Abstract
During meiosis, crossover events generate new allelic combinations, yet the abundance of these genetic exchanges in individual cells has not been measured previously on a genomic level. To perform a genome-wide analysis of recombination, we monitored the assortment of genetic markers in meiotic tetrads from Arabidopsis. By determining the number and distribution of crossovers in individual meiotic cells, we demonstrated (i) surprisingly precise regulation of crossover number in each meiosis, (ii) considerably reduced recombination along chromosomes carrying ribosomal DNA arrays, and (iii) an inversely proportional relationship between recombination frequencies and chromosome size. This use of tetrad analysis also achieved precise mapping of all five Arabidopsis centromeres, localizing centromere functions in the intact chromosomes of a higher eukaryote.
Keywords: meiosis, tetrad analysis, crossover interference
In the 1940s, the ability to analyze all four products of meiosis (tetrad analysis) revolutionized the understanding of genetic exchange (1, 2). This powerful approach identified regions conferring centromere functions and revealed crossover (CO) interference, recombination hot spots, and gene conversion in several fungi including Neurospora crassa and Saccharomyces cerevisiae (3–8). However, in multicellular organisms, meiotic products typically dissociate, restricting the use of tetrad analysis to exceptional cases in which two meiotic products can be examined, such as the Drosophila attached X chromosomes (9). This report presents the use of complete meiotic tetrads to survey genetic exchange in a higher eukaryote.
The Arabidopsis quartet1 mutation causes the four products of pollen meiosis to remain attached, making analysis of complete tetrads possible in a higher eukaryote (10). Marker assortment within these pollen tetrads can be monitored conveniently: Pollinating virgin flowers with individual tetrads typically yields four seeds, each sired by one member of the tetrad. Arabidopsis is amenable to genomic surveys of recombination. Only a small set of markers is required to detect all of the COs that occur during meiosis across its genetic map of ≈500 cM. Recombination across portions of the S. cerevisiae genome has been studied extensively, but its larger genetic map of >4,000 cM would require analysis of 8-fold more markers for a genome-wide survey of genetic exchange. Finally, because the common Arabidopsis laboratory strains Landsberg and Columbia differ in ≈1% of their genomic sequence (11), DNA sequence variation can be used to identify numerous polymorphic markers. Currently, over 600 DNA polymorphisms have been assembled into a dense genetic map (ref. 12 and http://nasc.nott.ac.uk/new_ri_map.html). In this study, we generated pollen tetrads segregating all of these molecular markers by crossing quartet1 mutants independently isolated in the Landsberg and Columbia backgrounds.
Genomic levels of recombination during meiosis have been estimated by (i) using cytological features such as chiasmata and recombination nodules to infer the genomic level of genetic exchange (13–16), (ii) using tetrad analysis to characterize recombination over limited regions or single chromosomes (3, 17), or (iii) generating genetic maps of an entire genome based on the average recombination frequencies in populations (18). However, these methods have not measured, at a genomic level, the variation in the number of genetic exchanges in individual meioses, and thus it is not clear whether the activity of the recombination machinery varies considerably between individual meiotic cells. Performing tetrad analysis in an organism with a relatively small genetic map presents an opportunity to overcome this limitation, making identification of all COs that occur during meiosis feasible. We monitored individual meiotic tetrads and assessed the segregation of markers, spaced at an average of 10.2 cM, on all five Arabidopsis chromosomes. This approach made it possible to examine variation in CO frequency and distribution, the number of DNA strands that undergo genetic exchange, and the frequency of gene conversion.
Conveniently, the data generated in this survey also revealed a precise map location for each of the five regions conferring centromere function in Arabidopsis. Centromeres are required for faithful chromosome transmission; in multicellular organisms, several methods have been used to define their genetic or physical map locations. Visualizing the primary chromosomal constriction (19–21) yields megabase resolution of centromere positions but cannot identify the DNA sequences essential for centromere functions. Chromosome fragmentation techniques, including treating cells with ionizing radiation, can define genetically stably transmitted segments (22, 23). Precise mapping of centromeres with this technique relies on the chance recovery of breakpoints near centromeres and can yield erroneous results when cryptic centromere functions become activated in fragmented chromosomes (24). Finally, repetitive sequences often reside in the vicinity of the cytologically defined centromeres of higher eukaryotes, and physical mapping of these arrays has been used to identify centromeric regions. Although juxtaposed to centromeres, repetitive arrays may not be required for centromere functions (25). In lower eukaryotes, including S. cerevisiae (4), tetrad analysis was instrumental in defining precisely centromere functions in intact chromosomes (Fig. 1). Here, we used this technique to identify the Arabidopsis chromosomal domains that contain the functional centromeres.
