Abstract
The process by which translation is initiated has long been considered similar in Bacteria and Eukarya but accomplished by a different unrelated set of factors in the two cases. This not only implies separate evolutionary histories for the two but also implies that at the universal ancestor stage, a translation initiation mechanism either did not exist or was of a different nature than the extant processes. We demonstrate herein that (i) the “analogous” translation initiation factors IF-1 and eIF-1A are actually related in sequence, (ii) the “eukaryotic” translation factor SUI1 is universal in distribution, and (iii) the eukaryotic/archaeal translation factor eIF-5A is homologous to the bacterial translation factor EF-P. Thus, the rudiments of translation initiation would seem to have been present in the universal ancestor stage. However, significant development and refinement subsequently occurred independently on both the bacterial lineage and on the archaeal/eukaryotic line.
The three major cellular information processing systems (replication, transcription, and translation) differ greatly in the degree to which their componentry is universally conserved (1, 2). At the one extreme is genome replication, where not even the central DNA polymerase is orthologous between the Bacteria and the Archaea/Eukarya (1, 3). At the other is translation, where most of the componentry is universal in distribution: ribosomal RNAs are strongly conserved in both primary and secondary structure among all organisms (1, 4); the majority of the ribosomal proteins are as well, as are most of the elongation factors, the tRNAs, and aminoacyl-tRNA synthetases (1, 2). The only major exception appears to be translation initiation.
Although translation initiation is functionally similar in all organisms, the underlying componentry seems to be quite dissimilar between the Bacteria and the Archaea/Eukarya. The bacterial mechanism seems simple; three (single subunit) proteins are involved: initiation factors IF-1, IF-2, and IF-3 (5). By contrast, eukaryotic initiation is complex, involving a larger number of protein factors, many of which comprise multiple subunits (6, 7).
The relationship between these two translation initiation systems addresses a central evolutionary question: How advanced in their evolutionary development were the various cellular information processing systems at the time the universal ancestor gave rise to the primary lines of organismal descent; more generally, what was the nature of this entity we call the universal ancestor? Was it more rudimentary than the cells we study today—and, if so, in what ways (2, 8, 9)?
The publication of the Methanococcus jannaschii genome (10–12) allowed the first complete (comparative) examination of the componentry of archaeal information processing systems. Before this, our understanding of archaeal translation initiation was at best rudimentary (13). The fact that archaeal mRNAs are polycistronic, uncapped, lack long poly(A) tails, and have Shine–Dalgarno sequences suggested (erroneously) that the archaeal process resembled the bacterial one. Yet, the M. jannaschii genome showed that archaeal translation initiation is remarkably similar to that seen in eukaryotes (10). Homologs of eukaryotic factors eIF-1A, eIF-2 (all three subunits), two of the five eIF-2B subunits (α and δ), eIF-4A, and eIF-5A were reported (10), a list that covers most eukaryotic factors (except for those involved with mRNA cap recognition).
The three recognized bacterial translation initiation factors have functional counterparts among the eukaryotic factors—although function in all cases is to one degree or another poorly defined. Bacterial IF-1 is thought to enhance the rate of ribosomal subunit dissociation and to stimulate the IF-2-dependent fMet-tRNAiMet binding to the small ribosomal subunit (5), a functionality quite like that ascribed to eIF-1A (14, 15). Bacterial IF-2 is the “central player” in translation initiation, for it associates the initiator Met-tRNAiMet (and GTP) with the small ribosomal subunit (5). Eukaryotic eIF-2, a more complex trimeric protein acts similarly (6), although the two functions differ somewhat in their mechanistic details (e.g., the stage at which GTP or mRNA participates into the complex) (5, 6). Finally, IF-3 (16) acts as a subunit antiassociation factor and promotes the selection of the initiator Met-tRNAiMet, a function quite similar to that of the eukaryotic SUI1 (17).
In Eukarya, the subsequent binding of the 43S preinitiation complex to the mRNA and the scanning for the start codon is mediated by eIF-4 (eIF-4A, eIF-4B, eIF-4E, eIF-4G, and eIF-4F) (6, 7). Finally, eIF-5 [whose N-terminal domain is homologous to the C-terminal domain of the eIF-2β (N.C.K., unpublished data)] is used to promote the hydrolysis of GTP bound to the preinitiation complex, the release of eIF-2⋅GDP, and the joining of the 60S ribosomal subunit (19). [In Bacteria, GTP hydrolysis is performed by IF-2 (20)].
