Abstract
Each day, approximately 0.5–0.9 l of water diffuses through (primarily) aquaporin-1 (AQP1) channels in the human choroid plexus, into the cerebrospinal fluid of the brain ventricles and spinal cord central canal, through the ependymal cell lining, and into the parenchyma of the CNS. Additional water is also derived from metabolism of glucose within the CNS parenchyma. To maintain osmotic homeostasis, an equivalent amount of water exits the CNS parenchyma by diffusion into interstitial capillaries and into the subarachnoid space that surrounds the brain and spinal cord. Most of that efflux is through AQP4 water channels concentrated in astrocyte endfeet that surround capillaries and form the glia limitans. This report extends the ultrastructural and immunocytochemical characterizations of the crystalline aggregates of intramembrane proteins that comprise the AQP4 “square arrays” of astrocyte and ependymocyte plasma membranes. We elaborate on recent demonstrations in Chinese hamster ovary cells of the effects on AQP4 array assembly resulting from separate vs. combined expression of M1 and M23 AQP4, which are two alternatively spliced variants of the AQP4 gene. Using improved shadowing methods, we demonstrate sub-molecular cross-bridges that link the constituent intramembrane particles (IMPs) into regular square lattices of AQP4 arrays. We show that the AQP4 core particle is 4.5 nm in diameter, which appears to be too small to accommodate four monomeric proteins in a tetrameric IMP. Several structural models are considered that incorporate freeze-fracture data for submolecular “cross-bridges” linking IMPs into the classical square lattices that characterize, in particular, naturally occurring AQP4.
Keywords: cross-bridges, furrows, FRIL, plastic deformation, water homeostasis
Precise control of water homeostasis is critical in the brain and spinal cord because even minor changes in water metabolism may result in brain edema and rapid development of potentially fatal compressive forces within the rigid encasement of the skull (King and Agre, 1996). Moderate hyponatremia may result in failure of synaptic transmission or in altered neuronal excitability (Kandel et al., 1992; Jefferys, 1995; Nielsen et al., 1997) and is a contributing factor in the pathogenesis of epilepsy (Dudek et al., 1990; Roper et al., 1992), whereas severe hyponatremia contributes to medical complications that arise from brain and spinal cord trauma, brain tumors, neonatal hydrocephalus, acute water intoxication, and drowning (Johnson and Loewy, 1990; King and Agre, 1996; Nielsen et al., 1997).
Rates of secretion vs. resorption of cerebrospinal fluid (CSF) is a major controlling factor in osmoregulation of the CNS. The aqueous component of CSF passes primarily through aquaporin-1 (AQP1) in the choroid plexus epithelium (Nielsen et al., 1993), with the remainder arising from metabolism of glucose, from simple transmembrane diffusion through cell membrane lipid bilayers [reviewed by Agre et al. (2002)], from facilitated transport via coupled ion exchangers [reviewed by Amiry-Moghaddam and Ottersen (2003)], and possibly by diffusion through as-yet-unidentified water channels. In turn, an equal volume of CSF is transported across astrocyte endfeet and into CNS parenchymal capillaries and subarachnoid space by molecular mechanisms that, until recently, were completely unknown.
Identification of AQP4 in “square arrays” of astrocytes and ependymocytes
Early freeze-fracture studies revealed abundant “square arrays” of intramembrane particles (IMPs) in freeze-fractured plasma membranes of astrocyte endfeet (Landis and Reese, 1974; Hatton and Ellisman, 1981, 1982; Landis, 1981) and in fast-twitch skeletal myofibers (Rash and Ellisman, 1974; Ellisman et al., 1976, 1978; Hudson et al., 1982; Frigeri et al., 1995) [These distinctive IMP arrays were originally called “rectilinear arrays” (Kreutziger, 1968), but subsequently have been called “assemblies” (Landis and Reese, 1974), “square arrays” (Rash and Ellisman, 1974; Friend, 1987), “rectangular arrays” (Rash et al., 1974), “orthogonal arrays” (Rash and Ellisman, 1974; Hatton and Ellisman, 1981, 1982), “orthogonal assemblies” (Anders and Brightman, 1979), “orthogonally arranged particles” (or “OAPs”; Verbavatz et al., 1994; Wolburg, 1995), “tetragonally arranged particles” (Zampighi et al., 1989), and even “type III gap junctions” (Staehelin, 1972 [subsequently retracted (Rash et al., 1974)]). Because “rectilinear” means “straight lines,” and “orthogonal,” “tetragonal,” and “rectangular” are terms equally applicable to any rectangular lattice, regardless of relative lengths of the orthogonal sides, we continue to use the geometric term denoting an array composed of equilateral rectangles, i.e. “square arrays.”]. Following initial identification of the class of proteins now called “aquaporins” (Agre et al., 1993, 2002; Crispeels and Agre, 1994), light and electron microscopic immunocytochemical studies (Verbavatz et al., 1994; Frigeri et al., 1995) suggested that the “mercurial insensitive water channel” (later renamed AQP4) closely paralleled the distribution of “OAPs” that were visualized in freeze-fracture replicas of astrocytes and ependymal cells in the mammalian CNS, as well as in skeletal muscle sarcolemmas and the basolateral plasma membranes of glandular epithelial cells. Nielsen et al. (1997) developed a series of antibodies to AQP4, which they used to show that AQP4 has a highly polarized distribution in the CNS, with greatest abundance in astrocyte endfeet that surround parenchymal capillaries and that form the glia limitans. They suggested that this polarized distribution of AQP4 provided a major pathway for equilibration of water in the CNS. However, unambiguous correlation of AQP4 with square arrays was by no means certain because those same membranes also contained abundant potassium “leak channels” [including particularly Kir4.1 (Nagelhus et al., 1999; Higashi et al., 2001)], as well as numerous unidentified 8 nm “tubular” IMPs in the same membranes (Rash et al., 1997). Conversely, apparently identical square arrays (by whatever name) also have been found in several tissues that either lacked AQP4 or that had not been shown to contain AQP4, including “tetragonally arranged particles” in lens fibers (Zampighi et al., 1989), “orthogonal arrays” in tracheal pneumocytes (Gordon, 1985; Bartels and Miragall, 1986) and gastric parietal cells (Bordi et al., 1986), and continuous sheets of square lattices in the plasma membranes of cells that comprise the filter chambers of sap-sucking and blood-sucking insects (Hubert et al., 1989; Duchesne et al., 2003). Thus, it was essential to determine if AQP4 is localized to square lattices of IMPs in astrocyte plasma membranes, to the tubular IMPs, or to an unrecognized class of IMPs near the square arrays (Warth et al., 2004).
The hypothesis that AQP4 is the primary component of square arrays is supported by:
expression of AQP4 cDNA in Chinese hamster ovary (CHO) cells, which resulted in the de novo appearance of abundant and unusually large square lattices of IMPs (Yang et al., 1996; Van Hoek et al., 1998);
freeze-fracture immunocytochemistry showing immunogold labeling of OAPs in membrane vesicles from AQP4-expressing CHO cells (Yang et al., 1996);
mice with targeted deletion of the AQP4 gene (“AQP4 knockout mice”) lacked square arrays in the plasma membranes of astrocyte endfeet, sarcolemmas of striated myofibers, and proximal tubules and descending loops of Henle in the kidney (Verbavatz et al., 1997), places which normally have abundant square arrays; and
freeze-fracture replica immunogold labeling (FRIL) of individual square arrays in astrocyte endfeet and ependymocyte lateral plasma membranes (Rash et al., 1998; Rash and Yasumura, 1999).
However, such immunogold localizations have recently been challenged by Warth et al. (2004) based on their unexpected detection of AQP4 immunoreactivity in plasma membranes of parenchymal astrocytoma cells, where they assumed square arrays to be absent, and their inability to detect immunofluorescence labeling for AQP4 above “background” in astrocyte and astrocytoma plasma membranes that they presumed to contain square arrays. They concluded that AQP4 is often “dissociated from the orthogonal arrays” and stated, “In our opinion, there is only one possible interpretation of these conflicting observations: in gliomatous membranes AQP4 must occur outside the OAPs.” In possibly related observations, Kobayashi et al. (2001) reported reverse transcription-polymerase chain reaction (RT-PCR) evidence for low but detectable levels of AQP4 messenger RNA (mRNA) in endothelial cells, and Amiry-Moghaddam et al. (2004) reported immunocytochemical evidence for weak AQP4 immunogold labeling of endothelial cell plasma membranes in cerebellum, a cell type in which square arrays have not been detected. If confirmed, those data would constitute evidence that AQP4 has an alternate state in which AQP4 protein is not assembled into square arrays.
In this report, we provide additional ultrastructural and immunocytochemical characterizations confirming that AQP4 is the primary or sole intramembrane constituent of square arrays, both in astrocyte and ependymocytes of normal adult rat CNS, as well as in CHO cells transfected with two alternatively spliced variants of AQP4. In addition, we confirm that one variant of AQP4 (M1), when expressed alone and in abundance, does not form stable square arrays (Furman et al., 2003; Silberstein et al., 2004). Stereoscopic freeze-fracture images made with improved methods reveal new ultra-structural details of normal AQP4 square arrays and of arrays formed following single-expression of AQP4 variant M23 and double-expression of M1 plus M23. These arrays exhibited submolecular cross-bridges linking most or all AQP4 IMPs and evidence for a centrally located water-filled channel or “vestibule” within each AQP4 particle. Substantial interstitial “node” material was demonstrated between each AQP4 particle within the arrays, thereby reducing the cross-sectional area within each AQP4 IMP. Finally, based on constraints imposed by the smaller size of AQP4 IMPs seen in replicas made with thinner platinum films, additional ultrastructural models for AQP4 assembly are proposed and compared.
EXPERIMENTAL PROCEDURES
Preparation of samples for freeze fracture and FRIL
Three adult male and one adult female Sprague–Dawley rats and one adult male Wistar rat (104–550 g) were anesthetized with ketamine (90 mg/kg) and xylazine (8 mg/kg) administered by i.p. injection. All experiments were approved by the Colorado State University Animal Care and Use Committee (ACUC) and were conducted according to the Principles of Laboratory Animal Care (NIH publication No. 86-23, Rev. 1985). Experiments were designed and conducted so as to minimize numbers of animals used and stress to animals. For conventional ultrastructural analysis, rats were chemically fixed via transcardiac perfusion with 2.5% glutaraldehyde in 0.15 M Sorensen’s phosphate buffer (pH 7.4; Hudson et al., 1981; Rash et al., 1995, 1996). For FRIL, animals were chemically fixed by whole-body perfusion (10 min) with 1% or 2% formaldehyde in phosphate buffer. [The irreversible cross-linking of proteins in glutaraldehyde-fixed tissues prevents effective tissue solubilization by sodium dodecyl sulfate (SDS) detergent, whereas formaldehyde fixation allows SDS washing, as is required in FRIL (Fujimoto, 1995, 1997; Rash and Yasumura, 1999).] Immediately following perfusion fixation, the animals were decapitated, consistent with recommendations of the Capital Panel on Euthanasia of the American Veterinary Association. After laminectomy, the brains and spinal cords were removed, and selected regions (suprachiasmatic, supraoptic nucleus, cerebellum and cervical spinal cord) were cut into 100 μm-thick slices using a Lancer Vibratome 3000 (Technical Products International, St. Louis, MO, USA) equipped with a refrigerated stage operated at 4 °C.
Freeze fracture and freeze etch
Tissue slices from rat CNS were either frozen without cryoprotection (for freeze etch) or infiltrated with 30% glycerol (cryoprotectant for conventional freeze fracture and for FRIL), mounted on aluminum “Slammer” supports (Heuser et al., 1981) and frozen by contact with a −195 °C “metal mirror” (Heuser et al., 1981; Phillips and Boyne, 1984). CHO cell cultures that had been transfected with AQP4 variants M1 and M23 cDNA [transfected cells provided by Dan Gorelick and Peter Agre, as described (Furman et al., 2003)] were pipetted onto gold specimen supports and frozen by plunging into a slurry of propane/ethane. For improved resolution, samples were fractured at −170 °C to −180 °C in a JEOL/RMC RFD 9010c freeze-fracture machine (RMC, Inc., Tucson, AZ, USA) and replicated by pre-coating with 0.5–1 nm of vaporized carbon (Winkler et al., 2002) and 1–1.5 nm of platinum/carbon (i.e. six to nine atom layers thick), followed by coating with 20–30 nm of carbon (Rash et al., 1997). For freeze-etch experiments, non-cryoprotected samples were fractured at −100 °C, etched for 1 min at −100 °C, cooled rapidly to −150 °C, and shadowed with 2–2.5 nm of platinum, and coated with carbon as above. Replicated (but still frozen) samples were bonded to gold 200-mesh “index” grids using a drop of 2–3% Lexan polycarbonate plastic dissolved in dichloroethane (Rash et al., 1995, 1996). Replicated samples were thawed and photomapped with a Molecular Dynamics Sarastro 2001 inverted laser scanning confocal microscope (Sunnyvale, CA, USA).