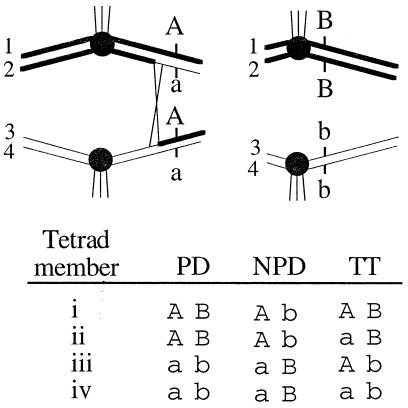
Figure 1.
Genetic analysis of marker assortment in tetrads. (Top) Two homologous pairs of chromosomes and their four chromatids (1–4) are shown at metaphase of meiosis I. Spindle fiber attachments (vertical lines) to the centromeres (filled circles) result in the separation of homologous centromeres. Two genetic loci (A and B) with Columbia (upper case) and Landsberg (lower case) alleles are shown. Recombination events (X) frequently separate distal markers (such as A) from the centromere. (Bottom) The results of scoring DNA markers in each of the members of a tetrad (i–iv) reveal the genotype. COs and the orientation of homologs at meiosis I dictate whether marker pairs assort into one of three categories: parental ditype (PD), nonparental ditype (NPD), or TT. TT patterns result only when recombination between a markers and its centromere occurs (as in the example shown at Top).
MATERIALS AND METHODS
Strain Construction.
Arabidopsis quartet1 (qrt1) mutants isolated independently from the Landsberg (qrt1–1, PLA 167) and Columbia (qrt1–2, PLA 606) strains (10) were crossed together to form F1 plants. Pollen tetrads from these F1 plants were placed on glass slides and individually manipulated onto stigmas by using a single-hair brush. These tetrad pollinations often yielded four seeds, and the plants derived from these seeds yielded permanent seed stocks (PLA 391–PLA 592, available from the Arabidopsis Resource Center, Ohio State University), as well as tissue that was used for the analysis of marker segregation.
To simplify genetic analysis, tetrads from the Landsberg/Columbia F1 plants preferably are deposited on females from “pure” stocks that carry a known allele at every locus. We constructed such a female by crossing CS64 (Landsberg, glabra1) to CS75 (Landsberg, male-sterile1) from the Arabidopsis Resource Center. This yielded male sterile plants that could be pollinated without requiring emasculation; incorporation of the visible glabra1 marker ensured identification of contaminants from rare self-pollinations. We subsequently discovered that CS64, although homozygous Landsberg at nearly every locus, has Columbia DNA in a few regions (on chromosome I at g2395 and T27K12, on chromosome III between GAPA and NIT1, and on chromosome V at nga139, nga76, and PHYC). In a small number of assays (26 of 10,146), this caused difficulty in assigning tetrad genotypes (six cases at g2395, four at T27K12, and five at NIT1), and for three tetrads, this caused difficulty in determining the total number of COs. For these ambiguous tetrads, we assumed that single rather than double COs occurred, yielding 508 (rather than 513) total COs.
Marker Analysis.
Single cauline leaves were crushed in 200 μl of 50 mM Tris⋅HCl (pH 8.0), 200 mM NaCl, 0.2 mM EDTA, 0.5% SDS, and 100 μg/ml proteinase K by using a power drill with a Teflon bit. Leaf slurries were incubated at 37°C for 30 min and then extracted with phenol and then chloroform. After the addition of 1/10 vol of sodium acetate, nucleic acids were precipitated with 2 vol of 100% ethanol. DNA pellets were washed twice with 70% ethanol and suspended in 100 μl of TE buffer (10 mM Tris⋅HCl, pH 8.0/1 mM EDTA).
