Abstract
Many genes expressed in the human genome have not been identified despite intensive efforts. We observed that the presence of long poly(dA/dT) sequences in the 3′ end of cDNA templates contributes significantly to this problem, because the hybrids formed randomly between poly(dA) and poly(dT) sequences of unrelated cDNA templates lead to loss of many templates in the normalization/subtraction reactions. The low abundant copies, which account for the majority of the expressed genes, are affected in particular by this phenomenon. We have developed a strategy called screening poly(dA/dT)− cDNAs for gene identification to overcome this obstacle. Applying this strategy can significantly enhance the efficiency of genome-wide gene identification and should have an impact on many functional genomic studies in the postgenome era.
Functional genomic studies of a particular species depend on the identification of all of the expressed genes from its genome. The difficulty of genomewide gene identification is proportional to the number of genes expressed in a particular genome. In the human genome, the number of expressed genes is estimated at between 60,000 and 150,000 (1–4). The EST (expressed sequence tag) project and CGAP (Cancer Genome Anatomy Project) are two major efforts to identify all of the expressed human genes (5, 6). These efforts have resulted in the identification of 38,039 human genes from 886,936 human EST sequences through the EST project and 44,391 human genes from 804,804 EST sequences through the CGAP (ref. 7; http://www.ncbi.nlm.nih.gov/UniGene/gene_discovery.html, January 5, 2000). However, the rate of novel gene identification through the EST project declined dramatically from 10.6% of EST sequences in 1996 (36,000 novel sequences from 340,000 EST sequences) (7) to only 2.7% of EST sequences collected in 1998 (638 novel sequences identified from 23,038 EST sequences, UniGene and dbEST databases), despite the fact that many expressed genes still were unidentified. Most of the procedures in the current CGAP are similar to the EST project except for the difference in the tissue sources (http://www.ncbi.nlm.nih.gov//ncicgap/). Therefore, the pattern of gene identification in the CGAP should be similar to that of the EST project. This implies that the rate of novel gene identification in the CGAP should decline at some point from its current rate (5.4%), leaving many expressed human genes unidentified.
There are several possible explanations for this situation. One is that genes expressed at a low level have a lower probability of being identified than those expressed at a higher level. By applying normalization or subtraction to reduce the redundancy, and by increasing the sequencing scale, one could identify most of these genes (8). However, analysis of the human EST sequence data does not fully support this explanation, because the large number of human EST sequences from various resources through the EST project has not resulted in a significant increase in the identification of novel human genes. Another explanation is that most of the expressed human genes have been identified (9). This conflicts with the current experimental data that 90,310 unique human sequences have been identified (Unigene Build 101, http://www.ncbi.nlm.nih.gov/UniGene/Hs.stats.shtml). A third explanation is that serious systematic flaws may exist in the current approaches, leading to difficulties in identifying novel genes. Our analysis of current technologies for genome-wide gene identification indicates that the existence of poly(dA/dT) sequences in cDNA clones causes the problem in large measure.
All cDNA libraries currently used for the genome-wide gene identification are generated exclusively through oligo(dT) priming for reverse transcription (ref. 8; http://genome.wustl.edu/est/est_protocols/libraries.html; http://www.ncbi.nlm.nih.gov/ncicgap). Because human mRNAs contain an average of 200 adenosine (A) residues at their 3′ end (10), the oligo(dT) priming in reverse transcription results in the inclusion of various lengths of poly(dA/dT) sequences at the 3′ end of cDNA templates. In a given cell, the majority of genes are expressed at lower levels and they constitute only a small portion of the total transcripts, whereas a small number of genes expressed at a high level constitutes a large portion of the total transcripts (1, 11). Therefore, direct screening of standard cDNA libraries will only identify highly expressed genes (12). Normalization and subtraction are needed to reduce the high-abundance copies and to increase the representation of the low-abundance copies to identify the genes expressed at low level (8). However, because of the presence of 3′ poly(dA/dT) sequences in cDNA templates, random hybridization can occur anywhere along the poly(dA) and poly(dT) sequences during the normalization/subtraction process. This would result in the formation of tangled poly(dA)/poly(dT) double-strand hybrids, independent of the sequence specificity (Fig. 1). Because double-stranded hybrids are removed, copies of many genes inappropriately annealed to the hybrids could be lost. The genes expressed at low levels will be particularly affected. This phenomenon may contribute directly to the low efficiency of novel gene identification in the current efforts of genome-wide gene identification. We have proven our hypothesis by various means. We have developed a strategy named “screening poly(dA/dT)− cDNA templates for gene identification” to overcome this problem. We have demonstrated that through applying our strategy, the rate of novel gene identification can be increased significantly.