FRIL
After photo-mapping, conventional (unlabeled) freeze-fracture and freeze-etch replicas were cleaned with Clorox bleach, followed by chrome-sulfuric acid. FRIL samples were washed for ca. 29 h at 35°–50 °C in 2.5% SDS detergent (pH 8.5) to remove bulk tissue components not directly absorbed to the platinum/carbon replica (Fujimoto, 1995). Samples were rinsed in blocking buffer (Dinchuk et al., 1987) and immunogold labeled (Fujimoto, 1995; Rash and Yasumura, 1999; Furman et al., 2003) using affinity-purified antibodies to AQP4 [provided by Søren Nielsen, University of Aarhus, DK (Nielsen et al., 1997) or obtained from Chemicon (Temecula, CA, USA)] and to connexin-26 (Cx26) and Cx30 [anti-Cx26 from Zymed Laboratories Incorporated (South San Francisco, CA, USA; Cx30; courtesy of James Nagy, University of Manitoba (Nagy et al., 2001)]. The anti-Cx26 antibody (catalogue no. 51–2800) does not cross-react with Cx30 (Nagy et al., 2001). Secondary goat anti-rabbit IgG conjugated to 10 nm gold beads (originally from Chemicon but no longer available from them) and goat anti-rabbit IgG conjugated to 12- and 18-nm gold beads (Jackson ImmunoResearch Laboratories, Westgrove, PA, USA) were used for FRIL. Replicas serving as “controls” were incubated with buffer containing no primary antibody to AQP4 but containing antibodies to other membrane proteins, including Cx30 and Cx26 (Rash and Yasumura, 1999; Rash et al., 2001). To permit examination of labeled samples by TEM, the electron-opaque Lexan support film was removed by immersion of replicas in dichloroethane for 2–4 h.
Electron microscopy
Replicas were examined at 100 kV in a JEOL 2000 EX-II TEM. Selected areas were photographed as stereoscopic pairs having an 8° included angle, and all images were analyzed stereoscopically. TEM magnifications were calibrated using “Mag*i*Cal” (Electron Microscopy Sciences, Fort Washington, PA, USA) and conventional diffraction grating magnification calibration standards. (Some images were not used for measurements because the square arrays were tilted with respect to the image plane, or because the z-axis compensation was not properly adjusted.)
Locations of areas photographed by TEM were plotted on the original light microscopic photomaps of the same slices (Rash et al., 1995, 1996, 1997). Data in this report are derived from > 1000 electron micrographs containing > 10,000 AQP4 square arrays, including many hundreds of immunogold-labeled square arrays. In labeled samples, “labeling efficiency” [defined as the number of gold beads vs. the number of subunit particles in all arrays in the quantified area (Rash and Yasumura, 1999)] was from approximately 1:100 in early samples to approximately 1:3 in our more recent samples. (At 1:100, ca. 35% of arrays are labeled, whereas at 1:3, >90% of arrays are labeled.) Electron microscope images were captured using an ArtixScan 2000f digital scanning device (Microtek, Carson, CA, USA), and prepared using Photoshop 7.01 (Adobe systems, San Jose, CA, USA).
Replica interpretation
By convention, most low magnification freeze-fracture images of tissue are printed with “white shadows” oriented up, generating the impression that non-shadowed “white” areas constitute the essential elements of the image. However, in high magnification images of replicated transmembrane proteins, “white shadows” generate the false presumption that the IMPs correspond to the bright, uncoated “shadow cap” rather than to the dark, platinum-coated particles that cast the white shadows. Viewing white-shadow images is equivalent to viewing photographic “negatives,” which are notoriously difficult to interpret. This can be particularly misleading when, for example, the local shadowing angle is low, which results in long white shadows that may be misinterpreted as elongate or “string-like” particles. (See comparison of “black” vs. “white shadow” images in Results.) Consequently, for proper interpretation of molecular detail, selected high magnification images are also presented with black shadows (i.e. platinum is white), according to the original convention of Steere (1957) and Steere et al. (1980), which yields replicated IMPs as if illuminated with white light and, therefore, having “lifelike” black shadows. Thus, in these unidirectionally shadowed images, the white platinum coat delineates (and possibly enlarges by up to 1 nm) the true outline of the molecule or molecular subunit.
To facilitate more accurate image interpretation, many of the images are presented as stereoscopic pairs, which allows three-dimensional details to be discriminated from phase contrast artifacts of near-focus in electron micrographs (Sjostrand, 1967; Steere et al., 1980; Rash and Yasumura, 1992), as well as to confirm that the gold labels are on the tissue-side of the replica, thereby representing specific labeling or “signal.” Gold labels on the carbon-coated side (none shown) necessarily represent non-specific labeling or “noise.” To facilitate interpretation and description, “colorization” of selected images was done by using the “MagicWand” (“tolerance”= 35) and “PaintBucket” functions in Photoshop to identify and to color areas of the replica having similar gray scale values (i.e. extent of colorized areas determined by Photoshop program, rather than by investigator bias and direct micro-area selection).
Image enhancement by rotary reinforcement
Selected examples of AQP4 P-face IMPs and E-face pits were enlarged to 1,850,000× and printed with four-fold “Markham rotational reinforcement” (Markham et al., 1963) using Photoshop. To “average out” and thereby reduce random photographic and replica “noise,” four identical images were successively superimposed and rotated 0°, 90°, 180°, and 270°. For summation, successive transparency values were 100%, 50%, 33% and 25%; the final image was merged; and image quality was adjusted using “levels” and “brightness/contrast.” Separate images were rotated around the central “dimple” found in P-face IMPs, the “pegs” in E-face pits, and the “nodes” separating E-face pits. [This report uses the internationally recognized nomenclature (Branton et al., 1975) for designating the two fracture “faces” created when membranes are split: “P-face” designates the “protoplasmic” leaflet (i.e. the replicated membrane leaflet remaining in contact with cyto-plasm, nucleoplasm, or mitochondrial plasm); whereas “E-face” designates the extraplasmic leaflet (i.e. the leaflet remaining in contact with the extracellular space, extranuclear space, or extra-mitochondrial space)]. To demonstrate relative complementarity of P-face IMPs and E-face pits, E- and P-face images from each of these reinforcement techniques were compared using reversed photographic contrast. Rotational reinforcement at three-fold, five-fold and six-fold rotation resulted in almost complete loss of image detail outside the central pixels (blurred images not shown). We chose not to use either computer software image averaging programs or fast Fourier transform analysis because the steps in those methods are not transparent to the viewer. Rather, the classical “Markham rotational reinforcement” method (Markham et al., 1963) was used because all image details are retained and the final image is directly comparable to the initial image.
RESULTS
FRIL of AQP4 in astrocyte endfeet
In our initial FRIL experiments to identify the one or more classes of IMP corresponding to AQP4 (Rash et al., 1998), analysis of astrocyte endfoot plasma membranes revealed abundant “square arrays” of P-face IMPs in regular square lattices having periodicities of 6.5–6.8 nm. Immunogold beads were present almost exclusively near square arrays in plasma membranes of astrocytes [Fig. 1A, modified from Rash et al. (1998)] and ependymocytes (Fig. 1B). Gold labels were not detected above the low nonspecific “background” on plasma membranes of any other cell type, including neurons, oligodendrocytes, or vascular endothelial cells. Less than 5% (i.e. the practical limit of detectability) of immunogold labels were dispersed elsewhere within the astrocyte plasma membranes, suggesting that > 95% of AQP4 protein was present in square arrays. (An alternative explanation for low labeling in areas devoid of square arrays may be that monomers and small arrays of AQP4 are more readily solubilized by SDS washing than the larger arrays, and hence, are not retained for FRIL labeling.) Where the arrays were small and densely packed (Fig. 1A), antibodies to AQP4 directly labeled ca. 35% of square arrays. However, in areas of high array density, it was not possible by FRIL to assign immunogold labels to individual IMPs or to nearby arrays because of the “radius of uncertainty” of immunogold labeling (Dinchuk et al., 1987).
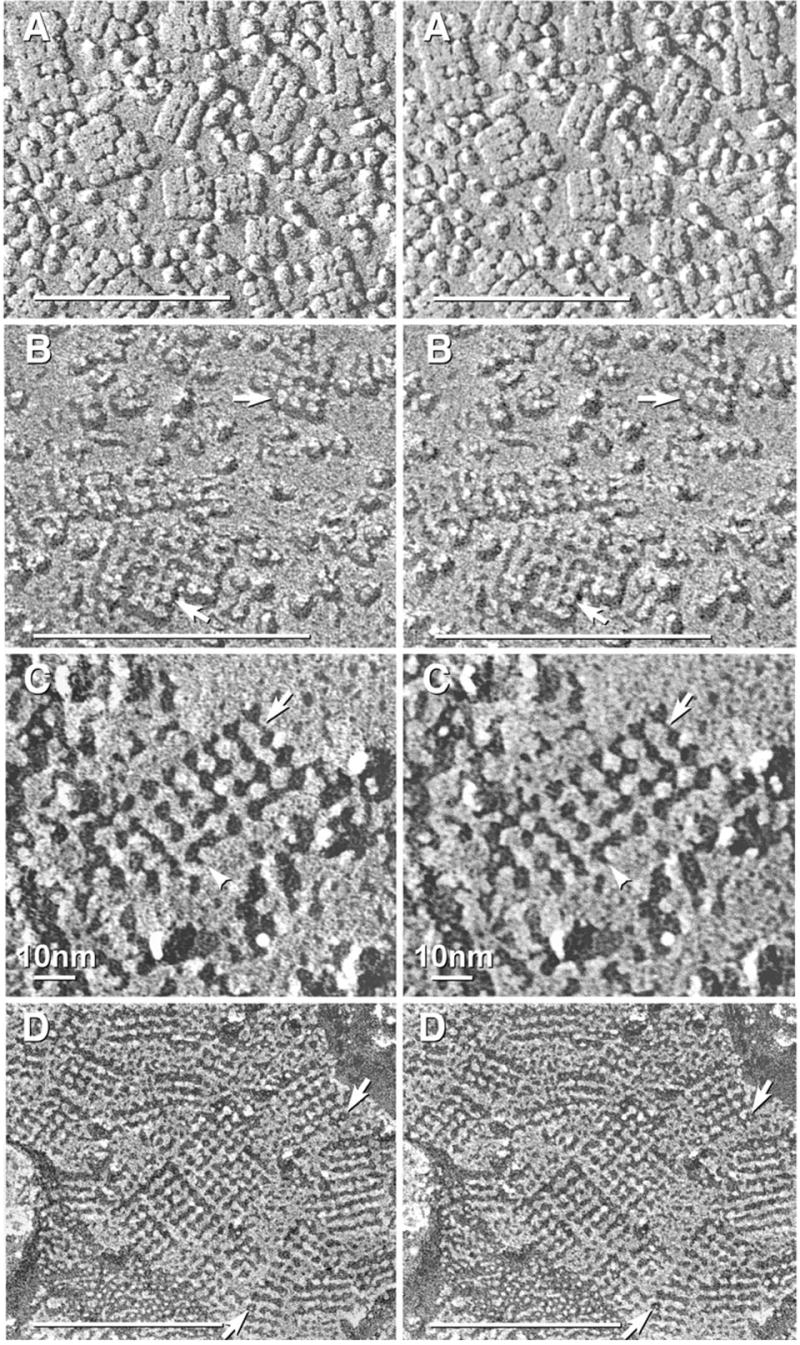
Fig. 1.
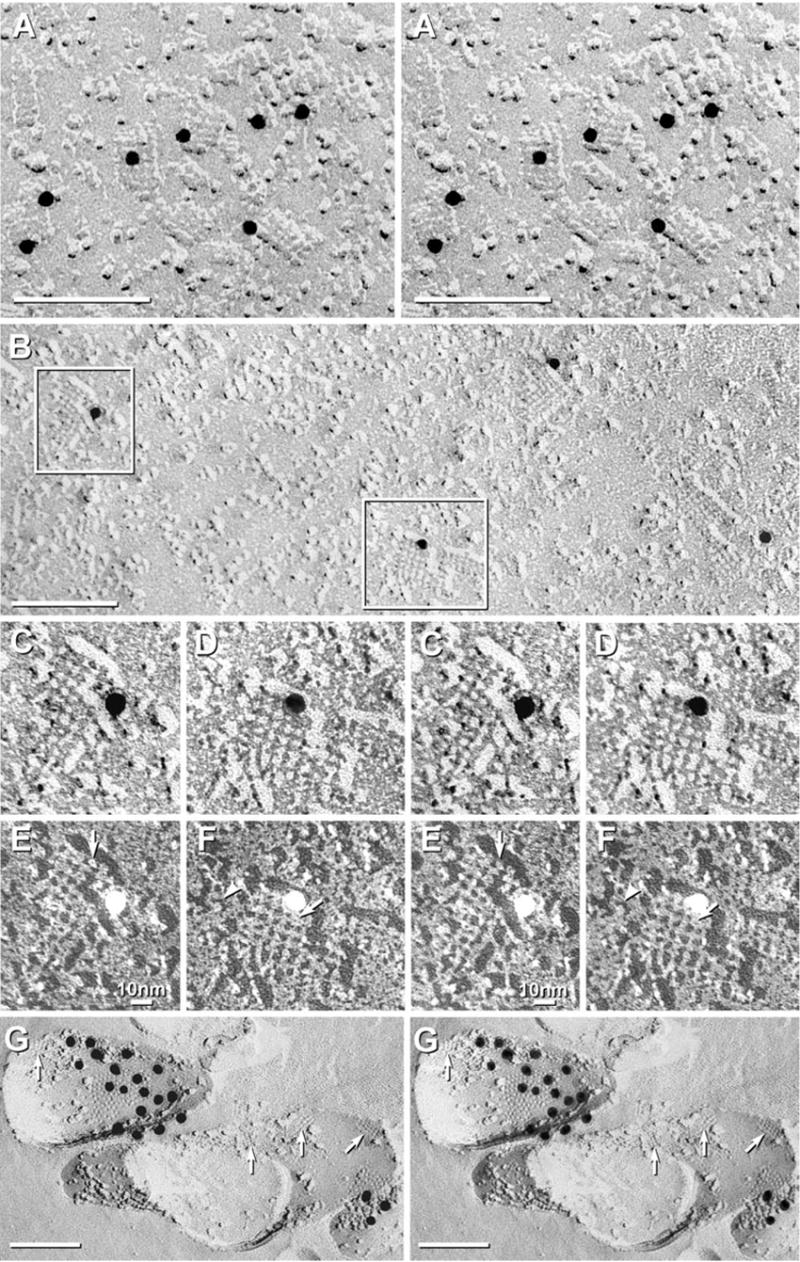
Square arrays in astrocyte endfeet of the glia limitans adjacent to the optic chiasm (A), in ependymocytes of the third ventricle near the suprachiasmatic nucleus (B–F), and in astrocyte processes in neuropil of adult rat spinal cord (G). (A) Stereoscopic image of area in which square arrays were in high density. Approximately 35% of arrays were immunogold labeled (10 nm gold beads). (B) Larger square arrays found in ependymocytes were most often labeled within the array perimeters, but occasionally, labels were 20–30 nm outside arrays (B, right side). (C–F) Higher magnification stereoscopic images of individual immunogold-labeled square arrays. The constituent IMPs within the 6.5 nm square lattices were 4.5 nm in diameter, with 1 × 2 nm cross-bridges linking adjacent IMPs. Many P-face IMPs had distinctive 1 nm central depressions or “dimples” (E, F, arrows). (G) Small-diameter astrocyte processes deep within neuropil in spinal cord of adult rat typically had multiple AQP4 square arrays. Immunogold beads labeled Cx30 (18 nm gold) and Cx26 (12 nm gold) in three gap junctions (middle gap junction is not visible at this tilt angle). In all images, unlabeled scale bars= 0.1 μm; labeled scale bars= 10 nm.
