Abstract
In Saccharomyces cerevisiae, most mutations induced by a wide range of mutagens arise during translesion replication employing the REV1 gene product and DNA polymerase ζ. As part of an effort to investigate mammalian mutagenic mechanisms, we have identified cDNA clones of the human homologs of the yeast REV genes and examined their function in UV mutagenesis. Previously, we described the isolation of a human homolog of yeast REV3, the catalytic subunit of pol ζ, and here report the identification and sequence of a human homolog of yeast REV1. This gene was isolated by identifying an expressed sequence tag encoding a peptide with similarity to the C terminus of yeast Rev1p, followed by sequencing of the clone and retrieval of the remaining cDNA by 5′ rapid amplification of cDNA ends. The human gene encodes an expected protein of 1,251 residues, compared with 985 residues in the yeast protein. The proteins share two amino-terminal regions of ≈100 residues with 41% and 20% identity, a region of ≈320 residues with 31% identity, and a central motif in which 11 of 13 residues are identical. Human cells expressing high levels of an hREV1 antisense RNA grew normally, and were not more sensitive to the cytotoxic effect of 254 nm UV radiation than cells lacking antisense RNA. However, the frequencies of 6-thioguanine resistance mutants induced by UV in the cells expressing antisense hREV1 RNA were significantly lower than in the control (P = 0.01), suggesting that the human gene has a function similar to that of the yeast homolog.
Information about the mechanisms that generate mutations in eukaryotes is likely to be useful for understanding human health concerns, such as genotoxicity and cancer (1). At the present, these mechanisms have been investigated most intensively in budding yeast, Saccharomyces cerevisiae, and in this organism almost all induced mutations arise during translesion replication, a process that promotes elongation past sites of unrepaired lesions that might otherwise block this event (1, 2). The major pathway for translesion replication in yeast employs DNA polymerase (pol) ζ (3) together with Rev1 protein (4), an enzyme that has two functions. These include a deoxycytidyl transferase activity that incorporates dCMP opposite abasic sites in the template and a second, as yet poorly defined, activity that is required for replication past a wide variety of lesions as well as abasic sites (J. R. Nelson, P.E.M.G., A. M. Nowicka, D. C. Hinkle and C.W.L., unpublished observations). In addition to this general pathway, yeast also possesses a specialized pathway for translesion replication employing pol η (5), a distant homolog of Rev1 protein that is encoded by RAD30 (6, 7). Although the substrate range of this enzyme has not yet been fully defined, it is likely that it entails only a few types of lesion, which pol η appears to bypass with relatively high accuracy. Pol η appears to contribute very little to overall mutagenesis compared with pol ζ/Rev1 both because RAD30 mutants have little effect on mutagenesis and because mutants lacking the pol ζ pathway are substantially deficient in mutagenesis induced by almost all mutagens.
DNA repair and damage tolerance mechanisms in yeast often provide good models for these processes in mammals, and evidence of several kinds suggests that this is likely to be the case with translesion replication. We (8) and others (9, 10) have identified and sequenced cDNA clones of a human homolog of the yeast REV3 gene, which encodes the catalytic subunit of Pol ζ, and a very similar gene has been described in the mouse (11). More particularly, we have further shown that UV-induced mutagenesis is markedly reduced in human cells expressing high levels of a REV3 antisense RNA, suggesting that the human gene is also likely to be used in translesion replication (8). In addition, human cells have been shown to possess two homologs of the yeast RAD30 gene (12–15). As further evidence for a yeast-like mechanism for translesion replication in humans, we report here the isolation and sequence of a cDNA clone of a human homolog of yeast REV1, together with evidence that it too is used in UV mutagenesis. A recent report also describes this gene, and provides evidence indicating that it possesses a deoxycytidyl transferase activity (16).
Materials and Methods
cDNA Synthesis, 5′ Rapid Amplification of cDNA Ends (RACE), and Cloning.