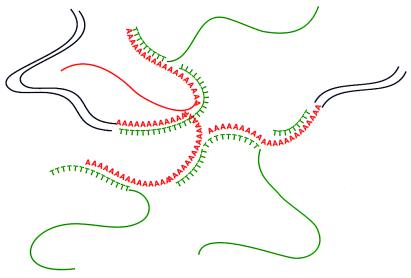
Figure 1.
Model for the formation of double-strand poly(dA)/poly(dT) complex during hybridization. Oligo(dT) priming in reverse transcription generates cDNAs with poly(dA/dT) sequences at the 3′ end. During the normalization or subtraction reaction, hybridization between poly(dA)/poly(dT) sequences among cDNA templates causes the formation of tangled double-strand hybrids. The removal of these hybrids causes the loss of many unrelated templates.
Materials and Methods
Analysis of EST Sequences.
To facilitate the analysis, the number of EST sequences in dbEST collected from the EST project and CGAP was separated. The number of EST sequences, 886,936, derived from the EST project was obtained by removal of the CGAP EST sequences, 804,804 (http://www.ncbi.nlm.nih.gov//ncicgap, January 5, 2000) from the total human EST sequences, 1,691,740 (dbEST release 113199, http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html). The number of human Unigene sequences from the EST project, 38,039, was calculated by removal of the human Unigene sequences derived from CGAP EST sequences, 44,391 (http://www.ncbi.nlm.nih.gov/ncicgap, January 5, 2000), and known genes, 10,501, from the total human Unigene sequences, 92,931 (http://www.ncbi.nlm.nih.gov/UniGene/Hs.stats.shtml, UniGene Build 105).
Subtraction of Poly(dT)+ Template.
A single-strand DNA template was synthesized as the tester. It contained 100 dA residues, an M13 sequence at its 5′ end, and a T3 promoter sequence at its 3′ end (5′-GTAAAACGACGGCCAGTACGN*B** (A)100 CTTTAGTGAGGGTTAATTTC-3′; N* = A, G, C, T; B** = A, G, C). Single-strand poly(dT)+ cDNAs used as the driver were converted from HL60 cell mRNA by oligo(dT) priming and MMLV reverse transcriptase. Tester DNA and driver cDNA were mixed, and a hybridization reaction was performed at 98°C for 2 min and 68°C for 10 h (13). Hydroxyapatite absorption followed the procedures (8). Controls without driver cDNA were set for each reaction. Quantitative PCR was used for quantification (13), in which a homologous control template with only 40 dA was used as the internal control for coamplification in quantitation PCR. The amplicons were fractionated on a 4% denaturing gel, exposed, and scanned by a PhosphorImager system (Molecular Dynamics).
Determination of the Specificity of Anchored Oligo(dT) in Reverse Transcription.
A double-strand DNA was generated by PCR with 5′ primer (T7-M13) and 3′ primer (T3) from the single-strand DNA template described above. This created a double-strand DNA with the T7 promoter at its 5′ end. In vitro transcripts were prepared from the templates with T7 RNA polymerase (Promega). cDNAs were synthesized with each anchored oligo(dT) primer tailed with SP6 sequences at their 5′ end and MMLV reverse transcriptase (Promega) or AMV reverse transcriptase (Invitrogen). The resulting cDNA was PCR-amplified with M13 and SP6 primers. PCR controls were set in which the DNA templates were amplified directly by Taq polymerase with M13 primer and each anchored oligo(dT) primer. The PCR products were cloned into a pCR2.1 vector (Invitrogen), sequenced with M13 reverse primer and dRhodamine sequencing kits, and analyzed with an ABI 377 Automatic Sequencer (Applied Biosystems).