All of the genetic markers used in this study have been described (http://genome-www.stanford.edu/Arabidopsis) and include, in order, for chromosome I: nga59, nga63, m59, g2395, m235, ZFPG, SO392, UFO, 7G6, T27K12, nga280, ETR, TAG1, ATHATPase, nga692; chromosome II: nga1145, m246, mi310, THY1, nga1126, nga361, nga168; chromosome III: nga32, nga162, ARLIM15, GAPA, GL1, NIT1, AFC1, nga112; chromosome IV: GA1, nga12, nga8, nga1111, DET1, COP9, SC5, g4539, AG, nga1139, nga1107; and chromosome V: CTR1, ca72, nga106, nga139, SO262, nga76, PHYC, SO191, DFR, ASB2, LFY3. Primers corresponding to these markers were purchased from Research Genetics (Huntsville, AL) and used for PCR without further purification as described (26, 27). Amplification was performed in a 96-well format by using an MJ Research (Cambridge, MA) PTC-200. PCR reactions were initiated at 95°C followed by 40 cycles of 15-s denaturation (94°C), 15-s annealing, and 30-s extension (72°C). Depending on the primer pair, annealing temperatures ranged from 52 to 57°C (first 10 cycles) followed by 54 to 61°C (final 30 cycles). Polymorphisms were detected by DNA electrophoresis on 1% agarose gels or 14% native polyacrylamide gels; in some cases, polymorphisms were revealed by digestion of the amplified product with an appropriate restriction enzyme (http://genome-www.stanford.edu/Arabidopsis).
Linkage Analysis.
Marker segregation patterns were compared with each other to produce a genetic map and to determine the location of the centromeres. To facilitate algorithmic manipulation of segregation data, Landsberg alleles were assigned a value of 1, and Columbia alleles were assigned a value of 0; the complete data set generated in this study has been deposited with the Arabidopsis Resource Center, Ohio State University. By using Microsoft Excel software, a program was designed to distinguish parental ditype, nonparental ditype, and tetratype (TT) patterns of assortment between every marker pair (Fig. 1). Distances (in centimorgans) between linked markers were calculated by the function: 100[(1/2 TT + 3 nonparental ditype)/n] and centromere linkage by: 100[(1/2 TT)/n], where n is the number of tetrads examined. The number of COs expected in a given interval was determined by (n)(interval size in cM/50), and the number of COs in adjacent intervals by (n)(size of first interval in cM/50)(size of second interval in cM/50).
RESULTS AND DISCUSSION
Pollinating stigmas with individual pollen tetrads from a qrt1–1/qrt1–2, Landsberg/Columbia plant yielded three or four viable seeds in 40% of the crosses. This produced 57 sets of plants used for DNA preparation and tetrad analysis, including 25 complete, four-member tetrads and 32 tetrads with only three surviving members. In each tetrad, we scored 52 PCR-based markers that spanned the genome with gaps no greater than 24 cM and an average spacing of 10.2 cM (http://nasc.nott.ac.uk/new_ri_map.html). With respect to the paternal (pollen) contribution, genetic markers assorted in a 2:2 pattern for Landsberg (L) and Columbia (C) alleles, and the maternal pattern yielded Landsberg alleles (Fig. 2A).
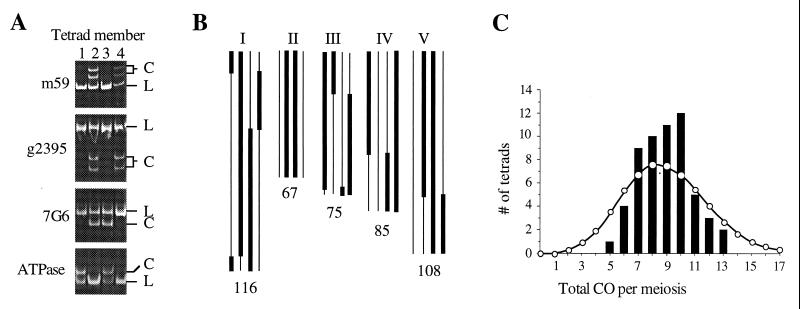
Figure 2.
(A) PCR amplification products for four polymorphic Landsberg (L) or Columbia (C) markers on chromosome I for each of the members (1–4) of the tetrad shown in B. (B) Map of recombination in one tetrad. The genotype of the four chromatids (bar = Columbia, line = Landsberg) for chromosomes I–V and approximate CO positions are shown. The genetic distance sampled is indicated at the bottom of each chromosome and totals 451 cM. (C) Histogram of the number of COs in each meiosis (bars) and a Poisson distribution with the same mean of 8.9 (circles). Deviation of the observed distribution from the Poisson was determined by testing χ squared for frequencies divided into ≥−2 classes (33).