Despite functional resemblance, the three bacterial factors are thought to be unrelated (specifically) in sequence to their eukaryotic counterparts. The two systems are seen as merely analogous (i.e., each evolved separately), with the strong attached implication that translation initiation at the universal ancestor stage was either nonexistent or very different (presumably much simpler) than the extant processes.
Herein, we show what the knowledge of archaeal translation initiation has made obvious, i.e., that there exists far more homology between these bacterial and archaeal/eukaryotic processes than had previously been thought—which dramatically changes the way we look at the evolution of that process and, for that matter, at the nature of the universal ancestor.
In the present communication, we consider the relationship and the phylogenetic distribution of the initiation factors IF-1/eIF-1A, eIF-1/SUI1, and EF-P/eIF-5A; the more complex IF-2/eIF-2/eIF-2B relationships will be considered separately.
MATERIALS AND METHODS
Databases.
The nonredundant protein sequence database at the National Center of Biotechnology Information was used for all of the sequence similarity searches. The complete database of Methanobacterium thermoautotrophicum (22) gene products was obtained from the web page of Genome Therapeutics, at http://www.cric.com/htdocs/sequences/methanobacter/abstract.html. The Archaeoglobus fulgidus (23) complete sequence database was retrieved from The Institute for Genomic Research at http://www.tigr.org. A query for the mere presence or absence of a sequence from the unpublished genomes of Deinococcus radiodurans, Treponema pallidum, and Borrelia burgdorferi was done through the blast server for The Institute for Genomic Research at http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-tigrbl. (A list of genome databases is available at http://geta.life.uiuc.edu/~nikos/genomes.html.)
Database searches were performed with blast (24) and wu-blast 2.0 (25) programs, by using the BLOSUM62 substitution matrix and default parameters, at http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-newblast?Jform=1.
Access to bibliographical databases was greatly facilitated by using entrez (26) (at http://www.ncbi.nlm.nih.gov/Entrez/), and srs (27) (at http://srs.ebi.ac.uk:5000/).
Sequence Analysis.
Multiple sequence alignments were performed by clustalw (28) and the pileup program of the GCG package, version 8.1 from the University of Wisconsin (29). Visualization of the conserved residues was facilitated by the boxshade (version 3.21) program, at http://ulrec3.unil.ch/software/BOX_form.html. Sequence profiles (30) were generated from the multiple sequence alignment of individual families and used to search protein sequence databases. Motif identification searches were used with the meta-MEME motif search tool (31) (see also http://www.sdsc.edu/MEME/meme/website/).
RESULTS AND DISCUSSION
Bacterial Translation Factor IF-1 and Eukaryotic eIF-1A Are Homologs.
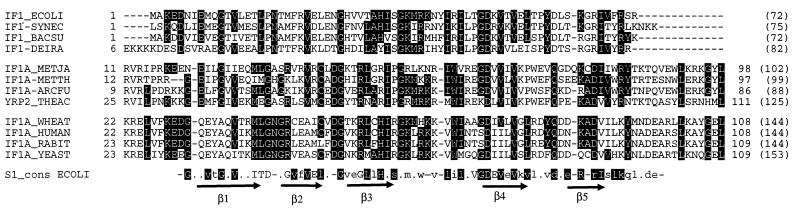
As stated above, despite their general functional similarity—i.e., each facilitates ribosomal subunit dissociation and stabilizes Met-tRNAiMet and mRNA binding to the small ribosomal subunit (5, 14, 15)—bacterial IF-1 and eukaryotic eIF-1A have been considered analogs, not homologs (32). However, this is not the case. Homology between the IF-1 and eIF-1A sequences is immediately apparent from profile searches (data not shown) and the alignment of Fig. 1. The archaeal and eukaryotic sequence are on average 38% identical, whereas archaeal and bacterial are 30% and bacterial and eukaryotic are 21%. However, within each of the three major groups, identities are greater than 50%.
Figure 1.