Determination of the Quantitative Pattern of cDNA Synthesis with Anchored Oligo(dT) Primers.
mRNA from HL60 cells was converted into single-strand poly(dT)+ or poly(dT)− cDNA with either oligo(dT)12–18 primers or the optimal set of anchored oligo(dT) primers by MMLV reverse transcriptase. Each cDNA was then purified and quantified. Two hundred nanograms of each cDNA was loaded side by side in an agarose gel and used for Southern blotting. As a control, 250 ng of mRNA was loaded in a denaturing gel for a Northern blotting. A group of genes representing high abundant and low abundant copies in HL60 cells was selected as the candidate genes (13). Probes for each selected gene were prepared with a random primer labeling kit (Ambion, Austin, TX). Three membranes containing each poly(dT)+ cDNA, poly(dT)− cDNA, and mRNA from both Southern and Northern blots were hybridized in the same tube with each probe. The membranes were then washed, exposed, and quantified with a PhosphorImager system (Molecular Dynamics). The signals from each Northern blot were set as 1. The signals of the corresponding genes in poly(dT)+ cDNA and poly(dT)− cDNA from the Southern blots were normalized to that number for comparison.
Comparison of the Level of cDNA Templates Generated by Oligo(dT) and Anchored Oligo(dT) Primers upon Subtraction.
A group of serial analysis of gene expression (SAGE) tag sequences was selected. These tag sequences were detected by SAGE in primary colon cells at three to nine copies per cell, but not in DLD1, a human colorectal adenocarcinoma cell line. The EST sequences corresponding to these SAGE tags were identified through a blast search. PCR primers were designed based on these matched EST sequences. The expression of these selected sequences in these two RNA samples was confirmed further by reverse transcription–PCR with these primers. Five genes (AI193160, AA435717, X03747, AA448394, and AA297150) amplified only in the colon cancer cells but not in DLD1 cells finally were selected, with the corresponding SAGE tag TGATCCCAAG, CTAGGATGAT, TTCTAACATA, CGGTGGGACC, and GAACAGCTCA. The sense primers were CCGGATGTAACACTGAGCAC, AGTGGCCAGGCCTGTGTCAT, CTGGAGGCATCACATGCTGG, GGCTGCCATGCGGTGGGAC, and ACCATGGAACAGCTCACAAG; the antisense primers were TCCTTGGGATCTCATGGTTG, ACATCGTCTCTTCCCTACTG, ACCTGACTGAATACAAGATC, GCCAGGAAAGTGAAGAGCTG, and AAGATACTCGTGCAATGTTG. Control templates for each sequence also were generated for quantitative PCR analysis (13). First-strand poly(dT)+ and poly(dT)− cDNA populations were generated as the tester with mRNA from the colon cells by either the oligo(dT)12–18 primer or with the optimal combination, and double-strand poly(dA/dT)+ cDNA and poly(dA/dT)− cDNA were generated as the drivers from DLD1 cell mRNA with a cDNA synthesis kit (Life Technologies, Gaithersburg, MD), except the anchored primers were used for the generation of poly(dA/dT)− cDNA. The subtraction reaction contained 160 ng of driver and 12.5 ng of tester. Five thousand nanograms of oligo(dT)20 DNA was used in the blocking reactions. In the subtraction reactions, the poly(dT)+ single-strand tester was combined with the poly(dA/dT)+ driver, and the poly(dT)− single-strand tester was combined with the poly(dA/dT)− driver.
Identification of Novel Sequences from Poly(dA/dT)− cDNA Libraries.
Poly(dA/dT)− cDNAs were generated from mRNA of normal primary colon cells with the optimal combination. To increase the specificity of the normalization reaction, facilitate the alignment of identified sequences with 3′ EST sequences in dbEST, and collect SAGE tag sequences for SAGE analysis, we collected and cloned the 3′ cDNAs (13), resulting in the 3′ poly(dA/dT)− colon cDNA library. A normalization reaction was performed by following method 3 (8). Each collected sequence was used for a blast search in databases and was defined as a known gene, an EST sequence, or a novel sequence. SAGE tags also were collected from sequences and matched in the SAGE database from normal colon cells (http://www.ncbi.nlm.nih.gov/SAGE/).