Total RNA or polyadenylated RNA from human brain (Stratagene) was used as a template for cDNA synthesis, using Superscript II Reverse Transcriptase (Life Technologies, Gaithersburg, MD) and primers specific for the candidate hREV1 cDNA clone (pHIBAC55; American Type Culture Collection). After removal of residual primer, an oligo(dC) tail was added to the cDNA with terminal transferase, and the cDNA selectively amplified by PCR, using elongase (Life Technologies) or Taq polymerase, a nested hREV1-specific primer, and a commercial primer with 3′ complementarity to the oligo(dC) tail. The PCR products were separated on agarose gels, and material from the trailing edge of the DNA smear was purified for use as a substrate for a second amplification with Taq polymerase; in this instance, both primers had 5′ sequences containing multiple dU residues. The products were again separated on agarose gels, and the largest visible DNA species were purified and treated with uracil N-glycosylase in the presence of the pAMP-1 vector (Life Technologies). The resulting annealed DNAs were used to transform Escherichia coli DH5α cells. For assembling clones with a nearly complete insert sequence, PCR used two hREV1-specific primers, each of which contained a restriction enzyme recognition site in the 5′ sequence. Subsequent digestion with the appropriate restriction nucleases enabled ligation with similarly digested vector DNA. A plasmid (pMlNo-10) containing the entire ORF and 3′-untranslated region, but with a truncated 5′ end, was constructed in pSPORT1 (Life Technologies).
DNA Purification and Sequence Analysis.
Plasmid DNAs were isolated by alkaline lysis, and in many cases further purified on midi columns (Qiagen, Chatsworth, CA) for sequence analysis. The initial clone, pHIBAC55, and clones from the first two rounds of 5′ RACE were sequenced entirely by the dideoxy method with Sequenase II. Later clones were sequenced by using dye terminator chemistries (Perkin–Elmer Applied Biosystems). Primers for sequencing and for PCR were developed as required from the known hREV1 sequence as it became available. Both strands of each clone were sequenced, and all regions of the cDNA were sequenced in at least three clones (except for the 5′ 26 nucleotides, where only one clone was available) to establish a consensus sequence for hREV1 free of reverse transcription or PCR-generated errors. Attempts to confirm the sequence at the extreme 5′ end of the mRNA with data from human expressed sequence tag (EST) sequences were unsuccessful, because none carried sequence at the 5′ end of the cDNA.
Preparation of Cells that Express hREV1 Antisense RNA.
A plasmid designed to express antisense hREV1 RNA was constructed by inserting a 4,117-bp REV1 fragment into pTet-Puro in the antisense orientation. The pTet-Puro plasmid, described previously (8), contains a puromycin-selectable marker and places the gene of interest under the control of the TetP promoter (17, 18), with the gene being transcribed in the absence of tetracycline (Tet OFF system). The 4,117-bp sequence contained the complete hREV1 ORF, the entire 3′ untranslated region, and a short oligo(A) sequence. Initial experiments, using a 972-bp fragment containing 75 bp of the hREV1 5′ untranslated sequence cloned in the antisense orientation into pTet-Puro, proved incapable of suppressing hREV1 function. This was probably caused by an interaction between the antisense RNA and 28S rRNA, and indeed the hREV1 5′ untranslated sequence has regions of strong similarity to part of the rRNA sequence. The XhoI and SalI fragments from pEcNo-4 containing the biologically effective 4,117-bp sequence were cloned into the SalI site of the vector pTet-Puro, to give the hREV1 anti-sense expressing plasmid, pR1P27-AS. The orientation of the insert in this clone was determined by restriction enzyme analysis and by sequencing across the ligation boundaries.
Plasmid pR1P27-AS was transfected into 7AGM cells, which were derived from MSU-1.2, a near diploid, nontumorigenic, karyotypically stable human fibroblast cell line obtained originally from foreskin material of a normal neonate (19). Strain 7AGM was engineered to contain the tetracycline-controlled transactivator (tTA), which can activate transcription of genes of interest controlled by the Tet-responsive element. The pTet-tTAk plasmid used to introduce the tTA element carried the gene for histidinol resistance (20). pR1P27-AS transfectants were selected for resistance to puromycin and screened for the level of expression of hREV1 antisense by Northern blot analysis.
Northern Blot Analysis.
