Abstract
Peptidoglycan is an essential polymer that forms a protective shell around bacterial cell membranes. Peptidoglycan biosynthesis is the target of many clinically used antibiotics, including the β-lactams, imipenems, cephalosporins, and glycopeptides. Resistance to these and other antibiotics has prompted interest in an atomic-level understanding of the enzymes that make peptidoglycan. Representative structures have been reported for most of the enzymes in the pathway. Until now, however, there have been no structures of any peptidoglycan glycosyltransferases (also known as transglycosylases), which catalyze formation of the carbohydrate chains of peptidoglycan from disaccharide subunits on the bacterial cell surface. We report here the 2.1-Å crystal structure of the peptidoglycan glycosyltransferase (PGT) domain of Aquifex aeolicus PBP1A. The structure has a different fold from all other glycosyltransferase structures reported to date, but it bears some resemblance to λ-lysozyme, an enzyme that degrades the carbohydrate chains of peptidoglycan. An analysis of the structure, combined with biochemical information showing that these enzymes are processive, suggests a model for glycan chain polymerization.
Keywords: antibiotic resistance, penicillin-binding protein, cell wall, transglycosylase
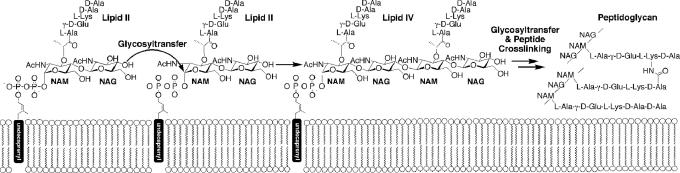
The major component of the bacterial cell wall is a cross-linked glycopeptide polymer called peptidoglycan. This polymer surrounds the cytoplasmic membrane of bacteria and functions as an exoskeleton, maintaining cell shape and stabilizing the membrane against fluctuations in osmotic pressure. Peptidoglycan is synthesized in an intracellular phase in which UDP-N-acetylglucosamine is converted to a diphospholipid-linked disaccharide-pentapeptide known as Lipid II, and in an extracellular phase in which the disaccharide (NAG-NAM) subunits of translocated Lipid II are coupled by peptidoglycan glycosyltransferases (PGTs; also known as transglycosylases) to form linear carbohydrate chains (Fig. 1), which are cross-linked through the attached peptide moieties by transpeptidases (1). A functioning peptidoglycan pathway is required for bacterial cell growth and division, and compounds that inhibit peptidoglycan biosynthesis have antibiotic activity (2). For example, the β-lactams, which have been used for decades to treat bacterial infections, irreversibly inhibit the transpeptidases. The emergence of resistance to β-lactams and other clinically used antibiotics has prompted intense interest in understanding peptidoglycan biosynthesis in detail. Major advances have been made in recent years with respect to the characterization of key biosynthetic enzymes (3–5). Several structures have been reported for enzymes that catalyze transpeptidation (6, 7), but no PGT structures have been described.
Fig. 1.
Peptidoglycan glycosyltransferases couple Lipid II to give higher-order glycan fragments that react with other Lipid II molecules. Transpeptidases cross-link the glycan products.
PGTs are defined by the presence of five conserved sequence motifs (Fig. 2A) and exist in two forms: (i) as N-terminal glycosyltransferase domains in bifunctional proteins that also contain a C-terminal transpeptidase domain [called class A penicillin-binding proteins (PBPs)], and (ii) as monofunctional proteins (MGTs) that do not contain transpeptidase domains (8). Different bacteria typically contain different numbers and types of PGTs, and it is thought that the different PGTs play different roles during the bacterial cell cycle, with some involved primarily in cell elongation and others recruited to the septal region during cell division (6). Regardless of their cellular roles, all PGTs catalyze glycosyltransfer from a polyprenyl-diphosphate moiety on the anomeric center of an N-acetyl muramic acid (NAM) unit to the C4 hydroxyl of an N-acetylglucosamine (NAG) moiety (Fig. 1). The mechanism of glycosyltransfer is not well understood, and it has taken considerable effort to identify soluble, well behaved PGT domains to use as model systems for detailed mechanistic and structural analysis (9–11). After screening a number of PGT domains from different organisms, we have identified the PGT domain from Aquifex aeolicus PBP1A as a good candidate for structural studies. We report here the 2.1-Å crystal structure of this PGT domain along with biochemical studies that suggest a model for processive glycosyltransfer.
Fig. 2.