Results and Discussion
Poly(dA/dT)+ cDNAs Cause the Loss of Templates.
To prove the validity of our hypothesis that the presence of poly(dA/dT) sequences at the 3′ end of cDNAs causes the loss of templates after subtraction, we designed an in vitro model. In this model, a single-strand synthetic DNA template containing 100 dA residues was subtracted with a cDNA sample generated by oligo(dT) priming and subsequently quantified by quantitative PCR after hydroxyapatite absorption. As shown in Fig. 2A, this template was lost after these procedures, indicating that the formation of poly(dA)/poly(dT) hybrids during subtraction indeed can result in the loss of templates.
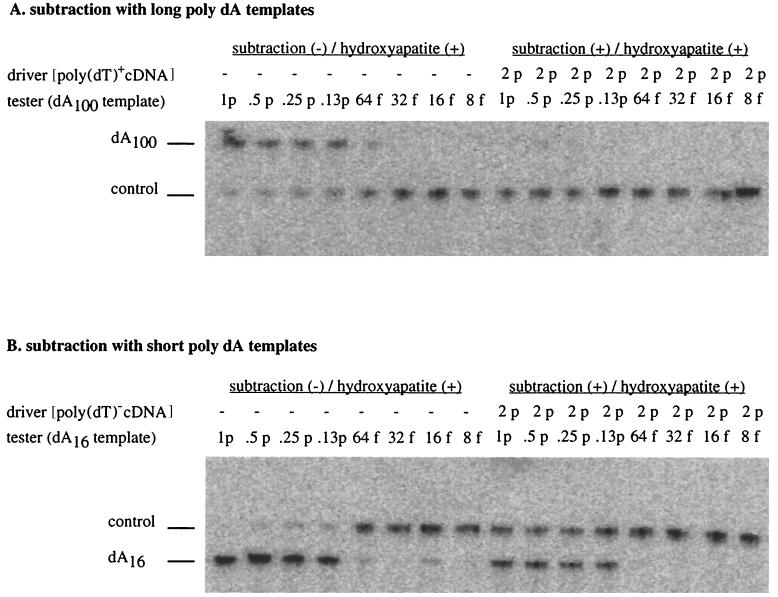
Figure 2.
Effects of the poly(dA/dT) sequence on the preservation of the templates upon subtraction. (A) Long poly(dA) sequences cause loss of the template. DNA templates with 100 dA were subtracted with poly(dT)+ cDNA and measured by quantitative PCR. Upper band, signal from the testing templates; lower band, signal from internal PCR control template with 40 dA. The left half was the control in which no driver cDNA was added. Note that in the right half containing driver, the 100 dA templates were lost. (B) Short poly(dA) sequences preserve the template; same as above except DNA templates contained 16 dA and were subtracted with poly(dT)− cDNA. Upper band, signal from internal PCR control template with 40 dA; lower band, signal from the testing templates. p, picograms; f, femtograms. Note that in the right half containing driver, the 16 dA templates were preserved.
Generation of Poly(dA/dT)− cDNA Templates.
We further reasoned that, if cDNA templates did not contain a long poly(dA/dT) sequence, these templates could be preserved after the subtraction. Such cDNA templates can be generated by use of 3′ anchored oligo(dT) primers instead of regular oligo(dT) primers for reverse transcription (14–17). The assumption is that only the primers annealed to the 5′ end of the mRNA poly(A) tail and its anchor nucleotide paired to the nucleotide immediately 5′ of the poly(A) sequence could result in extension by reverse transcriptase. Primers annealed to other positions along the poly(A) sequence should not be extended, because the unpaired anchors block the extension. These features should provide cDNA without long poly(dT) sequences. However, we frequently observed that many clones still contained long poly(dA/dT) sequences despite the use of anchored oligo(dT) primers (13).