The conditions used for comparing the level of expression of hREV1 antisense in the transfectant cell strains were described previously (21). Briefly, 15 μg total RNA was electrophoresed on a denaturing formaldehyde gel, transferred to a Hybond-N membrane by a downward capillary transfer technique, and fixed by UV crosslinking. The template DNA for preparing the hREV1 antisense DNA probe was excised from the pR1P27-AS plasmid using EcoRI restriction. A ≈2,900-bp fragment from the 5′ end of the insert hREV1 cDNA was radiolabeled by random-primer labeling (21). The probe to be used for the loading control was similarly prepared from PCR-amplified cDNA of the hypoxanthine phosphoribosyltransferase (HPRT) gene of human cells. Northern hybridization was performed at 42°C overnight in 50% formamide, containing SSPE [NaCl (0.75 M)/NaH2PO4⋅H2O (0.05 M)/EDTA (pH 8.0) (0.5 mM)] and the other components listed in ref. 22, and the blot was washed as described (21). Variation in RNA loading per lane was evaluated by probing with HPRT cDNA.
Determination of the Cytotoxic and Mutagenic Effects of UV Radiation.
Cells were routinely cultured in medium containing 10% supplemented calf serum (HyClone), hydrocortisone (1 μg/ml), penicillin (100 units/ml), and streptomycin (100 μg/ml). The medium used was McM medium (22) or Eagle's MEM, modified by addition of l-aspartic acid (0.2 mM), l-serine (0.2 mM), and pyruvate (1 mM). The cytotoxic effect of the 254-nm UV radiation was determined from the survival of colony-forming ability, as described (23). Briefly, cells were plated into a series of dishes at cloning densities (100–1,000 cells per 100-mm-diameter dish, depending on the cloning efficiency of the strain and the expected survival). When the cells had attached and flattened out, the medium was removed, and the cells were rinsed and irradiated as described (24), using the designated doses. The mutagenic effect of UV was determined from the frequency of HPRT-defective, 6-thioguanine-resistant cells. Briefly, for each experiment, cells in exponential growth were plated into a series of 150-mm-diameter dishes at a density of 0.5–2 × 106 cells per dish. For each dose of UV, sufficient dishes were used to ensure at least 1 × 106 surviving cells. At the same time, cells were plated at cloning density into a series of 100-mm-diameter dishes to be used for determining the cytotoxic effect of each dose. Again, when the cells had attached and flattened out, the medium was removed, and the cells were rinsed and irradiated. For each experiment, 1 × 106 cells were mock-treated as a control. Fresh medium containing serum was returned to the cells immediately after irradiation, and the cells were refed with culture medium after 24 h. The cells plated at cloning densities were allowed 14 days to form colonies, with one additional refeeding after 7 days. The cells irradiated at high density for mutation induction were allowed to replicate for 4–5 days. They were then detached from the dishes, pooled, and 1–2 × 106 plated again and allowed to continue replicating for 4 to 5 additional days to allow depletion of preexisting HPRT. The unused population for each dose was stored in liquid N2 for future use. After an 8- to 9-day expression period, at least 1 × 106 cells from each population were selected for resistance to 6-thioguanine at a density of 500 cells/cm2, as described (23). A portion of the cells from each population was also plated in nonselective medium at a density of 100 cells per 100-mm-diameter dishes to assay the colony-forming ability of the cells at the time of selection. This value was used to correct the observed frequency of mutants for the cloning efficiency of the cells at the time of selection (23).
Results and Discussion
Human Cells Possess a Homolog of Yeast REV1.
A candidate human REV1 cDNA clone was identified by screening the dbEST database of the National Center for Biotechnology Information, using the blast algorithm. This screen yielded a single EST (GenBank T08134) among the approximately 52,000 entries listed at that time, and the corresponding clone pHIBAC55 was obtained from the American Type Culture Collection. The 2.13-kb human DNA insert in this plasmid was fully sequenced and found to consist of 1,813 bp of the 3′ end of an ORF, 296 bp of 3′ untranslated DNA, and 20 bp of poly(A). The remainder of the ORF, together with at least some of the 5′ untranslated region, was obtained by using five rounds of 5′ RACE, with the fifth round extending the sequence to a point beyond which no further elongation was obtained. To eliminate possible errors of reverse transcription and amplification, a consensus sequence was established from at least three independent clones at each round of 5′ RACE.
