Abstract
The mouse p locus encodes a gene that functions in normal pigmentation. We have characterized a radiation-induced mutant allele of the mouse p locus that is associated with a failure-to-thrive syndrome, in addition to diminished pigmentation. Mice homozygous for this mutant allele, p100H, show delayed growth and die within 2 wk after birth. We have discovered that the mutant mice develop progressive atrioventricular heart block and significant ultrastructural changes in both cardiac and skeletal muscle cells. These observations are common characteristics described in human myopathies. The karyotype of p100H chromosomes indicated that the mutation is associated with a chromosome 7 inversion. We demonstrate here that the p100H chromosomal inversion disrupts both the p gene and the Sox6 gene. Normal Sox6 gene expression has been examined by Northern blot analysis and was found most abundantly expressed in skeletal muscle in adult mouse tissues, suggesting an involvement of Sox6 in muscle maintenance. The p100H mutant is thus a useful animal model in the elucidation of myopathies at the molecular level.
The mouse pink-eyed dilution (p) locus on chromosome 7 was one of the earliest known pigmentation loci identified in mice (1). In addition to known spontaneous p mutations, there is a series of radiation-induced p mutations that often are associated with additional (nonpigmentation) phenotypes caused by chromosome deletions or rearrangements of adjacent genes (2–4). These radiation-induced p alleles have been extremely useful for the identification of the genes in the proximal region of mouse chromosome 7 (3, 5, 6). The studies of these mutant alleles have led to discoveries such as involvement of the β3 subunit gene of the γ-aminobutyric acid type A receptor (Gabrb3) in palate development (7–9) and important roles for the HERC2 gene during development (10–12).
As part of our continuing effort to identify genes disrupted in radiation-induced p mutant alleles, we characterized the phenotype and chromosomal structure of the p100H allele (pink-eyed dilution 100 Harwell). p100H was first recognized as a postnatal lethal recessive mutation that did not fall into the complementation groups defined by other known p alleles. Using both cytological and molecular analysis, we found that the p100H mutation is caused by a chromosomal inversion, with one breakpoint in the p gene, the other in the Sox6 gene. Sox6 is a member of the Sox [Sry-related high mobility group (HMG) box] DNA binding protein family and initially was isolated from an adult testis cDNA library (13). Previous studies have suggested that Sox6 plays a role in the development of the central nervous system (CNS) (13) and chondrogenesis (14). However, our analysis of the p100H phenotype revealed that the p100H mutant develops myopathy and an atrioventricular (AV) heart block, a cardiac conduction defect that is a main cause of death in human cardiac myopathies (15). Electronmicroscopic evaluation of the mutant cardiac and skeletal muscle demonstrated significant change in ultrastructure. Thus, the phenotype of the p100H mutation suggests that the Sox6 protein also may be involved in maintaining normal physiological functions of muscle tissue, including the heart.
Materials and Methods
p Mutation and Karyotyping.
The p100H mutation was generated in the course of p locus mutagenesis conducted at the Medical Research Council Radiobiology Unit (Harwell, United Kingdom). A wild-type male was irradiated with 3 + 3 Gy at 24-h intervals, and mutant p alleles were uncovered by the specific locus test (16). Because homozygous neonates did not survive beyond 2 wk, the p100H allele was first recognized as a postnatal lethal mutation. This is a recessive mutation, and heterozygotes are normal and fertile.
Chromosomes of p100H/+ and wild-type mice were stained with Giemsa by using standard procedures (17). The banding pattern of the mutant chromosome 7 was compared with its wild-type homologue to detect the chromosomal rearrangement.
Reverse Transcription–PCR (RT-PCR) and Rapid Amplification of cDNA Ends (RACE).