Immunogold beads were often within the perimeter of the lattices (Fig. 1A, left center), but many were at the margins of the arrays (Figs. 1C, D) or within 20–40 nm. [Displacement of membrane proteins and their labels by up to 50 nm has been traced to the formation of lipid/protein “blebs” during SDS solubilization (Rash and Yasumura, 1999).] Similar concentrations of immunogold labels also were seen at the edges of gap junctions that had been labeled for connexins (Fig. 1G; also see Rash and Yasumura, 1999; Rash et al., 2000), and this “edge effect” (whatever its cause) may account for similar peripheral distributions near isolated square arrays.
AQP4 labeling of isolated square arrays in ependymocyte plasma membranes
To address the issues of reported lack of AQP4 immunofluorescence labeling in non-endfoot regions of astrocytes (Warth et al., 2004), we employed three independent approaches: 1) FRIL examination of ependymocyte plasma membranes, which have a lower density but larger size of square arrays (Fig. 1B–F); 2) analysis of astrocyte processes in areas not containing endfeet, including astrocyte processes in regions adjacent to myelin, nerve cell bodies, and nerve terminals (Fig. 1G); and 3) freeze-fracture and FRIL examination of CHO cells in which expression of one of the two alternatively-spliced variants of AQP4 (M23) results in the formation of unusually large aggregates of square arrays (composite “rafts”) of AQP4 protein (see Fig. 2, below).
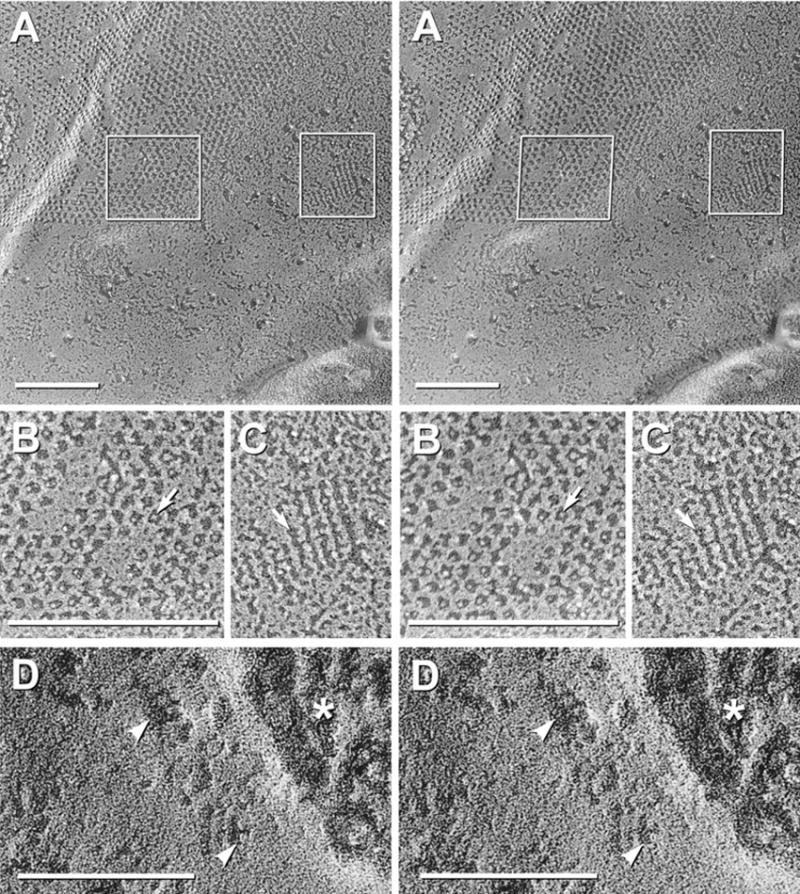
Fig. 2.
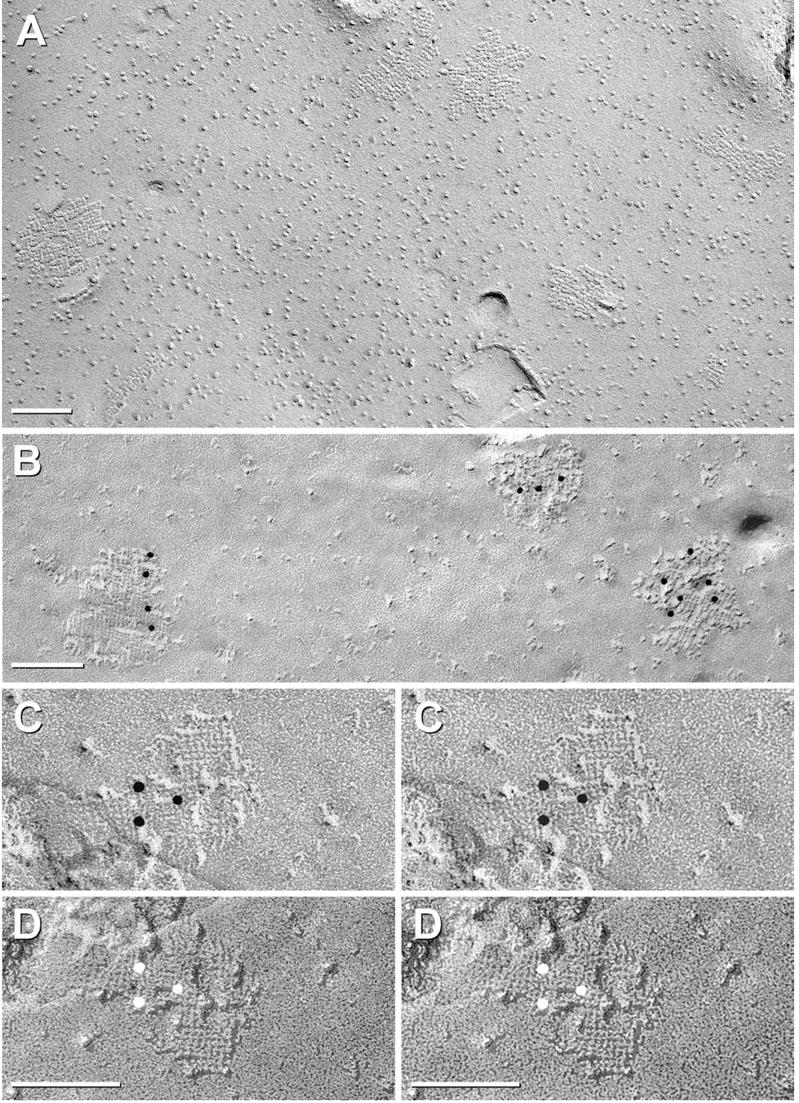
AQP4-M23 arrays in CHO cells. (A) Glutaraldehyde-fixed CHO cells expressing M23. Rafts represent large lattices of 4.5 nm-diameter IMPs, with many containing > 100 IMPs. (B–D) Formaldehyde-fixed CHO cell membranes that were immunogold labeled for AQP4. (B) Three “rafts” are labeled by four, three, and six 10 nm gold beads. (C, D) Stereoscopic images showing cross-bridges linking IMPs into a grid-like pattern. (D) Black shadow image more clearly reveals cross-bridges; IMPs are at intersections. Finer details are resolvable only at higher magnification (see Figs. 5–7, below).
In ependymocyte plasma membranes, where AQP4 immunofluorescence was low or absent, and where square arrays were few or absent (Hatton and Ellisman, 1981, 1982; Wolburg, 1995), immunogold labeling was essential for finding the isolated square arrays (Rash et al., 1997). In those areas, the square arrays frequently were larger than in astrocyte endfeet, and by FRIL, immunogold labels were found almost exclusively in close association with those large arrays (Fig. 1B–F). In black-shadow versions of the same images, the square arrays were resolved as assemblies of 4.5 nm IMPs linked by 1 × 2 nm “cross-bridges” (Fig. 1E, F), thereby forming the classical 6.5–6.8 nm square lattice spacing. The central IMP core and the cross-bridges represent those portions of AQP4 molecules projecting above/through the central plane (i.e. fracture plane) of the lipid bilayer. Occasionally, isolated 4.5 nm IMPs with one or two short lateral extensions, possibly representing unattached cross-bridges, were seen near but not incorporated into square arrays (Fig. 1F, arrowhead). Thus, FRIL allowed detection of AQP4-labeled square arrays in areas of ependymocyte membranes that light microscopic immunofluorescence and conventional freeze-fracture imaging had suggested were devoid of AQP4 or square arrays.
On a particle-by-particle basis, AQP4 arrays were labeled with approximately the same labeling efficiency as glial gap junctions labeled for connexins (Rash and Yasumura, 1999). However, exact quantification of labeling efficiency was not possible because preservation of square arrays was not optimal in formaldehyde-fixed samples, wherein some P-face arrays were so distorted as to be difficult to recognize and, hence, were unquantifiable (see Fig. 1A, upper right corner; Fig. 2B, right side). This “plastic distortion” (Steere et al., 1980) of IMPs within P-face arrays was confirmed to be excessive in some samples (and sometimes in adjacent areas of the same cell) based on comparison of P-face arrays to corresponding E-face imprints in the same replicas. In all cases, E-face imprints retained their distinctive square lattice morphology. Thus, the number of square arrays remained indirectly quantifiable based on examination of nearby E-faces (Fig. 1G). (For further details regarding plastic deformation of P-face IMPs but not E-face imprints, see section entitled “Complementarity of E-face impressions of AQP4 arrays”, below.)
In studies of more than 1000 replicas from 10 regions of brain and spinal cord, square arrays were relatively abundant in many of the fine tubular processes linking adjacent astrocytes (Fig. 1G, arrows; image obtained by Michio Morita in our laboratory), as well as in astrocyte somatic plasma membranes (see Fig. 6 in Rash et al., 1997). These processes were confirmed to be astrocytic based on: a) the presence of E-face imprints of square arrays, b) gap junctions that were immunogold labeled for Cx30 (18 nm gold beads) and Cx26 (12 nm gold beads), and c) glial fibrillary acidic protein filaments in the cytoplasm (not shown). These definitive immunocytochemical and structural markers for astrocytes are not present in any other cell type in CNS parenchyma (Rash et al., 1997; Nagy et al., 2001, 2003). Although a moderate level of square arrays was detected in many astrocyte processes in gray matter areas of brain and spinal cord, some areas of some astrocyte processes were devoid of arrays. Despite this large variability in density of square arrays, the relative amount and location of immunogold labeling of astrocyte plasma membranes correlated almost exactly with presence or absence of and relative local abundance of the arrays.
Fig. 6.