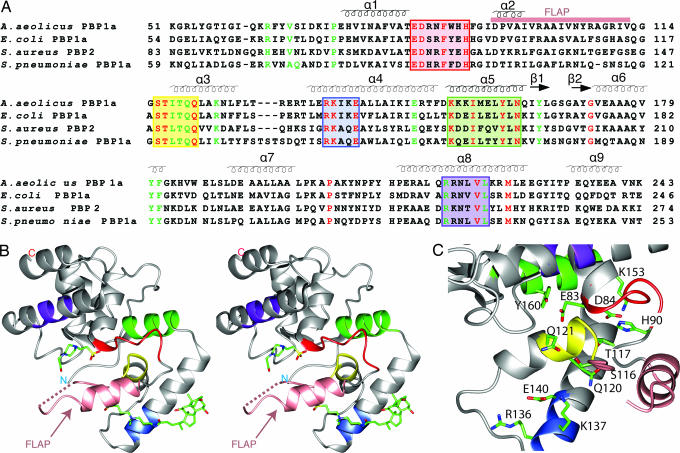
Overall structure of the PGT domain. (A) Sequence alignment of A. aeolicus ΔPBP1A[51–243] with PGTs from other class A penicillin-binding proteins (4 of 200 aligned sequences shown). Secondary structural elements of ΔPBP1A[51–243] are shown over the sequences. Conserved residues are highlighted (red, invariant; green, highly conserved). The conserved motifs that typify PGTs are colored in red, yellow, blue, green, and purple, and the flap region is marked by a pink bar. (B) Stereoview of the overall structure of PGT in a ribbon representation with the active site cleft facing left. The structure is shown in gray with the conserved motifs colored as in A and the flap region colored in pink with a dotted line representing the missing residues. The CHAPS molecule and Hepes molecule are rendered and colored by atom type. (C) View looking into the cleft (90° rotation from B) in a ribbon representation with the same color scheme as in B. Conserved residues are shown and colored by atom (carbon, green; nitrogen, blue; oxygen, red). All figures were created by CCP4 MG, Pymol, and Insight II.
Results
Overall Structure of the PGT Domain.
PBP1A from the hyperthermophile A. aeolicus is a class A PBP containing a short cytoplasmic region and a transmembrane helix followed by an N-terminal PGT domain and a C-terminal transpeptidase domain. Expression of the full-length enzyme was poor. An N-terminal thioredoxin (Trx)-His6 fusion protein lacking the transmembrane helix could be expressed in the cytoplasm but underwent proteolysis in the transpeptidase domain. Because crystal structures already exist for the C-terminal transpeptidase domain of orthologs of A. aeolicus PBP1A (7, 12, 13), we focused on identifying a well behaved PGT domain. Several PGT constructs of varying lengths were prepared and analyzed to establish the minimum size of the PGT domain. The shortest active fragment began at S67 and terminated at K243, producing a 177-residue polypeptide centered around the five conserved motifs that typify PGTs (Fig. 2A). This construct, ΔPBP1A[67–243], as well as two longer constructs, ΔPBP1A[29–243] and ΔPBP1A[51–243], was selected for crystallization trials. High quality crystals were obtained for ΔPBP1A[51–243]. The structure was determined by using phasing information from a single wavelength anomalous dispersion (SAD) data set of a selenomethionine-substituted crystal and refined by using a native data set acquired to a 2.1-Å resolution. There is one polypeptide in each asymmetric unit with a solvent content of 60%. The experimental electron density was of sufficient quality to trace the polypeptide chain from T57 to the C terminus except for a segment spanning amino acids 106–111. Because the shortest active A. aeolicus PGT domain begins at residue S67, the missing N-terminal residues (K51-Y56) are not part of the catalytic domain. The other missing residues are part of a mobile loop which is discussed further below. We observed additional density in the Fo − Fc map that was not part of the polypeptide, and a molecule of Hepes, as well as one of CHAPS (both used in the sample buffer), fits this density.
The A. aeolicus PGT domain is a globular protein composed of nine α-helices organized into two lobes that are separated by a cleft that spans the width of the protein (Fig. 2B). The larger lobe is formed from a long N-terminal coil of 16 residues, five helices (α1, α6, α7, α8, and α9) and two short antiparallel β strands that form a loop between α5 and α6; the smaller lobe is formed from helices α2, α3, and α4. Helix α5 is sandwiched between the two lobes and, along with part of the N-terminal coil, forms the back wall of the cleft. Many of the conserved amino acids that typify bacterial PGT domains line the cleft, which contains the active site (Fig. 2C). The bound Hepes molecule is located in the cleft near two invariant residues that have been implicated in the glycosyl transfer reaction (see The Active Site Cleft).
