Abstract
The major mutagenic base lesion in DNA caused by exposure to reactive oxygen species is 8-hydroxyguanine or 7,8-dihydro-8-oxoguanine (8-OH-G). Products of the human MMH/OGG1 gene are known to catalyze in vitro the reactions repairing this DNA lesion. To analyze the function of Mmh in vivo, we generated a mouse line carrying a mutant Mmh allele by targeted gene disruption. Mmh homozygous mutant mice were found to have a physically normal appearance, but to have lost nicking activity in liver extracts for substrate DNA containing 8-OH-G, exhibiting a 3-fold increased accumulation of this adduct at 9 weeks of age compared with wild-type or heterozygous mice. Further elevation to 7-fold was observed in 14-week-old animals. Substantial increase of spontaneous mutation frequencies was clearly identified in Mmh mutant mice bearing transgenic gpt genes. These results indicate that exposure of DNA to endogenous oxidative species continuously produces the mutagenic adduct 8-OH-G in mice, and Mmh plays an essential role in repair of this DNA damage.
Damage to genomic DNA is considered to be involved in aging and age-associated degenerative diseases, with 8-hydroxyguanine or 7,8-dihydro-8-oxoguanine (8-OH-G) induced by reactive oxygen species and ionizing radiation as one of the most critical mutagenic lesions. G → T transversion mutations found in mammalian cells appear to be caused by mispairing of this oxidized base with A (1–3). In bacteria and Saccharomyces cerevisiae, 8-OH-G is excised by a DNA glycosylase (MutM and OGG1, respectively) with associated lyase activity for chain cleavage. A mammalian homologue of 8-OH-G glycosylase/apurinic, apyrimidinic lyase (AP lyase; MutM homologue, MMH) has been identified and cloned (4–10). We recently showed, by using a human MMH type 1a-specific antibody, that human MMH (OGG1) type 1a protein (one isoform derived from the MMH gene) is a major enzyme for repair of 8-OH-G lesions in human cells (11). By employing an anti-human OGG1 antibody, which presumably recognizes all isoforms of the OGG1 product, S. Mitra and his coworkers (12) also reported a 38-kDa OGG1, identical to the MMH gene product, found in HeLa cell extract to be a major enzyme for 8-OH-G-specific DNA strand incision. It has remained unclear, however, whether this mammalian homologue works in vivo to repair oxidation-associated lesions in genomic DNA, and how deletion of this enzyme may affect cells in mammals.
Materials and Methods
Generation of Mmh-Deficient Mice.
For construction of a targeting vector, we isolated five λ phage clones from a genomic library of J1 embryonic stem (ES) cells by using mouse Mmh cDNA, mapped the clones in detail by restriction digestion, and used DNA fragments of two clones for vector construction. An approximately 8.3-kb HindIII–XbaI fragment containing exons 4–7 and a 2-kb HindIII–ScaI fragment containing exon 1 and the 5′ half of exon 2 were isolated and applied for construction of a targeting vector as long and short homologous sequences, respectively.
According to the procedure described previously (13), to generate mutant mice, J1 ES cells were electroporated with a targeting vector, and cells were selected in the presence of G418. To screen homologous recombinant ES cells, genomic DNA of G418-resistant clones was extracted, digested by XbaI, and subjected to Southern blot analysis with a 1-kb PstI–SacI fragment as a probe (probe 1 in Fig. 1A). Expected sizes of hybridized bands for wild-type and mutant Mmh alleles were 15 kb and 6.8 kb, respectively. Mutant ES cells were injected into C57BL/6J blastocysts, and resulting male chimeras were mated with female C57BL/6J mice. Germ-line transmission of mutant Mmh allele to F1 mice was confirmed by Southern-blot analysis. Genotyping of F2 offspring was performed by PCR amplification using tail lysates as templates with the primers shown in Fig. 1A (M2, CTCACTGGAGTGGCGTGCTGGCAGA; PGK-3, CCTGAAGAACGAGATCAGCAGCCTC; M5, CCATCCTGGTGGCCCTGTATCTGCA).