In tetrads with four surviving members, COs were always reciprocal, nondisjunction was not observed, and markers always segregated in a 2:2 ratio (n = 1247). However, aberrant chromosome segregation resulting in aneuploidy could have escaped detection in this study if nondisjunction events resulted in pollen lethality. The striking absence of 3:1 or 4:0 segregation patterns suggests that gene conversion events at each of the 52 loci we monitored are infrequent. For comparison, gene conversion has been detected at every locus examined in S. cerevisiae, ranging in frequency from 0.5 to 30% (3). Although previous observations of gene conversion have relied primarily on intraconversion of wild-type and mutant alleles with a single nucleotide difference, the DNA polymorphisms used in this study were in principle no different and should be subject to the same conversion events. Thus, Arabidopsis appears to undergo relatively low levels of gene conversion although examination of additional markers in our tetrads, or examination of additional tetrads, may reveal small tracts of gene conversion similar to those found near recombination break-points in yeast.
We constructed maps of the breakpoints in all four chromatids of every chromosome in each of the 57 meioses (10,146 assays) by scoring the segregation of molecular markers spanning 451 cM, or 78%, of the genome (Fig. 2B). Because aberrant segregation patterns were not observed in the tetrads with four surviving members, we included in this analysis those tetrads with only three surviving members and inferred the genotype of the fourth plant. A summary of the distribution and frequency of COs for each chromosome, chromatid, and chromosome arm is presented in Table 1. Our data indicate that nearly every chromosome recombined at least once (278/285, 98%), lending support to the suggestion, based on cytological observations (29), that at least one CO is required for proper chromosome assortment. At a low frequency, the small, rDNA-containing chromosomes (II and IV) were transmitted in the absence of COs, and in these instances their segregation was normal. This could be explained by our inability to detect COs near the ends of these chromosomes, due to the lack of convenient telomeric markers, or by the existence of an achiasmate segregation mechanism (30). Apparently, the absence of detectable COs did not affect pollen viability because some tetrads with four viable members lacked COs on chromosomes II or IV. Finally, in many cases, one arm of a chromosome lacked COs, indicating that every arm need not undergo recombination to achieve proper chromosome assortment.
Table 1.
CO occurrence and distribution in 57 meioses by chromosome
| Parameter | Chromosome
|
||||
|---|---|---|---|---|---|
| I | II | III | IV | V | |
| Meioses with 0 CO observed, n | 0 | 5 | 0 | 2 | 0 |
| 1 CO | 4 | 38 | 9 | 43 | 19 |
| 2 COs | 26 | 14 | 36 | 12 | 32 |
| 3 COs | 19 | 0 | 9 | 0 | 5 |
| 4 COs | 3 | 0 | 2 | 0 | 1 |
| 5 COs | 4 | 0 | 1 | 0 | 0 |
| 6 COs | 1 | 0 | 0 | 0 | 0 |
| Meioses without COs (upper arm), n* | 2 | 47† | 9 | 56† | 12 |
| Meioses without COs (lower arm), n* | 3 | 13 | 11 | 6 | 10 |
| Maximum separation of COs, cM‡ | 84 | 58 | 81 | 58 | 77 |
| COs observed (total), n | 151 | 66 | 121 | 67 | 103 |
| CO per megabase (Mb)§ | 0.092 | 0.058 | 0.162 | 0.055 | 0.107 |
| Average no. COs per meiosis | 2.7 | 1.2 | 2.1 | 1.2 | 1.8 |
| Meioses with COs on only two chromatids, n | 11 | 41 | 17 | 47 | 24 |
| Meioses with COs on only three chromatids, n | 24 | 7 | 26 | 7 | 19 |
| Meioses with COs on all four chromatids, n | 22 | 4 | 14 | 1 | 14 |
| Average no. recombinant chromatids per meiosis | 3.2 | 2.1 | 2.9 | 2.1 | 2.8 |
| COs observed, n/COs expected, n (12) | 1.14 | 0.87 | 1.41 | 0.69 | 0.84 |
COs were defined as reciprocal exchanges between two homologous chromatids.
*Arms were defined by the calculated centromere positions (Fig. 3).
Markers used to measure COs on chromosomes II and IV did not span the rDNA (12).
Map distances were derived from a male-specific meiotic map (Fig. 3).