Structural Similarities to λ-Lysozyme.
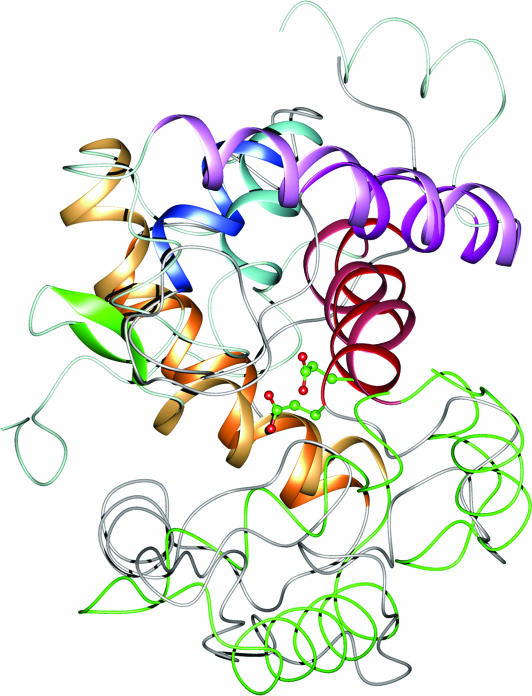
A structure similarity search using the Dali server did not return any structures with significant similarity to the PGT domain. However, a secondary structure matching (SSM) search returned bacteriophage λ-lysozyme (λR; also known as LaL), which breaks the β(1,4) glycosidic linkages between NAM and NAG residues of peptidoglycan, the same bonds that the PGTs make (14, 15). Terrak et al. (16) have previously suggested that the PGTs might resemble the lysozymes. The rmsd of 3.8 Å over 84 aligned residues between PGT and λR did not initially suggest significant similarity between the two structures; when superimposed, however, it is clear that the structures have a similar overall topology (Fig. 3). Both λR and the PGT domain are composed of a large and a small lobe separated by a cleft. The small lobe in λR contains a β sheet whereas the small lobe of the PGT domain is almost entirely α-helical, but the size and relative orientation of the small lobe to the larger lobe are similar for both proteins. The larger lobes share more similarities. Both consist largely of α-helical segments having similar connectivities and relative orientations (Fig. 3). Helix α1 of the PGT domain superimposes with helix α1 of λR but is one turn shorter; two helices, α5 and α6 of the PGT domain, which are interrupted by a nine-residue β sheet (β1 and β2), superimpose with the long helix α3 of λR. The relative orientation of α1 and α3 is among the most conserved structural features of the lysozyme family. In addition, helix α7 of the PGT domain superimposes with α5 of λR, whereas α8 superimposes with α6, the C-terminal helix of λR. Another conserved feature of the lysozyme family is the presence of a glutamic acid residue at the C terminus of helix α1. This residue, E19 in λR, is located in the active site cleft and functions as a general acid catalyst, protonating the leaving group during glycosidic bond cleavage by lysozyme. There is an invariant glutamate residue (E83) in the same location in the PGT domain. This glutamate residue is essential for catalysis and is proposed play a central role in the glycosyltransfer reaction (see The Active Site Cleft).
Fig. 3.
A superposition of the PGT domain and λR (chain B, PDB code 1D9U) with the clefts facing front. The four superimposable helices are shown in a ribbon representation and colored in red, orange, blue, and purple (colors in the PGT structure are lighter). The β1 and β2 strands separating helices α5 and α6 in the PGT domain are colored green. Nonsuperimposable regions in the big lobes are shown in a worm representation and colored gray for λR and light blue for PGT. The small lobes are shown in a worm representation and colored gray for λR and green for PGT. Glu-19 in λR and Glu-83 in PGT are shown as ball and stick and colored by atom (red, oxygen; green, carbon).
Despite the similarities, the β sheet that interrupts α5 and α6 in the PGT domain alters the orientation of the active site cleft substantially compared with that of λR. Therefore, the elongating peptidoglycan chain cannot bind in the same manner as the degrading peptidoglycan chain does in λR, as judged by a superposition of the PGT domain with the λR structure complexed to a PG substrate mimic, (GlcNAc)6.
Dimer Interface.