To determine the inversion break point within the p locus of the p100H mutation, RT-PCR was performed. First, 5 μg of total RNA from the eyes of p100H/p100H or C57BL/6 (+/+) mice was used to synthesize random-primed cDNA using standard procedures in a total volume of 20 μl. PCR was performed by using 10 sets of p gene-specific primer pairs that together cover all 24 exons. After determining which exon boundaries were not amplified from the mutant cDNA, RACE was carried out to isolate a possible chimeric transcript in the mutant. Poly(A)+ RNA was isolated from postnatal day-12 p100H/p100H brain by using the Fast-Track poly(A)+ RNA isolation kit (Invitrogen). cDNA synthesis and RACE were performed by using the Marathon cDNA Amplification Kit (CLONTECH), following the manufacturer's instructions. To obtain the sequence upstream of exon 24 in putative chimeric cDNA, 5′RACE was carried out by using a gene-specific reverse primer in exon 24 of the p gene (MHB329: 5′-TCTTCGGAGGTGCTGTGGGGAAGCCTTC-3′, nucleotides 2749–2722; GenBank accession no. M97900), followed by nested PCR using the primer MHB310 (5′-CTTCCACCTGATGCTGATGCTGA-3′, nucleotides 2725–2703). PCR products were analyzed on 1.2% agarose gel, and distinct bands were gel-purified and cloned into the TA cloning vector pCR 2.1 (Invitrogen). cDNA clones were sequenced by an Applied Biosystems automated sequencer, and the sequences were analyzed with the sequencher and macvector programs. After obtaining a chimeric cDNA sequence (Sox6-p), PCR primers were designed to amplify 5′p-3′Sox6 and 5′Sox6-3′p reciprocal cDNAs from the mutant: MHB346 (5′-TGGGACACTTATCGGAGCATCCAC-3′, exon 23 of the p gene, nucleotides 2458–2481), MHB329 (exon 24 of the p gene nucleotides 2749–2722, sequence above), MHB345 (5′-TCGGCAGGGGCAGTCTCACCTAC-3′, nucleotides 1353–1375 in Sox6: GenBank accession no. U32614), and MHB347 (5′-GCAGAGCCATTCATTGCTTTGCTTCC-3′, nucleotides 1927–1902 in Sox6: GenBank accession no. U32614). Mixed gene pairs (MHB346 and MHB347, MHB345 and MHB329) as well as the same gene pairs (MHB346 and 329, MHB345 and MHB347) were used for PCR using wild-type C57BL/6 and homozygous p100H brain cDNA as templates. The products were analyzed on 2.0% agarose gels, TA-cloned in pCR2.1, and their sequence was analyzed as described above.
Northern Blot Hybridization.
Mouse multiple tissue Northern and embryonic Northern filters were purchased from CLONTECH. The filters were hybridized with Sox6 cDNA sequences (nucleotides 1353–1927; GenBank accession no. U32614) following the manufacturer's protocols and exposed to a X-Omat AR film (Kodak) at −80°C with two intensifying screens for 18 h.
Electrocardiogram.
p100H homozygous mice and wild-type littermates (postnatal 3–13 days old) were anesthetized with inhaled Halothane (Halocarbon Laboratory, River Edge, NJ) before electrocardiogram (ECG) recordings. We recorded ECGs from a total of six p100H homozygotes along with two wild-type littermates for each as controls. Recording electrodes were placed on the right axilla and lower left abdomen, and a grounding electrode was placed on the abdominal right lower quadrant. ECG recordings were collected until voluntary movements were observed. The 12 QRS complexes immediately preceding the initiation of voluntary movements were chosen for analysis of each mouse to minimize anesthetic effects. ECG signals were amplified ×100 (Grass RPS107; Grass Instruments, Quincy, MA) and displayed on an oscilloscope. The signals were sampled at a rate of 500 Hz and stored digitally using an Instrutech ITC-16 digital computer interface (Instrutech, Greatneck, NY) and axodata 1.2.2 software (Axon Instruments, Foster City, CA) and later analyzed by using axograph 3.0 software (Axon Instruments).
Electron Microscopy of Skeletal and Heart Muscle.
Skeletal muscle from hind leg (gastrocnemius) and heart muscle (ventricle) were dissected from p100H homozygote and a wild-type littermate (postnatal day 4) and fixed in 3% glutaraldehyde in cacodylate (0.1 M, pH 8.0) buffer. Tissues were embedded in Spurr resin and sectioned at the University of Arizona core facility. Then, 1-μm sections were stained with Trichrome to define the orientation of the tissue. Thin sections (80 nm) were stained with uranyl acetate and lead citrate. Grids were viewed on an electron microscope (Philips CM12) at an accelerating voltage of 80 kV at the core facility.
Results
Determination of the Inversion Breakpoint.