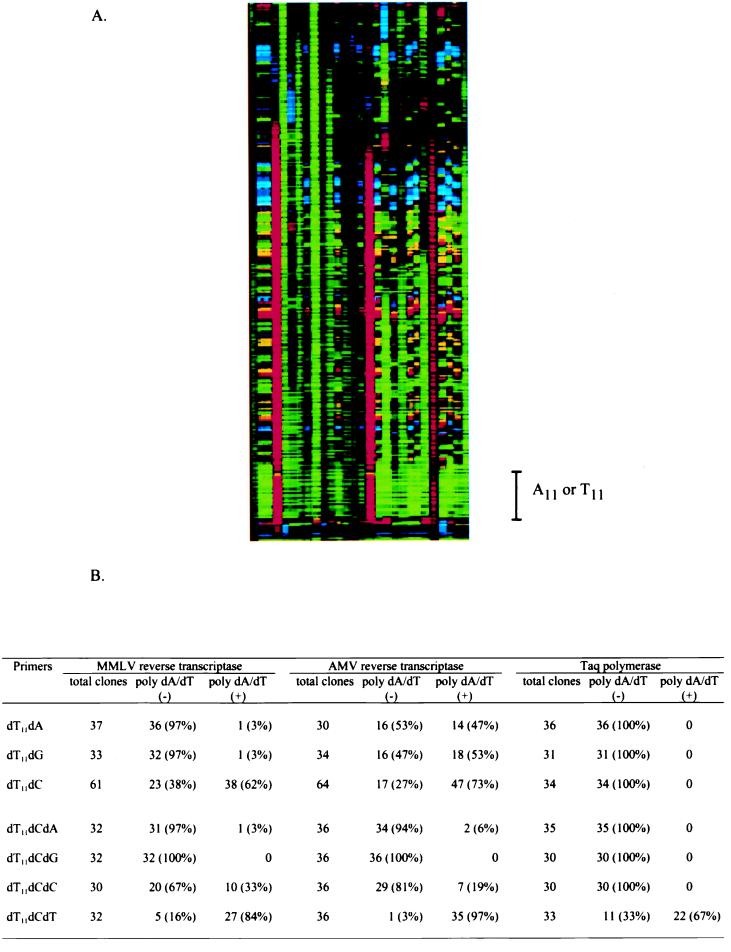
We systematically examined the pattern of cDNA synthesis with various anchored oligo(dT) primers and reverse transcriptases. An in vitro transcript was synthesized to mimic mRNA templates. It contained 100 adenosine residues, randomized nucleotides of A, G, or C at the first position 5′ of the poly(A) sequences, and randomized nucleotides of A, G, C, or T at the second position 5′ of the poly(A) sequences to reflect all of the possible combinations at these two positions within natural mRNA populations. After reverse transcription, a given cDNA clone either could contain 11 dA/dTs at its 3′ end, derived from the primer annealed to the 5′ end of the poly(A) sequences, or it could contain a longer poly(dA/dT) sequence at its 3′ end, extended from the primer annealed randomly along the poly(A) sequences. We classified the former as a poly(dA/dT)− clone and the latter as a poly(dA/dT)+ clone. To our surprise, the results showed that the lengths of the poly(dA/dT) sequences in the cDNA clones are anchor nucleotide-dependent and reverse transcriptase-dependent (Fig. 3B). For example, most clones generated with a dC-anchored primer were poly(dA/dT)+ (Fig. 3A). Apparently, the dC-anchored primer does not provide a discriminatory function for the synthesis of the poly(dA/dT)− clone in reverse transcription. This is due to the fact that the reverse transcriptases from retroviruses have high mis-pairing extension capacity during RNA-dependent DNA synthesis in order to maintain high mutation rates for retrovirus integration and replication (18–20). This feature contributes directly to the inherent problem of high false-positive rates of gene identification in the differential display technique (21). Because of the random length of poly(dA/dT) sequences at the 3′ end of cDNA templates resulting from the dC-anchored primer, the size of cDNA templates varies even for the cDNAs from the same mRNA. This makes gene identification through gel fractionation highly unreliable. The addition of a second anchor (dA, dG, dC) to the dC-anchor primer corrected this problem, except for the dCdT anchor, in which most of clones were poly(dA/dT)+. Further addition of nucleotides to the dCdT anchors will not improve this situation because of the nonspecificity of dCdT itself. Through this analysis, we determined that the combination of dA-, dG-, dCdA-, dCdG-, dCdC-anchored oligo(dT) primers and MMLV reverse transcriptase provides the optimal condition for the generation of poly(dA/dT)− cDNAs with both simplicity and specificity. The coverage of the total expressed sequences with these primers should be 91.7%, assuming a random distribution of A, G, C, and T in the last and second-to-last positions before the poly(A) sequences in the mRNA population.