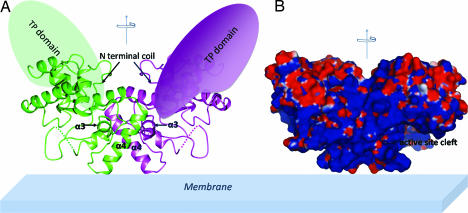
The A. aeolicus PGT domain elutes as a dimer on a size exclusion column [supporting information (SI) Fig. 6]. It also forms a covalent dimer in the presence of cross-linking agents. Several class A PBPs containing PGT domains, including Escherichia coli PBP1A and PBP1B, are reported to form dimers (17, 18), but the location of the dimer interface has not been established for any of these proteins. Packing analysis of the A. aeolicus PGT crystal structure reveals an extensive interface between the external surfaces of α3 and α4 as well as part of the N-terminal coil of two PGT monomers that are related by a crystallographic 2-fold axis of symmetry (Fig. 4A). The buried surface area of ≈1,200 Å2 per monomer is twice as large as the next largest crystal-packing interface in the lattice, making it the likely candidate for the dimer interface. There are extensive hydrophobic contacts as well as a network of polar interactions on the periphery of the interface. A surface conservation analysis of the interface based on an alignment of 40 class A PBPs shows a conserved central hydrophobic region consisting of residues L126, F127, L128, L142, and L143 (SI Fig. 7). On the periphery, R63, which is highly conserved, is within hydrogen bonding distance of the backbone carbonyls of E148 (also highly conserved) and R149 located at the C terminus of helix α4. The conserved residue analysis suggests that the dimer interface identified in the A. aeolicus PGT domain is conserved in other PGT domains.
Fig. 4.
The PGT domain of PBP1A forms a dimer. (A) The PGT dimer (green or purple for each monomer) and the proposed orientation with respect to the membrane. The approximate locations of the TP domains are indicated (green in back, purple in the front). Helices α3 and α4 and the N-terminal coil involved in the dimer interface are labeled for each monomer. Dotted lines in each monomer represent missing loop residues. (B) Solvent accessible surface of the PGT dimer colored according to electrostatic potential (blue, positive; red, negative) as calculated by APBS in Pymol. Active site clefts are indicated with arrows.
The physiological relevance of dimerization is not clear from the crystal structure because the active site clefts in each monomer are located on opposite sides of the dimer and are presumed to be functionally separate. It has been speculated for other class A PBPs that dimerization somehow facilitates transpeptidation of nascent glycan strands. A crystal structure for the TP domain of an ortholog of A. aeolicus PBP1A complexed to a tryptic fragment of a peptide that lies N-terminal to the catalytic portion of the PGT domain has been reported (13) and provides information about the locations of both the N and C termini of the PGT domain. This structure makes it possible to estimate the approximate locations of the TP domains relative to the A. aeolicus PGT domains (Fig. 4A). Because of the 2-fold symmetry of the active site clefts in the dimer, the growing glycan strands would emerge in opposite directions, making unlikely a mechanism in which two elongating glycan chains are fed directly from the PGT domains into the active site of one of the TP domains. The topology instead suggests that each elongating strand must be cross-linked to other glycan chains, which may lie within the existing framework of the bacterial sacculus. Bidirectional peptidoglycan chain synthesis also suggests that if PGTs are found in multiprotein complexes, as proposed (19), then the complexes remain stationary at biosynthetic loci rather than sliding along the membrane surface. Recent observations that new peptidoglycan is incorporated in patches throughout the bacterial sacculus (20) are consistent with such a model. It is not yet known whether catalytically active monomers can be produced if the dimerization interface is disrupted, but the structure reported here provides a basis to guide the design of experiments to address the functional role, if any, of dimerization by class A PBPs.
Proposed Orientation of the PGT Domain with Respect to the Membrane.
PGTs are anchored to bacterial membranes by N-terminal transmembrane helices. The A. aeolicus PGT domain described here lacks the TM helix and ≈25 additional N-terminal residues, but it is reasonable to assume that it must be oriented with respect to the membrane in a manner that enables membrane-bound substrates access to the active site. The orientation shown in Fig. 4, in which the small lobes of both monomers are directed toward the membrane, would allow access of the membrane-anchored substrates to the active site clefts. In this orientation, the 2-fold axis of the dimer is perpendicular to the membrane, and the calculated dipole of each monomer, which is parallel to the 2-fold axis, is strongly positive near the membrane (Fig. 4B). Both the bacterial membrane and the PGT substrates are negatively charged and would be expected to interact favorably with the positive field. The positively charged residues around the cleft, which include the conserved residues K124, R136, K137, and K153, may play roles in orienting the negatively charged substrates in the active site.