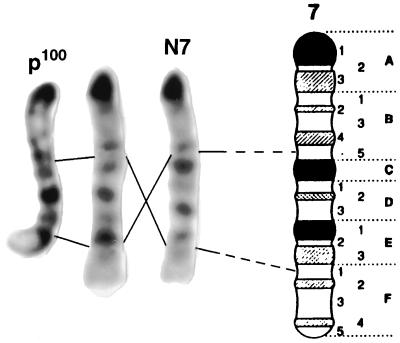
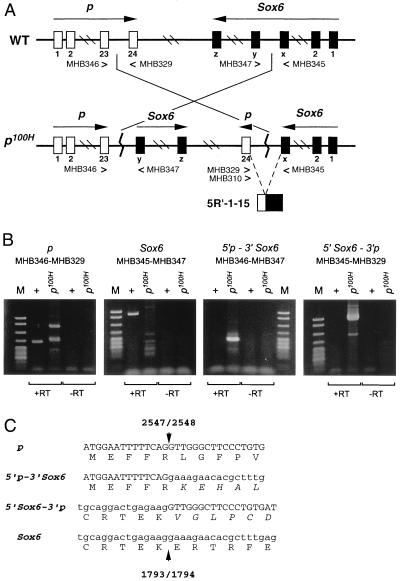
The p100H phenotype was unique among previously isolated p alleles and did not fall in any of the previously identified complementation groups (3, 6) of the series of p-deletions (data not shown). Based on this observation, a chromosomal rearrangement was suspected. Preliminary evidence of an inversion was provided by crossover-suppression between the loci of p and Tyr. In a backcross of p100H +/+ Tyrc-ch × p Tyrc-ch/p Tyrc-ch (Tyrc-ch = chinchilla), with 10–15% recombination expected, no crossovers were found among 74 mice. Karyotyping confirmed the presence of an inversion with breakpoints at 7B5 and 7F1 (Fig. 1), the former corresponding to the map location of the p locus (18). RT-PCR was performed first to determine the inversion breakpoint of p100H in the p gene. Primer sets were chosen to cover all 24 exons of the p gene. Among the primer sets tested, only one primer pair that spans the 23rd and 24th exons failed to amplify from p100H cDNA (data not shown). This result led us to speculate that the p100H mutation had an inversion breakpoint in the 23rd intron of the p gene. However, primer pairs located within exons 1–23 as well as those within exon 24 amplified the expected size fragments in both wild-type and the mutant cDNA with equivalent intensity (data not shown). Because both the mutant and wild-type cDNA gave equally robust signals with the primer pairs within the 24th exon, we hypothesized that the 24th exon was part of a chimeric transcript of the p gene and an unknown gene fused by the inversion. Based on this assumption, 5′ RACE was performed to isolate this chimeric cDNA using the reverse primers MHB329 and MHB310 in exon 24 of the p gene. A clone, 5′R-1–15, isolated from this assay supported this hypothesis (Fig. 2A). The clone 5′R-1–15 contained part of the Sox6 coding sequence (nucleotides 1275–1793: GenBank accession no. U32614), followed immediately by the sequence of exon 24 of the p gene. The Sox6 gene was previously mapped near D7Mit40 on chromosome 7 (13), corresponding to the distal breakpoint of p100H, band 7F1.
Figure 1.
Karyotype and idiogram of chromosome 7 from p100H showing the presence of paracentric inversion with breakpoints in bands 7B5 and 7F1. The central chromosome is a composite made by cutting and inverting the appropriate segment from the normal chromosome 7 (N7). The banding pattern of this composite chromosome then resembles that from a p100H/+ heterozygote (p100H).
Figure 2.
(A) A schematic diagram of the p locus and the Sox6 locus on mouse chromosome 7. WT, wild type; p100H, p100H mutant. Primers used for RT-PCR are listed underneath each exon (23, 24, x, y, z). Nucleotide sequences of the primers are described in Materials and Methods. 5′R-1–15 is the first chimeric cDNA clone isolated and was used to identify the inversion breakpoints in the p100H mutation (see text). (B) RT-PCR products amplified from wild-type (+) and the p100H homozygote (p100H) brain cDNA using primers listed. +RT and −RT represent synthesis with or without RT, respectively in a cDNA reaction. Sizes of the marker standard fragments (M: pBR322 MspI digest; New England Biolabs) are from the top: 622 bp, 527 bp, 404 bp, 307 bp, 238/242 bp, 217 bp. The expected sizes of the RT-PCR products are: 292 bp for p, 575 bp for Sox6, 224 bp for 5′p-3′Sox6, and 643 bp for 5′Sox6-3′p. The two fragments seen in p100H (+RT) lane of p were cloned and sequenced. Both fragments (327 bp and 480 bp) have no significant sequence matches with the p gene except for the primer sequences and are therefore artifactual. (C) Amino acid sequence in the vicinity of the breakpoints. The predicted amino acid sequences of the chimeric transcripts are shown below the nucleotide sequence. Arrowhead indicates the breakpoint, and the numbers above and below it correspond to nucleotide numbers of the breakpoint in the p gene cDNA (GenBank accession no. M97900) or the Sox6 gene cDNA (GenBank accession no. U32614).
To confirm the site of the inversion in the Sox6 gene, primer sets (shown in Fig. 2A) were designed to detect the reciprocal chimeric transcripts, 5′p-3′Sox 6 or 5′Sox6-3′p in p100H. As shown in Fig. 2B, wild-type cDNA fragments of the p and Sox6 genes were observed only in wild type; however, the mutant cDNA fragments (5′p-3′Sox6, 5′Sox6-3′p) were observed only in p100H. In the 5′Sox6-3′p transcript, the switch to the p coding sequence leads to a frame shift (Fig. 2C) that introduces a premature termination codon 11 aa downstream of the break point. This truncation eliminates the HMG DNA binding domain of Sox6 that is downstream of the breakpoint (HMG BOX is nucleotides 2022–2261 in Sox6). Similarly, the frame shift of the 5′p-3′Sox6 transcript (Fig. 2C) results in a 78-aa substitution downstream of the breakpoint replacing the normal 27 aa encoded by exon 24 in the wild-type p protein. This frameshift eliminates the last transmembrane domain of the p protein. The frameshifts of both p and Sox6 most likely generate nonfunctional proteins because of loss of functionally critical domains or instability of the mutant proteins.
