. The peptide antibiotic LL-37/hCAP-18 is expressed in epithelia of the human lung where it has broad antimicrobial activity at the airway surface. Proc Natl Acad Sci U S A. 1998;95:9541–9546. doi: 10.1073/pnas.95.16.9541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bals R, Weiner DJ, Meegalla RL, Wilson JM. Transfer of a cathelicidin peptide antibiotic gene restores bacterial killing in a cystic fibrosis xenograft model. J Clin Invest. 1999;103:1113–1117. doi: 10.1172/JCI6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman P, Johansson L, Asp V, Plant L, Gudmundsson GH, Jonsson AB, Agerberth B. Neisseria gonorrhoeae downregulates expression of the human antimicrobial peptide LL-37. Cell Microbiol. 2005;7:1009–1017. doi: 10.1111/j.1462-5822.2005.00530.x. [DOI] [PubMed] [Google Scholar]
- Bergman P, Termen S, Johansson L, Nystrom L, Arenas E, Jonsson AB, Hokfelt T, Gudmundsson GH, Agerberth B. The antimicrobial peptide rCRAMP is present in the central nervous system of the rat. J Neurochem. 2005;5:1132–1140. doi: 10.1111/j.1471-4159.2005.03081.x. [DOI] [PubMed] [Google Scholar]
- Brogden KA, Kalfa VC, Ackermann MR, Palmquist DE, McCray PB, Jr, Tack BF. The ovine cathelicidin SMAP29 kills ovine respiratory pathogens in vitro and in an ovine model of pulmonary infection. Antimicrob Agents Chemother. 2001;45:331–334. doi: 10.1128/AAC.45.1.331-334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassell GH. The Role of Research. Washington, D.C: 1997. New and Reemerging Infectious Diseases: A Global Crisis and Immediate Threat to the Nation’s Health; pp. 1–11. [Google Scholar]
- Cassell GH, Archer GL, Beam TR, Gilchrist MJ, Goldmann D, Hooper DC, Jones RN, Kleven SH, Lederberg J, Levy SB, Lein DH, Moellering RC, O’Brien TF, Osburn B, Osterholm M, Shlaes DM, Terry M, Tolin SA, Tomasz A. Report of the ASM Task Force on Antibiotic Resistance. Washington D.C: 1994. [Google Scholar]
- Chen C, Brock R, Luh F, Chou Ping-Jung, Larrick JW, Huang Rong-Fong, Huang Tai-huang. The solution structure of the active domain of CAP18-a lipopolysaccharide binding protein from rabbit leukocytes. FEBS Letters. 1995;370:46–52. doi: 10.1016/0014-5793(95)00792-8. [DOI] [PubMed] [Google Scholar]
- Davidson DJ, Currie AJ, Reid GS, Bowdish DM, MacDonald KL, Ma RC, Hancock RE, Speert DP. The cationic antimicrobial peptide LL-37 modulates dendritic cell differentiation and dendritic cell-induced T cell polarization. J Immunol. 2004;172:1146–1156. doi: 10.4049/jimmunol.172.2.1146. [DOI] [PubMed] [Google Scholar]
- De Y, Chen Q, Schmidt AP, Anderson GM, Wang JM, Wooters J, Oppenheim JJ, Chertov O. LL-37, the neutrophil granule- and epithelial cell-derived cathelicidin, utilizes formyl peptide receptor-like 1 (FPRL1) as a receptor to chemoattract human peripheral blood neutrophils, monocytes, and T cells. J Exp Med. 2000;192:1069–1074. doi: 10.1084/jem.192.7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschner RA, Pestonjamasp VK, Tamakuwala S, Ohtake T, Rudisill J, Nizet V, Agerberth B, Gudmundsson GH, Gallo RL. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J Invest Dermatol. 2001;117:91–97. doi: 10.1046/j.1523-1747.2001.01340.x. [DOI] [PubMed] [Google Scholar]
- Duits LA, Nibbering PH, van Strijen E, Vos JB, Mannesse-Lazeroms SP, van Sterkenburg MA, Hiemstra PS. Rhinovirus increases human beta-defensin-2 and -3 mRNA expression in cultured bronchial epithelial cells. FEMS Immunol Med Microbiol. 2003;38:59–64. doi: 10.1016/S0928-8244(03)00106-8. [DOI] [PubMed] [Google Scholar]