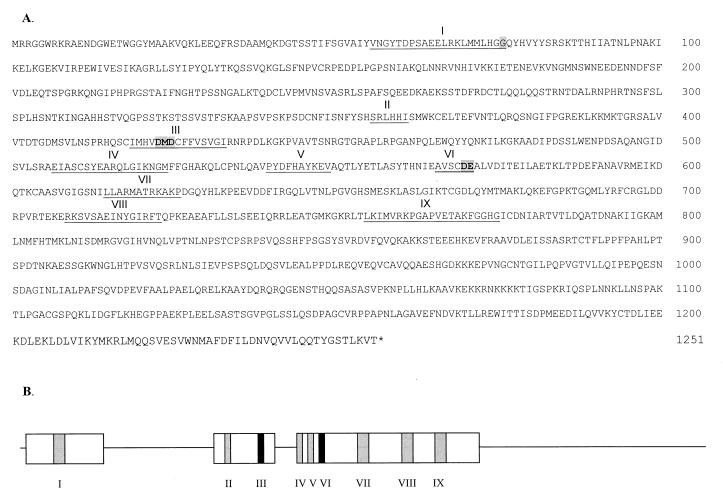
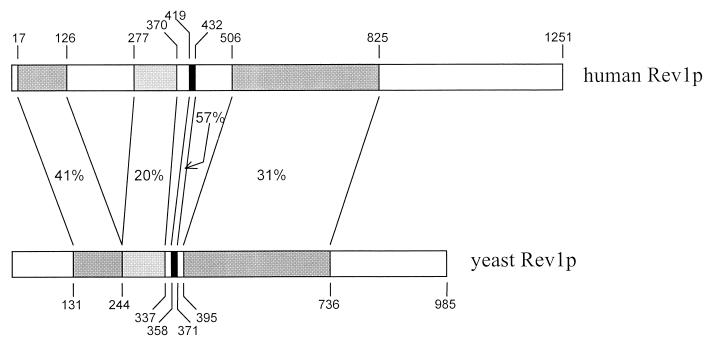
The hREV1 ORF (GenBank accession no. AF206019) encodes an expected protein of 1,251 residues (Fig. 1A) with significant identity to yeast REV1 (Fig. 2) and to candidate REV1 genes from other species, including Caenorhabditis elegans, Arabidopsis thaliana, Schizosaccharomyces pombe, and Drosophila melanogaster (data not shown). Significant identity exists in two amino-terminal regions, each ≈100 residues in length, that exhibit 41% and 20% identity with their corresponding yeast sequences, a region of ≈330 residues with 31% identity, and a centrally located motif that is highly conserved in Rev1 proteins and also found in E. coli dinB. A total of perhaps nine sequence motifs are suggested by alignments with the Rev1 proteins from S. cerevisiae, S. pombe, C. elegans, D. melanogaster, and A. thaliana (Fig.1B). Motif I lies within a region that shows weak homology to the terminal region of the BRCA1 gene (BRCT), although the similarity is greater with the Rev1 protein from budding yeast than with the human protein and the possible functional significance is unclear. However, of greater functional significance, motif I in the human protein contains G76 (highlighted in Fig. 1A), homologous to G193 in yeast and the site of the G193R mutation found in the yeast rev1-1 mutant (25); the Rev1-1 protein retains a significant fraction of its deoxycytidyl transferase activity, but has lost almost all of its general bypass function (J. R. Nelson, P.E.M.G., A. M. Nowicka, D. C. Hinkle and C.W.L., unpublished observations). Conversely, the highly conserved DXD and DE sequences found in motifs III and VI, respectively, appear to be concerned with catalysis (5, 26), and a pol η D156A, E157A tandem double mutation lacks polymerase activity (5).
Figure 1.