To confirm the exon–intron boundaries in the Sox6 gene assessed by the RT-PCR clones shown in Fig. 2, genomic clones containing the inversion breakpoint were isolated. The exon–intron boundaries were confirmed from the genomic sequence to be 1793/1794 (data not shown).
Expression of the Sox6 Gene.
Northern blot hybridization was performed to examine the expression of the Sox6 gene in both adult and embryonic tissues. As shown in Fig. 3, two (≈3 kb and ≈8–9 kb) transcripts hybridize to the Sox6 cDNA probe (nucleotides 1353–1927). In adult tissues, the large transcript is most abundantly expressed in skeletal muscle; however, it also is expressed in a variety of other tissues at moderate levels. In contrast, the ≈3-kb transcript is expressed predominantly in testis (Fig. 3A). Similar expression profiles (sizes and abundance) were obtained when Northern blot analyses of human tissues were probed with Sox6 cDNA (data not shown).
Figure 3.
(A) Mouse multiple tissue Northern blot. Each lane contains 2 μg of poly(A)+ RNA from the tissues indicated hybridized with 32P-labeled 575 bp Sox6 cDNA fragment (nucleotides 1353–1927). Control hybridization with β-actin was performed to confirm equal loading of RNAs (data not shown). sk, skeletal. (B) Mouse embryonic multiple tissue Northern blot hybridized with the same probe as A. Each lane contains 2 μg of poly(A)+ RNA isolated from embryos whose ages are listed above each lane as embryonic day (ED). Numbers on left are sizes of standard marker fragments.
Temporal expression of the two transcripts in developing embryos is shown in Fig. 3B. The large transcript appears by embryonic day 11, preceding the expression of the ≈3-kb transcript. The signal of the large transcript remains elevated throughout the later stages of embryogenesis (embryonic day 17). It has been reported that, in the developing embryonic nervous system, Sox6 expression is diminished by embryonic day 12.5 (13). These results, together with the present data, suggest that the expression of the large transcript during later stages of embryogenesis is most likely from nonneural tissues.
It has been previously reported that the large and small transcripts are generated by alternative splicing from the Sox6 gene, and the 3′ untranslated region (UTR) unique to the large form has been determined (14). We isolated multiple Sox6 cDNA clones by 5′ RACE from adult mouse skeletal muscle as well as by screening an embryonic limb buds cDNA library (gift of David J. Goldhamer, Univ. of Pennsylvania, Philadelphia), and identified large transcript-specific 5′ UTR sequences. Interestingly, all of the clones share the same 5′ coding sequence previously reported (13); however, analysis of the 5′ UTR sequences indicated complex alternative splicing in the 5′ noncoding region of the Sox6 gene (data not shown).
Phenotype Analysis of the p100H Mutation.
To better characterize the postnatal lethal phenotype of p100H, the number of p100H homozygous newborns were recorded from 16 timed p100H/+ intercrosses. Of 134 newborns, 26 were p100H homozygotes (19.4% of total births); therefore, the p100H allele is not an embryonic lethal mutation. One-half of the newborn mutant homozygotes died within 24 h after birth. The surviving mutant homozygotes exhibited runting, and by postnatal day 14, typically weighed one-third of the normal littermates. In the postnatal period, mutant mice died suddenly and often unexpectedly (several homozygotes died while being handled). We examined the major organs of both male and female p100H homozygotes at postnatal day 14. The only gross anomaly observed in near-death homozygotes was a bloated intestine, which may be a secondary effect of overall declining health. At the light microscopic level, no apparent structural anomalies were found in brain, heart, lung, thymus, stomach, kidney, liver, testis, ovary, intestine, or spleen (data not shown).