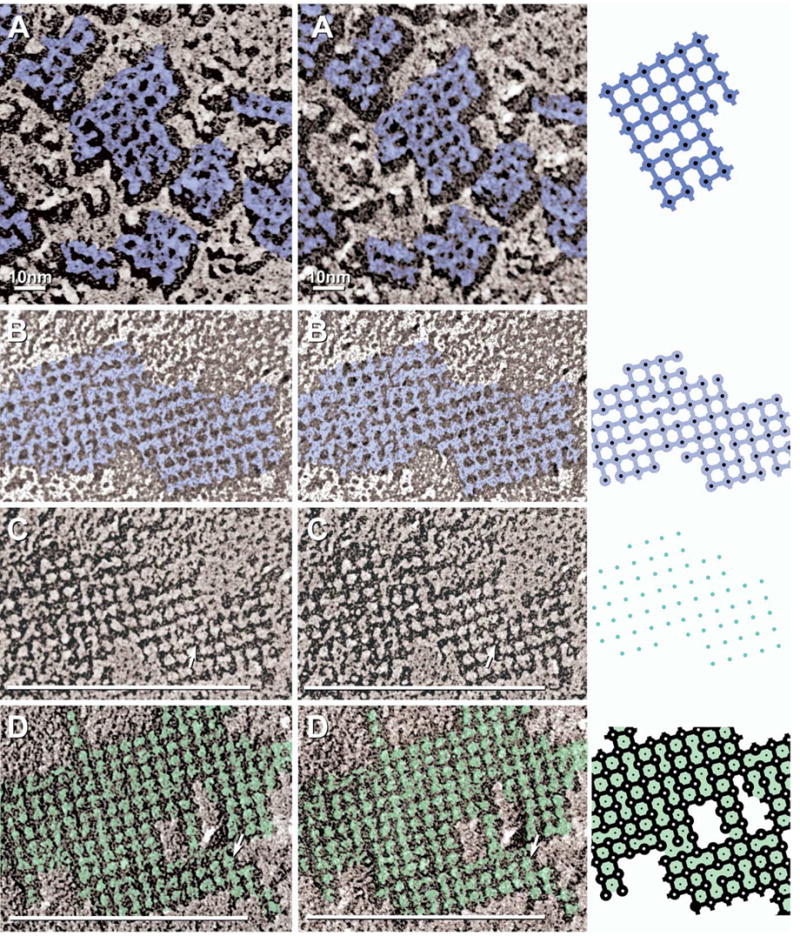
High magnification stereoscopic images of AQP4 IMPs (A) and E-face pits (B–D), with corresponding drawings (right column). Color is used to delineate structures with similar electron opacities. (A) Black-shadow image of P-face AQP4 square array IMPs (dark blue) linked by 1×2 nm cross-bridges. (B, C) White-shadow (B) and black-shadow images (C) of square array E-face imprints (light blue in B) showing furrows linking membrane pits. Most pits contain a 0.3–0.7 nm “peg” (C; arrow; blue dots in diagram). (D) E-face image of M23 raft in CHO cell. Pits are linked by furrows and are separated by 4.5–5.0 nm “nodes” (arrow; green shading).
FRIL labeling of IMP “rafts” in CHO cells expressing the M23 variant of AQP4
As an independent method for investigating the possibility that AQP4 corresponds to IMPs other than those in square arrays, we recently analyzed AQP4 expression in CHO cells, which do not normally form square arrays (Fig. 3A, described below). Using the M23 variant of AQP4, we showed that transfection of CHO cells with M23 AQP4 resulted in the de novo formation of abundant and extremely large conglomerate square lattices of IMPs (Fig. 2A; modified from Furman et al., 2003). Others have shown similar images from CHO cells transfected with “the” gene for AQP4 (Yang et al., 1996; Van Hoek et al., 1998), but at that time, the M1 AQP4 variant had not been isolated. Because only cells with large arrays were seen in those replicas (Yang et al., 1996; Van Hoek et al., 1998), the M1 variant presumably was not expressed in those cells. Using antibodies against the carboxy terminus of AQP4, these large arrays were labeled by multiple gold beads within the perimeter of individual rafts (Fig. 2B–D), with virtually no gold beads on membrane areas that were devoid of rafts. Thus, restriction of immunogold beads to rafts demonstrated that AQP4 is the primary or sole trans-membrane constituent of the newly-created rafts.
Fig. 3.

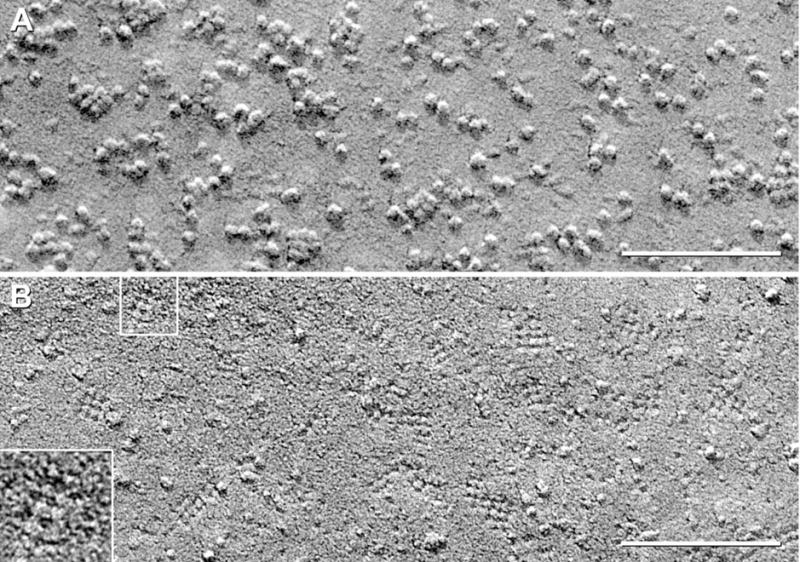
Black shadow images of E-face of control CHO cells (A) and P- and E-face images of CHO cells expressing M1 variant of AQP4 (B–G). (A) E-face image of control CHO cell has “baseline” number of large particles but very few 4.5 nm diameter pits. (B) P-face image of M1 expressing cell has abundant 4–6 nm diameter IMPs, and in this area, only one square array (arrow). (C) E-face of CHO cell expressing M1. A large increase in 4.5–5 nm pits is observed above “control” value. Selected areas are shown stereoscopically at higher magnification (D–G). (D–G) Pegs are seen in most pits (white arrowheads). Many pits exhibit 1×2 nm “furrows,” frequently oriented at right angles (D, F; black arrowheads).
One form of AQP4 (M1) does not form “rafts” or stable square arrays
In non-transfected “control” CHO cells, plasma membrane E-faces (Fig. 3A) and P-faces (not shown) have a low level of 4–6 nm-diameter P-face IMPs and E-face pits. It is upon this “baseline” that changes in relative numbers of particles and pits were measured after expression of AQP4 M1. Additional larger (6–8 nm) IMPs/pits are also present in P-and E-faces. In low-magnification images, some of the 6–8 nm IMPs were difficult to discriminate from the isolated 4.5 nm particles/pits; consequently, all images are shown at high magnification (i.e. > 200,000× ).
Normal astrocytes express M1 and M23 at a ratio of 1:4 (Neely et al., 1999). Thus, it was appropriate to examine CHO cells expressing M1 and M23 separately and together. However, technical limitations of the CHO cell expression system permitted co-expression of M1 and M23 only at a near 1:1 ratio. Transfection of CHO cells with the full-length M1 variant of AQP4 resulted in a large increase in the number of irregularly shaped, small-diameter (< 6 nm) P-face IMPs (Fig. 3B, arrow) and E-face pits (Fig. 3D–G, arrowheads). Most of the 4.5 nm E-face pits had a central “peg” (Fig. 3D–G, arrowheads), and many E-face pits had 1 × 2 nm lateral “furrows” that usually extended in orthogonal directions (Fig. 3D–F, black arrowheads). The relevance of E-face pits with orthogonal furrows is described below.
Most of the small P-face IMPs and E-face pits were randomly distributed in the plasma membrane. Occasionally, however, the small P-face IMPs and E-face pits formed “incipient arrays” having irregularly spaced IMPs/pits in approximate square lattice (Fig. 3B, arrow). In E-faces, some pits possibly were linked by indistinct furrows (Fig. 3E, below the arrowhead). The few incipient arrays contained two to 12 IMPs/pits, with most having six or fewer subunits. Overall, therefore, > 85% of 4.5 nm IMPs/pits in M1 samples were singlets (Fig. 3C–G), and <15% of 4.5 nm IMPs/pits were present within incipient arrays. This is to be contrasted with both M23-expressing CHO cells (Fig. 2), wherein > 98% of 4.5 nm IMPs/pits were in rafts, and with normal astrocytes, which had > 95% of 4.5 nm IMP/pits in square arrays (Furman et al., 2003).
Co-expression: M1 down-regulates square array assembly by M23
Simultaneous co-expression of M1+M23 in CHO cells did not result in the combination of large rafts (as with M23-expressing cells) intermixed with dispersed IMPs/pits and incipient arrays (as with M1-expressing cells). Rather, co-expression resulted in the formation of abundant but small square arrays (Fig. 4A, B; modified from Furman et al., 2003). However, these arrays were less than half the size of square arrays in normal astrocyte endfeet (average < 12 IMPs/array in double-expressing CHO cells vs. > 25 IMPs/array in astrocytes). Thus, the smaller square arrays in CHO cells co-expressing M1+M23, like the square arrays in normal astrocyte plasma membranes, represent assemblages formed in the presence of and presumably containing variable mixtures of both M23 and M1 AQP4. In the five cases investigated (normal astrocytes, normal CHO cells, CHO cells expressing M1, CHO cells expressing M23, and CHO cells expressing both M1 and M23), square array size was inversely related to the ratio of M23:M1, with higher ratios of M1 associated with progressively down-regulated array size. (For additional details regarding relative ratios of M1:M23 in each of these cell types, see Discussion.) Further tests of the hypothesis that M1 down-regulates square array size will require analysis of cells expressing a wider range of ratios of M1 to M23, from 10:1 to 1:10.
Fig. 4.
P- and E-face views of CHO cells co-expressing M1+M23. (A) P-face images reveal abundant but small square arrays. (B) E-face images of square array imprints. Pegs are visible in almost all pits. Many pits are linked by furrows, but up to 50% of furrows are missing. Inset shows array of seven pits linked by about four cross-bridges, but excessive granularity of this replica prevented more detailed analysis.
Improved freeze fracture replicas revealed substructure in AQP4 P-face particles: cross-bridges and “dimples”
In our early freeze fracture electron micrographs prepared with “conventional” (i.e. 1.5–2.5 nm-thick) platinum films (Fig. 5A), as well as in virtually all previously published images by other laboratories, square arrays appeared to consist of 6.5 nm globular P-face IMPs in regular 6.5 nm square lattice, with little or no apparent separation of particles. Those artifactually enlarged particles provided an apparent ultrastructural basis to accommodate the known molecular weights and proposed polymeric assembly of AQP4. However, improvements in high vacuum technology and shadow coating methods (Rash et al., 1990; Rash and Yasumura, 1992) resulted in reduced or absent water vapor contamination on IMPs, reduced thickness of platinum coating layers, and (usually) virtually grainless platinum films, allowing finer details to be resolved in the structural components within square arrays. By analogy, a deep snowfall may obscure entire automobiles, but a thin layer of snow may coat and clearly delineate even small structures, such as the handlebars on a bicycle.
Fig. 5.
Stereoscopic images comparing “conventional” (A) and higher-resolution replicas of AQP4 square arrays (B–D). (A) In “old-style” replicas, IMPs are enlarged by excess platinum and by water vapor “contamination.” (B) In replicas made with ca. 1 nm of platinum, P-face images of AQP4 square arrays reveal 4.5 nm diameter IMPs linked by indistinct cross-bridges (at lower arrow). Some IMPs have indistinct “dimples” (arrows). (C) In E-face images, imprints of square arrays consist of 4.0–4.5 nm membrane “pits,” with most pits containing a central 0.7 nm central “peg” (faintly resolved). Each pit is linked to its nearest neighbors by 1×2 nm grooves or “furrows” (arrow). Some furrows at the edge of arrays (arrowhead) extend outward for possible attachment of additional IMPs/pits. Stereoscopic imaging reveals that the replicated P-face cross-bridges and E-face furrows are co-planar with the platinum replica and thus do not represent artifacts of underfocus or overfocus, which otherwise appear as a superimposed three-dimensional “fog.” (D) E-face imprints of square arrays in astrocyte endfoot in adult rat spinal cord. Pegs (arrows) are present in most E-face pits, and most pits are linked by 1×2 nm furrows.
Improved replicas revealed that each astrocytic square array consisted of IMPs in conventional 6.8 nm square lattices, but the constituent IMP cores were 4.5 nm in diameter (Fig. 5B, lower arrow) and more clearly delineated than in previous replicas. Likewise, E-face pits were larger and also more clearly delineated (Fig. 5C; also see Fig. 1B–F), with E-face pits often identical in diameter to their corresponding P-face IMPs, suggesting that IMPs were not detectably enlarged by the platinum coat and likewise, that E-face pits were not partially filled up by the platinum. Interestingly, IMPs/pits at the edge of square arrays often had cross-bridges/furrows extending to where an additional particle might be expected to attach (Fig. 5C, arrowhead). At high magnification, a few of the component IMPs resembled “Maltese crosses,” with each 4.0–4.5 nm diameter central core linked to its nearest neighbors by 1 nm × 2 nm cross-bridges (Fig. 5B, lower arrow; Fig. 6A; also see Fig. 2D). Approximately 25–50% of IMPs exhibited a 0.3 nm-0.7 nm diameter central depression (Fig. 5B, lower arrow). However, up to 75% of the other constituent IMPs were clearly altered by “plastic deformation” (stretching and inelastic rebound; Steere et al., 1980) during the fracturing event (Fig. 5B, upper arrow; Fig. 6A), making those deformed IMPs of little value for ultrastructural analysis.