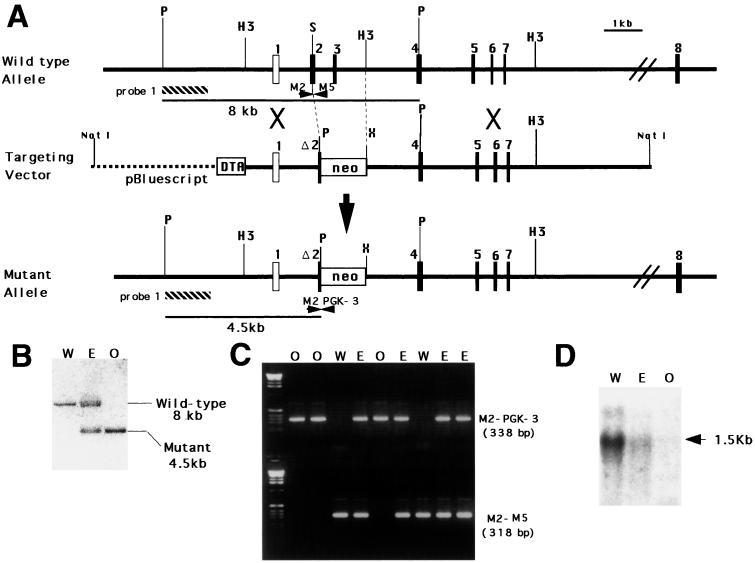
Figure 1.
Targeted disruption of mouse Mmh. (A) Restriction map of wild-type Mmh and structures of the targeting vector and the mutant allele generated by homologous recombination. Coding exons are represented by filled boxes and the noncoding exon by a white box. The genomic fragment used as a probe (probe 1) for Southern-blot analysis is shown by a hatched box. neo, neomycin phosphotransferase gene flanked by the phosphoglycerate kinase (Pgk) promoter and a polyadenylation signal; DTA, diphtheria toxin A gene driven by the MC1 promoter; P, PstI; H3, HindIII; S, ScaI; X, XbaI. (B) Southern-blot analysis of liver genomic DNA extracted from F2 mice obtained by F1 heterozygous intercrosses. PstI digestion generated hybridized bands of the expected sizes for wild-type and mutant Mmh of 8 kb and 4.5 kb respectively. W, wild-type; E, heterozygote; O, homozygous mutant. These notations are also used in C and D. (C) Genotyping of F2 offspring by PCR amplification. Tail lysates were used as the templates. Amplification of fragments from the mutant and wild-type allele was achieved by using M2/PGK-3 and M2/M5 primer pairs, respectively. The positions of the primers are indicated in A. (D) Northern-blot analysis of Mmh expression in liver RNA extracted from F2 mice. Each lane contained 2 μg of poly(A)-RNA.
Northern-blot analysis was carried out as described (14). Poly(A)-RNA was extracted from wild-type and Mmh mutant mouse liver by using FastTrack reagents (Invitrogen), fractionated by agarose gel electrophoresis with 1% formaldehyde, transferred to nylon membrane, and hybridized with a Mmh cDNA probe that corresponds to the sequence from exons 5–7.
AP Lyase Activity Toward 8-OH-G-Containing DNA.
Mouse tissues were homogenized in lysis buffer (50 mM Hepes/250 mM NaCl/0.1% Nonidet P-40/1 mM phenylmethylsulfonyl fluoride) with a Potter-type homogenizer and then centrifuged at 18,000 × g for 15 min. The supernatants thus obtained were used as the crude enzyme extracts. The 21-base oligonucleotide containing a single 8-OH-G (Goh) was synthesized by the method described previously (15), and the other oligonucleotides (without 8-OH-G) were synthesized with an Expedite DNA synthesizer model 8900 (PerSeptive Biosystems). The oligonucleotides were purified by HPLC (C18-MS column, 0.46 × 25 cm, Nacalai Tesque, Kyoto) and developed with a linear acetonitrile gradient (5–30%) in 24 ml of 0.1 M triethylamine acetate (pH 7.0) at a flow rate of 1.2 ml/min. The sequences of oligonucleotides used as substrates are shown below:
oligo 1: 5′-CAGCCAATCAGohTGCACCATCC-3′
oligo 2: 5′-CAGCCAATCAGTGCACCATCC-3′
oligo 3: 5′-GGATGGTGCACTGATTGGCTG-3′
oligo 4: 5′-GGATGGTGCAGTGATTGGCTG-3′
oligo 5: 5′-GGATGGTGCAATGATTGGCTG-3′
oligo 6: 5′-GGATGGTGCATTGATTGGCTG-3′.
An 8-OH-G-containing oligonucleotide (oligo 1) or merely G-containing oligonucleotide (oligo 2) was 5′-end labeled with [γ-32P]ATP and T4 polynucleotide kinase, and annealed with complementary oligonucleotides (oligos 3–6). Crude extracts or purified recombinant enzyme was incubated with 100 fmol of the end-labeled double-stranded oligonucleotide in 25 μl of 50 mM Tris⋅HCl, pH 7.5/50 mM KCl/5 mM EDTA at 37°C for 1 hr. After the incubation, labeled oligonucleotide fragments in the reaction mixture were precipitated with ethanol and isolated by electrophoresis in a 20% polyacrylamide gel containing 7 M urea. The radioactivity of the nicked products was quantified with a bioimaging analyzer (BAS2000, Fuji Photo Film, Tokyo).