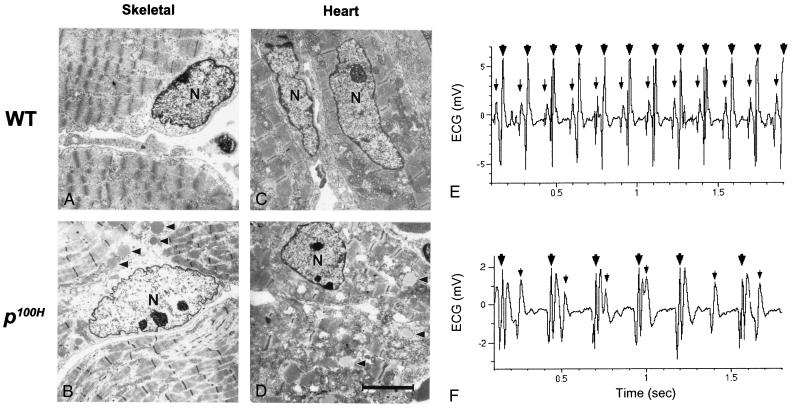
The expression pattern of Sox6 (Fig. 3) suggested that it may play a critical role in muscle tissue. We examined the ultrastructures of skeletal and cardiac muscle using transmission electron microscopy (Fig. 4 A–D). As represented in Fig. 4B, mutant skeletal muscle fibers are significantly thinner than those of wild type (Fig. 4A) and appear liquefied in some places that may indicate degeneration. Curiously, in addition to the abnormal fiber organization, many mutant nuclei are irregularly shaped, often jagged and pleated. In mutant cardiac muscle cells, vacuole structures are obvious, and increased number of mitochondria is observed in many fields (data not shown). Mutant muscles, both skeletal and cardiac, accumulate intracellular deposits, possibly lipid droplets, as indicated by arrows in Fig. 4 B and D. These observations are typical structural changes associated with myopathies.
Figure 4.
Electron micrographs of skeletal and cardiac muscle cells (A–D) and ECG of wild-type (E) and the mutant (F). WT, the wild-type littermate (A and C), p100H, the p100H homozygote (B and D). N, nucleus. Arrow heads in B and D indicate deposits found in the mutant muscle cells. The scale bar in D is equal to 4 μm. Representative ECG of the wild type (E) and the mutant (F) are shown. The small arrow indicates P-wave, activation of the atria, and the large arrow indicates QRS-complex, activation of the ventricles. The ECG of F shows the third degree (complete) heart block observed in a postnatal day-12 p100H mouse less than 24 h before death.
To assess the physiological function of the mutant heart, ECG were recorded in mutant mice and their wild-type littermates. In wild type (Fig. 4E), a normal sinus rhythm was observed in each with consistent intervals between P wave and QRS activation. In p100H, however, variable degrees of AV conduction defects, including prolongation of the PR interval (first-degree AV block) and complete atrial-ventriclar dissociation (complete heart block) (Fig. 4F), were observed. As represented in Fig. 4 E and F, the mutants exhibited significantly slower average heart rates than wild-type littermates (wild type 418 ± 73; p100H 320 ± 64; heart rate per min ± SD, P = 0.004). These findings are characteristic of abnormalities of the cardiac system. One p100H homozygote did not exhibit heart block on initial ECG recording at postnatal day 6, but subsequently demonstrated complete heart block at postnatal day 10, and died on postnatal day 13. These observations suggest that p100H mutants have progressive AV heart block that is a likely cause of the early and sudden postnatal lethality.
Discussion
We have characterized both the phenotype and genotype of the x-ray-induced p100H postnatal lethal mutation. This mutation is caused by an intrachromosomal inversion: one breakpoint in the p gene (that functions in pigmentation) and the other in the Sox6 gene. The Sox gene family is defined by the well-conserved SRY-related HMG box domain that mediates sequence-specific DNA binding (19, 20). The known functions of the Sox proteins during development are both diverse and complex: Sry has a well-described role in sex determination (21), Sox1–3 in lens development (22), Sox4 in endocardial ridge development in the heart and pro-B cell expansion (23), Sox9 in chondrogenesis and sex determination (24, 25), and Sox10 in neural crest cell differentiation (26, 27).
The function of the Sox6 protein in early development is not yet fully understood. It has been reported that Sox6 is expressed in the CNS during embryogenesis (13). The Sox6 protein also has been associated with the activation of the type II collagen gene (Col2a1) during chondrogenesis. It has been suggested that the Sox6 protein, along with Sox5 and Sox9, cooperatively activate the Col2a1 gene (14). Type II collagen is the major component of cartilage and when the Col2a1 gene is disrupted, the homozygous null mouse suffers from severe skeletal anomalies (28). However, p100H homozygotes, although small in size, do not show any of the visible skeletal anomalies reported in Col2a1 null mice, and we were able to detect Col2a1 transcripts in p100H mice by RT-PCR and Northern blot analysis (data not shown). Additionally, we did not detect gross anomalies in the CNS of p100H homozygotes. These observations could imply that Sox6 may play a minor role in Col2a1 expression and CNS development, or it may reflect a functional redundancy of Sox proteins in the development of these tissues. For example, Sox1 null mice exhibit only subtle CNS abnormalities (29), despite the high expression of the gene in the developing CNS (30). This observation could be explained by the overlapping expression pattern of Sox2 and Sox3 in the nervous system (20, 29, 30). Similarly, for the p100H mutant, the loss of Sox6 expression in cartilage and the CNS may be compensated for by other Sox proteins with overlapping expression.