Figure 3.
Patterns of cDNA sequences generated with anchored oligo(dT) primers and reverse transcriptases. (A) Sequence ladder generated with dC-anchored oligo(dT) primer and MMLV reverse transcriptase. The poly(dA/dT)− clone contained only 11 dA (green) or 11 dT (red) residues, depending on the cloning orientation. Poly(dA/dT)+ clones contained longer dA or dT residues. (B) Summary of the results for different anchored oligo(dT) primers and reverse transcriptases.
cDNAs Generated by Anchored Oligo(dT)s Maintain the Quantitative Pattern in the Original mRNAs.
It is critical to determine whether the anchored oligo(dT)s would selectively convert different mRNA templates into cDNAs because of the presence of the anchors in the primers. We generated poly(dT)− cDNAs with the anchored oligo(dT)s. We also generated poly(dT)+ cDNAs with regular oligo(dT) primers. We then compared the level of different genes within these two cDNAs by Southern blot analysis and compared the level of these genes in the original mRNA determined by Northern blot analysis. As shown in Fig. 4, the quantitative pattern of the cDNAs synthesized with anchored oligo(dT)s is equivalent to the cDNAs generated with regular oligo(dT) primer. The quantitative patterns of both cDNAs are consistent with those in the original mRNAs for most of the genes. This indicates that cDNAs generated with anchored oligo(dT)s largely maintain the quantitative pattern in the original mRNAs.
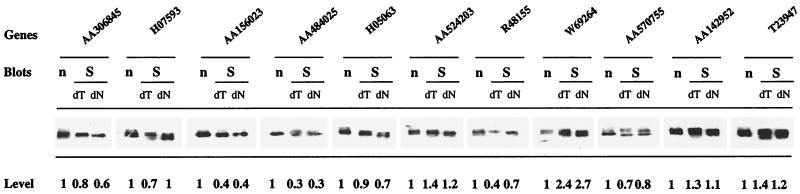
Figure 4.
Quantitative pattern of cDNAs generated by anchored oligo(dT)s. Poly(dT)− and poly(dT)+ cDNAs were generated by anchored oligo(dT) or regular oligo(dT) primers and used for Southern blot analysis. The original mRNA was used for Northern blot analysis. The relative levels of a group of genes in the cDNAs and mRNA were determined by hybridization. The quantity for each gene in Northern blot analysis was set as 1, and the quantity for each gene from the Southern blot analysis was normalized to that for each corresponding gene. n, Northern blot; S, Southern blot; dT, oligo(dT) priming in reverse transcription; dN, anchored oligo(dT) priming in reverse transcription.
Poly(dA/dT)− cDNAs Reserve the Templates upon Subtraction.
We next performed an experiment similar to that illustrated in Fig. 2A to test whether the poly(dA/dT)− templates would be preserved upon subtraction. The driver used was poly(dT)− cDNAs generated with the optimal combination, and the tester template contained only 16 dA. As shown in Fig. 2B, the tester was largely retained after the procedures. This indicates that the exclusion of long poly(dA/dT) in the cDNA indeed preserves templates upon subtraction.
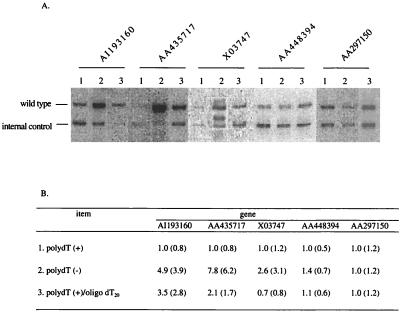
We further compared our strategy with the current approaches used in the genome-wide gene analysis to determine whether our strategy would provide a higher efficiency for gene identification. An mRNA sample from normal colon epithelium cells was chosen for this comparison. This sample has been analyzed extensively by using the SAGE technique. Of 14,721 genes identified from 62,168 SAGE tags, more than 70% were expressed at 5 copies or fewer per cell (22). The relative quantities of five sequences expressed at less than five copies per cell in this sample were compared after subtraction (Fig. 5). The results showed that the levels of these sequences in the poly(dA/dT)− reactions were between 1.4- and 7.8-fold higher in four of the five genes than in the poly(dA/dT)+ samples. The addition of a large excess of oligo(dT)20, used routinely in normalization or subtraction in an attempt to block the poly(dA)/poly(dT) hybridization (8), only resulted in a minor increase in two samples. This indicates that that approach does not adequately preserve the low abundance copies. That the level of the AA297150 sequence showed no change among all three reactions might be due to the absence of a long poly(A) sequence in its original mRNA template.