(A) Sequence of the translation product of hREV1 mRNA. The underlined sequences numbered I–IX are possible sequence motifs, and the highlighted residues in motifs I, III, and VI are known to be important for function. (B) Depiction of regions of similarity to C. elegans and D. melanogaster sequences (open boxes) together with the location of the predicted motifs. The darker filled boxes indicate the location of motifs III and VI.
Figure 2.
Schematic representation of the alignment of hREV1 and yeast REV1 protein sequences. Regions with significant identity are shaded, and the percent identity is indicated.
The sequence 5′ to the ATG codon at the start of the main ORF contains an out-of-frame ATG sequence at nucleotide −35, initiating a small reading frame that terminates at a TGA stop codon overlapping the main ORF ATG. Further, the sequence context of the out-of frame ATG is almost as good as that of the ORF ATG. This feature suggests that the hREV1 message is translated very inefficiently, and predicts that hRev1 protein levels are likely to be low (27). A number of the clones analyzed encoded a protein of 1,250, rather than 1,251, residues, in which the sequence CAGCAG at nucleotides 1041–1046 was replaced by CAG. A similar pair of sequences was also found in the homologous region of the mouse REV1 sequence (data not shown); in both cases, they may result from slippage at the 3′ splice site of an intron. The protein sequence given in Fig. 1A is identical with that of ref. 16, but differs by one synonymous mutation (Ile1150, ATT vs. ATC), that may represent a polymorphism. Finally, a sequence yielding an exact match to the 3′-terminal 60% of the ORF of hREV1 is described in GenBank as encoding a protein that interacts with α3A integrin (28); the significance of this observation is unclear.
Although determining the sequence of the ORF presented few problems, establishing the sequence of the 5′ untranslated region of the mRNA was less straightforward, and it is unlikely that it is complete. Because the 5′ sequence is GC-rich, attempts to extend primers into this region resulted in multiple premature termination events in clones with intron sequences, as identified both by the presence of canonical 3′ splice junction sequences and by the loss of the ORF. Other attempts recovered clones showing rearrangements of the cDNA sequence that presumably occurred during either synthesis or amplification. To confirm that the GC-rich sequence indeed constituted the 5′ untranslated sequence, sense-strand primers were designed specific to each of the possible upstream sequences, together with a common antisense primer. Only the GC-rich specific primer yielded an appropriate amplification product (data not shown). Additional evidence supporting this conclusion is provided by the sequences of three mouse ESTs (accession nos. AA4202230, AI019222, and AI481088), which show strong similarity to the region of hREV1 flanking the putative initiation codon. In contrast, sequence identified by an alternative upstream primer was clearly an artifact, because it matched sequence of an mRNA from human cortex (HUMMRNAC, accession no. L10374).
UV-Induced Mutagenesis Is Much Reduced in Human Cells Expressing hREV1 Antisense RNA.
In yeast, the REV1 gene is required for ≈95% of the base pair substitutions induced by UV (19), and rev1 mutants are more readily killed by this radiation, although this hypersensitivity is relatively modest (30). Unlike REV3 however, the REV1 gene appears to be required to a much smaller extent for the production of frame-shift mutations induced by UV. The extent varies among the different genetic sites investigated, however, and at three of them >80% of the UV-induced frameshifts depended on REV1 function (29, 31). Results with other mutagens, although less extensive, appear to be similar to those with UV, both with respect to base pair substitutions and frameshifts (32). To examine whether human REV1 is also required for UV mutagenesis, we have used the antisense method used previously with hREV3 (8), in which high levels of an antisense RNA are expressed under the control of the TetP promoter. To this end, a series of independent puromycin-resistant colonies obtained by transfection of parental 7AGM cells with hREV1 antisense-expressing plasmid, pR1P27-AS, were isolated and expanded to ≈10 × 106 cells. A portion was used for Northern blot analysis for antisense expression, and the rest were cryopreserved. Two unequivocally independent clones, designated 7AGM-12B-R1 and 7AGM-17C-R1, found to express antisense RNA at a level much higher than the level of natural transcript from the endogenous hREV1 gene (Fig. 3), were chosen for study. The level of expression of antisense REV1 RNA in these two strains, corrected for slight differences in RNA loading per lane, was very similar (Fig. 3).