Analysis of 8-OH-G in DNA with an HPLC-Electrochemical Detector (ECD) (16).
Genomic DNA was extracted from 100–200 mg of tissues with a DNA Extractor WB Kit (Wako, Japan) and digested with nuclease P1 (Sigma) and acid phosphatase type XA (Sigma) at 37°C for 30 min in 100 μl of 10 mM sodium acetate solution (pH 4.5) containing 1 mM EDTA. The resulting deoxynucleoside mixture was treated with the ion-exchange resin Muromac (Muromachi Kagaku, Tokyo), to remove the NaI and centrifuged at 18,000 × g for 5 min. The supernatant was filtered with Microcon 10 (Amicon) and analyzed with an HPLC-ECD system: pump, L-6000 (Hitachi); UV detector, L-4000 (Hitachi); ECD, Coulochem (ESA); column, Beckman Ultrasphere ODS (0.46 × 25 cm); eluent, 10 mM NaH2PO4 buffer containing 8% methanol; flow rate, 1 ml/min. The amounts of deoxyguanosine (dG) and 8-hydroxydeoxyguanosine (8-OH-dG) in the DNA samples were measured with UV at 290 nm and ECD (guard cell, +350 mV; detector I, +150 mV; detector II, +300 mV), respectively. Ten micrograms of dG (Sigma) and 100 pg of 8-OH-dG (Sigma) were injected as standards. The ratio of 8-OH-dG to dG in the DNA sample was determined from the peak areas of the standards.
All measurements of AP lyase activity and 8-OH-G amount in this study were performed on a blind-test basis.
Spontaneous Mutation Assay in Vivo.
gpt transgenic mice carrying ecogpt genes for detection of mutations introduced in vivo were used to estimate spontaneous mutation frequency in Mmh homozygous mutant mice compared with wild-type mice. gpt transgenic male mice of C57BL6 background were mated with Mmh F0 chimera to obtain F1 (gpt/Mmh+/−) mice. Mmh homozygous mutant (Mmh−/−) and wild-type (Mmh+/+) mice hemizygous for the gpt transgene were obtained by mating F1 (Mmh+/−) males with F1 (gpt/Mmh+/−) females. The copy number of gpt genes serving as targets of mutagenesis was estimated as approximately 80 per hemizygous allele in autosomes. High molecular weight DNAs of mice carrying the gpt transgene were extracted from liver according to the method of Suzuki et al. (17) and used for rescuing λ phage containing a linear plasmid. The gpt gene is included in this plasmid, and it was excised from the λ phage vector in host Escherichia coli expressing Cre recombinase to give a circularized form that eventually confers sensitivity to 6-thioguanine (6-TG). To rescue λ phages from the DNA, an in vitro packaging reaction was performed with TransPack extracts (Stratagene). Four micrograms of genomic DNA from an Mmh mutant mouse carrying the gpt transgene was used for each packaging reaction, and the resultant lysate was suspended in 500 μl of SM buffer (50 mM Tris⋅HCl, pH 7.5/10 mM MgSO4/100 mM NaCl/0.01% gelatin). All aliquots of the 500-μl phage suspension were infected into E. coli strain YG6020 expressing Cre recombinase to determine mutant frequencies for the gpt reporter gene. Infected cells were poured on plates containing 1.5% agar, 1× M9 salts, 1% glycerol, 2 mM MgSO4, 0.1 mM CaCl2, 5 μg/ml thiamin, 50 μg/ml proline, 50 μg/ml leucine, 50 μg/ml isoleucine, 25 μg/ml chloramphenicol, and 25 μg/ml 6-TG (18). For estimating the total rescue efficiencies of phages from mouse genomic DNA, diluted suspensions of infected cells were poured on plates containing chloramphenicol without 6-TG. Plates were incubated for 3 days at 37°C for selection of colonies harboring the plasmids carrying mutated gpt genes. To confirm the results of 6-TG selection, surviving colonies were replated on fresh 6-TG plates and incubated for another 3 days. The gpt mutant frequency was calculated as M/(W × d) where M, W, and d stand for total number of mutant colonies on a 6-TG plate, number of colonies on a plate without 6-TG, and the fold dilution, respectively.
To analyze the mutation spectrum, the entire gpt gene of each mutant was amplified by the PCR procedure with a primer pair as follows; Gpt-1seq: TACCACTTTATCCCGCGTCAGG; Gpt-2seq: ACAGGGTTTCGCTCAGGTTTGC. Obtained PCR products were purified and sequenced directly with a ABI Prism 210 DNA Sequencer (Applied Biosystems).