- Faden H. The microbiologic and immunologic basis for recurrent otitis media in children. Eur J Pediatr. 2001;160:407–413. doi: 10.1007/s004310100754. [DOI] [PubMed] [Google Scholar]
- Frohm Nilsson M, Sandstedt B, Sorensen O, Weber G, Borregaard N, Stahle-Backdahl M. The human cationic antimicrobial protein (hCAP18), a peptide antibiotic, is widely expressed in human squamous epithelia and colocalizes with interleukin-6. Infect Immun. 1999;67:2561–2566. doi: 10.1128/iai.67.5.2561-2566.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo RL, Kim KJ, Bernfield M, Kozak CA, Zanetti M, Merluzzi L, Gennaro R. Identification of CRAMP, a cathelin-related antimicrobial peptide expressed in the embryonic and adult mouse. J Biol Chem. 1997;272:13088–13093. doi: 10.1074/jbc.272.20.13088. [DOI] [PubMed] [Google Scholar]
- Giebink GS, Berzins IK, Marker SC, Schiffman G. Experimental otitis media after nasal inoculation of Streptococcus pneumoniae and influenza A virus in chinchillas. Infect Immun. 1980;30:445–450. doi: 10.1128/iai.30.2.445-450.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giebink GS, Ripley ML, Wright PF. Eustachian tube histopathology during experimental influenza A virus infection in the chinchilla. Ann Otol Rhinol Laryngol. 1987;96:199–206. doi: 10.1177/000348948709600212. [DOI] [PubMed] [Google Scholar]
- Gitiban N, Jurcisek JA, Harris RH, Mertz SE, Durbin RK, Bakaletz LO, Durbin JE. Chinchilla and murine models of upper respiratory tract infections with respiratory syncytial virus. J Virol. 2005;79:6035–6042. doi: 10.1128/JVI.79.10.6035-6042.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough M, Hancock RE, Kelly NM. Antiendotoxin activity of cationic peptide antimicrobial agents. Infect Immun. 1996;64:4922–4927. doi: 10.1128/iai.64.12.4922-4927.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson GH, Agerberth B. Neutrophil antibacterial peptides, multifunctional effector molecules in the mammalian immune system. J Immunol Methods. 1999;232:45–54. doi: 10.1016/s0022-1759(99)00152-0. [DOI] [PubMed] [Google Scholar]
- Hancock RE, Scott MG. The role of antimicrobial peptides in animal defenses. Proc Natl Acad Sci U S A. 2000;97:8856–8861. doi: 10.1073/pnas.97.16.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RH, Wilk D, Bevins CL, Munson RS, Jr, Bakaletz LO. Identification and characterization of a mucosal antimicrobial peptide expressed by the chinchilla (Chinchilla lanigera) airway. J Biol Chem. 2004;279:20250–20256. doi: 10.1074/jbc.M400499200. [DOI] [PubMed] [Google Scholar]
- Harrison A, Dyer DW, Ray WC, Mungur R, Carson MB, Zhong H, Gipson J, Gipson M, Johnson LS, Lewis L, Bakaletz LO, Munson RS., Jr Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. infleunzae serotype d, strain KW20. J Bacteriol. 2005;13:4627–36. doi: 10.1128/JB.187.13.4627-4636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausdorff WP, Yothers G, Dagan R, Kilpi T, Pelton SI, Cohen R, Jacobs MR, Kaplan SL, Levy C, Lopez EL, Mason EO, Syriopoulou V, Wynne B, Bryant J. Multinational study of penumococcal serotypes causing acute otitis media in children. Pediatr Infec Dis J. 2002;21:1008–16. doi: 10.1097/00006454-200211000-00007. [DOI] [PubMed] [Google Scholar]
- Heikkinen T, Chonmaitree T. Importance of respiratory viruses in acute otitis media. Clin Microbiol Rev. 2003;16:230–241. doi: 10.1128/CMR.16.2.230-241.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam D, Bandholtz L, Nilsson J, Wigzell H, Christensson B, Agerberth B, Gudmundsson G. Downregulation of bactericidal peptides in enteric infections: a novel immune escape mechanism with bacterial DNA as a potential regulator. Nat Med. 2001;7:180–185. doi: 10.1038/84627. [DOI] [PubMed] [Google Scholar]