Average number of COs observed/chromosome size, calculated by using 28.7, 20, 13.1, 21.5, and 16.9 Mb for chromosomes I–V, respectively (28). For chromosomes II and IV, elimination of the rDNA from the calculation yields 0.068 and 0.063 CO/Mb, respectively.
Examining CO dispersal showed that chromosomes I, III, and V, which lack rDNA arrays, undergo recombination at a frequency (COs per megabase) inversely proportional to their physical size; a similar pattern is observed for the 15 non-rDNA containing S. cerevisiae chromosomes (31). Arabidopsis chromosomes II and IV, which contain ≈3.7 Mb of rDNA, do not fit this trend (Table 1). Although small in size, both chromosomes exhibit the lowest observed number of COs per megabase, even when the presence of 3 Mb of rDNA is accounted for (Table 1). This discrepancy raises the possibility that the CO suppression observed previously in the rDNA (32) might extend throughout the chromosome. Thus, recombination levels on individual chromosomes may be mediated by both chromosome size and content.
We determined the total number of COs in each meiosis and found a surprising disparity between our observations and the number predicted by models that assume COs are independent on a genomic level. The number of COs in each meiosis varied <3-fold, ranging from 5 to 13, with an average of 8.9 (Fig. 2C). It was striking to note that, on a genomic scale, CO frequency deviated significantly from a Poisson distribution (P < 0.025); there were more COs near the mean and fewer at the extremes than expected if CO events are independent (Fig. 2C). This deviation is unlikely caused by a small data set; performing the same analysis on the subset of 25 tetrads with four surviving members also yielded a significant difference from a Poisson distribution. In addition, similar comparisons of the number of COs observed on each chromosome (Table 1) also deviated significantly from the Poisson (not shown). These data implicate a mechanism that ensures that each Arabidopsis chromosome participates in the COs essential for assortment, while limiting exchange in the genome as a whole.
Control of CO frequency might result from the cumulative effect of interference between COs (34). We measured interference across the Arabidopsis genome, determining that the number of double COs in adjacent intervals was 65% lower than that expected from the genetic map (12); 33 double COs were observed and 93 were predicted (see Materials and Methods). However, although CO interference was detected in adjacent intervals across entire chromosomes, COs were distributed randomly among the four homologous DNA strands (Table 1). The distribution of COs on the chromatids did not deviate significantly (P > 0.05) from a random distribution of 1:2:1 for 2-strand-to-3-strand-to-4-strand double COs (chromosome I, 3:16:7; chromosome II, 3:7:4; chromosome III, 6:23:7; chromosome IV, 4:7:1; and chromosome V, 5:17:10). Thus, this study revealed that chromatid interference in Arabidopsis is not a significant factor in the regulation of CO distribution.
Unlike previous surveys that relied on recombination in both male and female gametogenesis (12), our analysis selectively examined male meiosis. The overall level of exchange in male meioses was equivalent to that measured in recombinant inbred lines derived from a Columbia/Landsberg F1 (12), with 508 COs observed in 57 meioses vs. 514 expected. However, we detected striking differences when comparing individual chromosomes. In males, chromosome III displayed a 41% increase in recombination, and chromosome IV manifested a 31% decrease (P < 0.005, Table 1). Although a direct comparison of male recombination frequencies with recombination during female meiosis could reveal even larger differences, little data on map distances in females are currently available.
We localized sex-specific meiotic differences to 10 small, randomly distributed intervals that were clustered at neither telomeres nor centromeres (Fig. 3). Measuring recombination between adjacent markers yielded a male-specific genetic map (Fig. 3) with distances in cM calculated as described in Materials and Methods. This map contains four intervals with enhanced, and six with reduced, recombination during male meiosis, with deviations as large as 5-fold (significant by χ2 test, P < 0.05) from the expected frequencies. Although the recombinant inbred map represents the accumulated data from several laboratories, the differences we observed are unlikely to have arisen from errors in marker placement. First, the regions we identified confirmed and more precisely localized several previously noted male-specific regions of increased recombination (37). Second, we verified that the genetic distances measured from male meioses were significantly different than those determined from the combination of male and female meioses for two intervals. In an independent F2 population derived from a Columbia/Landsberg F1, we found 12.8 cM between nga63 and nga59 (n = 94) and 11 cM between nga1139 and nga1107 (n = 90) whereas these intervals measure 27 and 4 cM, respectively, in male meiosis. Finally, although on average a 1-cM interval in Arabidopsis measures 200 kb (12, 38), comparisons with the physical map (39, 40) revealed ≈160 kb/cM for one of the male-specific hot spots (THY1-nga1126) and ≈450 kb/cM for a cold spot (nga1139-nga1107).