Figure 3.
Northern blot analysis of the level of expression of hREV1 antisense RNA in the cell strains. RNA extracted from the two derivative cell strains, 7AGM-17C and 7AGM-12B, which had been transfected with plasmid pR1P27-AS containing the 4,117-bp sequence of the hREV1 RNA in an antisense orientation, and from their nontransfected parental cell strain, MSU-1.2-7AGM, and analyzed for expression of hREV1 antisense RNA (hREV1 AS RNA) and/or the endogenous hREV1 sense RNA. The latter mRNA, which is ≈4.4 kbp in length, can be seen as a faint band just above the antisense band in the first two lanes, and in the third lane. The lower band, approximately 1.4 kbp in length, is the endogenous HPRT mRNA, which was used to normalize the amount of RNA loaded per lane. There was no significant difference between the two derivative cell strains in the level of expression of the antisense RNA.
The frequency of 254-nm UV-induced mutants and the sensitivity to UV cell killing in these two cell strains was compared with that in their nontransfected parental cell strain, 7AGM. In all but one experiment, the parental cell strain and one or other or both of the derivative strains 7AGM-12B-R1 and 7AGM-17C-R1 were compared simultaneously. As shown in Fig. 4A, these two cell strains expressing hREV1 antisense RNA were not more sensitive than their parental strain to the cytotoxic effect of UV. The line with the slightly steeper slope is a least squares fit to the data from the parent strain 7AGM and also from strain 7AGM-17C-R1; the other line represents the least squares fit to data from strain 7AGM-12B-R1. Although the survival curves did not show a significant difference between these two strains and their parental strain, the frequency of 6-thioguanine-resistant mutants induced in the two strains expressing hREV1 antisense RNA was significantly lower (P = 0.01) than that seen with their parental cell strain (Fig. 4B). The 7AGM-17C-R1 cells showed a ≈64% decrease; the 7AGM-12B-R1 cells showed a ≈94% decrease in frequency.
Figure 4.
Percent survival (A) and frequencies of UV-induced 6-thioguanine resistant mutants (B) in the parent cell strain 7AGM and the hREV1 antisense RNA-expressing cell strains 7AGM-17C-R1 and 7AGM-12B-R1, plotted against fluence of 254 nm UV. The data are the average of results from four experiments with the parent strain, 7AGM; three experiments with 7AGM-17C-R1 cells; and two experiments with 7AGM-12B-R1cells. The background frequencies of mutants, which have been subtracted to obtain the frequencies induced above the background frequency by UV (23), are cited in the text. The mutant frequencies observed for untreated control and each UV fluence were corrected for the cloning efficiency of the cells determined after an 8-day expression period, i.e., at the time they were plated into selection medium containing 6-thioguanine. The average cloning efficiencies for control and UV-irradiated cells were: 7AGM cells, 33.3% ± 1.4% (SEM for four independent experiments); 7AGM-17C-R1 cells, 26.7 ± 1.3% (SEM for three independent experiments); and 7AGM-12B-R1 cells, 31.8% ± 0.9% (SEM for two independent experiments).
The results shown in Fig. 4B for the latter strain were obtained from two independent experiments, using 0, 11, 13, and 15 J/m2 of UV in one experiment, and 0, 12, and 14 J/m2 of UV in the other. No mutants were seen in the unirradiated control population. The slope of the line for this strain in Fig. 4B was fitted to the five data points from these two experiments. Although the slope is virtually parallel with the x axis, the line cannot, in reality, intersect the y axis above zero because the data represent the increase in frequency of mutants above the background frequency in the population. Cell strain 7AGM-17C-R1 was tested in three experiments. In each, the UV fluences used were 0, 11, 13, and 15 J/m2. The values shown for each UV fluence represent the average of the three determinations. Nevertheless, the slope of the line shown in Fig. 4B for this strain was fitted to the nine individual data points. The background frequencies of mutants per 106 cells in the unirradiated population for these three experiments, i.e., 0, 0, and 5, were almost as low as those observed with cell strain 7AGM-12B-R1. The values shown in Fig. 4B for parental cell strain 7AGM were taken from four experiments. Except for a single determination at 10 J/m2, the data are the average from two or three independent determinations in which the same UV fluence was used. Again, the values shown for each fluence, except 10 J/m2 UV, represent the average of the multiple determinations. Nevertheless, the slope of the line was fitted to the ten individual determinations obtained. The background frequency of mutants per 106 cells in the experiments with this strain were 4, 10, 13, and 17. To allow a comparison of the surviving fractions, the number of mutants seen, the total cells assayed for mutants, and their corresponding cloning efficiency at the time of selection for 6-thioguanine resistance, an example of the data obtained from an experiment for each strain is shown in Table 1.