Results
Genetic Inactivation of the Mmh Gene in Mice.
Using homologous recombination in ES cells, we obtained two independent ES clones (cl 132 and cl 165) carrying mutant alleles of Mmh, in which the 3′ half of exon 2 and the entire sequence of exon 3 were replaced by the pgk-neo gene cassette (Fig. 1A). Chimeric mice were generated and germ-line transmission of mutant Mmh alleles to F1 offspring was confirmed for chimeric mice generated from both ES clones (Fig. 1B). To produce F2 progeny for the analysis, interbreeding of F1 heterozygotes originating from the cl 165 ES cell was performed. The results described in this paper were obtained from the analysis of the resultant F2 mice. Genotype analysis of 86 F2 offspring was performed by PCR (Fig. 1C): 22 were wild type (25%), 41 were heterozygous (48%), and 23 were homozygous mutants (27%), consistent with the ratio expected from Mendelian inheritance. Mmh expression was not detected in the liver of the Mmh homozygous mutant by Northern-blot analysis (Fig. 1D). Clearly the Mmh gene product does not play an essential role in embryonic development, and both homozygous and heterozygous mutants appeared healthy with no obvious differences from wild-type littermates in survival and gross appearance until 50 weeks old. No tumor formation has been observed thus far. Both male and female homozygotes proved fertile, and F3 homozygous offspring obtained by homozygous intercrosses were also found to be healthy.
AP Lyase Activity in Mmh Mutant Liver Cells.
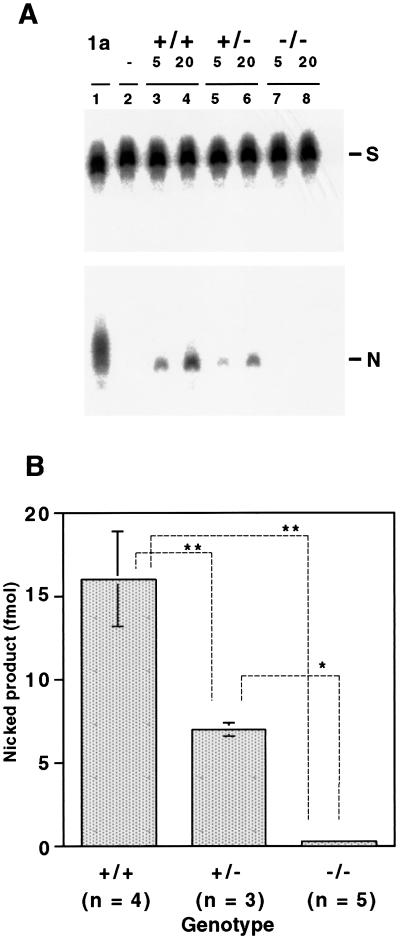
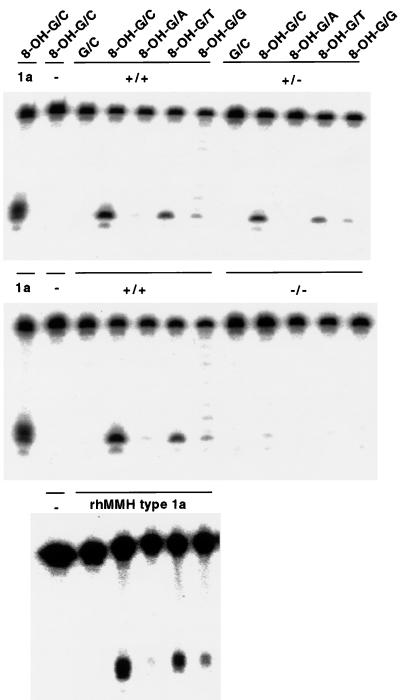
Because mammalian MMH/OGG1 products have been shown to have glycosylase/AP lyase activity that may specifically excise 8-OH-G in DNA, AP lyase activity in liver extracts of Mmh mutant mice was analyzed. Nicking activity toward substrate DNA with 8-OH-G⋅C pairs was measured by titrating the radioactivity of nicked products on gels. As shown in Fig. 2, the results showed a clear decrease of activity consistent with the gene dosage; 16 fmol/25 μg of protein for the wild type, 7.0 for heterozygous mutants, and 0.36 for homozygous mutants. Although not totally zero when compared with negative control (Figs. 2A and 3), the residual activity of unknown origin in homozygotes was at most less than 5% of that for wild-type mice. The substrate specificity of nicking activity in liver extracts was also determined (Fig. 3). In wild-type mice it was in the following order: 8-OH-G/C > 8-OH-G/T > 8-OH-G/G > 8-OH-G/A ≫ G/C, identical to findings for recombinant human MMH type 1a (4, 19). Although reduced to half of the wild type, extracts obtained from heterozygotes showed exactly the same spectrum of specificity. Activity against all substrates almost disappeared in liver extracts of homozygotes. From these observations we conclude that major repair of 8-OH-G is carried by the Mmh gene product, at least in mouse liver cells.