- Johansson J, Gudmundsson GH, Rottenberg ME, Berndt KD, Agerberth B. Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J Biol Chem. 1998;273:3718–3724. doi: 10.1074/jbc.273.6.3718. [DOI] [PubMed] [Google Scholar]
- Kaplan B, Wandstrat TL, Cunningham JR. Overall cost in the treatment of otitis media. Pediatr Infect Dis J. 1997;16:S9–11. doi: 10.1097/00006454-199702001-00003. [DOI] [PubMed] [Google Scholar]
- Kennedy BJ, Novotny LA, Jurcisek JA, Lobet Y, Bakaletz LO. Passive transfer of antiserum specific for immunogens derived from a nontypeable Haemophilus influenzae adhesin and lipoprotein D prevents otitis media after heterologous challenge. Infect Immun. 2000;68:2756–2765. doi: 10.1128/iai.68.5.2756-2765.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ST, Cha HE, Kim DY, Han GC, Chung YS, Lee YJ, Hwang YJ, Lee HM. Antimicrobial peptide LL-37 is upregulated in chronic nasal inflammatory disease. Acta Otolaryngol. 2003;123:81–85. doi: 10.1080/0036554021000028089. [DOI] [PubMed] [Google Scholar]
- Kurosaka K, Chen Q, Yarovinsky F, Oppenheim JJ, Yang D. Mouse cathelin-related antimicrobial peptide chemoattracts leukocytes using formyl peptide receptor-like 1/mouse formyl peptide receptor-like 2 as the receptor and acts as an immune adjuvant. J Immunol. 2005;10:6257–65. doi: 10.4049/jimmunol.174.10.6257. [DOI] [PubMed] [Google Scholar]
- Larrick JW, Hirata M, Balint RF, Lee J, Zhong J, Wright SC. Human CAP18: a novel antimicrobial lipopolysaccharide-binding protein. Infect Immun. 1995;63:1291–1297. doi: 10.1128/iai.63.4.1291-1297.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrick JW, Morgan JG, Palings I, Hirata M, Yen MH. Complementary DNA sequence of rabbit CAP18--a unique lipopolysaccharide binding protein. Biochem Biophys Res Commun. 1991;179:170–175. doi: 10.1016/0006-291x(91)91350-l. [DOI] [PubMed] [Google Scholar]
- Lehrer RI, Ganz T. Cathelicidins: a family of endogenous antimicrobial peptides. Curr Opin Hematol. 2002a;9:18–22. doi: 10.1097/00062752-200201000-00004. [DOI] [PubMed] [Google Scholar]
- Lehrer RI, Ganz T. Defensins of vertebrate animals. Curr Opin Immunol. 2002b;14:96–102. doi: 10.1016/s0952-7915(01)00303-x. [DOI] [PubMed] [Google Scholar]
- Lysenko ES, Gould J, Bals R, Wilson JM, Weiser JN. Bacterial phosphorylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expressed in the upper respiratory tract. Infect Immun. 2000;68:1664–1671. doi: 10.1128/iai.68.3.1664-1671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA, Rehg JE. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J Infect Dis. 2002;186:341–350. doi: 10.1086/341462. [DOI] [PubMed] [Google Scholar]
- Monto AS, Ullman BM. Acute respiratory illness in an American community, The Tecumseh study. JAMA. 1974;227:164–169. [PubMed] [Google Scholar]
- Murakami M, Ohtake T, Dorschner RA, Gallo RL. Cathelicidin antimicrobial peptides are expressed in salivary glands and saliva. J Dent Res. 2002;81:845–850. doi: 10.1177/154405910208101210. [DOI] [PubMed] [Google Scholar]
- Mygind PH, Fischer RL, Schnorr KM, Hansen MT, Sonksen CP, Ludvigsen S, Raventos D, Buskov S, Christensen B, De Maria L, Taboureau O, Yaver D, Elvig-Jorgensen SG, Sorensen MV, Christensen BE, Kjaerulff S, Frimodt-Moller N, Lehrer RI, Zasloff M, Kristensen H. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature. 2005;437:975–980. doi: 10.1038/nature04051. [DOI] [PubMed] [Google Scholar]