Table 1.
Example of UV cytotoxicity and mutagenicity in parental cell strain MSU-1.2-7AGM and two derivative cell strains expressing hREV1 antisense RNA
| Cell strain | UV (J/m2) | Percent survival | Cells selected (×106) | Mutants observed | Cloning efficiency (%) | Mutant frequency (×106) | Induced frequency (×106) |
|---|---|---|---|---|---|---|---|
| 7AGM | 0 | 100 | 1 | 4 | 40 | 10 | 0 |
| 10 | 62 | 1 | 18 | 36 | 50 | 40 | |
| 12 | 52 | 1 | 22 | 40 | 55 | 45 | |
| 7AGM | 0 | 100 | 1.8 | 2 | 24 | 5 | 0 |
| 17C- | 11 | 47 | 1.8 | 6 | 24 | 14 | 9 |
| R1 | 13 | 33 | 1.8 | 12 | 21 | 32 | 27 |
| 15 | 20 | 1.8 | 7 | 21 | 19 | 14 | |
| 7AGM | 0 | 100 | 1 | 0 | 33 | 0 | 0 |
| 12B- | 11 | 57 | 1 | 0 | 32 | 0 | 0 |
| R1 | 13 | 35 | 1.5 | 1 | 32 | 2 | 2 |
| 15 | 26 | 1.5 | 0 | 30 | 0 | 0 |
The data in Fig. 4B indicate that, in cells expressing of hREV1 antisense, the frequency of UV-induced mutants was significantly reduced. Unlike what is found with yeast, however, there was no evidence for increased sensitivity to the cytotoxic effect of UV radiation in either of the two cell strains expressing the hREV1 antisense, and in which, presumably, the level of Rev1 protein is reduced by virtue of the antisense RNA. Such a lack of hypersensitivity may well result from the redirection of DNA damage into another pathway, one in which mutations are less likely to occur, or it may reflect a decreased importance of translesion replication for survival in these human cells, compared with yeast cells.
Acknowledgments
We thank Dr. Dennis Gilliland of Michigan State University, Department of Statistics & Probability, for statistical analysis of the mutagenicity data, and Beatrice Tung for advice and technical assistance with the human cell studies. This work was supported by U.S. Public Health Service Grants GM21858 (to C.W.L.), ES09822, and CA56796 (to V.M.M.), and CA73984 (to W.G.M.) from the National Institutes of Health.
Abbreviations
- RACE
rapid amplification of cDNA ends
- pol
DNA polymerase
- EST
expressed sequence tag
- HPRT
hypoxanthine phosphoribosyltransferase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF206019).