Figure 2.
AP lyase activity in mouse liver crude extracts. A 5′-end-labeled oligonucleotide containing an 8-OH-G hybridized with the complementary oligonucleotide was incubated with the crude extracts, and then nicked products produced were analyzed by 20% PAGE. (A) Representative AP lyase assay: lane 1, 1 ng of purified recombinant human MMH type 1a protein as a positive control; lane 2, the lysis buffer as a negative control; lanes 3 and 4, 5 and 20 μg of crude extract of liver from an Mmh wild-type mouse; lanes 5 and 6, 5 and 20 μg of crude extract of liver from an Mmh heterozygous mutant mouse; lanes 7 and 8, 5 and 20 μg of crude extract of liver from an Mmh homozygous mutant mouse. S, substrate oligonucleotide; N, nicked product. (B) Quantification of the nicked products by analyzing radioactivity with a bioimaging analyzer BAS2000. Numbers of mice used are indicated under the genotype. Results are mean and SD. *, P < 0.0003; **, P < 0.0001 (Fisher statistical analysis)
Figure 3.
Substrate specificity of AP lyase activity in crude liver extracts of Mmh wild-type, Mmh heterozygous mutant, and Mmh homozygous mutant mice. The 8-OH-G- or G-containing oligonucleotide was labeled and annealed with one of the four complementary oligonucleotides having C, A, T, or G opposite the 8-OH-G or G. The crude liver extracts, purified recombinant human MMH type 1a protein (rhMMH type 1a or 1a) or lysis buffer (−) were incubated with the substrates listed above and then the nicked products were analyzed by 20% PAGE.
Accumulation of 8-OH-G in DNA of Mmh Mutant Mice.
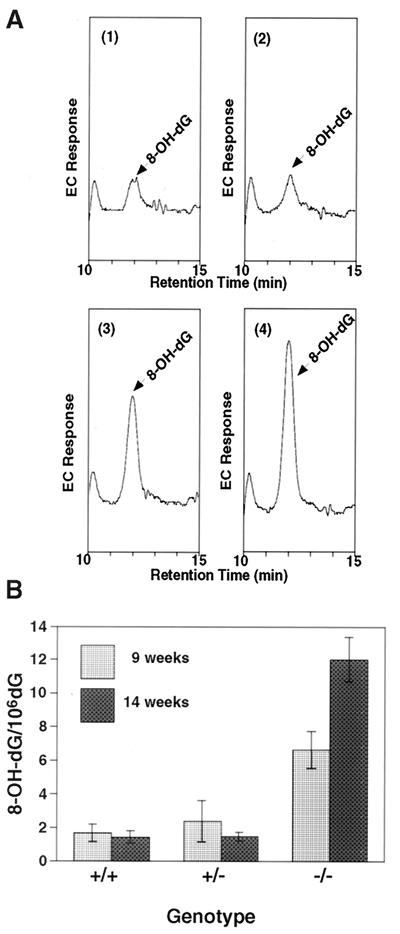
To analyze the effects of Mmh deficiency on oxidative DNA lesion accumulation in mice, the amount of 8-OH-G in genomic DNA was measured by a HPLC–electrochemical detection method, which is highly selective, with a sensitivity at the fmol level (Fig. 4). In DNA of wild-type mice the amount was measured to be 1.0–2.0 in 106 dG, in good agreement with our previous report (20). A significant increase was observed in the liver DNA of Mmh homozygous mutants (Fig. 4B): at 9 weeks of age, the level of 8-OH-G in liver DNA was 6.6 in 106 dG, about 4.2-fold the wild-type value, and further increase to 12.0 in 106 dG was observed in 14-week-old animals. This indicated accumulation of 5.4 8-OH-G in 106 dG within 5 weeks, or approximately 3,900 8-OH-Gs every week per diploid DNA of liver cells that are deficient in the Mmh gene product. Although heterozygotes possessed only half the AP lyase activity of wild-type mice, this seemed to be enough to process the usual 8-OH-G load, since no statistically significant elevation was apparent.
Figure 4.