Figure 5.
Increased level of low-abundance copies with poly(dA/dT)− cDNAs. (A) Quantitative PCR showing the signals in different samples. Lanes: 1, poly(dA)+ tester and poly(dA/dT)+ driver; 2, poly(dA)− tester and poly(dA/dT)− driver; 3, poly(dA)+ tester, poly(dA/dT)+ driver plus oligo(dT)20 blocking primer. (B) Comparison of relative levels. The numbers within parentheses are the original ratio between wild-type and control amplicons. The number from the reaction of the poly(dA)+ tester and poly(dA/dT)+ driver (line 1) was set at 1.0. Numbers from other samples were normalized to this value.
Screening Poly(dA/dT)− cDNAs Increases the Rate of Novel Gene Identification.
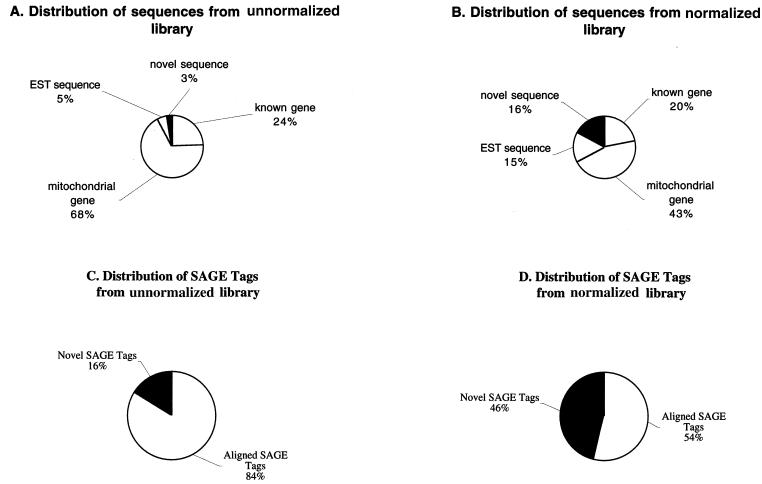
To verify whether our strategy indeed can yield a higher rate of novel gene identification, we screened a normalized poly(dA/dT)− colon cDNA library directly. As shown in Fig. 6, the rate of novel sequences identified in the normalized poly(dA/dT)− cDNA library increased to 16%, compared with 3% in the control sample. As a second validation, SAGE tags collected from these sequences showed that the rate of novel SAGE tags in the normalized poly(dA/dT)− cDNA library was 43%, compared with 16% in the control sample. These data clearly indicate that screening normalized/subtracted poly(dA/dT)− cDNAs can provide a much higher degree of novel gene identification than can the current approaches.
Figure 6.
Distribution of sequences collected in poly(dA/dT)− colon cDNA library. A total of 109 clones from a nonnormalized library and 193 clones from a normalized library were sequenced. All of the sequences were aligned with databases. SAGE tags containing 10 nt were also collected from these clones and searched in the SAGE database. (A) Distribution of sequences from nonnormalized library. (B) Distribution of sequences from normalized library. (C) Distribution of SAGE tags from nonnormalized library. (D) Distribution of SAGE tags from normalized library.
In summary, we have identified and corrected a fundamental flaw in the current genome-wide gene studies. Applying our strategy of screening poly(dA/dT)− cDNAs should substantially accelerate the rate of genome-wide novel gene identification in many eukaryotic species. In the postgenome era, although most of the genes in many genomes will be known, the identification of genes expressed under various particular conditions will become a challenge. The principles we described here also should be readily applicable for the genome analysis in the postgenome stage.
