Abstract
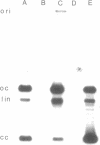
Resistance to tetracycline and lincosamide antibiotics was transferred en bloc from a strain of Bacteroides fragilis (V503) to a plasmidless strain of Bacteroides uniformis (V528) during in vitro filter matings. Resistance transfer was detected at frequencies of 10(-5) to 10(-6) drug-resistant progeny per input donor cell and was dependent on cell-to-cell contact of donors and recipients. Transfer was insensitive to DNase and was not mediated by chloroform- or filter-sterilized donor broth cultures. A determinant for resistance to cefoxitin in V503 was not transferred in this system. V503 contained a 3.7 x 10(6)-dalton plasmid (pVA503). Drug-resistant progeny of V503 x V528 matings usually contained pVA503, but up to 20% of the total progeny of such crosses were plasmid free. Filter blot DNA hybridization studies (Southern method) confirmed that pVA503 was not integrated into the host chromosome of the plasmidless progeny. Drug-resistant progeny from V503 x V528 matings (with or without pVA503) conjugally transferred clindamycin resistance an tetracycline resistance to a suitable recipient strain. None of the drug resistance determinants of V503 were affected by treatment with standard plasmid curing regimens, and methods designed to detect very large plasmid molecules failed to suggest the involvement of extrachromosomal DNA in this resistance transfer system. The well-characterized Bacteroides R plasmid, pBF4 (conferring clindamycin resistance), was found to share hybridizing sequences with bulk cellular V503 DNA when examined by filter blot hybridization. Similarly sized sequences were found in drug-resistant progeny recovered from matings. Neither of the two pBF4 derivatives carrying deletions that abolished clindamycin resistance hybridized with V503 DNA.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Brefort G., Magot M., Ionesco H., Sebald M. Characterization and transferability of Clostridium perfringens plasmids. Plasmid. 1977 Nov;1(1):52–66. doi: 10.1016/0147-619x(77)90008-7. [DOI] [PubMed] [Google Scholar]
- Franke A. E., Clewell D. B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of "conjugal" transfer in the absence of a conjugative plasmid. J Bacteriol. 1981 Jan;145(1):494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbach S. L., Bartlett J. G. Anaerobic infections. 1. N Engl J Med. 1974 May 23;290(21):1177–1184. doi: 10.1056/NEJM197405232902106. [DOI] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina F. L., Jones K. R., Wood P. H. Chimeric streptococcal plasmids and their use as molecular cloning vehicles in Streptococcus sanguis (Challis). J Bacteriol. 1980 Sep;143(3):1425–1435. doi: 10.1128/jb.143.3.1425-1435.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina F. L., Kopecko D. J., Jones K. R., Ayers D. J., McCowen S. M. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. 1978 Jun;1(3):417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- Macrina F. L., Reider J. L., Virgili S. S., Kopecko D. J. Survey of the extrachromosomal gene pool of Streptococcus mutans. Infect Immun. 1977 Jul;17(1):215–226. doi: 10.1128/iai.17.1.215-226.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina F. L., Weatherly G. G., Curtiss R., 3rd R6K plasmid replication: influence of chromosomal genotype in minicell-producing strains of Escherichia coli K-12. J Bacteriol. 1974 Dec;120(3):1387–1400. doi: 10.1128/jb.120.3.1387-1400.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore W. E., Cato E. P., Holdeman L. V. Some current concepts in intestinal bacteriology. Am J Clin Nutr. 1978 Oct;31(10 Suppl):S33–S42. doi: 10.1093/ajcn/31.10.S33. [DOI] [PubMed] [Google Scholar]
- Privitera G., Sebald M., Fayolle F. Common regulatory mechanism of expression and conjugative ability of a tetracycline resistance plasmid in Bacteroides fragilis. Nature. 1979 Apr 12;278(5705):657–659. doi: 10.1038/278657a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roberts M. C., Smith A. L. Molecular characterization of "plasmid-free" antibiotic-resistant Haemophilus influenzae. J Bacteriol. 1980 Oct;144(1):476–479. doi: 10.1128/jb.144.1.476-479.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker N. B., Smith M. D., Guild W. R. DNase-resistant transfer of chromosomal cat and tet insertions by filter mating in Pneumococcus. Plasmid. 1980 Jan;3(1):80–87. doi: 10.1016/s0147-619x(80)90036-0. [DOI] [PubMed] [Google Scholar]
- Smith C. J., Markowitz S. M., Macrina F. L. Transferable tetracycline resistance in Clostridium difficile. Antimicrob Agents Chemother. 1981 Jun;19(6):997–1003. doi: 10.1128/aac.19.6.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tally F. P., Snydman D. R., Gorbach S. L., Malamy M. H. Plasmid-mediated, transferable resistance to clindamycin and erythromycin in Bacteroides fragilis. J Infect Dis. 1979 Jan;139(1):83–88. doi: 10.1093/infdis/139.1.83. [DOI] [PubMed] [Google Scholar]
- Thayer R. E. An improved method for detecting foreign DNA in plasmids of Escherichia coli. Anal Biochem. 1979 Sep 15;98(1):60–63. doi: 10.1016/0003-2697(79)90705-x. [DOI] [PubMed] [Google Scholar]
- Varel V. H., Bryant M. P. Nutritional features of Bacteroides fragilis subsp. fragilis. Appl Microbiol. 1974 Aug;28(2):251–257. doi: 10.1128/am.28.2.251-257.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch R. A., Jones K. R., Macrina F. L. Transferable lincosamide-macrolide resistance in Bacteroides. Plasmid. 1979 Apr;2(2):261–268. doi: 10.1016/0147-619x(79)90044-1. [DOI] [PubMed] [Google Scholar]
- Welch R. A., Macrina F. L. Physical characterization of Bacteroides fragilis R plasmid pBF4. J Bacteriol. 1981 Feb;145(2):867–872. doi: 10.1128/jb.145.2.867-872.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga M. C., Durham D. R., Welch R. A. Plasmid- and chromosome-mediated dissimilation of naphthalene and salicylate in Pseudomonas putida PMD-1. J Bacteriol. 1981 Sep;147(3):836–843. doi: 10.1128/jb.147.3.836-843.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]