- Nagaoka I, Tsutsumi-Ishii Y, Yomogida S, Yamashita T. Isolation of cDNA encoding guinea pig neutrophil cationic antibacterial polypeptide of 11 kDa (CAP11) and evaluation of CAP11 mRNA expression during neutrophil maturation. J Biol Chem. 1997;272:22742–22750. doi: 10.1074/jbc.272.36.22742. [DOI] [PubMed] [Google Scholar]
- Nakamura A, DeMaria TF, Lim DJ, van Blitterswijk CA. Primary culture of chinchilla middle ear epithelium. Ann Otol Rhinol Laryngol. 1991;100:774–782. doi: 10.1177/000348949110000916. [DOI] [PubMed] [Google Scholar]
- Nakamura A, DeMaria TF, van Blitterswijk C, Lim DJ. Effect of endotoxin on cultured chinchilla middle ear epithelium. Ann Otol Rhinol Laryngol. 1992;101:607–611. doi: 10.1177/000348949210100712. [DOI] [PubMed] [Google Scholar]
- Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- Plotkowski MC, Puchelle E, Beck G, Jacquot J, Hannoun C. Adherence of type I Streptococcus pneumoniae to tracheal epithelium of mice infected with influenza A/PR8 virus. Am Rev Respir Dis. 1986;134:1040–1044. doi: 10.1164/arrd.1986.134.5.1040. [DOI] [PubMed] [Google Scholar]
- Polack FP, Irusta PM, Hoffman SJ, Schiatti MP, Melendi GA, Delgado MF, Laham FR, Thumar B, Hendry RM, Melero JA, Karron RA, Collins PL, Kleeberger SR. The cysteine-rich region of respiratory syncytial virus attachment protein inhibits innate immunity elicited by the virus and endotoxin. Proc Natl Acad Sci U S A. 2005;102:8996–9001. doi: 10.1073/pnas.0409478102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud D, Sanders SP, Wiehler S. Human rhinovirus infection induces airway epithelial cell production of human beta-defensin 2 both in vitro and in vivo. J Immunol. 2004;172:4637–4645. doi: 10.4049/jimmunol.172.7.4637. [DOI] [PubMed] [Google Scholar]
- Ritonja A, Kopitar M, Jerala R, Turk V. Primary structure of a new cysteine proteinase inhibitor from pig leucocytes. FEBS Lett. 1989;255:211–214. doi: 10.1016/0014-5793(89)81093-2. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2001. [Google Scholar]
- Sorensen OE, Cowland JB, Theilgaard-Monch K, Liu L, Ganz T, Borregaard N. Wound healing and expression of antimicrobial peptides/polypeptides in human keratinocytes, a consequence of common growth factors. J Immunol. 2003a;170:5583–5589. doi: 10.4049/jimmunol.170.11.5583. [DOI] [PubMed] [Google Scholar]
- Sorensen OE, Follin P, Johnsen AH, Calafat J, Tjabringa GS, Hiemstra PS, Borregaard N. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood. 2001;97:3951–3959. doi: 10.1182/blood.v97.12.3951. [DOI] [PubMed] [Google Scholar]
- Sorensen OE, Gram L, Johnsen AH, Andersson E, Bangsboll S, Tjabringa GS, Hiemstra PS, Malm J, Egesten A, Borregaard N. Processing of seminal plasma hCAP-18 to ALL-38 by gastricsin: a novel mechanism of generating antimicrobial peptides in vagina. J Biol Chem. 2003b;278:28540–28546. doi: 10.1074/jbc.M301608200. [DOI] [PubMed] [Google Scholar]
- Starner TD, Agerberth B, Gudmundsson GH, McCray PB., Jr Expression and activity of beta-defensins and LL-37 in the developing human lung. J Immunol. 2005;174:1608–1615. doi: 10.4049/jimmunol.174.3.1608. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Bakaletz LO. Synergistic effect of adenovirus type 1 and nontypeable Haemophilus influenzae in a chinchilla model of experimental otitis media. Infect Immun. 1994;62:1710–1718. doi: 10.1128/iai.62.5.1710-1718.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K, Gorodetsky S, Good L, Davis S, Musgrave D, Stelwagen K, Farr V, Molenaar A. Expression of a beta-defensin mRNA, lingual antimicrobial peptide, in bovine mammary epithelial tissue is induced by mastitis. Infect Immun. 2004;72:7311–7314. doi: 10.1128/IAI.72.12.7311-7314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack BF, Sawai MV, Kearney WR, Robertson AD, Sherman MA, Wang W, Hong T, Boo LM, Wu H, Waring AJ, Lehrer RI. SMAP-29 has two LPS-binding sites and a central hinge. Eur J Biochem. 2002;269:1181–1189. doi: 10.1046/j.0014-2956.2002.02751.x. [DOI] [PubMed] [Google Scholar]