Complementarity of E-face impressions of AQP4 arrays; “furrows” and “pegs”
An independent way of verifying molecular structure for AQP4 particles was to compare the relative “complementarity of fit” (Steere and Moseley, 1969; Challcroft and Bullivant, 1970; Rash and Yasumura, 1992) of P-face particles and their corresponding E-face pits. Because the E-face pits are not altered by the shearing forces that distort the proteins as they are removed from the pits (Steere et al., 1980), the shapes and sizes of pits more reliably preserve the original shapes of extracted protein molecules (Rash and Yasumura, 1992). Stated differently, plastic deformation of IMPs occurs as an artifact of tensile fracture (IMP stretching) rather than brittle fracture (Steere et al., 1980), whereas the E-face pits are not subject to plastic deformation.
In high-magnification, near-focus stereoscopic images (Fig. 5C; replica made with <1 nm Pt, but at conventional, non-optimum 45° local shadow angle), the 6.8 nm lattice spacings of the arrays were clearly discerned. As above, however, each E-face pit (i.e. AQP4 imprint) within the lattice was 3.8 nm to 4.5 nm in diameter, and each pit was linked to its nearest neighbors by 1 × 2 nm “furrows” (Figs. 5C, D, 6B–D). The precise complementarity of the E-face furrows with at least some P-face cross-bridges suggests that the furrows represent the impressions remaining where protein cross-bridges had been extracted by fracturing.
In areas shadowed at >50° angle, most E-face pits contained a 0.3–0.7 nm central “peg” (Figs. 5D, 6C, arrows) that was complementary to the “dimple” seen in many of the P-face IMPs (Figs. 5B, 6A), suggesting that the E-face pegs represent material removed from the dimples in the P-face particles. Pegs were clearly discerned within most pits in E-face images of square arrays from normal astrocytes (Fig. 5D, arrow; Fig. 6C, arrow), in M23 arrays (Fig. 6D), and in both incipient arrays and isolated E-face pits in M1 samples (Fig. 3D–G, arrowheads). Because pegs were not replicated where the local shadow angle was <45° (Rash and Yasumura, 1999), but were usually visible when shadowed at a higher angle, trigonometric considerations imply that the E-face pegs extend ca. 1–1.5 nm above the bottom of each pit, or one-third to halfway through the plasma membrane, from the extracellular surface into each IMP. Moreover, when gap junctions and AQP4 arrays were in close proximity (Fig. 7A), the pegs in their respective E-face pits were resolved to have two different diameters: ca. 1.0–1.4 nm in connexon imprints (Fig. 7B) vs. 0.3–0.7 nm in AQP4 imprints (Fig. 7C), thereby documenting a reproducible two-fold difference in the sizes of the pegs in connexon vs. AQP4 imprints. Slight variability in the sizes of the AQP4 pegs may be due to slight differences in the height of the fracture plane within the material that makes up the pegs, as well as to random differences in the small number of replicating platinum atoms at these near atomic dimensions.
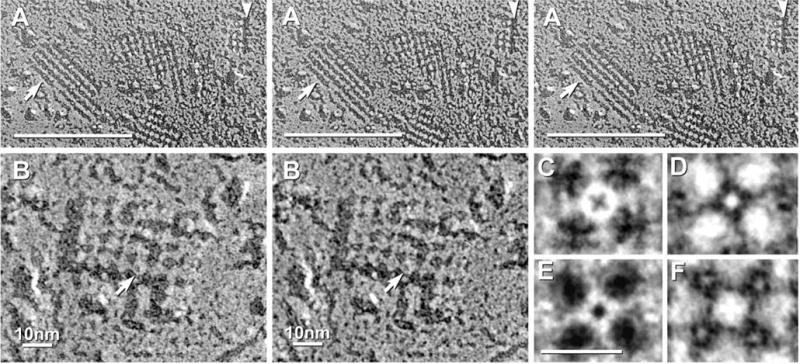
Fig. 7.
Comparison of two-fold difference in diameter of pegs in E-face pits of gap junctions and AQP4 arrays; and evidence for disappearance of pegs following shallow “etching.” (A) Stereoscopic image of gap junction and several square arrays in ependymocyte of third ventricle in adult rat suprachiasmatic nucleus. Boxed areas are enlarged in B and C. (B) Pegs in gap junction E-face pits (arrow) are 1.0–1.5 nm in diameter. (C) Pegs in square array E-face pits (arrow) are 0.3–0.7 nm in diameter, or about half the size of pegs in adjacent connexon imprints. (D) E-face image of square arrays after brief freeze etching. Etching is evident in extracellular space (asterisk). Where E-face pits are wider and deeper, pegs are missing (arrowheads), but nodes are intact.
E-face “pegs” removed by freeze etching
In images from ultrarapidly frozen, non-glycerinated samples, brief freeze etching (60 s at − 100 °C) resulted in lowering of the frozen water (ice) table in the cytoplasm and extracellular space by 20–30 nm. In the narrow range of conditions in which the delicate, unsupported plasma membrane E-faces were not perforated by excess etching (Dinchuk et al., 1987), square array E-face pegs disappeared (Fig. 7D, arrowheads), as did the pegs in gap junction E-faces (not shown). The presence of pegs in almost 100% of E-face pits in non-etched specimens (Figs. 5D, 6C, D), and their disappearance after gentle etching (Fig. 7D) suggests that the pegs represent a component that sublimes away under conditions of vacuum and temperature in which only frozen water (i.e. ice) is volatile (Böhler, 1979). Thus, we conclude that each peg represents the frozen water core extracted from within each AQP4 IMP. Moreover, the dimple in each etched P-face IMP was deeper and more distinct after etching (not shown), suggesting that the ice pegs originally extended deeper into each AQP4 IMP but were sheared off part way into the IMP, perhaps at a narrow constriction.
Comparison of bridges and furrows
The cross-bridges linking many P-face particles (Figs. 1E, F, 2B–D, 5B, 6A) are complementary to the furrows linking most E-face pits (Figs. 5C, D, 6B–D). In “black-shadow” images, furrows are black and pegs are white. However, for clear delineation of the several types of structural components having sharp reversals of molecular contour, the same images are also printed with reversed photographic contrast (i.e. with white shadows), thereby allowing the furrows to be delineated in “false color.” Computer-determined areas of colorization emphasize the precise correspondence of the P-face cross-bridges (Fig. 6A, dark blue) and E-face furrows (Fig. 6B, light blue).
The presence of furrows linking essentially 100% of E-face pits in M23 arrays (Fig. 6D) and of cross-bridges linking 100% of P-face particles in M23 arrays (Fig. 2B–D) confirmed that cross-bridges were never separated from P-face particles in arrays composed solely of M23, suggesting that the cross-bridges represent lateral covalent extensions of the AQP4 molecules. Likewise, the furrows linking almost all E-face pits (Fig. 6D) revealed that the P-face cross-bridges were present within and removed from within the entire 1.5 nm thickness of the external membrane leaflet. Thus, the cross-bridges are parallel to and at the interface of the external leaflet membrane lipid with the aqueous extracellular surface.
In notable contrast to the extremely regular and complete lattices that characterize M23 arrays (Fig. 2D, Fig. 6D), P-face cross-bridges linked only about 70 – 80% of IMPs in astrocyte square arrays (Figs. 5B, 6A; also see Fig. 1E, F); and in E-faces, furrows linked ca. 70 – 80% of E-face pits (Figs. 5C, D, 6B, C). Thus, in astrocytes and ependymocytes, we estimated that ca. 20 –30% of P-face cross-bridges and E-face furrows were missing. With M1 constituting ca. 20% of the AQP4 molecules in astrocyte endfeet (Neely et al., 1999), it appears that the number of M1 variant molecules in an array prevents formation of a comparable number of cross-bridges. Similar partial blocking of cross-bridge formation may also occur in both normal astrocyte square arrays (Fig. 5B, C), as well as in square arrays formed in CHO cells doubly expressing M1+M23 (Fig. 4B). Moreover, single expression of M1 precluded stable cross-bridge formation between 4.5 nm IMPs in M1-expressing CHO cells and resulted in almost complete absence of furrows in E-faces (Fig. 3C–G) and failure to form stable arrays. Thus, in normal astrocytes and in M1+M23 co-expressing cells, the presence of IMPs in regular lattices that are, nonetheless, missing cross-bridges between some P-face IMPs may provide a clue regarding mechanisms of AQP4 lattice assembly (see Discussion).
P-face IMPs separated by 4.5–5 nm E-face “nodes”
In P-face images of square arrays, the IMPs were somewhat variable in shape and size, ranging from 4.0–4.5 nm in diameter. In corresponding E-face images of astrocyte and ependymocyte square arrays, and more abundantly in M23 rafts, the complementary E-face pits were 3.8 nm-4.0 nm in diameter. However, each pit was uniformly separated from each adjacent pit by a 4.5–5 nm-diameter “node” (Fig. 6D, light green). In replicas shadowed at a higher angle and with slightly more platinum, smaller diameter (3.8 nm) pits were observed, surrounded by 5–6 nm nodes, indicating that even these very thin platinum layers partially obscure fine details. Thus, the smaller pits may have been reduced in diameter by up to 1 nm and the nodes increased in diameter by up to 1 nm based on the thickness of the Pt/C “cap.” If so, the P-face IMPs and E-face nodes are approximately equal in cross-sectional area.
The “node” material was not removed by etching (Fig. 7D). Therefore, the nodes represent etch-resistant, relatively stable E-leaflet molecules (lipid or protein) that surrounded and separated the P-face IMPs prior to fracture. In non-etched specimens, the relative height of the nodes vs. surrounding E-leaflet lipid molecules was assessed by stereoscopic viewing of conventional and “intaglio” (reversed) stereoscopic images (Fig. 8A). In intaglio views (right pair of images), the nodes consistently extended ca. 1 nm above the relief of the surrounding replicated E-face lipids. This relative difference in elevation of nodes vs. the lipids of the surrounding E-faces was also reflected in the increased width of the black shadows (arrowhead) in the direction of the shadowing source, as well as in an increased platinum thickness at the edge of the array (arrow) opposite the shadowing source. By comparing intaglio (right pair) vs. conventional stereoscopic images (left pair), the nodes were ca. 1 nm shorter than the surrounding lipid, i.e. extended from the extracellular surface to less than halfway through the membrane bilayer (for interpretive drawing, see Fig. 9 in Discussion). Thus, the nodes represent a substantial intramembrane volume not occupied by the AQP4 IMP, and this volume must be taken into account in any model of AQP4 structure and assembly in intact biological membranes. (See Discussion.)
Fig. 8.
(A) Stereoscopic (left pair) and reverse stereoscopic images (right pair) of square arrays in E-face showing that the “nodes” are fractured at a lower level than the surrounding membrane E-face. In “intaglio” images (right pair), this relative elevation is reversed so that the “nodes” artificially appear as raised “waffles.” (B) High-magnification stereoscopic image of one AQP4 square array. Several IMPs have central depressions or “dimples” (arrow). (C) Rotational reinforcement image of the IMP indicated in “B.” The central depression appears as an “X.” (D, E) Rotational reinforcement images of E-face pits printed with black shadows (D) and white shadows (E). In the black-shadow image (D), the peg (bright spot) is smaller than the dimple in the adjacent P-face IMP (C). In the white shadow image (E), the (now white) pit appears almost identical to the IMP in the black shadow image (C). Four nodes around the central peg (D, E) are approximately the same diameter as P-face IMPs, indicating that both nodes and IMPs each occupy approximately 50% of the cross-sectional area within a square array. (F) Rotational reinforcement of “node” showing reinforcement of four surrounding “pegs” (bright spots within darker E-face pits).
Fig. 9.