In the p100H mutation, the inversion breakpoints fall into the introns of the p gene and the Sox6 gene, creating reciprocal chimeric transcripts without loss of exons (Fig. 2A). The reciprocal nature of the chimeric transcripts also implies that only two genes, p and Sox6, are directly disrupted by the inversion. Deduced amino acid sequence of both chimeric transcripts predicts a frameshift downstream of the breakpoints (Fig. 2C). In the 5′Sox6-3′p transcript, the frameshift creates a premature termination codon, causing a truncation before the HMG DNA-binding domain of the Sox6 gene. Therefore, the Sox6 protein in p100H is likely to be a null form without sequence-specific DNA binding activity. The fact that the lack of the p protein only leads to a pigmentation defect (2–8, 10–12) implies that the other phenotypes of the p100H mutation described here are caused by the disruption of the Sox6 gene. However, it is also formally possible that expression of another unidentified gene (or genes) is altered in p100H, the result of a position effect or yet uncovered (linked) mutation.
Alternative splicing has been reported for several Sox genes including Sox6 (20). In the case of the human SOX5 gene expressed in testis, alternative promoter usage has been suggested for generating the unique 5′ untranslated sequences (31). As shown in Fig. 3A, of the multiple forms observed, the large Sox6 transcripts are expressed in a variety of adult tissues. This result suggests a possible function of Sox6 in these tissues. A similar expression pattern was obtained in human tissues (data not shown). A Sox6 ortholog isolated in nonmammalian organisms (e.g., trout) includes a ≈10-kb message as well as the testis-specific 3-kb message (32). These conserved expression patterns in evolutionarily distant species imply a functional significance of the large Sox6 transcript in a variety of tissues.
The observation that the large form of the Sox6 transcript is most abundantly expressed in skeletal muscle in adult (Fig. 3A) prompted us to examine muscle tissues of the mutant in detail. Comparison of the ultrastructures of skeletal and cardiac muscles of the mutant with those of wild type showed that p100H develops a widespread myopathy that affects skeletal and cardiac myocytes (Fig. 4 A–D). Because the p100H homozygote dies suddenly within the first two postnatal weeks, we suspected compromised cardiac function in the mutant. In fact, ECG recordings of the mutant mice showed a slower heart rate and progressive AV heart block (Fig. 4F). These mutants die within a few days after developing a heart block, indicating that the direct cause of early postnatal lethality is likely cardiac in origin.
While analyzing the phenotype of the p100H mutation, we were intrigued by its similarities to the Emery–Dreifuss muscular dystrophy (EDMD) in humans (OMIM310300, 181350). Patients with EDMD suffer from both skeletal and cardiac myopathy, the latter leading to heart block in adulthood. This cardiac conduction defect is the leading cause of death in these patients. Two genes are associated with EDMD: the nuclear membrane proteins emerin and lamin A/C (15). We examined the expression of these two genes in the postnatal p100H mutants by RT-PCR and by Northern hybridization; however, both transcripts were found at equivalent levels in the mutant and wild-type skeletal muscle (data not shown). Furthermore, the phenotype of the p100H mutant is more severe than human EDMD. Given the likely roles of Sox6 as a transcription factor, the p100H mutation may have more wide-ranging effect on muscle tissues. Detailed examination of differences in gene expression between the p100H mutant and wild type will lead to elucidation of the downstream targets of the Sox6 transcription factor in muscle development.
Many genes associated with heart impairments in humans have been discovered in the last decade. Most of these are muscle structural proteins or ion channels, but transcription factors, including NKX2–5 and TBX5, also have been implicated. Mutations in these genes cause both structural and conduction defects in human (33–35). In the case of NKX2–5 mutations, the severity of atrial septal defect differs among individual patients, but these individuals all develop progressive AV conduction delays that are potentially life-threatening and require pacemaker implantation (33). Based on this observation, it is suggested that NKX2–5 expression in the adult may be required for normal electrophysiological function of the heart (33).
Analyses of Sox4 and Sox9 mutations have demonstrated the importance of these genes in heart development. In mice, a Sox4 null mutation is embryonic lethal because of defects in the formation of the cardiac outflow tract (23). In humans, Campomelic syndrome caused by SOX9 mutations includes a congenital heart defect in some cases (36). The observation that Sox9 is expressed in heart during mouse embryogenesis (37, 38) corroborates the phenotype seen in humans. If Sox6 is required for normal cardiac function, it differs from Sox4 and Sox9 primarily in the absence of gross morphological defects of the heart.
The cardiac conduction defect detected in p100H mutant homozygotes may suggest a role for the Sox6 protein in postnatal heart function. The phenotype of the p100H mutation (early postnatal lethality associated with progressive atrioventricular heart block and myopathy) makes this mutant a useful model for studying early onset myopathies. Identifying candidate genes whose expression is regulated by Sox6 will help us to understand the pathways leading to some heart function disorders.
