Abstract
This study reports the radiation-chemical yields for DNA single strand breaks (ssb) in crystals of CGCACG:CGTGCG (I) and CACGCG:CGCGTG (II) duplexes, induced by direct ionization using X-rays. The DNA fragmentation products, consisting of 3’- and 5’-phosphate-terminated fragments, were quantified by ion-exchange chromatography using a set of reference compounds. The yields of single strand breaks in I and II are 0.16±0.04 μmol/J and 0.07±.02 μmol/J, respectively. The probability of cleavage at a given site is relatively independent of which of the four bases is at that site. For the very small sample of base sequences studied to date, there is no obvious dependence on base sequence. However, there appears to be an increased frequency of strand breaks at the non-phosphorylated termini of the oligodeoxynucleotides. These results show that direct ionization is efficient at producing single strand breaks in DNA and its action is relatively indiscriminate with respect to base sequence.
Introduction
It has been known for some time that there are two different pathways by which ionizing radiation damages DNA, direct-type and indirect-type (1). It is primarily reactions of the products of water radiolysis with DNA that makes up the indirect-type pathway. Much is known about this pathway, both from the mechanistic and qualitative standpoints. In particular, the indirect-type’s propensity for causing single strand breaks (ssb) in DNA has been extensively studied (2–4).
Direct-type damage to DNA is caused by the direct interaction of ionizing radiation with the bases or sugar-phosphate backbone or by holes and dry electrons transferred to the DNA from the hydration waters surrounding the DNA (5–7). The direct pathway is particularly important in cells, where the amount of unbound water surrounding DNA is relatively small. Work on cellular DNA has implicated direct-type damage as significant in causing strand breaks (8). Recently we reported the yields of ssb in crystals of two Z-form DNAs. An HPLC study on crystals of d(CGCG)2 and d(CGCGCG)2 duplexes, x-irradiated at 4 K and room temperature, showed that the ssb yield is relatively insensitive to the irradiation temperature and it is about 10% of the free radical yield measured at 4 K (9).
In the current study, we extended this approach to the non-palindromic crystalline d(CGCACG:CGTGCG) (I) and d(CACGCG:CGCGTG) (II); both duplexes are Z-form. Based on our previous results on unaltered base release from several DNA crystals, we hypothesized that the precursor for ssb in directly ionized DNA is the sugar radical cation, which rapidly undergoes deprotonation to give a neutral sugar radical (10). Deprotonation of the radical cation competes with hole transfer from the sugar to the DNA bases, the rates of these two reactions being comparable. One question that needs addressed is to what extent this process depends on the local environment of the primary radical cation, and in particular, on the base sequence. There is a possibility, for example that guanine being the most oxidizable DNA base could “quench” the sugar radical cation more efficiently than the other bases (11). The positive correlation between energetics and rates of electron transfer is valid, however, only for moderately exothermic processes (12) while electron transfer from the sugar radical cation to any DNA base is expected to be highly energetically favorable. In order to test what is the actual situation in DNA, we extended our investigation to sequences containing a single AT base pair. The single adenine in these structures has no adjacent guanines. This should render the adenine site more susceptible to strand breakage if the rate of hole transfer is controlled by thermodynamics.
Materials/Methods
Reference 3’- and 5’-phosphorylated oligodeoxynucleotides for each of the possible strand-break products were purchased from Midland Certified Reagent and purified by ion-exchange chromatography. Crystals of CGCACG:CGTGCG (I) and CACGCG:CGCGTG (II) were grown from oligodeoxynucleotides purchased from Ransom Hill Bioscience (used without further purification) following published procedures (13). Both I and II are Z-form DNA. In the crystals, there are about 4.2 water molecules per nucleotide (14). The base stacking is continuous; e.g., the hexamers sit on top of one another so that the base stacking distance between the hexamer ends is the same as the internal stacking distance.
Single crystal and polycrystalline samples were removed from the sitting drop by capturing in a quartz capillary. Then the excess mother liquor was removed by the application of fine paper wicks and the samples were transferred to thin-walled quartz capillaries (Charles Supper). The sample size, weighed to ±1 μg accuracy on a Cahn-60 microbalance, was typically around 200 μg with a range of 90–400 μg. X-irradiation was performed with a Varian/Eimac OEG-76H tungsten-target tube operated at 70 keV and 20 mA. For room temperature (RT) irradiation, the capillaries were positioned at the face of the x-ray tube, and for 4 K irradiation, the capillaries were in a Janis Cryostat (15). The dose rates were 26 kGy/hr and 21.5 kGy/min, respectively. Most samples were irradiated at RT since our previous study has revealed only a minor influence of irradiation temperature on the yields (9).
The irradiated samples were dissolved, under air, in a 20 mM phosphate buffer (50 μl buffer per 100 μg of crystals) containing a known concentration of thymidine-5’-monophosphate (TMP) employed as an internal standard for quantification purposes. The solutions were held at 70° C for 30 minutes to ensure the complete conversion of heat-labile lesions to strand breaks (10) (9).
Samples were run on a Waters Alliance™ HPLC system equipped with a 2690 solvent delivery system and a 996 PDA detector. The products were separated on a Dionex DNAPac PA-100 4.6 mm×250 mm strong anion-exchange column at 60°C using 50 mM Tris (pH 10.22) as a mobile phase and applying a linear NaCl gradient. The products were detected by their absorbance at 260 nm and quantified by comparison with the standard containing reference oligonucleotides and TMP at known concentrations. The concentrations of reference oligonucleotides (in μM) were calculated from the optical densities by using the conversion factors provided by Midland Certified Reagent Company.