Acknowledgments
We acknowledge B. Vogelstein and K. W. Kinzler (John Hopkins University) for helpful comments. We thank L. Wagner and C. Tolstoshev (National Center for Biotechnology Information) for providing EST and CGAP database information. We thank J. Jessee (Life Technologies) for providing Gene II enzyme. We also thank Mr. X. Xu for assistance. This work was supported by National Cancer Institute Grants CA42557 (J.D.R) and CA78862-01 (J.D.R and S.M.W.), American Cancer Society Grant IRG-41-40 (S.M.W.), and the G. Harold and Lelia Y. Mathers Foundation (S.M.W.).
Abbreviations
- SAGE
serial analysis of gene expression
- EST
expressed sequence tag
- CGAP
Cancer Genome Anatomy Project
Footnotes
References
- 1.Cohen J. Science. 1997;275:769. [Google Scholar]
- 2.Bishop J O, Morton J G, Rosbach M, Richardson M. Nature (London) 1974;250:199–204. doi: 10.1038/250199a0. [DOI] [PubMed] [Google Scholar]
- 3.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D. In: Molecular Biology of the Cell. Robertson M, editor. New York: Garland; 1994. p. 369. [Google Scholar]
- 4.Lewin B. In: Gene VI. Lewin B, editor. New York: Oxford Univ. Press; 1997. p. 687. [Google Scholar]
- 5.Boguski M S. Trends Biochem Sci. 1995;20:295–296. doi: 10.1016/s0968-0004(00)89051-9. [DOI] [PubMed] [Google Scholar]
- 6.Strausberg R L, Dahl C A, Klausner R D. Nat Genet. 1997;15:415–416. doi: 10.1038/ng0497supp-415. [DOI] [PubMed] [Google Scholar]
- 7.Gerhold D, Caskey C T. BioEssays. 1996;18:973–981. doi: 10.1002/bies.950181207. [DOI] [PubMed] [Google Scholar]
- 8.Bonaldo M F, Lennon G, Soares M B. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- 9.Fields C, Adams M D, White O, Venter J C. Nat Genet. 1994;7:345–346. doi: 10.1038/ng0794-345. [DOI] [PubMed] [Google Scholar]
- 10.Lewin B. In: Gene VI. Lewin B, editor. New York: Oxford Univ. Press; 1997. p. 170. [Google Scholar]
- 11.Velculescu V E, Madden S L, Zhang L, Lash A E, Yu J, Rago C, Lal A, Wang C J, Beaudry G A, Ciriello K M, et al. Nat Genet. 1999;23:387–388. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 12.Adams M D, Dubnick M, Kerlavage A R, Moreno R, Kelley J M, Utterback T R, Nagle J W, Fields C, Venter J C. Nature (London) 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- 13.Wang S M, Rowley J D. Proc Natl Acad Sci USA. 1998;95:11909–10914. doi: 10.1073/pnas.95.20.11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan A S, Wilcox A S, Hopkins J A, Silela J M. Nucleic Acids Res. 1991;19:1715. doi: 10.1093/nar/19.7.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiriangkum J, Vainshtein I, Elliott J F. Nucleic Acids Res. 1992;20:3793–3794. doi: 10.1093/nar/20.14.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang P, Pardee A B. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 17.Liang P, Zhu W, Zhang X, Guo Z, O'Connell R P, Averboukh L, Wang F, Pardee A B. Nucleic Acids Res. 1994;22:5763–5764. doi: 10.1093/nar/22.25.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abbotts J, Jaju M, Wilson S H. J Biol Chem. 1991;266:3937–3943. [PubMed] [Google Scholar]
- 19.Bakhanashvili M, Hizi A. FEBS Lett. 1993;319:201–205. doi: 10.1016/0014-5793(93)80067-5. [DOI] [PubMed] [Google Scholar]
- 20.Yu H, Goodman M F. Biol Chem. 1992;267:10888–10896. [PubMed] [Google Scholar]
- 21.Sun Y, Hegamyer G, Colburn N H. Cancer Res. 1994;54:1139–1144. [PubMed] [Google Scholar]
- 22.Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]