Acknowledgments
We thank our colleagues, Drs. Ray Runyan, Carol Gregorio, John Gardner, J Newton, and Richard Gniewek for helpful discussions, and Roderick Bronson for initial histology. We also thank Carol Sacks for technical support. We are grateful to Dr. E. P. Evans for the karyotyping and Fig. 1. We dedicate this paper to Dr. Noboru Sueoka on the occasion of his retirement. The animal studies at Harwell were carried out under the guidance issued by the Medical Research Council in “Responsibility in the use of animals in medical research” (July 1993) and Home Office Project license 30/00875. This work was supported by National Institutes of Health Grant GM43840 (to M.H.B.).
Abbreviations
- AV
atrioventricular
- HMG
high mobility group
- RT-PCR
reverse transcription–PCR
- RACE
rapid amplification of cDNA ends
- UTR
untranslated region
- ECG
electrocardiogram
- CNS
central nervous system
Note Added in Proof
No putative open reading frames identified by grail 1.3 analysis (Oak Ridge National Laboratory) in the sequenced portion (26 kb) of the Sox6 breakpoint-containing intron (27 kb) are significantly homologous to any known gene sequence in the nonredundant blast database (National Institutes of Health).
References
- 1.Haldane J B S, Sprunt A D, Haldane N M. J Genet. 1915;5:133–135. [Google Scholar]
- 2.Brilliant M H. Mamm Genome. 1992;3:187–191. doi: 10.1007/BF00355717. [DOI] [PubMed] [Google Scholar]
- 3.Lyon M F, King T R, Gondo Y, Gardner J M, Nakatsu Y, Eicher E M, Brilliant M H. Proc Natl Acad Sci USA. 1992;89:6968–6972. doi: 10.1073/pnas.89.15.6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doolittle D P, Davisson M T, Guidi J N, Green M C. In: Genetic Variants and Strains of the Laboratory Mouse. 3rd ed. Lyon M F, Rastan S, Brown S D M, editors. New York: Oxford Univ. Press; 1996. pp. 17–854. [Google Scholar]
- 5.Johnson D K, Stubbs L J, Culiat C T, Montgomery C S, Russell L B, Rinchik E M. Genetics. 1995;141:1563–1571. doi: 10.1093/genetics/141.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell L B, Montgomery C S, Cacheiro N L A, Johnson D K. Genetics. 1995;141:1547–1562. doi: 10.1093/genetics/141.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Culiat C T, Stubbs L, Nicholls R D, Montgomery C S, Russell L B, Johnson D K, Rinchik E M. Proc Natl Acad Sci USA. 1993;90:5105–5109. doi: 10.1073/pnas.90.11.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakatsu Y, Tyndale R F, DeLorey T M, Durham-Pierre D, Gardner J M, McDanel H J, Nguyen Q, Wagstaff J, Lalande M, Sikela J M, et al. Nature (London) 1993;364:448–450. doi: 10.1038/364448a0. [DOI] [PubMed] [Google Scholar]
- 9.Homanics G E, DeLorey T M, Firestone L L, Quinlan J J, Handforth A, Harrison N L, Krasowski M D, Rick C E, Korpi E R, Makela R, et al. Proc Natl Acad Sci USA. 1997;94:4143–4148. doi: 10.1073/pnas.94.8.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehman A L, Nakatsu Y, Ching A, Bronson R T, Oakey R J, Keiper-Hrynko N, Finger J N, Durham-Pierre D, Horton D B, Newton J M, et al. Proc Natl Acad Sci USA. 1998;95:9436–9441. doi: 10.1073/pnas.95.16.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji Y, Walkowicz M J, Buiting K, Johnson D K, Tarvin R E, Rinchik E M, Horsthemke B, Stubbs L, Nicholls R D. Hum Mol Genet. 1999;8:533–542. doi: 10.1093/hmg/8.3.533. [DOI] [PubMed] [Google Scholar]
- 12.Walkowicz M, Ji Y, Ren X, Horsthemke B, Russell L B, Johnson D, Rinchik E M, Nicholls R D, Stubbs L. Mamm Genome. 1999;10:870–878. doi: 10.1007/s003359901106. [DOI] [PubMed] [Google Scholar]
- 13.Connor F, Wright E, Denny P, Koopman P, Ashworth A. Nucleic Acids Res. 1995;17:3365–3372. doi: 10.1093/nar/23.17.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefebvre V, Li P, de Crombrugghe B. EMBO J. 1998;19:5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris G E, Manilal S. Hum Mol Genet. 1999;8:1847–1851. doi: 10.1093/hmg/8.10.1847. [DOI] [PubMed] [Google Scholar]