Accumulation of 8-OH-G in DNA of Mmh-deficient mice. (A) Electrochemical chromatograms of 8-OH-dG in Mmh mutant mouse liver genomic DNAs. The HPLC patterns were traced by an electrochemical (EC) detector as described in Materials and Methods. Patterns are for 8-OH-dG level, in 1, a wild-type mouse; 2, an Mmh heterozygous mutant mouse; 3, an Mmh homozygous mutant mouse at 9 weeks of age; and 4, an Mmh homozygous mutant mouse at 14 weeks of age. Arrows indicate 8-OH-dG peaks, the areas of those for homozygotes being markedly increased. (B) The measured amounts of 8-OH-G in liver tissues from Mmh wild-type, heterozygous mutant, and homozygous mutant mice at 9 and 14 weeks of age are summarized. Values are mean ± SD (n = 3). The 8-OH-G levels in Mmh homozygous mutant mice were significantly higher than those in Mmh wild-type mice at both 9 and 14 weeks of age at P < 0.0001 (Fisher's test). The 8-OH-G levels in 14-week-old homozygotes were significantly higher than those in 9-week-old Mmh homozygous mutant mice at P < 0.0001.
Increased Mutation Frequencies in Mmh Mutants.
Mmh homozygous mutant (Mmh−/−) and wild-type (Mmh+/+) mice hemizygous for the gpt transgene, serving as a target of mutagenesis, were established to assess spontaneous mutation frequencies in the mutant mice. Liver DNA of six Mmh homozygous mutants 16–20 weeks old and six wild-type mice of the same age, all carrying the gpt gene, were used independently for the assay. The number of gpt mutant colonies (6-TG-resistant and chloramphenicol-resistant), the total number of colonies plated (chloramphenicol-resistant), and the calculated mutation frequencies for each mouse are shown in Table 1. Consistent with the previous report (18), the mutation frequencies for gpt/Mmh+/+ transgenic mice were under 1 × 10−5, although the values fluctuated among these six mice. On the other hand, in liver DNAs obtained from six Mmh homozygous mutants, the mutation frequencies tended to increase and, on average, 2.3-fold elevation of the frequencies was noted. This increase was considered to be statistically significant (Fisher's test, P < 0.05). To analyze the spectrum of mutations introduced spontaneously in vivo, plasmids were recovered from gpt mutant colonies and the ORFs of the gpt gene were sequenced. Mutations thus found were all single-base substitutions. In Mmh-deficient mice, 26 out of a total of 46 base substitutions (57%) were G → T or C → A transversions, and the rest were 15 G → A transitions and 5 T → A transversions. In the wild type, 8 G → T or C → A transversions out of 23 single-base-pair substitutions (35%) were found. This mutation spectrum implies that more than 70% of the increase in mutations in Mmh homozygotes was accounted for by G → T or C → A transversions. Because 8-OH-G has been shown to mispair with A, resulting in G → T transversions (1–3), these results suggest that the majority of increased mutation in vivo was caused by the 8-OH-G formation in Mmh-deficient mice.
Table 1.
Mutation frequencies observed in gpt/Mmh+/+ (W) and gpt/Mmh−/− (O) mouse liver cells
| Mouse no. | Mmh | CmR colonies | 6-TGR and CmR colonies | Mutation frequency |
|---|---|---|---|---|
| 1 | W | 1.3 × 106 | 4 | 3.1 × 10−6 |
| 2 | W | 1.1 × 106 | 3 | 2.7 × 10−6 |
| 3 | W | 1.2 × 106 | 5 | 4.2 × 10−6 |
| 4 | W | 1.8 × 106 | 3 | 1.7 × 10−6 |
| 5 | W | 3.9 × 105 | 4 | 10.0 × 10−6 |
| 6 | W | 6.0 × 105 | 4 | 6.4 × 10−6 |
| Mean | (4.7 ± 3.1) × 10−6 | |||
| 7 | O | 1.0 × 106 | 11 | 11.0 × 10−6 |
| 8 | O | 1.7 × 106 | 8 | 4.7 × 10−6 |
| 9 | O | 3.1 × 105 | 6 | 19.0 × 10−6 |
| 10 | O | 8.6 × 105 | 7 | 8.1 × 10−6 |
| 11 | O | 1.1 × 106 | 9 | 8.2 × 10−6 |
| 12 | O | 3.9 × 105 | 5 | 13.0 × 10−6 |
| Mean | (11.0 ± 5.0) × 10−6 |
6-TGR and CmR stand for 6-TG-resistant and chloramphenicol-resistant, respectively. Numbers of colonies are for six independent wild-type mice 16–20 weeks old or six homozygous mice of the same age. On average, Mmh homozygotes shows a 2.3-fold elevation of mutation frequency relative to the wild-type counterparts (P < 0.03, Fisher's test).