- Tarver AP, Clark DP, Diamond G, Russell JP, Erdjument-Bromage H, Tempst P, Cohen KS, Jones DE, Sweeney RW, Wines M, Hwang S, Bevins CL. Enteric beta-defensin: molecular cloning and characterization of a gene with inducible intestinal epithelial cell expression associated with Cryptosporidium parvum infection. Infect Immun. 1998;66:1045–1056. doi: 10.1128/iai.66.3.1045-1056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teele DW, Klein JO, Rosner B. Epidemiology of otitis media during the first seven years of life in children in greater Boston: a prospective, cohort study. J Infect Dis. 1989;160:83–94. doi: 10.1093/infdis/160.1.83. [DOI] [PubMed] [Google Scholar]
- Termen S, Tollin M, Olsson B, Svenberg T, Agerberth B, Gudmundsson GH. Phylogeny, processing and expression of the rat cathelicidin rCRAMP: a model for innate antimicrobial peptides. Cell Mol Life Sci. 2003;60:536–549. doi: 10.1007/s000180300045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjabringa GS, Rabe KF, Hiemstra PS. The human cathelicidin LL-37: a multifunctional peptide involved in infection and inflammation in the lung. Pulm Pharmacol Ther. 2005;18:321–327. doi: 10.1016/j.pupt.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Turner J, Cho Y, Dinh N, Waring AJ, Lehrer RI. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob Agents Chemother. 1998;42:2206–2214. doi: 10.1128/aac.42.9.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehkamp J, Schmidt K, Herrlinger KR, Baxmann S, Behling S, Wohlschlager C, Feller AC, Stange EF, Fellermann K. Defensin pattern in chronic gastritis: HBD-2 is differentially expressed with respect to Helicobacter pylori status. J Clin Pathol. 2003;56:352–357. doi: 10.1136/jcp.56.5.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wezyk MT, Makowski A. pH of fluid collected from middle ear in the course of otitis media in children. Otolaryngol Pol. 2000;54:131–133. [PubMed] [Google Scholar]
- Yang YH, Zheng GG, Li G, Zhang XJ, Cao ZY, Rao Q, Wu KF. Expression of bioactive recombinant GSLL-39, a variant of human antimicrobial peptide LL-37, in Escherichia coli. Protein Expr Purif. 2004;37:229–235. doi: 10.1016/j.pep.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zaiou M, Nizet V, Gallo RL. Antimicrobial and protease inhibitory functions of the human cathelicidin (hCAP18/LL-37) prosequence. J Invest Dermatol. 2003;120:810–816. doi: 10.1046/j.1523-1747.2003.12132.x. [DOI] [PubMed] [Google Scholar]
- Zanetti M. Cathelicidins, multifunctional peptides of the innate immunity. J Leukoc Biol. 2004;75:39–48. doi: 10.1189/jlb.0403147. [DOI] [PubMed] [Google Scholar]
- Zanetti M, Gennaro R, Romeo D. Cathelicidins: a novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett. 1995;374:1–5. doi: 10.1016/0014-5793(95)01050-o. [DOI] [PubMed] [Google Scholar]
- Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Nguyen T, Boo LM, Hong T, Espiritu C, Orlov D, Wang W, Waring A, Lehrer RI. RL-37, an alpha-helical antimicrobial peptide of the rhesus monkey. Antimicrob Agents Chemother. 2001;45:2695–2702. doi: 10.1128/AAC.45.10.2695-2702.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