Multiple sequence alignment of bacterial IF-1, eukaryotic eIF-1A, and their archaeal homologs (aIF-1A). Positions in which sequence conservation is >50% identity are highlighted in black. The last line is a consensus of the six S1 motifs found in E. coli ribosomal protein S1: highlighted uppercase type denotes those residues that are also highly conserved in the IF-1/eIF-1A/aIF-1A family, whereas highlighted lowercase type denotes the residues for which a related amino acid occurs in the family. (Dots denote nonconserved positions in the S1 consensus, and dashes denote gaps/insertions of more than one residue.) The horizontal arrows display the positions of the β-strands according to the three-dimensional structure (33). Protein names (and accession number—when different) are as follows: IF1_ECOLI, E. coli IF-1; IF1-SYNEC, Synechocystis sp. IF-1 (EMBL:D90905_47); IF1_BACSU, Bacillus subtilis IF-1; IF1-DEIRA, unpublished ORF from Deinococcus radiodurans; IF1A_METJA, Methanococcus jannaschii aIF-1A; IF1A-METTH, Methanobacterium thermoautotrophicum unpublished ORF; IF1A-ARCFU, Archaeoglobus fulgidus unpublished ORF; YRP2_THEAC, Thermoplasma acidophilum hypothetical protein; IF1A_WHEAT, Triticum aestivum eIF-1A; IF1A_HUMAN, human eIF-1A; IF1A_RABIT, Oryctolagus cuniculus eIF-1A; IF1A_YEAST, Saccharomyces cerevisiae eIF-1A.
Given that the archaeal IF-1A sequences are unique but more similar to their eukaryotic than to their bacterial counterparts, we support the previously proposed renaming of them to archaeal IF-1A (aIF-1A) (32).
A well-known sequence motif, characteristic of bacterial ribosomal protein S1, also has been reported in IF-1 and eIF-2α, as well as in a number of other RNA-binding proteins (34, 35). The (solution) structures for Escherichia coli IF-1 (36) and the S1 domain (33) are built around a five-stranded antiparallel β-barrel, a structure that displays striking resemblance to proteins belonging to the OB (oligonucleotide/oligosaccharide binding) family, many members of which are single-stranded nucleic acid-binding proteins (37). However, the composition of the S1 motif correlates only weakly with the conservation patterns of the IF-1/eIF-1A alignment (see Fig. 1), not only among Bacteria, Archaea, and Eukarya but even within the Bacteria themselves. These conservation patterns would seem, then, to suggest functionality over and above simple nucleic acid binding.
Eukaryotic Initiation Factor eIF-1/SUI1 Occurs in Archaea and Some Bacteria.
Although mammalian eIF-1 is a single subunit factor (38) and yeast SUI1 is one of the eight eIF-3 subunits (17), the two are very similar at the sequence level (59% identity), and both are reported to have similar functions, i.e., stabilizing mRNA and initiator tRNA binding to the 40S ribosomal subunit (16).
As the alignment of Fig. 2 shows, all known archaeal genomes contain homologs of eIF-1/SUI1 (11). A few bacterial examples are known as well [i.e., enteric bacteria (39, 40) and cyanobacteria (41)], although the majority of the known bacterial genomes [i.e., Helicobacter pylori (42), Mycoplasma genitalium (43), M. pneumoniae (44), Treponema pallidum, Borrelia burgdorferi, and Deinococcus radiodurans] contain none.
Figure 2.
Multiple sequence alignment of the eukaryotic SUI1/eIF-1 protein family with their archaeal and bacterial homologs. Protein names (and accession number—when different) are as follows: YCIH_ECOLI, E. coli hypothetical protein; YCIH_SALTY, Salmonella typhimurium hypothetical protein; YCIH_HAEIN, Haemophilus influenzae hypothetical protein; YCIH-SYNEC, Synechocystis sp. hypothetical protein (European Molecular Biology Laboratory accession no. D64003_48); SUI1-METJA, M. jannaschii hypothetical ORF MJ0463 (Protein Information Resource accession no. G64357); YRP1_METVA, Methanococcus vannielii hypothetical protein; SUI1-METTH, M. thermoautotrophicum unpublished ORF; SUI1-ARCFU, A. fulgidus unpublished ORF; SUI1_HUMAN, human SUI1; SUI1_ANOGA, Anopheles gambiae SUI1; SUI1_YEAST, Saccharomyces cerevisiae SUI1; SUI1_ARATH, Arabidopsis thaliana SUI1.