Discussion
The results of this study provide evidence of an essential role of Mmh in the mouse in repair of 8-OH-G, the major mutagenic base lesion in DNA caused by exposure to reactive oxygen species. Naturally these species may be derived from both exogenous and endogenous sources. Our findings indicate that oxidative adducts in mouse liver cells are constantly produced by oxygen species of endogenous origin, resulting in age-dependent accumulation of 8-OH-G in the absence of the Mmh gene product in mice. Mmh deficiency further caused increase of spontaneous mutation frequency in vivo largely because of the mispairing of A with 8-OH-G in DNA.
Previous reports have suggested the presence of two distinct enzymes to repair 8-OH-G in human cells (12, 21). A mutation of the Msh2 gene, one of the mismatch repair genes, has also be found to cause accumulation of 8-OH-G in mouse ES cells (23). Although there is a clear discrepancy between these results and ours with respect to basal levels of 8-OH-G in DNA from wild-type controls, DeWeese et al. (22) reported that ES cells from Msh2−/− mice accumulated much larger amounts of 8-OH-G in DNA compared with Msh2+/+ mice, either with or without low-level radiation treatment in tissue culture system. The results of our study described here, however, clearly indicate that the Mmh gene product may be solely responsible for repair of 8-OH-G in liver cells of mice. Bessho et al. (23) previously showed that mammalian N-methylpurine-DNA glycosylase releases 8-OH-G from DNA in vitro as well as in vivo when transfected into E. coli. The excision of 8-OH-G from synthetic DNA in vitro by a human nucleotide excision repair system was also reported by Reardon et al. (24). On the basis of our results, it is likely that none of these enzymatic systems plays a major role in repair of 8-OH-G in the mouse liver.
In this study, significantly increased levels of 8-OH-G in Mmh-deficient mice were observed. Levels of 8-OH-G detected in liver DNA from wild-type mice are approximately 1.5 8-OH-G per 106 dG in the system we used in the present study. It is possible that this is an over-estimation because of artificial generation of 8-OH-G during the course of isolation of DNA. In that case, deletion of Mmh must have exerted a devastating effect on increase of 8-OH-G in cellular DNA.
A relatively low spontaneous mutation rate and absence of tumors does not appear consistent with the marked accumulation of potentially miscoding 8-OH-G residues in the genome, as observed here for Mmh-deficient liver tissues. There are several possible explanations of this discrepancy. First, it may be merely that fixation of mutations is limited because only a small fraction of adult liver cells are proliferating. Second, it could be that the human homologue of the MutY DNA glycosylase provides repair of the opposite strand. It can excise A misincorporated opposite 8-OH-G in dividing cells. Such removal would result in possible reversion to a correct C. Third, there could be another complementary repair of 8-OH-G by the nucleotide excision-repair (NER) pathway. The NER system was shown to repair 8-OH-G lesions in vitro (24). 8-OH-G could be removed by relatively acute phase transcription-coupled NER activities which would suppress the level of the miscoding 8-OH-G in livers of Mmh-null mice. On the other hand, the genome-wide NER could be coupled with DNA replication, and in the Mmh-null liver cells this might not work effectively, because cell turnover is limited in the liver of adult mice, but genome-wide NER could still prevent the fixation of the mutation caused by the 8-OH-G lesion in proliferating cells. This would be consistent with the large increase of 8-OH-G in DNA and the observed relatively small increase in spontaneous mutations in livers of Mmh-deficient mice. Further study on the appropriate synthetic mutant mice is required to clarify this functional overlapping of the Mmh gene with other repair systems in mammals.
During revision of this paper, Klungland et al. (25) reported on independently obtained Mmh/Ogg1-deficient mice. They noted accumulation of 8-OH-G and increase of spontaneous mutation frequency in Mmh/Ogg1-deficient mice, consistent with our results.
Acknowledgments
We thank Eiko Ohtsuka for valuable reagents. We also express our appreciation to Hitomi Yamanaka, Yoshinobu Sugitani, Akiko Kawakami, Shioko Ito, and Hiromi Takahashi for their expert technical support and to Toshiyuki Kobayashi for valuable discussions. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Sports, Science and Culture of Japan (O.M. and T.N.).