References
- 1.Lawrence C W, Hinkle D C. Cancer Surv. 1996;28:21–31. [PubMed] [Google Scholar]
- 2.Morrison A, Christensen R B, Alley J A, Beck A K, Bernstine E G, Lemontt J F, Lawrence C W. J Bacteriol. 1989;171:5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson J R, Lawrence C W, Hinkle D C. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 4.Nelson J R, Lawrence C W, Hinkle D C. Nature (London) 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 5.Johnson R E, Prakash S, Prakash L. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 6.McDonald J P, Levine A S, Woodgate R. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roush A A, Suarez M, Friedberg E C, Radman M, Siede W. Mol Gen Genet. 1998;257:686–692. doi: 10.1007/s004380050698. [DOI] [PubMed] [Google Scholar]
- 8.Gibbs P E M, McGregor W G, Maher V M, Nisson P, Lawrence C W. Proc Natl Acad Sci USA. 1998;95:6876–6881. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morelli C, Mungall A J, Negrini M, Barbanti-Brodano G, Croce C M. Cytogenet Cell Genet. 1998;83:18–20. doi: 10.1159/000015157. [DOI] [PubMed] [Google Scholar]
- 10.Lin W, Wu X, Wang Z. Mutat Res. 1999;433:89–98. doi: 10.1016/s0921-8777(98)00065-2. [DOI] [PubMed] [Google Scholar]
- 11.Van Sloun P P H, Romeijn R J, Eeken J C J. Mutat Res. 1999;433:109–116. doi: 10.1016/s0921-8777(98)00067-6. [DOI] [PubMed] [Google Scholar]
- 12.Matsutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. Nature (London) 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 14.Johnson R E, Kondratick C M, Prakash S, Prakash L. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 15.McDonald J P, Rapic-Otrin V, Epstein J A, Broughton B C, Wang X, Lehman A R, Wolgemuth D J, Woodgate R. Genomics. 1999;60:20–30. doi: 10.1006/geno.1999.5906. [DOI] [PubMed] [Google Scholar]
- 16.Lin W, Xin H, Zhang Y, Wu X, Yuan F, Wang Z. Nucleic Acids Res. 1999;27:4468–4475. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shockett P, Difilippantonio M, Hellman N, Schatz D G. Proc Natl Acad Sci USA. 1995;92:6522–6526. doi: 10.1073/pnas.92.14.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin C C, Wang Q, Maher V M, McCormick J J. Cell Growth Differ. 1994;5:1381–1387. [PubMed] [Google Scholar]
- 20.Hartmann S C, Mulligan R C. Proc Natl Acad Sci USA. 1988;85:8047–8051. doi: 10.1073/pnas.85.21.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qing J, Maher V M, Tran H, Argraves W S, Dunstan R W, McCormick J J. Oncogene. 1997;15:2159–2168. doi: 10.1038/sj.onc.1201385. [DOI] [PubMed] [Google Scholar]
- 22.Ryan P A, Maher V M, McCormick J J. Exp Cell Res. 1987;172:318–328. doi: 10.1016/0014-4827(87)90390-9. [DOI] [PubMed] [Google Scholar]
- 23.Maher V M, McCormick J J. In: Technologies for Detection of DNA Damage and Mutations. Pfeifer G P, editor. New York: Plenum; 1996. pp. 381–390. [Google Scholar]
- 24.Patton J D, Rowan L A, Mendrala A L, Howell J N, Maher V M, McCormick J J. Photochem Photobiol. 1984;39:37–42. doi: 10.1111/j.1751-1097.1984.tb03401.x. [DOI] [PubMed] [Google Scholar]
- 25.Larimer F W, Perry J R, Hardigree A A. J Bacteriol. 1989;171:230–237. doi: 10.1128/jb.171.1.230-237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerlach V L, Aravind L, Gotway G, Schultz R A, Koonin E V, Friedberg E C. Proc Natl Acad Sci USA. 1999;96:11922–11927. doi: 10.1073/pnas.96.21.11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak M. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 28.Wixler V, Laplantine E, Geerts D, Sonnenberg A, Petersohn D, Eckes B, Paulsen M, Aumailley M. FEBS Lett. 1999;445:351–355. doi: 10.1016/s0014-5793(99)00151-9. [DOI] [PubMed] [Google Scholar]
- 29.Lawrence C W, Christensen R B. J Mol Biol. 1978;122:1–21. doi: 10.1016/0022-2836(78)90104-3. [DOI] [PubMed] [Google Scholar]
- 30.Lemontt J F. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence C W, O'Brien T, Bond J. Mol Gen Genet. 1984;195:487–490. doi: 10.1007/BF00341451. [DOI] [PubMed] [Google Scholar]
- 32.McKee R H, Lawrence C W. Genetics. 1979;93:375–381. doi: 10.1093/genetics/93.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]