According to Table 1, percent identities between domains are more or less the same, in the 25–30% identity range, a break with the now-familiar pattern of the highest similarity being between archaeal and eukaryotic components of the translation components. Within each major group, the percent identities are greater than 55%, except for the bacterial examples (see Table 1). The 38% identity between the enteric Bacteria and the cyanobacterium questions their orthology. Although it is reasonable to assume that Archaea possess a eIF-1/SUI1 type of function (based on the overall similarity of archaeal and eukaryotic translation initiation), one wonders about the functional significance of the few phylogenetically scattered bacterial examples. Given that the bacterial versions of eIF-1/SUI1 are no more diverged from the archaeal and eukaryotic versions than these are from one another, the idea that this molecule is involved in translation initiation in some Bacteria (despite its being paralogous therein) must be seriously considered and subject to experimental test.
Table 1.
Pairwise amino acid percentage identity between the eukaryotic SUI1 family and its archaeal and bacterial homologs
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|
| 1 (YCIH_ECOLI) | — | 38.7 | 34.0 | 29.4 | 32.2 | 35.0 | 31.0 | 26.0 | 30.7 |
| 2 (YCIH-SYNEC) | 38.7 | — | 30.9 | 30.6 | 25.2 | 29.0 | 30.7 | 35.5 | 31.3 |
| 3 (SUI1-METJA) | 34.0 | 30.9 | — | 78.4 | 67.0 | 60.4 | 31.9 | 30.5 | 28.4 |
| 4 (YPRI_METVA) | 29.4 | 30.6 | 78.4 | — | 61.0 | 55.4 | 35.4 | 29.3 | 32.6 |
| 5 (SUI1-METTH) | 32.2 | 25.2 | 67.0 | 61.0 | — | 54.5 | 26.3 | 27.0 | 23.7 |
| 6 (SUI1-ARCFU) | 35.0 | 29.0 | 60.4 | 55.4 | 54.5 | — | 29.1 | 25.5 | 28.0 |
| 7 (SUI1_HUMAN) | 31.0 | 30.7 | 31.9 | 35.4 | 26.3 | 29.1 | — | 59.4 | 58.5 |
| 8 (SUI1_YEAST) | 26.0 | 35.5 | 30.5 | 29.3 | 27.0 | 25.5 | 59.4 | — | 53.7 |
| 9 (SUI1_ARATH) | 30.7 | 31.3 | 28.4 | 32.6 | 23.7 | 28.0 | 58.5 | 53.7 | — |
Protein names and their references are as in Fig. 2. Internal lines separate bacterial, archaeal, and eukaryal sequences.
It has been suggested that SUI1 functions in concert with eIF-2 to confine Met-tRNAiMet recognition to the AUG initiator codon, because mutants of SUI1 allow tRNAiMet to initiate protein synthesis at UUG codons as well (45). Because it is believed that there may be a functional relationship between the eukaryotic and the bacterial start-site selection processes (46), the presence of SUI1 homologs in some bacterial genomes supports a similar start-site recognition mechanisms among Archaea, Eukarya, and some Bacteria.
Archaeal/Eukaryotic Initiation Factor eIF-5A Is Related to Bacterial Elongation Factor EF-P.
Eukaryotic translation initiation factor eIF-5A appears to promote the synthesis of the initial peptide bond in mRNA translation (47) and is unique in being the only known cellular protein that contains the unusual amino acid hypusine, formed by a posttranslational modification of a specific lysine residue (48). This (modified) protein has been identified in all Eukarya and Archaea so far analyzed, but it appears to be absent from Bacteria (49). However, a similar facilitating function may be performed in Bacteria by translation factor EF-P (50).