Abbreviations
- 8-OH-G
8-hydroxyguanine or 7,8-dihydro-8-oxoguanine
- 8-OH-dG
8-hydroxydeoxyguanosine
- AP lyase
apurinic, apyrimidinic lyase
- ES cells
embryonic stem cells
- 6-TG
6-thioguanine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050404497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050404497
References
- 1.Kasai H, Nishimura S. In: Oxidative Stress: Oxidants and Antioxidants. Sies H, editor. London: Academic; 1991. pp. 99–116. [Google Scholar]
- 2.Cheng K C, Cahill D S, Kasai H, Nishimura S, Loeb L A. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 3.Moriya M. Proc Natl Acad Sci USA. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M, et al. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- 5.Rosenquist T A, Zharkov D O, Grollman A P. Proc Natl Acad Sci USA. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roldan-Arjona T, Wei Y F, Carter K C, Klungland A, Anselmino C, Wang R P, Augustus M, Lindahl T. Proc Natl Acad Sci USA. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radicella J P, Dherin C, Desmaze C, Fox M S, Boiteux S. Proc Natl Acad Sci USA. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arai K, Morishita K, Shinmura K, Kohno T, Kim S R, Nohmi T, Taniwaki M, Ohwada S, Yokota J. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- 9.Lu R, Nash H M, Verdine G L. Curr Biol. 1997;7:397–407. doi: 10.1016/s0960-9822(06)00187-4. [DOI] [PubMed] [Google Scholar]
- 10.Bjoras M, Luna L, Johnsen B, Hoff E, Haug T, Rognes T, Seeberg E. EMBO J. 1997;16:6314–6322. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monden Y, Arai T, Asano M, Ohtsuka E, Aburatani H, Nishimura S. Biochem Biophys Res Commun. 1999;258:605–610. doi: 10.1006/bbrc.1999.0649. [DOI] [PubMed] [Google Scholar]
- 12.Hazra T K, Izumi T, Maidt L, Floyd R A, Mitra S. Nucleic Acids Res. 1998;26:5116–5122. doi: 10.1093/nar/26.22.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakai S, Kawano H, Yudate T, Nishi M, Kuno J, Nagata A, Jishage K, Hamada H, Fujii H, Kawamura K, Shiba K, Noda T. Genes Dev. 1995;9:3109–3121. doi: 10.1101/gad.9.24.3109. [DOI] [PubMed] [Google Scholar]
- 14.Shibata H, Toyama K, Shioya H, Ito M, Hirota M, Hasegawa S, Matsumoto H, Takano H, Akiyama T, Toyoshima K, et al. Science. 1997;278:120–123. doi: 10.1126/science.278.5335.120. [DOI] [PubMed] [Google Scholar]
- 15.Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Miura K, Ohtsuka E, Nishimura S. Nature (London) 1987;327:77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- 16.Asami S, Hirano T, Yamaguchi R, Tsurudome Y, Itoh H, Kasai H. Biochem Biophys Res Commun. 1998;243:678–682. doi: 10.1006/bbrc.1998.8166. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T, Itoh T, Hayashi M, Nishikawa Y, Ikezaki S, Furukawa F, Takahashi M, Sofuni T. Environ Mol Mutagen. 1996;28:348–353. doi: 10.1002/(SICI)1098-2280(1996)28:4<348::AID-EM8>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 18.Nohmi T, Katoh M, Suzuki H, Matsui M, Yamada M, Watamnabe M, Suzuki M, Horiya N, Ueda O, Shibuya T, et al. Environ Mol Mutagen. 1996;28:465–470. doi: 10.1002/(SICI)1098-2280(1996)28:4<465::AID-EM24>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto F, Kasai H, Bessho T, Chung M H, Inoue H, Ohtsuka E, Hori T, Nishimura S. Jpn J Cancer Res. 1992;83:351–357. doi: 10.1111/j.1349-7006.1992.tb00114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasai H, Crain P F, Kuchino Y, Nishimura S, Ootsuyama A, Tanooka H. Carcinogenesis. 1986;7:1849–1851. doi: 10.1093/carcin/7.11.1849. [DOI] [PubMed] [Google Scholar]
- 21.Bessho T, Tano K, Kasai H, Ohtsuka E, Nishimura S. J Biol Chem. 1993;268:19416–19421. [PubMed] [Google Scholar]
- 22.DeWeese T L, Shipman J M, Larrier N A, Buckley N M, Kidd L R, Groopman J D, Cutler R G, te Riele H, Nelson W G. Proc Natl Acad Sci USA. 1998;95:11915–11920. doi: 10.1073/pnas.95.20.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bessho T, Roy R, Yamamoto K, Kasai H, Nishimura S, Tano K, Mitra S. Proc Natl Acad Sci USA. 1993;90:8901–8904. doi: 10.1073/pnas.90.19.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reardon J T, Bessho T, Kung H C, Bolton P H, Sancar A. Proc Natl Acad Sci USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes D E. Proc Natl Acad Sci USA. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]