The Active Site Cleft.
Viewed from the front, half of the cleft that separates the large and small lobes of the PGT domain is open but the other half is occluded by a flap created by the polypeptide chain between the start of helix α2 and the long coil preceding helix α3 (Fig. 2B). The B factors for residues in the flap region are higher than average and the electron density for several residues was not adequate to place them, suggesting that this region is mobile and may undergo a conformational rearrangement when the substrates bind. Behind the flap lie several invariant residues, including E83 and D84. We prepared mutants of these and two other invariant residues in the occluded region of the cleft (H90, which is within hydrogen bonding distance of D84, and K153) to assess their importance in catalysis. All mutants could be expressed in soluble form at levels comparable with the WT parent enzyme, suggesting that the mutations do not interfere with protein folding. The kinetic parameters for the WT enzyme are kcat = 3.5 min−1 and kcat/Km = 6 × 105 M−1·min−1. This truncated PGT domain thus has comparable activity to the most efficient PGT studied to date, full length E. coli PBP1B (16, 21, 22), which contains the transmembrane helix and the TP domain. Activity was not detectable for either the E83 or the D84 mutant (E83A, E83Q, D84A, and D84N) under our standard assay conditions, which can only detect turnover greater than 0.04 min−1; however, turnover was observed for D84N, but not E83Q, at high enzyme concentrations and extended reaction times. In contrast, catalytic activity was readily detected for the H90A and K153S mutants under standard assay conditions, although the reaction rates for both were reduced ≈5-fold compared with the WT enzyme. These results suggest that E83, and to a lesser extent D84, play important roles in the catalytic mechanism. Consistent with our findings, Terrak et al. (16) have identified the invariant glutamate residue in conserved sequence motif 1 as critical for catalysis by the PGT domain of E. coli PBP1B.
Carboxylate residues play important roles in the hydrolysis of peptidoglycan by lysozymes and lytic transglycosylases. For example, E19 in λ-lysozyme is proposed to protonate the glycosidic bond oxygen of the NAG leaving group (15). The structural resemblance of the A. aeolicus PGT domain, which catalyzes the formation of glycosidic bonds in peptidoglycan, to λR, which catalyzes their breakdown, makes it tempting to speculate that E83 may function to deprotonate the C4 hydroxyl of the attacking NAG moiety. Because the precise positions of the substrates in the active site cleft are as yet unknown, it is possible that E83 could play other roles, e.g., helping to stabilize the incipient oxocarbenium ion intermediate.
Another feature of the structure that is worthy of comment is an exposed hydrophobic patch comprising residues in helix α4 and the loop preceding helix α2. We propose that this hydrophobic patch, which is conserved in PGTs, is an interaction site for the lipid chains of the substrates. Consistent with the suggestion that there is a recognition site for at least the first part of the lipid chain on the substrates, we have established that the A. aeolicus PGT domain, as well as other PGT domains, polymerizes only substrates that, like the natural substrate, contain cis-allylic polyprenyl groups (23). The hydrophobic patch leads into the active site, suggesting a mechanism in which substrates enter the active site with their lipid chains interacting with this patch. In the crystal, the side chain of the CHAPS molecule (a bile acid derivative) penetrates into the active site via an approach from the hydrophobic funnel (Fig. 2B). This side chain, which is hydrophobic, terminates in a negatively charged moiety and may mimic part of the diphospholipid chain on the substrate.
A Model for Processive Glycosyl Transfer.