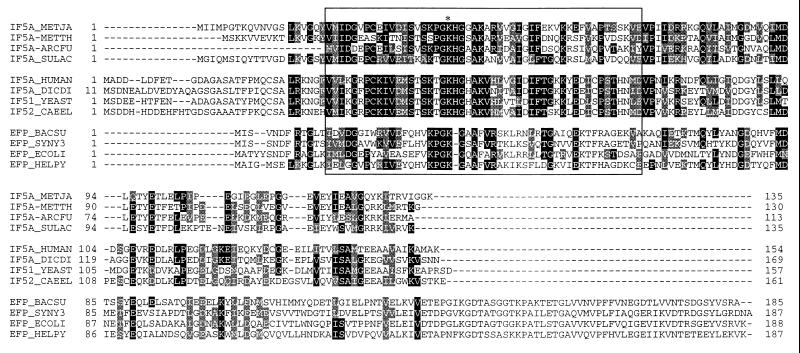
Fig. 3 shows that, despite the lack of hypusine in the bacterial case, bacterial EF-P and eukaryotic/archaeal eIF-5A are homologs. Although sequence similarity between the eukaryotic and bacterial sequences are relatively low (in the 20% range), both the alignment of Fig. 3 and profile searches (data not shown) generated with either the Bacteria or the archaeal/eukaryotic family make clear their relatedness.
Figure 3.
Multiple sequence alignment of the eukaryotic translation initiation eIF-5A protein family with their archaeal homologs and the bacterial translation elongation EF-P family. The minimum domain of eukaryotic IF-5A needed for hypusine modification (51) is boxed; the asterisk denotes the lysine residue that is posttranslationally modified to hypusine. Protein names (and accession number—when different) are as follows: IF5A_METJA, M. jannaschii aIF-5A; IF5A-METTH, M. thermoautotrophicum unpublished ORF; IF5A-ARCFU, A. fulgidus unpublished ORF; IF5A_SULAC, Sulfolobus acidocaldarius aIF-5A; IF5A_HUMAN, human eIF-5A; IF5A_DICDI, Dictyostelium discoideum eIF-5A; IF51_YEAST, S. cerevisiae eIF-5A; IF52_CAEEL, Caenorhabditis elegans eIF-5A; EFP_BACSU, B. subtilis EF-P; EFP_SYNY3, Synechocystis sp. EF-P; EFP_ECOLI, E. coli EF-P; EFP_HELPY, Helicobacter pylori EF-P.
The archaeal IF-5A family displays an average of 32% and 26% identity to the eukaryotic and bacterial families, respectively, whereas bacterial and eukaryotes display an average of only 20%. The highest density of sequence conservation is found in the vicinity of the minimal region of the protein that must be conserved for lysine → hypusine conversion to occur (see boxed area of Fig. 3) (50).
Although hypusine is not present in the bacterial examples, a lysine residue usually is found in the corresponding position in the sequence. Note in Fig. 3 that in all archaeal and eukaryotic examples, the modified lysine residue is followed by histidine but that in all bacterial examples, an amino acid residue is absent at the corresponding position. Note also that bacterial EF-P sequences have a well-conserved C-terminal section that is absent in both archaeal and eukaryotic eIF-5A sequences, which suggests functional differentiation of the two molecular types.
A number of detailed suggestions have been made regarding eIF-5A function: the essential hypusine modification is suggested to stabilize Met-tRNAiMet binding to the peptidyltransferase center of the ribosome because formylation of the initiator Met residue (which removes its positive charge) renders it less dependent upon the presence of eIF-5A function (18). Complete intracellular depletion of eIF-5A (by gene deletion) results in inhibition of cell growth, although protein synthesis seems to be only slightly reduced (∼25%) (21). This has been interpreted to mean that eIF-5A may participate only in the translation of a subclass of mRNAs required directly for cell growth (21).
CONCLUSIONS
The above analysis makes clear that, contrary to generally accepted understanding, parts of the componentry of the translation initiation system (i.e., SUI1 and its relatives, the IF-1/eIF1A group, and the eIF-5A/EF-P group) are indeed universally distributed; representatives occur in at least some members of each primary line of descent. Whether the various members of any one group are orthologs or merely paralogs is not clear and, in any case, is a matter to be decided by experimentation. However, in at least one instance, the SUI1 group, it seems likely that functional orthology will hold across the entire group: Molecular length is nearly constant throughout the group; the sequence conservation pattern covers the entire molecule; and the degree of similarity is about the same among all three major taxa. Thus, it would appear that at least some aspects of a translation initiation system existed at the universal ancestor stage, which suggests a more advanced ancestral translation function than would otherwise be the case.