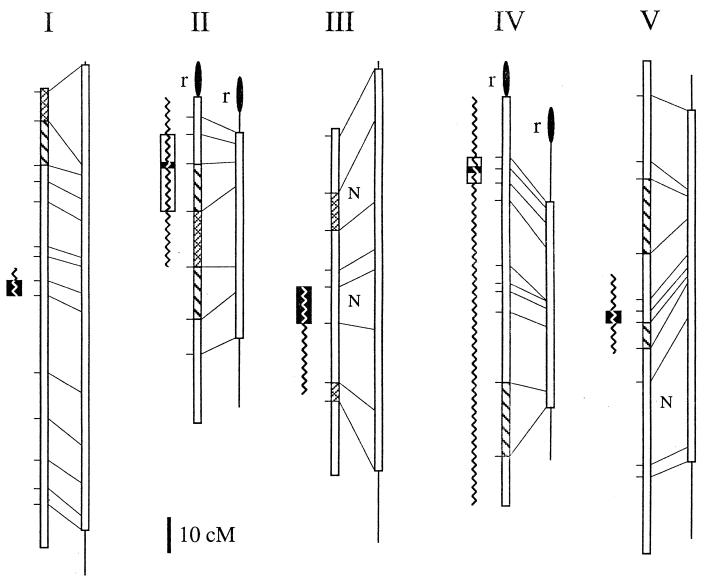
Figure 3.
Genetic maps of Arabidopsis chromosomes I–V. For each column, on the left is the published map from male and female meioses (ref. 12; http://nasc.nott.ac.uk/new_ri_map.html) and on the right is a male-specific meiotic map calculated by using the data presented here (the lines at the extremities of the male-specific map represent unsampled regions). Marker positions (horizontal lines), corresponding markers on both maps (diagonal lines), intervals with observed nonparental ditype tetrads (N), regions of enhanced (cross-hatched boxes) and reduced (hatched boxes) recombination in males, and rDNA (filled ovals) are indicated. Centromere positions as defined by recombinant individuals at the markers assayed (open boxes) and calculated by a mapping function (solid boxes; see Materials and Methods) are shown at the left of each chromosome map. Centromeres for chromosomes I–V were mapped between 7G6 and T27K12 (52–59 cM), m246 and THY1 (11–33 cM), GL1 and NIT1 (45–55 cM), GA1 and nga8 (17–29 cM), and nga76 and PHYC (71–74 cM), respectively. At each centromeric interval, there are 6, 14, 15, 7, and 2 (chromosomes I–V, respectively) recombinants remaining. On chromosomes 2 and 4, no recombinants were detected at mi310 and nga12, respectively. Previous centromere localizations derived from chromosome fragmentation experiments are displayed as jagged lines (35, 36).
By pair-wise comparison of marker assortment on each of the five chromosomes, we localized the regions that confer meiotic centromere functions to intervals that comprise only 4% of the Arabidopsis genome. This represents a dramatic improvement over chromosome fragment assays that previously mapped mitotic centromere functions to regions totaling 39% of the genome (Fig. 3; refs. 35 and 36). With tetrad analysis, centromere mapping uses the same principles as mapping any genetic locus. Linkage relationships can be analyzed by recombination frequency: Markers close to the centromere rarely are separated by crossing over, and markers far from the centromere yield tetratype assortment patterns (see Fig. 1). Thus, the frequency of tetratype tetrads approaches zero when both markers are near their centromeres (1, 4). Extending this analysis to markers along each chromosome (Fig. 4) revealed a discrete location for each meiotic centromere that overlapped the intervals previously shown to contain the mitotic centromeres (Fig. 3; refs. 35 and 36). Of interest, each of the centromeric intervals we defined was linked to an array of 180 bp repeats (refs. 39–41; EKR markers, E. J. Richards, personal communication). As new genetic markers are identified, it will be possible to use the recombinant tetrads to determine whether these repetitive DNA arrays cosegregate with centromere functions. It is curious that, although centromeric regions have been shown to be the least variable portion of the Drosophila genome (42), the centromeric intervals we defined appear to have a high rate of polymorphism. We found clustering in the centromeric intervals of DNA clones ≈1 kb in size that are unique to either the Columbia or Landsberg genomes (43). Between these strains, this degree of sequence variation is rare, suggesting that Arabidopsis centromeric regions are subject to rapid change.