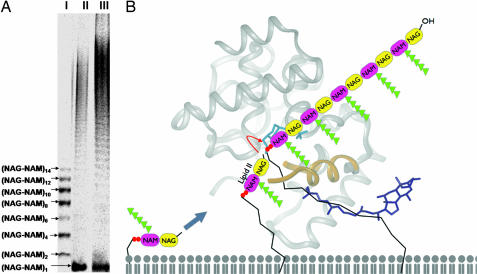
Glycosyl transfer by PGTs is thought to proceed by elongation at the reducing end of the growing polymer (24, 25). It has also been suggested that PGTs are processive, meaning that they catalyze multiple rounds of coupling without releasing the elongating product; however, no definitive evidence for processivity has been presented. Using a gel electrophoresis assay that enables separation of products to single disaccharide (NAG-NAM) resolution, we find that the A. aeolicus PGT domain and an E. coli ortholog, PBP1A, make glycan chains up to at least 40 disaccharides in length (Fig. 5A). Under steady-state conditions, a ladder of products is observed with no accumulation of short products. Because we have previously established that elongated glycan strands such as Lipid IV are poor substrates for PGTs compared with Lipid II (26), and thus would not be expected to rebind and react faster than Lipid II, this distribution is consistent with a processive mechanism in which elongation occurs without release of the growing polymer chain rather than a distributive mechanism in which product release occurs before the next reaction cycle. The topology of the active site cleft, combined with the location of the two carboxylates, allows us to propose a model in which the elongating glycan chain (the glycosyl donor) is bound so that the reacting end of the molecule is anchored near E83/D84 behind the flap that folds over the cleft, whereas the lipid chain extends down past the hydrophobic patch and into the membrane (Fig. 5B). The position of the bound Hepes molecule in the PGT structure is predicted to coincide approximately with the location of the reducing end NAM moiety of the growing chain. The glycosyl acceptor (Lipid II) binds in the more open side of the cleft with the NAG moiety directed toward the glycosyl donor. After coupling, the flap prevents dissociation of the glycan product, and the elongated glycan chain shifts in the cleft so that the new reducing terminus shuttles into the active site. It has been noted that clamps or flaps that prevent product release are one way in which enzymes can achieve a degree of processivity (27). Even so, we find it remarkable that such a small domain can processively couple Lipid II units to make long glycan chains.
Fig. 5.
The PGT domain acts as a processive glycosyltransferase. (A) Products of the PGT reaction separated by 9% tricine-SDS/PAGE. Lane I, marker lane showing (NAG-NAM)2n diphospholipid oligomers. Lane II, 0.8 μM E. coli PBP1A incubated with 8 μM ([14C]-GlcNAc)-heptaprenyl Lipid II (LII*, specific activity 288 μCi/μmol) for 5 min. Lane III, 0.03 μM A. aeolicus PBP1A (PGT domain only) incubated with 10 μM LII* at 55°C for 1 h. The LII* starting material is designated (NAG-NAM)1 in the figure. (B) A model for processive glycosyltransfer by PGT. The protein structure, represented as a ribbon and oriented to the membrane as described in the text, is shown in gray. The flap that occludes the active site is shown in yellow, with the broken ends of the chain indicating the missing residues. The Hepes and CHAPS molecules are shown in light and dark blue, respectively. The diphosphosphate moieties on Lipid II and the growing polymer chain are represented as two orange ovals, with the attached lipid represented by a black zig-zag.
Conclusion
We have reported here the previously undescribed structure of a peptidoglycan glycosyltransferase domain involved in forming the NAM-β(1,4)-NAG glycosidic linkages of bacterial peptidoglycan. PGTs use diphospholipid donors rather than nucleotide-sugar donors and the structure of this A. aeolicus PGT domain is quite different from the >100 nucleotide-sugar glycosyltransferase structures that have been reported (28), which all contain some variant of an α/β open sheet motif (a Rossmann-like fold). The PGT domain is almost completely α-helical and resembles the lysozyme-fold family of glycosidases more than it resembles any known glycosyltransferases. This resemblance may have implications for inhibitor design. The PGT structure reported here suggests a model for how processivity is achieved and can guide the design of experiments to test the role of various structural features in the glycosyltransfer reaction. Efforts to obtain cocomplexes with bound substrates are underway.
Materials and Methods
Sample Preparation.
The mrcA gene encoding PBP1A was PCR-amplified from purified A. aeolicus VF5 genomic DNA and subcloned into pET24b(+) at the NheI and XhoI restriction sites to produce pET24b:mrcA, which was used as a parent for subsequent cloning. Genes encoding only PGT domain constructs ΔPBP1A[29-243], ΔPBP1A[51-243], and ΔPBP1A[67-243] were PCR-amplified from pET24b:mrcA and subcloned into pET48b(+) at the BamHI and XhoI restriction sites. PGT fusion proteins containing N-terminal Trx-His6 tags were expressed in E. coli and purified by nickel affinity chromatography. Tags were cleaved by proteolysis with His6-tagged HRV3C (1 unit of protease per 30 μg of protein) at 4°C for 16 h while dialyzing into buffer A (20 mM Tris·HCl/500 mM NaCl, pH 7.0) containing 0.5% CHAPS, each protein solution was incubated with Ni-NTA resin for 2 h at 4°C and filtered, and the filtrate was loaded onto a Superdex 200 prep-grade size-exclusion column and eluted with buffer A containing 0.5% CHAPS. Tag-free protein was collected, concentrated to 7.5 mg/ml, and stored at −80°C.