The three initiation factors under discussion are seen as playing relatively “peripheral” roles in the process. The central role, that of introducing the initiator tRNA into the mechanism, falls either to IF-2 (in Bacteria) or eIF-2 (in Eukarya and Archaea). Yet, IF-2 and eIF-2 are not specifically related to one another. Herein lies a puzzle: One does not expect molecules that refine or embellish a process to have evolved as such (functionally) before the evolution of the fundaments of that process. Although our observations do not lend themselves to satisfying conclusions at this time, they do offer the promise of ultimately understanding a great deal about the evolution of translation initiation (and translation in general) and focus on the need for comparative (experimental) approaches to this process.
Acknowledgments
We thank C. Ouzounis (European Bioinformatics Institute/United Kingdom) and D. Graham (University of Illinois at Urbana-Champaign) for critical reading of the manuscript. This work was supported by grants from the National Aeronautics and Space Administration (NAG 54500) and Department of Energy (DEFG C02-95ER61963) to C.R.W.
ABBREVIATION
- IF
initiation factor
Note Added in Proof
After the submission of this paper, the complete genomes of A. fulgidus and M. thermoautotrophicum were published. Below are the ORF numbers of the genes mentioned above: IF1A-METTH, MTHI004; IF1A-ARCFU, AFO777; SUI1-METTH, MTHI0; SUI1-ARCFU, AFO914; IF5A-METTH, MTH869; and IF5A-ARCFU, AFO645.
References
- 1.Olsen G J, Woese C R. Cell. 1997;89:991–994. doi: 10.1016/s0092-8674(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 2.Ouzounis C, Kyrpides N. FEBS Lett. 1996;390:119–123. doi: 10.1016/0014-5793(96)00631-x. [DOI] [PubMed] [Google Scholar]
- 3.Edgell D R, Doolittle W F. Cell. 1997;89:995–998. doi: 10.1016/s0092-8674(00)80285-8. [DOI] [PubMed] [Google Scholar]
- 4.Dennis P P. Cell. 1997;89:1007–1010. doi: 10.1016/s0092-8674(00)80288-3. [DOI] [PubMed] [Google Scholar]
- 5.Gualerzi C O, Pon C L. Biochemistry. 1990;29:5881–5888. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- 6.Pain V M. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 7.Clark B, Grunberg-Manago M, Gupta N K, Hershey J, Hinnebusch A G, Jackson R J, Maitra U, Mathews M B, Merrick W C, Rhoads R E, Sonenberg N, Spremulli L L, Trachsel H, Voorma H O. Biochimie. 1996;78:1119–1122. [Google Scholar]
- 8.Woese C R. In: Organization and Control in Prokaryotic and Eukaryotic Cells. Charles H P, Knight B C J G, editors. Cambridge: University Press; 1970. pp. 39–53. [Google Scholar]
- 9.Woese C R, Fox G F. J Mol Evol. 1977;10:1–6. doi: 10.1007/BF01796132. [DOI] [PubMed] [Google Scholar]
- 10.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 11.Kyrpides N, Olsen G, Klenk H-P, White O, Woese C. Microbiol Comp Genomics. 1996;1:329–338. [PubMed] [Google Scholar]
- 12.Andrade M, Casari G, de Daruvar A, Sander C, Schneider R, Tamares J, Valencia A, Ouzounis C. Comput Appl Biosci. 1997;13:481–483. doi: 10.1093/bioinformatics/13.4.481. [DOI] [PubMed] [Google Scholar]
- 13.Keeling P J, Doolittle W F. Proc Natl Acad Sci USA. 1995;92:5761–5764. doi: 10.1073/pnas.92.13.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas A, Goumans H, Voorma H O, Benne R. Eur J Biochem. 1980;107:39–45. doi: 10.1111/j.1432-1033.1980.tb04621.x. [DOI] [PubMed] [Google Scholar]
- 15.Wei C-L, Kainuma M, Hershey J W B. J Biol Chem. 1995;270:22788–22794. doi: 10.1074/jbc.270.39.22788. [DOI] [PubMed] [Google Scholar]
- 16.Hershey J W B, Asano K, Naranda T, Vornlocher H P, Hanachi P, Merrick W C. Biochimie. 1996;78:903–907. doi: 10.1016/s0300-9084(97)86711-9. [DOI] [PubMed] [Google Scholar]
- 17.Naranda T, Macmillan S E, Donahue T F, Hershey J B. Mol Cell Biol. 1996;16:2307–2313. doi: 10.1128/mcb.16.5.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershey J W B, Smit-McBride Z, Schnier J. Biochim Biophys Acta. 1990;1050:160–162. doi: 10.1016/0167-4781(90)90159-y. [DOI] [PubMed] [Google Scholar]