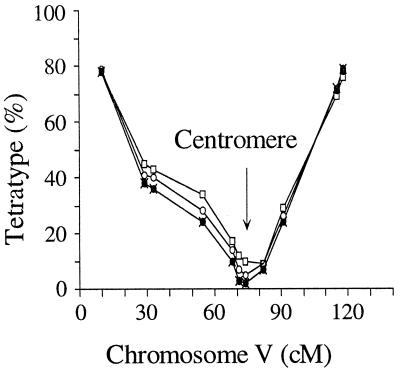
Figure 4.
%TT for markers on chromosome V calculated by comparison to centromere-linked markers. Four plots, using centromeric markers on chromosomes I-IV (filled and open symbols) provide a consensus location for the centromere (arrow).
The analysis of complete Arabidopsis tetrads, coupled with a high marker density, provides an excellent framework for dissecting genetic mechanisms in higher eukaryotes. Our characterization makes possible the identification of the sites of genetic exchange at base pair resolution and will allow screens for new mutants with enhanced or suppressed recombination. Moreover, the availability of tetrad analysis, combined with the relatively large chromosome size, makes it possible to resolve both centromere structure and function in Arabidopsis.
Acknowledgments
We thank L. Dupree and S. Kniss for technical assistance, J. Bender and N. Crawford for molecular markers, D. Bishop, B. Charlesworth, R. Esposito, J. A. Mayfield, L. Mets, L. K. Wilhelmi, and G. Zinkl for critical reading, and M. Jensen and R. Mauricio for helpful discussion. Seed stocks used for this study are available from the Arabidopsis Biological Resource Center. This work was funded in part by grants from the United States Department of Agriculture (G.P.C.) and the Consortium for Plant Biotechnology Research. Additional support was provided by the National Science Foundation, the Department of Energy, and the Searle Scholars Foundation.
ABBREVIATIONS
- CO
crossover
- TT
tetratype
- cM
centimorgan
References
- 1.Mather K, Beale G H. J Genetics. 1942;43:1–30. [Google Scholar]
- 2.Perkins D D. Genetics. 1949;34:607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petes T D, Malone R E, Symington L S. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Broach J R, Jones E S, Pringle J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1991. pp. 407–521. [Google Scholar]
- 4.Carbon J, Clarke L. New Biol. 1990;2:10–19. [PubMed] [Google Scholar]
- 5.Fogel S, Hurst D D. Genetics. 1967;57:455–481. doi: 10.1093/genetics/57.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fink G R, Styles C A. Genetics. 1974;77:231–244. doi: 10.1093/genetics/77.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White M A, Petes T D. Curr Genet. 1994;26:21–30. doi: 10.1007/BF00326300. [DOI] [PubMed] [Google Scholar]
- 8.Whitehouse H L K. J Genetics. 1942;41:23–62. [Google Scholar]
- 9.Anderson E G. Genetics. 1925;10:403–417. doi: 10.1093/genetics/10.5.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Preuss D, Rhee S Y, Davis R W. Science. 1994;264:1458–1460. doi: 10.1126/science.8197459. [DOI] [PubMed] [Google Scholar]
- 11.Chang C, Bowman J L, DeJohn A W, Lander E S, Meyerowitz E M. Proc Natl Acad Sci, USA. 1988;85:6856–6860. doi: 10.1073/pnas.85.18.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lister C, Dean C. Plant J. 1993;4:745–750. [Google Scholar]
- 13.Jones G H. In: Controlling Events in Meiosis. Evans C W, Dickinson H G, editors. Cambridge, U.K.: The Company of Biologists; 1984. pp. 293–320. [Google Scholar]
- 14.Sherman J D, Stack S M. Genetics. 1985;141:683–708. doi: 10.1093/genetics/141.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orr-Weaver T L. Proc Natl Acad Sci USA. 1995;92:10443–10449. doi: 10.1073/pnas.92.23.10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carpenter A T C. In: Genetic Recombination. Kucherlapati R, Smith G R, editors. Washington, DC: Am. Soc. Microbiol.; 1988. pp. 529–547. [Google Scholar]