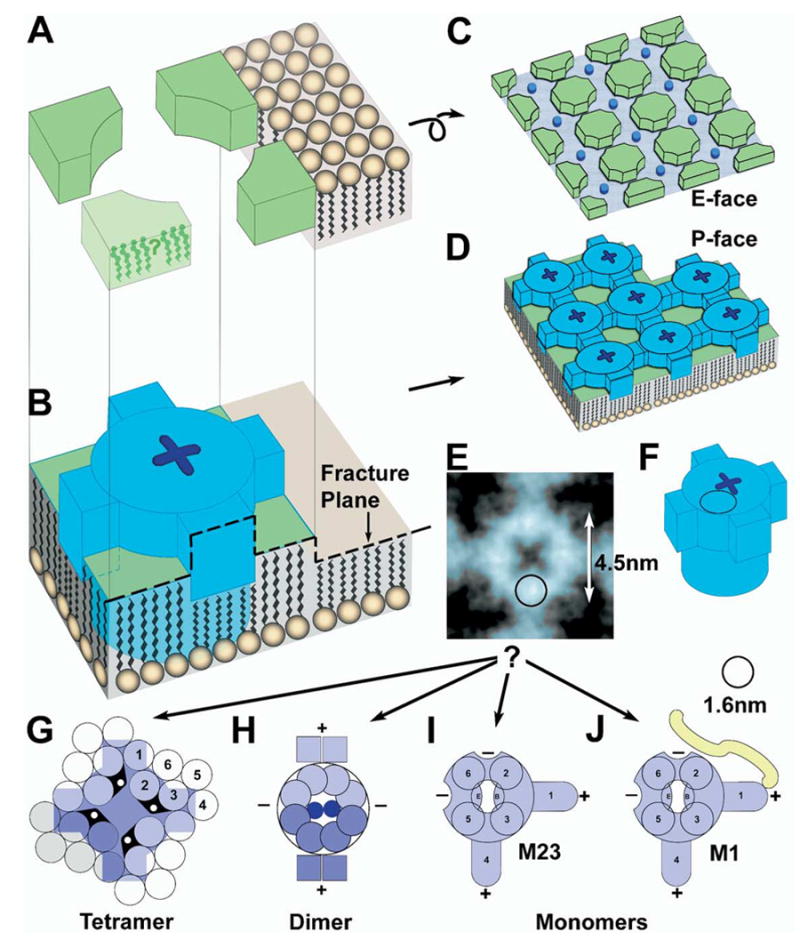
Interpretive drawings of components of naturally occurring square arrays and their constituent IMPs. (A, B) Fracture plane through an individual IMP, with the resulting formation of E-face (C) and P-face (D) images of a square array. Dark blue indicates an individual AQP4 IMP (B and D), with “X-shaped dimple.” (E, F) Approximate comparison of rotationally reinforced image on one IMP (E) with stylized AQP4 protein (F). Superimposed circles in remaining drawings are the approximate diameter of an α-helix, 1.6 nm, prior to platinum coating. A single α-helix is almost half the diameter of an AQP4 particle. (G–J) Comparison of tetrameric (G), dimeric (H), and monomeric (I) models of AQP4 IMP. The IMP seen by freeze fracture (blue in E) is much smaller than the 24 transmembrane α-helices required of a tetrameric IMP. (H) The dimeric model of AQP4 has conflicting positions for several α-helices. (I, J) Monomeric models for M23 (I) and M1 (J) AQP4. Two cross-bridges (“+”) are envisioned to arise from amphipathic α-helices (S1 and perhaps S4), while two additional cross-bridge binding sites (“−”) are envisioned as part of the core IMP, thereby providing for up to four cross-bridge binding sites per IMP. The additional 23 amino acid sequence on M1 (J) is proposed to interfere with close approach of M1 monomers to other monomers, and perhaps also to block additional cross-bridge binding sites.
Rotational reinforcement
Selected P-face IMPs, E-face nodes, and E-face pits were enlarged to 1,850,000× and photographically printed (using Photoshop) by four-fold Markham rotary reinforcement superposition (Markham et al., 1963). This procedure enhances substructures having four-fold symmetry and reduces the contribution of random “noise” that arises from platinum granularity, photographic noise, and artifacts of defocus in the electron microscope. In enhanced white-shadow images of IMPs that had minimal plastic deformation, the core of each IMP within an array was 4.5 nm in diameter (Fig. 8C) and had four 1 nm×2 nm lateral projections (i.e. cross-bridges). Selected IMPs with obvious x-shaped dimples (unshadowed central depressions) prior to four-fold rotary reinforcement (Fig. 8B, arrow) were enhanced especially well by rotary reinforcement (Fig. 8C). Because of differences in height of various substructures, shadowing at an oblique angle resulted in apparent slight lateral displacements of different array components having different topographic relief. Consequently, separate rotational reinforcement was conducted on nodes rotated around pegs (Fig. 8D, E) and pegs rotated around nodes (Fig. 8F). (Fig. 8D and E are white-shadow and black-shadow versions of the same image.) Apparent substructure in nodes was enhanced, but the surrounding pegs were somewhat diminished in their relative intensity due to incomplete rotational reinforcement of asymmetrically shadowed structures having different topographic elevations. Regardless of image processing method, however, IMP cores were consistently 4–4.5 nm in diameter (including thickness of platinum cap); cross-bridges were 1 × 2 nm; pegs from images shadowed at > 45°angle were 0.3 nm–0.7 nm (3–7Å) in diameter; and IMP dimples were 1–1.5 nm in width.
DISCUSSION
FRIL of AQP4 in normal astrocyte square arrays and multiple labeling of individual AQP4 “rafts” formed in the in vitro CHO cell expression system provided compelling evidence that AQP4 is the primary transmembrane protein of square arrays in these naturally occurring biological membranes. More important, freeze-fracture replicas made with improved replication methods revealed new details of the molecular substructure of square arrays, including:
In P-faces, each constituent protein particle is significantly smaller than in “old-style” replicas made with excess platinum. The central IMP core is 4.0–4.5 nm in diameter, and most of these core IMPs are linked to their closest neighbors by 1×2 nm cross-bridges, thereby creating the classical 6.5–6.8 nm lattice spacing (Fig. 9 is an interpretive drawing; core IMPs are indicated as blue barrels).
In E-faces, square arrays are resolved as 3.8 nm–4.5 nm diameter pits linked by 1×2 nm–1×3 nm furrows (Fig. 9C). The complementarity of P-face cross-bridges and E-face furrows demonstrates that the cross-bridges and furrows represent different ways of demonstrating the same structural component.
The furrows in E-faces indicate that the P-face cross-bridges extend laterally within the plane of the external lipid layer, at or close to the interface between membrane lipid and extracellular aqueous matrix (Fig. 9B, blue arms extending from barrels), consistent with a simple amphipathic α-helix at the lipid-aqueous interfaces (Hristova et al., 1999).
In astrocytes and ependymocytes, most images of square arrays lacked one or more cross-bridges between IMPs, leaving only the central 4.5 nm IMP core. It is the diameter of this core IMP that must accommodate all trans-membrane α-helices.
In CHO cells expressing M1, cross-bridges are not detectable (or were rarely detected), suggesting that the additional 23 amino acid sequence in the N terminus interferes with the formation of cross-bridges (for conceptual representation of bridge blocking, see Fig. 9J; yellow segment).
Adjacent IMPs were either linked by a complete cross-bridge, or the cross-bridge was missing entirely (i.e. no half-length bridges were detected), suggesting that a complete cross-bridge is contributed by only one of the two adjacent IMPs.
We propose that cross-bridges provide a molecular cross-linking mechanism for assembling and imparting structural stability to the arrays.
The large arrays established by the M23 variant of AQP4 were constructed from IMPs that presumably had four orthogonal binding sites available for lateral attachment and lattice growth. However, when M1 was present in an array, the additional cytoplasmic segment appeared to interfere with close approach by other AQP4 IMPs or to prevent cross-bridge formation, thereby effectively interfering with lattice assembly. At a ratio for M1:M23 of 1:1 (as in CHO cells), arrays were small (ave.= 12 IMPs/pits); at 1:4 (as in astrocytes), arrays were larger (ave.= 25 IMPs/pits); and at 0:1 (as in M23-expressing CHO cells), areas were larger still (ave.= 75; with up to 600 IMPs/pits per array). Accordingly, a few M1 monomers appear to be capable of interfering with AQP4 lattice formation by M23. However, the hypothesis that M1 down-regulates M23 AQP4 lattice formation has yet to be tested by freeze-fracture analysis of cells expressing more extreme ratios of M1:M23. [Note: After submission of this manuscript, Silberstein et al. (2004) reported that M23-expressing LLC-PK1 (pig kidney) cells had an eight-fold higher water permeability [Pf] than M1-expressing cells, even though the M1-expressing cells had a five-fold greater content of AQP4 protein. Although contradictory to a previous report (Neely et al., 1999), the data from Silberstein and coworkers provides evidence that assembly into arrays may be essential for opening water channels.]
Functionally, cross-bridges may allow for either “assembly-based osmoregulation” or “stretch-sensitive” gating of the AQP4 channel in intact biological membranes. If osmoregulation occurs by assembly or by stretch-sensitive gating (neither has been demonstrated), AQP4 might be more easily regulated (activated or inactivated) when arrayed into complex lattices of IMPs. If either hypothetical regulatory mechanism occurs, the number of functional channels would increase geometrically with the size of the square lattices (Fig. 6). For example, if only those IMPs having all four cross-bridges occupied were functional, the minimum functional array would be five IMPs arrayed as a “+,” with only the central IMP functional. Linear assembly would allow concentration of AQP4 molecules within an area while retaining low water permeability, whereas rearrangement into square lattices would result in rapid increase in permeability. Rapid regulation of water flux might also occur by disrupting cross-bridges between adjacent IMPs within an array (see for example Fig. 5C), by removing a few constituent particles from an array, or by disassembling square arrays. Experiments involving extreme hyponatremia and hypernatremia are underway to test those conjectures.
Structural location of the water channel
In square arrays, a central 0.7 nm “dimple” was present in some P-face particles. A corresponding 0.3–0.7 nm ice “peg” occurred in almost all E-face pits. The removal of E-face pegs by brief etching at −100 °C suggests that the pegs represent the water/ice core removed from each IMP-whether a peg represents a single channel per functional aquaporin molecule (Gonen et al., 2004) or the “vestibule” formed by four composite water channels within each core particle (Jung et al., 1994). The “x-shaped” dimples seen in some P-face IMPs (Fig. 8B, arrow; rotational reinforcement in Fig. 8C) may constitute freeze-fracture evidence for four channels per IMP. However, at the current state of the art, freeze-fracture imaging is not capable of clearly resolving one vs. four 0.3–0.7 nm channels within a single 4.5 nm-diameter IMP.
Structure, possible composition, and potential significance of the “nodes”
E-face nodes represent unidentified “spacer” material between the AQP4 IMP cores. The low relief of the E-face nodes as compared with surrounding E-face lipid (Fig. 8A) requires that nodes represent either: a) a cluster of shorter membrane lipids, such as tightly packed cholesterol (illustrated diagrammatically in Fig. 9 A, green), or b) an un-identified class of membrane proteins that penetrates only about one-third the way through the bilayer, and less than the surrounding lipids. Based on the AQP1 and AQP0 assembly models of Engle, Walz and coworkers (Walz et al., 1997; Gonen et al., 2004), the nodes may represent additional AQP4 molecules oriented with opposite polarity in the membrane (i.e. the “interdigitating protein model”). At this time, however, the composition and function of the nodes are unknown.
Models of AQP4 square array assembly
Available sequence data suggest a common molecular configuration for aquaporin proteins that form square arrays, either in vivo [e.g. AQP4, AQP0 (Zampighi et al., 1989), AQPcic (Hubert et al., 1989), and AeaAQP (Duchesne et al., 2003)] or under artificial crystallization conditions [e.g. AQP1 (Walz et al., 1997)]. Although the 6.8 nm lattice spacing in freeze-fracture replicas is the same as that in artificial crystals of AQP1 (which are crystallized in artificial lipid films composed of only one or two kinds of lipid molecules), it has not been established that the square lattices of purified AQP1 occur in the same configuration and spacing as the square arrays in naturally occurring biological membranes. Also not clear is the relationship of the E-face “node” material to the intervening IMPs in each square array. This additional node material within an array occupies approximately 50% of the cross-sectional area of each lattice spacing, thereby in our estimation, leaving insufficient area to accommodate an IMP corresponding to a 24-pass (or 28-pass) AQP4 tetramer (Fig. 9G).
By way of comparison, gap junctions consist of individual 8.5 nm IMPs/8.5 nm E-face pits (Rash and Yasumura, 1992). Each connexon is composed of six connexin molecules, which are “four-pass” polypeptides strands (Makowski et al., 1977; Unwin and Zampighi, 1980). Thus, a connexon consists of 24 transmembrane α-helices that form a replicated 8.5 nm IMP having a cross-sectional area of 56 nm2. With the same (or fewer) transmembrane α-helices than a tetramer of AQP4, each 24-pass connexon is approximately twice the diameter and four times the cross-sectional area of the AQP4 core IMP, as described in this report. Moreover, each of the 24 replicated α-helices in a connexon is calculated to have an average cross-sectional area of ca. 2.5 nm2 and a diameter of 1.6 nm.
If the same values for cross-sectional area and diameter of a replicated α-helix are applied to a tetrameric “hourglass” model for AQP4 (Jung et al., 1994), and assuming that all α-helices in AQP4 span the bilayer, a tetramer consisting of 24 transmembrane spanning domains plus eight half membrane-spanning α-helices (i.e. the “B” and “E” loops; Jung et al., 1994; Ren et al., 2000) would have a cross-sectional area of 70 nm2 and create an IMP 9 nm in diameter. However, AQP4 particles have a 4.5 nm central IMP core (Fig. 8B, C and Figs. 9B, D, F; blue barrels), or one-fourth the required area. Using the upper and lower limits for the size of a replicated α-helix (also see Eskandari et al., 1998) and size constraints imposed by IMP measurements (this report), we compared the tetrameric IMP model (Fig. 9G) with a “dimeric IMP” model (Fig. 9H), and a “monomeric IMP” model (Fig. 9I, J). Although all three models are derived from freeze-fracture images, they are based on the “hourglass” model for aqua-porin monomers (Jung et al., 1994).
The 24-pass (or 28-pass) tetrameric IMP model (Fig. 9G; diagram modified from Jung et al., 1994) appears to be incompatible with the 4.5 nm IMPs seen in freeze-fracture replicas because the central core of each AQP4 IMP is too small to accommodate more than eight to 12 transmembrane α-helices. Consequently, in the 12-pass (or 14-pass) “dimeric” AQP4 IMP model (Fig. 9H), the central IMP is envisioned to contain a maximum of eight transmembrane α-helices, plus two B-loops and two E-loops. Moreover, the two structural cross-bridges are envisioned as being formed from two α-helices per monomer (Fig. 9H, labeled “+”). Two additional cross-bridge attachment sites (labeled “−”) provide for orthogonal bonding of additional cross-bridges from adjacent dimers. If two oppositely oriented dimers form an “interdigitating” tetramer, the dimeric IMP/node model could create the observed 6.8 nm lattice spacing. At present, however, there are no data from diffraction studies to support either of the dimeric IMP models over the tetrameric IMP model.