Mutagenesis.
Site-directed mutagenesis was performed by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) using the construct producing ΔPBP1A[29-243] as a template. The appropriate primers for introducing mutations E83A, E83Q, D84A, D84N, H90A, and K153S are included in SI Table 1.
Enzyme Assays.
Kinetic parameters were obtained for the WT PGT domain by measuring reaction rates in assay buffer (50 mM Hepes, pH 7.5/10 mM CaCl2/20% DMSO) at varying concentrations (0.5–20 μM) of ([14C]-GlcNAc)-heptaprenyl Lipid II (LII*, 288 μCi/μmol) (1 Ci = 37 GBq), as described by Chen et al. (21). Standard reactions for the mutants (5 μl each) were carried out in nonstick PCR tubes containing 60 nM enzyme, assay buffer, and 4 μM LII*. Reactions were kept on ice before initiation with a rapid temperature ramp to 55°C in a PCR cycler which is 12°C below the optimal reaction temperature for the PGT domain. If no turnover was observed under these conditions, reactions were repeated with 2 μM enzyme and reaction times of 30 min. Glycan chain sizes were evaluated after separation of products on a 9% SDS-polyacrylamide gel as described in SI Materials and Methods.
Crystallization, Data Collection, and Structure Determination.
Crystals of ΔPBP1A[51–243] were obtained by the hanging-drop vapor-diffusion method at 22°C by mixing 1 μl of protein sample (7.5 mg/ml protein in 20 mM Tris, pH 7.0/0.5 M NaCl/0.5% CHAPS) with 1 μl of well solution [6% (wt/vol) PEG 6,000/100 mM Hepes, pH 7.5]. Crystals of selenomethionine-labeled (SeMet) protein were grown under the same conditions. Crystals were cryoprotected by serial transfer in four steps to 25% (vol/vol) glycerol, 6% (wt/vol) PEG 6,000, 100 mM Hepes, pH 7.5, and flash-frozen in liquid nitrogen.
Complete data sets for both native and SeMet crystals were collected at 100 K at the ID-24 beamline of the Advance Photon Source (Argonne National Laboratories). SAD data were collected at the peak wavelength. Data were processed and scaled with HKL2000 (29). Both native and SeMet crystals belonged to space group I222. Two selenium sites were located, and SAD phases were calculated with BnP interface coupling Shake-and-Bake and PHASES (30). Density modification and automatic model building were carried out by RESOLVE (31) and provided a partial polyAla model. The model was completed by interactive rounds of manual fitting in COOT 0.2 (32) and refinement in CNS 1.2 (33). Data collection and refinement statistics are included in SI Table 2.
Supplementary Material
Acknowledgments
We thank Robert Huber (University of Regensburg, Regensburg, Germany) for providing A. aeolicus VF5 genomic DNA, Andrew Wang (Harvard University, Cambridge, MA) for providing E. coli PBP1A, and Bing Chen for useful discussions. This work was supported by National Institutes of Health Grant GM076710.
Abbreviations
- PGT
peptidoglycan glycosyltransferase
- PBP
penicillin-binding protein
- NAG
N-acetylglucosamine
- NAM
N-acetylmuramoyl-pentapeptide.
Footnotes
The authors declare no conflict of interest.
Data deposition: The coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 2OQO).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701160104/DC1.