- 19.Das K, Chevesich J, Maitra U. Proc Natl Acad Sci USA. 1993;90:3058–3062. doi: 10.1073/pnas.90.7.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laalami S, Sacerdot C, Vachon G, Mortensen K, Sperling-Petersen H U, Cenatiempo Y, Grunberg-Manago M. Biochimie. 1991;73:1557–1566. doi: 10.1016/0300-9084(91)90191-3. [DOI] [PubMed] [Google Scholar]
- 21.Kang H A, Hershey J W B. J Biol Chem. 1994;269:3934–3940. [PubMed] [Google Scholar]
- 22.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, et al. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klenk H-P, Clayton R A, Tomb J-F, White O, Nelson K E, et al. Nature (London) 1997;390:364–375. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 24.Altchul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Altchul S F, Gish W. Methods Enzymol. 1996;266:460–480. doi: 10.1016/s0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- 26.Schuler G D, Epstein J A, Ohkawa H, Kans J A. Methods Enzymol. 1996;266:141–162. doi: 10.1016/s0076-6879(96)66012-1. [DOI] [PubMed] [Google Scholar]
- 27.Etzold T, Argos P. Comput Appl Biosci. 1993;9:49–57. doi: 10.1093/bioinformatics/9.1.49. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devereux J, Haeberli P, Smithies N O. Nucleic Acids Res. 1984;12:387–396. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gribskov M, McLachlan A D, Eisenberg D. Proc Natl Acad Sci USA. 1987;89:10915–10919. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bailey T L, Elkan C. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. Menlo Park, CA: AAAI; 1994. pp. 28–36. [PubMed] [Google Scholar]
- 32.Keeling P J, Doolittle W F. Mol Microbiol. 1995;17:399–400. doi: 10.1111/j.1365-2958.1995.mmi_17020399.x. [DOI] [PubMed] [Google Scholar]
- 33.Bycroft M, Hubbard T J P, Proctor M, Freund S M V, Murzin A G. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 34.Gribskov M. Gene. 1992;119:107–111. doi: 10.1016/0378-1119(92)90073-x. [DOI] [PubMed] [Google Scholar]
- 35.Langer D, Lottspeich F, Zillig W. Nucleic Acids Res. 1994;22:694. doi: 10.1093/nar/22.4.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sette M, van Tilborg P, Spurio R, Kaptein R, Paci M, Gualerzi C O, Boelens R. EMBO J. 1997;16:1436–1443. doi: 10.1093/emboj/16.6.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murzin A G. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kasperaitis M A M, Voorma H O, Thomas A A M. FEBS Lett. 1995;365:47–50. doi: 10.1016/0014-5793(95)00427-b. [DOI] [PubMed] [Google Scholar]
- 39.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goedeu M A, Rose D J, Mau B, Shuo Y. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 40.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 41.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, et al. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 42.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, et al. Nature (London) 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 43.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 44.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B C, Herrmann R. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoon H, Donahue T F. Mol Cell Biol. 1992;12:248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon H, Donahue T F. Mol Microbiol. 1992;6:1413–1419. doi: 10.1111/j.1365-2958.1992.tb00861.x. [DOI] [PubMed] [Google Scholar]
- 47.Smit-McBride Z, Dever T E, Merrick W C, Hershey J W B. J Biol Chem. 1989;264:1578–1583. [PubMed] [Google Scholar]
- 48.Cooper H L, Park M H, Folk J E, Safer B, Braverman R. Proc Natl Acad Sci USA. 1983;80:1854–1857. doi: 10.1073/pnas.80.7.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Magdole V, Klier H, Wohl T, Klink F, Hirt H, Hauber J, Lottspeich F. Mol Gen Genet. 1994;244:646–652. doi: 10.1007/BF00282755. [DOI] [PubMed] [Google Scholar]
- 50.Joe Y A, Park M H. J Biol Chem. 1994;264:25916–25921. [PubMed] [Google Scholar]
- 51.Aoki H, Adams S L, Chung D G, Yaguchi M, Chuang S E, Ganoza M C. Nucleic Acids Res. 1991;19:6215–6220. doi: 10.1093/nar/19.22.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]