From freeze-fracture data, the “monomeric IMP” model (Fig. 9I, J) is preferred because it allows sufficient room for six transmembrane domains per IMP (or seven, with Band E-loops corresponding to a single α-helix). Each of the four circles in the diagram corresponds to a 1.4–1.7 nm-diameter α-helix, according to size estimates from other replicated transmembrane proteins (also see Eskandari et al., 1998). The additional 23 amino acid residues forming the M1 translated segment is indicated diagrammatically (Fig. 9J, yellow segment). The “monomeric IMP” model is consistent with the “hourglass” model by having one functional channel per monomer (Vidossich et al., 2004). If AQP4 is present as monomeric IMPs, the six α-helices may have been resolved in the freeze-fracture replicas described in this report. Four apparent structural subunits were detected surrounding the central “dimple” (Fig. 8C; the four bright spots in cloverleaf configuration; one indicated by circle in Fig. 9E). The two remaining α-helices (S1 and perhaps S4) would correspond to two of the four cross-bridges per monomeric IMP (Figs. 8C and 9I). Assembly of monomeric IMPs into square arrays would occur by orthogonal attachment of “+” ends of amphipathic intramembrane α-helices (Hristova et al., 1999) to “−” sites on adjacent monomers (Fig. 9I), and one or both of these “−” sites would be blocked by the additional segment on M1 (Fig. 9J).
The monomeric IMP model (Fig. 9I, J) appears to be inconsistent with current diffraction data because the lattice spacing of square arrays is twice that of monomers in artificial AQP1 crystals formed in pure lipid films. Moreover, limited evidence suggests that M1 is functional only when assembled into a tetramer (Neely et al., 1999), whereas functional M1 IMPs do not assemble into stable square arrays (Furman et al., 2003). Taken at face value, these two observations imply that M1 tetramers correspond to individual IMPs. However, a recent report suggests that AQP1 may be functional as an isolated monomer (Vidossich et al., 2004). If so, perhaps AQP4 may also be functional as a monomer, but this possibility has been questioned by Silberstein and coworkers (2004) based on measurements of high water permeability only when AQP4 is assembled into square arrays and rafts. We anticipate that diffraction data from AQP4 crystals formed in lipid films formed from naturally occurring complex lipids will provide compelling evidence for one or the other models.
CONCLUSIONS
There is compelling evidence from electron diffraction studies that AQP1 assembles as tetramers in artificial protein crystals in simple lipid bilayers, that the crystals have a 6.9 nm lattice spacing, and that the protein molecules completely fill the lattices, with no intervening voids. Moreover, in those crystals, 1×2 nm structural cross-bridges have not been identified. In contrast, freeze-fracture images of AQP4 arrays in biological membranes reveal 4.5 nm transmembrane particles, linked by 1×2 nm cross-bridges, and separated by 4.0–4.5 nm nodes, the latter of which at least superficially resembles the “interdigitating,” oppositely directed AQP1 and AQP0 tetramers in artificial crystals formed in pure lipid films. However, assembly of artificial crystals of AQP1 is not constrained by enzymatically regulated insertion of transmembrane proteins, as occurs during protein synthesis in vivo. Thus, the freeze-fracture images from intact biological membranes seem incompatible with the data from electron diffraction from artificial membranes. The significance of these structural differences is not established. To resolve these discrepancies, we suggest that artificial crystals of various aquaporins be examined in high-resolution freeze-fracture replicas to determine if the same morphology, size and lattice spacing of IMPs is observed as in naturally occurring AQP4 arrays. We also suggest electron diffraction studies be conducted on aquaporin crystals formed in the presence of complex lipids, including cholesterol, to determine the contributions of complex lipids to organization of AQP4 arrays. Clearly, it is essential to reconcile the data from these two distinct methodologies.
Acknowledgments
Funded by grants from NIH (R01NS 38121, R01NS44010, R21NS39040, and S10RR05831 to J.E.R.). We thank Aaron Magnie and Jennifer Sampson for assistance preparing stereoscopic images using Photoshop, and James L. Rash (NASA Goddard Space Flight Center) for assistance with computer image analysis.
Abbreviations
- AQP1
aquaporin-1
- AQP4
aquaporin-4
- CHO
Chinese hamster ovary
- CNS
central nervous system
- CSF
cerebrospinal fluid
- Cx26
connexin-26
- Cx30
connexin-30
- FRIL
freeze-fracture replica immunogold labeling
- IMP
intramembrane particle
- OAP
orthogonally arranged particle
- SDS
sodium dodecyl sulfate
- TEM
transmission electron microscopy
Footnotes
Available online 5 November 2004
References
- Agre P, King LS, Yasui M, Guggino WB, Ottersen OP, Fujiyoshi Y, Engel A, Nielsen S. Aquaporin water channels-from atomic structure to clinical medicine. J Physiol. 2002;542:3–16. doi: 10.1113/jphysiol.2002.020818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agre P, Preston GM, Smith BL, Jung JS, Raina S, Moon C, Guggino WB, Nielsen S. Aquaporin CHIP: the archetypal molecular water channel. Am J Physiol. 1993:F463–F476. doi: 10.1152/ajprenal.1993.265.4.F463. [DOI] [PubMed] [Google Scholar]
- Amiry-Moghaddam M, Ottersen OP. The molecular basis of water transport in the brain. Nat Rev Neurosci. 2003;4:991–1002. doi: 10.1038/nrn1252. [DOI] [PubMed] [Google Scholar]
- Amiry-Moghaddam M, Xue R, Haug F-M, Neely JD, Bhardwaj A, Agre P, Adams ME, Froehner SC, Mori S, Ottersen OP. Alpha-syntrophin deletion removes the perivascular but not endothelial pool of aquaporin-4 at the blood-brain barrier and delays the development of brain edema in an experimental model of acute hyponatremia. FASEB J. 2004;18:542–544. doi: 10.1096/fj.03-0869fje. [DOI] [PubMed] [Google Scholar]
- Anders JJ, Brightman MW. Assemblies of particles in the cell membranes of developing, mature, and reactive astrocytes. J Neurocytol. 1979;8:777–795. doi: 10.1007/BF01206676. [DOI] [PubMed] [Google Scholar]
- Bartels H, Miragall F. Orthogonal arrays of particles in the plasma membranes membranes of pneumocytes. J Submicrosc Cytol. 1986;18:637–646. [PubMed] [Google Scholar]
- Böhler S. Artifacts in defects of preparation in freeze-etch technique. In: Rash JE, Hudson CS, editors. Freeze fracture: methods, artifacts, interpretations. New York, NY: Raven Press; 1979. pp. 19–29. [Google Scholar]
- Bordi C, Amherdt M, Perrelet A. Orthogonal arrays of particles in the gastric parietal cell of the rat: differences between superficial and basal cells in the gland and after pentagastrin and metiamide treatment. Anat Rec. 1986;215:28–34. doi: 10.1002/ar.1092150105. [DOI] [PubMed] [Google Scholar]
- Branton D, Bullivant S, Gilula NB, Karnovsky MJ, Moor H, Northcote DH, Packer L, Satir B, Satir P, Speth V, Staehelin LA, Steere RL, Weinstein RS. Freeze-etching nomenclature. Science. 1975;190:54–56. doi: 10.1126/science.1166299. [DOI] [PubMed] [Google Scholar]
- Challcroft JP, Bullivant S. An interpretation of liver cell membrane and junction structure based on observation of freeze-fracture replicas of both sides of the fracture. J Cell Biol. 1970;47:49–60. doi: 10.1083/jcb.47.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispeels MJ, Agre P. Aquaporins: water channel proteins of plant and animal cells. Trends Biochem Sci. 1994;19:421–425. doi: 10.1016/0968-0004(94)90091-4. [DOI] [PubMed] [Google Scholar]
- Dinchuk JE, Johnson TJA, Rash JE. Postreplication labeling of E-leaflet molecules: membrane immunoglobulins localized in sectioned labeled replicas examined by TEM and HVEM. J Electron Microsc Tech. 1987;7:1–16. doi: 10.1002/jemt.1060070102. [DOI] [PubMed] [Google Scholar]
- Duchesne L, Hubert J-F, Verbavatz J-M, Pietrantonio PV. Mosquito (Aedes aegypti) aquaporin, present in tracheolar cells, transports water, not glycerol, and forms orthogonal arrays in Xenopus oocyte membranes. Eur J Biochem. 2003;270:422–429. doi: 10.1046/j.1432-1033.2003.03389.x. [DOI] [PubMed] [Google Scholar]
- Dudek FE, Obenaus A, Tasker JG. Osmolality-induced changes in extracellular volume alter epileptiform bursts independent of chemical synapses in the rat: importance of non-synaptic mechanisms in hippocampal epileptogenesis. Neurosci Lett. 1990;120:267–270. doi: 10.1016/0304-3940(90)90056-f. [DOI] [PubMed] [Google Scholar]
- Ellisman MH, Brooke MH, Kaiser KK, Rash JE. Appearance in slow muscle sarcolemma of specializations characteristic of a fast muscle after reinnervation by a fast muscle nerve. Exp Neurol. 1978;58:59–67. doi: 10.1016/0014-4886(78)90120-6. [DOI] [PubMed] [Google Scholar]
- Ellisman MH, Rash JE, Staehelin LA, Porter KR. Studies of excitable membranes: II. A comparison of specializations at neuromuscular junctions and nonjunctional sarcolemmas of mammalian fast and slow twitch muscle fibers. J Cell Biol. 1976;68:752–774. doi: 10.1083/jcb.68.3.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskandari S, Wright EM, Kreman M, Sterace DM, Zampighi GA. Structural analysis of cloned plasma membrane proteins by freeze-fracture electron microscopy. Proc Natl Acad Sci USA. 1998;95:11235–11240. doi: 10.1073/pnas.95.19.11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend DS. Loss of square (orthogonal) arrays from cultured airway epithelial cells. J Electron Microsc Tech. 1987;6:237–246. [Google Scholar]
- Frigeri A, Gropper MA, Umenishi F, Kawashima M, Brown D, Verkman AS. Localization of MIWC and GLIP water channels homologs in neuromuscular, epithelial and glandular tissues. J Cell Sci. 1995;108:2933–3002. doi: 10.1242/jcs.108.9.2993. [DOI] [PubMed] [Google Scholar]
- Fujimoto K. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins: application to the immunogold labeling of intercellular junctional complexes. J Cell Sci. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- Fujimoto K. SDS-digested freeze-fracture replica labeling electron microscopy to study the two-dimensional distribution of integral membrane proteins and phospholipids in biomembranes: practical procedure, interpretation and application. Histochem Cell Biol. 1997;107:87–96. doi: 10.1007/s004180050092. [DOI] [PubMed] [Google Scholar]
- Furman CS, Gorelick-Feldman DA, Davidson KGV, Yasumura T, Neely JD, Agre P, Rash JE. Aquaporin-4 square array assembly: opposing actions of M1 and M23 isoforms. Proc Natl Acad Sci USA. 2003;100:13609–13614. doi: 10.1073/pnas.2235843100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen T, Sliz P, Kistler J, Cheng Y, Walz T. Aquaporin-0 membrane junctions reveal the structure of a closed water pore. Nature. 2004;429:193–197. doi: 10.1038/nature02503. [DOI] [PubMed] [Google Scholar]
- Gordon RE. Orthogonal arrays in normal and injured respiratory airway epithelium. J Cell Biol. 1985;100:648–651. doi: 10.1083/jcb.100.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton JD, Ellisman MH. The distribution of orthogonal arrays and their relationship to intercellular junctions in neuroglia of the freeze-fractured hypothamo-neurohypophysial system. Cell Tissue Res. 1981;215:309–323. doi: 10.1007/BF00239117. [DOI] [PubMed] [Google Scholar]
- Hatton JD, Ellisman MH. The distribution of orthogonal arrays in freeze-fractured rat median eminence. J Neurocytol. 1982;11:335–349. doi: 10.1007/BF01258250. [DOI] [PubMed] [Google Scholar]
- Heuser JE, Reese TS, Dennis MJ, Jan Y, Jan L, Evans L. Synaptic vesicle exocytosis captured by quick freezing and correlated with quantal transmitter release. J Cell Biol. 1981;275:275–300. doi: 10.1083/jcb.81.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi K, Fujita A, Inanobe A, Tanemoto M, Doi K, Kubo T, Kurachi Y. An inwardly rectifying K(+) channel, Kir4.1, expressed in astrocytes surrounds synapses and blood vessels in brain. Am J Physiol. 2001;281:C922–C931. doi: 10.1152/ajpcell.2001.281.3.C922. [DOI] [PubMed] [Google Scholar]