- 17.Brown P. Curr Opin Genet Dev. 1994;4:366–373. doi: 10.1016/0959-437x(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 18.Sturtevant A H. Zeit Ind Abst Vererb. 1915;13:234–287. [Google Scholar]
- 19.Sunkel C E, Coelho P A. Curr Opin Genet Dev. 1995;5:756–767. doi: 10.1016/0959-437x(95)80008-s. [DOI] [PubMed] [Google Scholar]
- 20.Rattner J B. Bioessays. 1991;13:51–56. doi: 10.1002/bies.950130202. [DOI] [PubMed] [Google Scholar]
- 21.Earnshaw W C, Cooke C A. Genome. 1989;31:541–552. doi: 10.1139/g89-103. [DOI] [PubMed] [Google Scholar]
- 22.Murphy T D, Karpen G H. Cell. 1995;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyler-Smith C, Oakey R J, Larin Z, Fisher R, Crocker M, Affara N A, Ferguson-Smith M A, Muenke M, Zuffardi O, Jobling M A. Nat Genet. 1993;5:368–375. doi: 10.1038/ng1293-368. [DOI] [PubMed] [Google Scholar]
- 24.Sacchi N, Magnani I, Fuhrman-Conti A M, Monard S P, Darfler M. Cytogenet Cell Genet. 1996;73:123–129. doi: 10.1159/000134322. [DOI] [PubMed] [Google Scholar]
- 25.Kipling D, Warburton P. Trends Genet. 1997;13:141–145. doi: 10.1016/s0168-9525(97)01098-6. [DOI] [PubMed] [Google Scholar]
- 26.Bell C J, Ecker J R. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 27.Konieczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 28.Samoylova T I, Meister A, Miséra S. Plant J. 1996;10:949–954. doi: 10.1046/j.1365-313x.1996.10050949.x. [DOI] [PubMed] [Google Scholar]
- 29.Baker B S, Carpenter A T C, Esposito M S, Esposito R E, Sandler L. Annu Rev Genet. 1976;10:53–134. doi: 10.1146/annurev.ge.10.120176.000413. [DOI] [PubMed] [Google Scholar]
- 30.Wolf K W. Bioessays. 1994;16:107–114. doi: 10.1002/bies.950160207. [DOI] [PubMed] [Google Scholar]
- 31.Kaback D B, Guacci V, Barber D, Mahon J W. Science. 1992;256:228–232. doi: 10.1126/science.1566070. [DOI] [PubMed] [Google Scholar]
- 32.Copenhaver G P, Pikaard C S. Plant J. 1996;9:259–272. doi: 10.1046/j.1365-313x.1996.09020259.x. [DOI] [PubMed] [Google Scholar]
- 33.Sokal R R, Rohlf F J. Biometry. New York: Freeman; 1981. p. 703. [Google Scholar]
- 34.Muller H J. Am Nat. 1916;50:193–221. ; 284–305; 350–366; 421–434. [Google Scholar]
- 35.Sears L M S, Lee-Chen S. Can J Genet Cytol. 1970;12:217–223. [Google Scholar]
- 36.Koornneef M, van Eden J, Hanhart C J, Stam P, Braaksma F J, Feenstra W J. J Hered. 1983;74:265–272. [Google Scholar]
- 37.Vizir I Y, Corol A B. J Hered. 1990;65:379–383. [Google Scholar]
- 38.Hwang I, Kohchi T, Hauge B M, Goodman H M, Schmidt R, Cnops G, Dean C, Gibson S, Iba K, Lemieux B, Arondel V, Danhoff L, Somerville C. Plant J. 1991;1:367–374. doi: 10.1046/j.1365-313x.1991.t01-5-00999.x. [DOI] [PubMed] [Google Scholar]
- 39.Zachgo E A, Wang M L, Dewdney J, Bouchez D, Camilleri C, Belmonte S, Huang L, Dolan M, Goodman H M. Genome Res. 1996;6:19–25. doi: 10.1101/gr.6.1.19. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt R, West J, Love K, Lenehan Z, Lister C, Thompson H, Bouchez D, Dean C. Science. 1995;270:480–483. doi: 10.1126/science.270.5235.480. [DOI] [PubMed] [Google Scholar]
- 41.Maluszynska J, Heslop-Harrison J S. Plant J. 1991;1:159–166. [Google Scholar]
- 42.Begun D J, Aquadro C F. Nature (London) 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y-G, Mitsukawa N, Lister C, Dean C, Whittier R F. Plant J. 1996;10:733–736. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]