References
- 1.van Heijenoort J. Nat Prod Rep. 2001;18:503–519. doi: 10.1039/a804532a. [DOI] [PubMed] [Google Scholar]
- 2.Walsh CT. Antibiotics: Actions, Origins, Resistance. Washington, DC: Am Soc Microbiol; 2003. [Google Scholar]
- 3.Smith CA. J Mol Biol. 2006;362:640–655. doi: 10.1016/j.jmb.2006.07.066. [DOI] [PubMed] [Google Scholar]
- 4.Bugg TD, Lloyd AJ, Roper DI. Infect Disord Drug Targets. 2006;6:85–106. doi: 10.2174/187152606784112128. [DOI] [PubMed] [Google Scholar]
- 5.Hu Y, Chen L, Ha S, Gross B, Falcone B, Walker D, Mokhtarzadeh M, Walker S. Proc Natl Acad Sci USA. 2003;100:845–849. doi: 10.1073/pnas.0235749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. FEMS Microbiol Rev. 2006;30:673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 7.Lovering AL, De Castro L, Lim D, Strynadka NC. Protein Sci. 2006;15:1701–1709. doi: 10.1110/ps.062112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goffin C, Ghuysen JM. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostash B, Walker S. Curr Opin Chem Biol. 2005;9:459–466. doi: 10.1016/j.cbpa.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Terrak M, Nguyen-Disteche M. J Bacteriol. 2006;188:2528–2532. doi: 10.1128/JB.188.7.2528-2532.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Offant J, Michoux F, Dermiaux A, Biton J, Bourne Y. Biochim Biophys Acta. 2006;1764:1036–1042. doi: 10.1016/j.bbapap.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Contreras-Martel C, Job V, Di Guilmi AM, Vernet T, Dideberg O, Dessen A. J Mol Biol. 2006;355:684–696. doi: 10.1016/j.jmb.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 13.Macheboeuf P, Di Guilmi AM, Job V, Vernet T, Dideberg O, Dessen A. Proc Natl Acad Sci USA. 2005;102:577–582. doi: 10.1073/pnas.0407186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evrard C, Fastrez J, Declercq JP. J Mol Biol. 1998;276:151–164. doi: 10.1006/jmbi.1997.1499. [DOI] [PubMed] [Google Scholar]
- 15.Leung AK, Duewel HS, Honek JF, Berghuis AM. Biochemistry. 2001;40:5665–5673. doi: 10.1021/bi0028035. [DOI] [PubMed] [Google Scholar]
- 16.Terrak M, Ghosh TK, van Heijenoort J, Van Beeumen J, Lampilas M, Aszodi J, Ayala JA, Ghuysen JM, Nguyen-Disteche M. Mol Microbiol. 1999;34:350–364. doi: 10.1046/j.1365-2958.1999.01612.x. [DOI] [PubMed] [Google Scholar]
- 17.Charpentier X, Chalut C, Remy MH, Masson JM. J Bacteriol. 2002;184:3749–3752. doi: 10.1128/JB.184.13.3749-3752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertsche U, Breukink E, Kast T, Vollmer W. J Biol Chem. 2005;280:38096–38101. doi: 10.1074/jbc.M508646200. [DOI] [PubMed] [Google Scholar]
- 19.Holtje JV. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Pedro MA, Schwarz H, Koch AL. Microbiology. 2003;149:1753–1761. doi: 10.1099/mic.0.26125-0. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Walker D, Sun B, Hu Y, Walker S, Kahne D. Proc Natl Acad Sci USA. 2003;100:5658–5663. doi: 10.1073/pnas.0931492100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz B, Markwalder JA, Seitz SP, Wang Y, Stein RL. Biochemistry. 2002;41:12552–12561. doi: 10.1021/bi026205x. [DOI] [PubMed] [Google Scholar]
- 23.Ye XY, Lo MC, Brunner L, Walker D, Kahne D, Walker S. J Am Chem Soc. 2001;123:3155–3156. doi: 10.1021/ja010028q. [DOI] [PubMed] [Google Scholar]
- 24.van Heijenoort J. Glycobiology. 2001;11:25–36. doi: 10.1093/glycob/11.3.25r. [DOI] [PubMed] [Google Scholar]
- 25.Fraipont C, Sapunaric F, Zervosen A, Auger G, Devreese B, Lioux T, Blanot D, Mengin-Lecreulx D, Herdewijn P, Van Beeumen J, et al. Biochemistry. 2006;45:4007–4013. doi: 10.1021/bi051055m. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Fechter E, Wang T-S, Barrett D, Walker S, Kahne D. J Am Chem Soc. 2007 Feb 27; doi: 10.1021/ja069060g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breyer WA, Matthews BW. Protein Sci. 2001;10:1699–1711. doi: 10.1110/ps.10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breton C, Snajdrova L, Jeanneau C, Koca J, Imberty A. Glycobiology. 2006;16:29–37. doi: 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Methods in Enzymology. In: Carter CW, Sweet RM, editors. Macromolecular Crystallography, Part A. Vol 276. New York: Academic; 1997. pp. 307–326. [Google Scholar]
- 30.Weeks CM, Blessing RH, Miller R, Mungee R, Potter SA, Rappleye J, Smith GD, Xu H, Furey W. Z Kristallogr. 2002;217:686–693. [Google Scholar]
- 31.Terwilliger TC, Berendzen J. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.