- Hristova K, Wimley WC, Mishra VK, Anantharamiah GM, Segrest JP, White SH. An amphipathic alpha-helix at a membrane interface: a structural study using a novel X-ray diffraction method. J Mol Biol. 1999;290:99–117. doi: 10.1006/jmbi.1999.2840. [DOI] [PubMed] [Google Scholar]
- Hubert JF, Thomas D, Cavalier A, Gouranton J. Structural and biochemical observations on specialized membranes of the “filter chamber,” a water-shunting complex in sap-sucking homopteran insects. Biol Cell. 1989;66:155–163. [PubMed] [Google Scholar]
- Hudson CS, Dyas BK, Rash JE. Changes in number and distribution of orthogonal arrays during postnatal muscle development. Dev Brain Res. 1982;4:91–101. doi: 10.1016/0165-3806(82)90100-6. [DOI] [PubMed] [Google Scholar]
- Hudson CS, Rash JE, Shinowara N. In: Freeze-fracture and freeze-etch methods. In: Current trends in morphological techniques. Johnson JE, editor. II. Boca Raton: CRC Press; 1981. pp. 183–217. [Google Scholar]
- Jefferys JG. Nonsynaptic modulation of neuronal activity in the brain: electric currents and extracellular ions. Physiol Rev. 1995;75:689–723. doi: 10.1152/physrev.1995.75.4.689. [DOI] [PubMed] [Google Scholar]
- Johnson AK, Loewy AD. In: Central Regulation of Autonomic Functions. In: Circumventricular organs and their role in visceral functions. Loewy AD, Spyer KM, editors. New York: Oxford University Press; 1990. pp. 247–267. [Google Scholar]
- Jung JS, Preston GM, Smith BL, Guggino WB, Agre P. Molecular structure of the water channel through aquaporin CHIP: the hourglass model. J Biol Chem. 1994;269:14648–14654. [PubMed] [Google Scholar]
- Kandel ER, Schwartz JH, Jessell TM. Principles of neural science. New York: Elsevier; 1992. [Google Scholar]
- King LS, Agre P. Pathophysiology of the aquaporin water channels. Annu Rev Physiol. 1996;58:619–648. doi: 10.1146/annurev.ph.58.030196.003155. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Minami S, Itoh S, Shirashi S, Yokoo H, Yanagita T, Uezono Y, Mohri M, Wada A. Aquaporin subtypes in rat cerebral microvessels. Neurosci Lett. 2001;297:163–166. doi: 10.1016/s0304-3940(00)01705-5. [DOI] [PubMed] [Google Scholar]
- Kreutziger GO. Freeze-etching of intercellular junctions of mouse liver. Microsc Soc Am Proc. 1968;26:234–235. [Google Scholar]
- Landis DMD. Membrane structure in mammalian astrocytes: a review of freeze-fracture studies on adult, developing, reactive and cultured astrocytes. J Exp Biol (Br) 1981;95:35–48. doi: 10.1242/jeb.95.1.35. [DOI] [PubMed] [Google Scholar]
- Landis DMD, Reese TS. Arrays of particles in freeze-fractured astrocytic membranes. J Cell Biol. 1974;60:316–320. doi: 10.1083/jcb.60.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski L, Caspar DLD, Phillips WC, Goodenough DA. Gap junction structures: II. Analysis of the x-ray diffraction data. J Cell Biol. 1977;74:629–645. doi: 10.1083/jcb.74.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham R, Frey S, Hills GJ. Methods for the enhancement of image detail and accentuation of structure in electron microscopy. Virology. 1963;20:88–102. [Google Scholar]
- Nagelhus EA, Horio Y, Inanobe A, Fujita A, Haug F-M, Nielsen S, Kurachi Y, Ottersen OP. Immunogold evidence suggests that coupling of K+ siphoning and water transport in rat retinal Muller cells is mediated by coenrichment of Kir4.1 and AQP4 in specific membrane domains. Glia. 1999;26:47–54. doi: 10.1002/(sici)1098-1136(199903)26:1<47::aid-glia5>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Nagy JI, Ionescu A-V, Lynn BD, Rash JE. Coupling of astrocyte connexins Cx26, Cx30, Cx43 to oligodendrocyte Cx29, Cx32, Cx47: implications from normal and connexin32 knockout mice. Glia. 2003;44:205–218. doi: 10.1002/glia.10278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy JI, Li X, Rempel J, Stelmack GL, Patel D, Staines WA, Yasumura T, Rash JE. Connexin26 in adult rodent CNS: demonstration at astrocytic gap junctions and co-localization with connexin30 and connexin43. J Comp Neurol. 2001;441:302–323. doi: 10.1002/cne.1414. [DOI] [PubMed] [Google Scholar]
- Neely JD, Christensen BM, Nielsen S, Agre P. Heterotetrameric composition of aquaporin-4 water channels. Biochemistry. 1999;38:11156–11163. doi: 10.1021/bi990941s. [DOI] [PubMed] [Google Scholar]
- Nielsen S, Nagelhus EA, Amiry-Moghaddam M, Bourque C, Agre P, Ottersen OP. Specialized membrane domains for water transport in glial cells: high-resolution immunogold cytochemistry of aquaporin-4 in rat brain. J Neurosci. 1997;17:171–180. doi: 10.1523/JNEUROSCI.17-01-00171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S, Smith BL, Christensen EI, Agre P. Distribution of aquaporin CHIP in secretory and resorptive epithelia and capillary endothelia. Proc Natl Acad Sci USA. 1993;90:7275–7279. doi: 10.1073/pnas.90.15.7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TE, Boyne AF. Liquid nitrogen-based quick freezing: experiments with bounce-free delivery of cholinergic nerve terminals to a metal surface. J Electron Microsc Tech. 1984;1:9–29. [Google Scholar]
- Rash JE, Dillman RK, Bilhartz BL, Duffy HS, Whalen LR, Yasumura T. Mixed synapses discovered and mapped throughout mammalian spinal cord. Proc Natl Acad Sci USA. 1996;93:4235–4239. doi: 10.1073/pnas.93.9.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash JE, Dillman RK, Morita M, Whalen LR, Guthrie PB, Fay-Guthrie D, Wheeler DW. Grid-mapped freeze fracture: correlative confocal laser scanning microscopy and freeze-fracture electron microscopy of preselected cells in tissue slices. In: Severs NJ, Shotton DM, editors. Rapid freezing, freeze fracture, and deep etching. New York: Wiley-Liss, Inc; 1995. pp. 127–150. [Google Scholar]
- Rash JE, Duffy HS, Dudek FE, Bilhartz BL, Whalen LR, Yasumura T. Grid-mapped freeze-fracture analysis of gap junctions in gray and white matter of adult rat central nervous system, with evidence for a “panglial syncytium” that is not coupled to neurons. J Comp Neurol. 1997;388:265–292. doi: 10.1002/(sici)1096-9861(19971117)388:2<265::aid-cne6>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Rash JE, Ellisman MH. Studies of excitable membranes: I. Macromolecular specializations of the neuromuscular junction and the nonjunctional sarcolemma. J Cell Biol. 1974;63:567–586. doi: 10.1083/jcb.63.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash JE, Staehelin LA, Ellisman MH. Rectangular arrays of particles on freeze-cleaved plasma membranes are not gap junctions. Exp Cell Res. 1974;86:187–190. doi: 10.1016/0014-4827(74)90670-3. [DOI] [PubMed] [Google Scholar]
- Rash JE, Staines WA, Yasumura T, Patel D, Hudson CS, Stelmack GL, Nagy JI. Immunogold evidence that neuronal gap junctions in adult rat brain and spinal cord contain connexin36 (Cx36) but not Cx32 or Cx43. Proc Natl Acad Sci USA. 2000;97:7573–7578. doi: 10.1073/pnas.97.13.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash JE, Yasumura T. Improved structural detail in freeze-fracture replicas: high-angle shadowing of gap junctions cooled below −170° C and protected by liquid nitrogen-cooled shrouds. J Electron Microsc Tech. 1992;20:187–204. doi: 10.1002/jemt.1070200207. [DOI] [PubMed] [Google Scholar]
- Rash JE, Yasumura T. Direct immunogold labeling of connexins and aquaporin4 in freeze-fracture replicas of liver, brain and spinal cord: factors limiting quantitative analysis. Cell Tissue Res. 1999;296:307–321. doi: 10.1007/s004410051291. [DOI] [PubMed] [Google Scholar]
- Rash JE, Yasumura T, Dudek FE, Nagy JI. Cell-specific expression of connexins, and evidence for restricted gap junctional coupling between glial cells and between neurons. J Neurosci. 2001;21:1983–2001. doi: 10.1523/JNEUROSCI.21-06-01983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash JE, Yasumura T, Hudson CS, Agre P, Nielsen S. Direct immunogold labeling of aquaporin-4 in “square arrays” of astrocyte and ependymocyte plasma membranes in rat brain and spinal cord. Proc Natl Acad Sci USA. 1998;95:11981–11986. doi: 10.1073/pnas.95.20.11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash JE, Yasumura T, Walrond JP, Steere RL, Okutomi S. Gap junctions: a new level of detail revealed by shadowing at −170 to −182 C° in a JEOL JFD-9000 equipped with cryopump and liquid nitrogen-cooled shrouds. Int Cong Electron Microsc. 1990;12:800–801. [Google Scholar]
- Ren G, Cheng A, Reddy V, Melnyk P, Mitra AK. Three-dimensional fold of the human AQP1 water channel determined at 4 Å resolution by electron crystallography of two-dimensional crystals embedded in ice. J Mol Biol. 2000;301:369–387. doi: 10.1006/jmbi.2000.3949. [DOI] [PubMed] [Google Scholar]
- Roper SN, Obenaus A, Dudek FE. Osmolality and nonsynaptic epileptiform: the bursts in rat CA1 and dentate gyrus. Ann Neurol. 1992;31:81–85. doi: 10.1002/ana.410310115. [DOI] [PubMed] [Google Scholar]
- Silberstein C, Bouley R, Huang Y, Fang P, Pastor-Soler N, Brown D, Van Hoek AN. Membrane organization and function of M1- and M23-isoforms of aquaporin-4 in epithelial cells. Am J Physiol Renal Physiol. 2004;287:F501–F511. doi: 10.1152/ajprenal.00439.2003. [DOI] [PubMed] [Google Scholar]
- Sjostrand FS. Electron microscopy of cells and tissues. Vol. 1. New York: Academic Press; 1967. [Google Scholar]
- Staehelin LA. Three types of gap junctions interconnecting intestinal epithelial cells visualized by freeze-etching. Proc Natl Acad Sci USA. 1972;69:1318–1321. doi: 10.1073/pnas.69.5.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere RL. Electron microscopy of structural detail in frozen biological specimens. J Biophys Biochem Cytol. 1957;3:45–60. doi: 10.1083/jcb.3.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere RL, Erbe EF, Moseley JM. Prefracture and cold-fracture images of yeast plasma membranes. J Cell Biol. 1980;86:113–122. doi: 10.1083/jcb.86.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere RL, Moseley M. New dimensions in freeze-etching. Proc Elec Microsc Soc Am. 1969;27:202–203. [Google Scholar]
- Unwin PNT, Zampighi G. Structure of the junction between communicating cells. Nature. 1980;283:545–549. doi: 10.1038/283545a0. [DOI] [PubMed] [Google Scholar]
- Van Hoek AN, Yang B, Kirmiz S, Brown D. Freeze-fracture analysis of plasma membranes of CHO cells stably expressing aquaporins 1–5. J Membr Biol. 1998;165:243–254. doi: 10.1007/s002329900438. [DOI] [PubMed] [Google Scholar]
- Verbavatz J-M, Van Hoek AN, Ma T, Sabolic I, Valenti G, Ellisman MH, Ausiello DA, Verkman AS, Brown D. A 28 kDa sarcolemmal antigen in kidney principal cell basolateral membranes: relationship to orthogonal arrays and MIP26. J Cell Sci. 1994;107:1083–1094. doi: 10.1242/jcs.107.4.1083. [DOI] [PubMed] [Google Scholar]
- Verbavatz JM, Ma T, Gobin P, Verkman AS. Absence of orthogonal arrays in kidney, brain, and muscle from transgenic knockout mice lacking aquaporin-4. J Cell Sci. 1997;110:2855–2860. doi: 10.1242/jcs.110.22.2855. [DOI] [PubMed] [Google Scholar]
- Vidossich P, Cascella M, Carloni P. Dynamics and energetics of water permeation through the aquaporin channel. Proteins. 2004;55:924–931. doi: 10.1002/prot.10642. [DOI] [PubMed] [Google Scholar]
- Walz T, Hirai T, Murata K, Heymann JB, Mitsuoka K, Fujiyoshi Y, Smith BL, Agre P, Engel A. The three-dimensional structure of aquaporin-1. Nature. 1997;387:624–627. doi: 10.1038/42512. [DOI] [PubMed] [Google Scholar]
- Warth A, Kröger, Wolburg H. Redistribution of aquaporin-4 in human glioblastoma correlates with loss of agrin immunoreactivity from brain capillary basal laminae. Acta Neuropathol. 2004;107:311–318. doi: 10.1007/s00401-003-0812-0. [DOI] [PubMed] [Google Scholar]
- Winkler H, Wildhaber H, Gross H. Decoration on the surface of a regular protein layer. Ultramicroscopy. 2002;16:331–339. [Google Scholar]
- Wolburg H. Orthogonal arrays of intramembrane particles: a review with special reference to astrocytes. J Brain Res. 1995;36:239–258. [PubMed] [Google Scholar]
- Yang B, Brown D, Verkman AS. The mercurial-insensitive water channel (AQP-4) forms orthogonal arrays in stably transfected CHO cells. J Biol Chem. 1996;271:4577–4580. [PubMed] [Google Scholar]
- Zampighi GA, Hall JE, Ehring GR, Simon SA. The structural organization and protein composition of lens fiber junctions. J Cell Biol. 1989;108:2255–2275. doi: 10.1083/jcb.108.6.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]