- 16.Cattanach B M, Rasberry C. Mutat Res. 1994;311:77–84. doi: 10.1016/0027-5107(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 17.Evans E P. In: Genetic Variants and Strains of the Laboratory Mouse. 3rd ed. Lyon M F, Rastan S, Brown S D M, editors. New York: Oxford Univ. Press; 1996. pp. 1446–1449. [Google Scholar]
- 18.Brilliant M H, Williams R W, Holdner B C, Angel J M. Mamm Genome. 1996;6:S135–S150. [PubMed] [Google Scholar]
- 19.Pevny L H, Lovell-Badge R. Curr Opin Genet Dev. 1997;7:338–344. doi: 10.1016/s0959-437x(97)80147-5. [DOI] [PubMed] [Google Scholar]
- 20.Wegner M. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodfellow P N, Lovell-Badge R. Annu Rev Genet. 1993;27:71–92. doi: 10.1146/annurev.ge.27.120193.000443. [DOI] [PubMed] [Google Scholar]
- 22.Kamachi Y, Uchikawa M, Collignon J, Lovell-Badge R, Kondoh H. Development (Cambridge, UK) 1998;125:2521–2532. doi: 10.1242/dev.125.13.2521. [DOI] [PubMed] [Google Scholar]
- 23.Schilham M W, Oosterwegel M A, Moerer P, Ya J, de Boer P A, van de Wetering M, Verbeek S, Lamers W H, Kruisbeek A M, Cumano A, Clevers H. Nature (London) 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 24.Bi W, Deng J M, Zhang Z, Behringer R R, de Crombrugghe B. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 25.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli F D, Keutel J, Hustert E, et al. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 26.Southard-Smith E M, Kos L, Pavan W J. Nat Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- 27.Pingault V, Bondurand N, Kuhlbrodt K, Goerich D E, Prehu M O, Puliti A, Herbarth B, Hermans-Borgmeyer I, Legius E, Matthijs G, et al. Nat Genet. 1998;18:171–173. doi: 10.1038/ng0298-171. [DOI] [PubMed] [Google Scholar]
- 28.Li S-W, Prockop D J, Helminen H, Fassler R, Lapvetelainen T, Kiraly K, Peltarri A, Arokoski J, Lui H, Arita M, Khillan J S. Genes Dev. 1995;9:2821–2830. doi: 10.1101/gad.9.22.2821. [DOI] [PubMed] [Google Scholar]
- 29.Nishiguchi S, Wood H, Kondoh H, Lovell-Badge R, Episkopou V. Genes Dev. 1998;12:776–781. doi: 10.1101/gad.12.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pevny L H, Sockanathan S, Placzek M, Lovell-Badge R. Development (Cambridge, UK) 1998;125:1967–1978. doi: 10.1242/dev.125.10.1967. [DOI] [PubMed] [Google Scholar]
- 31.Wunderle V M, Critcher R, Ashworth A, Goodfellow P N. Genomics. 1996;36:354–358. doi: 10.1006/geno.1996.0474. [DOI] [PubMed] [Google Scholar]
- 32.Takamatsu N, Kanda H, Tsuchida I, Yamada S, Ito M, Kabeno S, Shiba T, Yamashita S. Mol Cell Biol. 1995;15:3759–3766. doi: 10.1128/mcb.15.7.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schott J J, Benson D W, Basson C T, Pease W, Silberbach G M, Moak J P, Maron B J, Seidman C E, Seidman J G. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 34.Basson C T, Bachinsky D R, Lin R C, Levi T, Elkins J A, Soults J, Grayzel D, Kroumpouzou E, Traill T A, Leblanc-Straceski J, et al. Nat Genet. 1997;15:30–35. doi: 10.1038/ng0197-30. [DOI] [PubMed] [Google Scholar]
- 35.Li Q Y, Newbury-Ecob R A, Terrett J A, Wilson D I, Curtis A R, Yi C H, Gebuhr T, Bullen P J, Robson S C, Strachan T, et al. Nat Genet. 1997;15:21–29. doi: 10.1038/ng0197-21. [DOI] [PubMed] [Google Scholar]
- 36.Houston C S, Optiz J M, Spranger J W, Macpherson R I, Reed M H, Gilbert E F, Herrmann J, Schinzel A. Am J Med Genet. 1983;15:3–28. doi: 10.1002/ajmg.1320150103. [DOI] [PubMed] [Google Scholar]
- 37.Ng L J, Wheatley S, Muscat G E, Conway-Campbell J, Bowles J, Wright E, Bell D M, Tam P P, Cheah K S, Koopman P. Dev Biol. 1997;183:108–121. doi: 10.1006/dbio.1996.8487. [DOI] [PubMed] [Google Scholar]
- 38.Wright E, Hargrave M R, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, Koopman P. Nat Genet. 1995;9:15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]