Results
Typical ion-exchange chromatograms of x-irradiated CGCACG:CGTGCG (I) and CACGCG:CGCGTG (II) are shown in Figure 1A and 1B along with the reference chromatograms. It is concluded, through the comparison of the irradiated crystal and reference spectra, that all the major damage products found in the crystal are present in the reference samples. These products are strand-break fragments phospohorylated at either the 3’- or 5’-end. The production of other strand-break species, i.e., not present in the reference sample, is minimal. In particular, there is no evidence of 3’-phosphoglycolates that are the products of C4’ hydrogen abstraction from DNA in the presence of oxygen (16).
Fig 1.
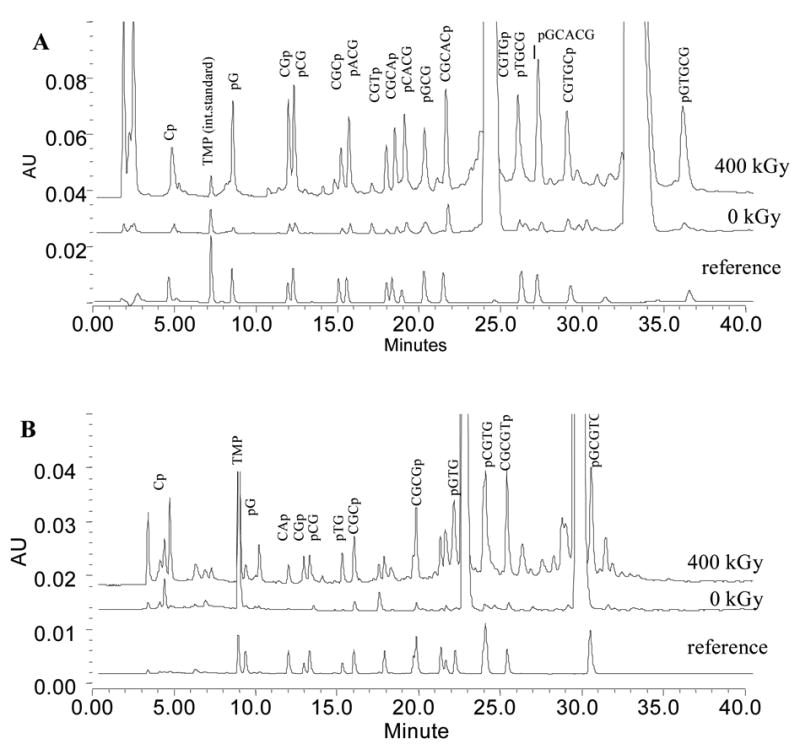
Ion-exchange chromatograms of I (A) and II (B) before and after irradiation to the dose of 400 kGy. The lower traces are reference chromatograms produced by mixtures of authentic oligonucleotides at known concentrations. Only the products used in further analysis are marked in Figure 1B.
Dose response plots for accumulation of DNA fragmentation products in I are shown in Figure 2. For the sake of clarity the data sets corresponding to each product are shifted vertically by 5 μM increments. As can be seen from the plots, dose saturation sets in above 500 kGy. Portions of the plots below 500 kGy were treated as linear dependencies and used to calculate the radiation-chemical yields. The yield of each DNA fragment was determined from the slope of the corresponding linear least squares fit and reported in Table 1.
Fig 2.
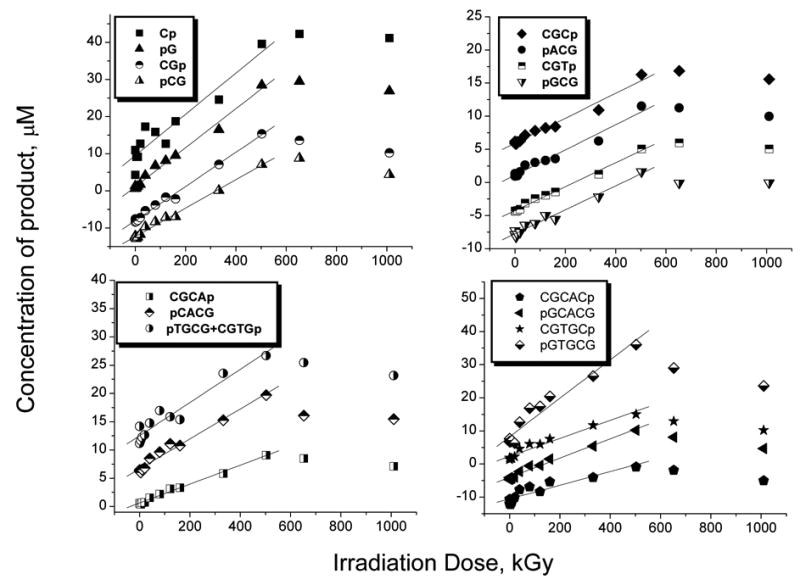
Dose response curves for product accumulation in I. For clarity, the curves in each graph are shifted vertically with respect to each other by 5 μM. The concentrations reported in the graph were obtained by dissolving x μg of the crystals in x/2 μl of phosphate buffer containing TMP.
Table 1.
Radiation-Chemical Yields of DNA Fragmentation Products in d(CGCACG:CGTGCG) (site numbering starts at the 5’-end in each strand)
| Product | Yield | Error | Cleavage Site
|
|
|---|---|---|---|---|
| (μmol/J) | (95% conf. level) | A-strand | T-strand | |
| Cp | 0.028 | 0.003 | G2 | G2 |
| pG | 0.026 | 0.001 | C5 | C5 |
| CGp | 0.022 | 0.001 | C3 | T3 |
| pCG | 0.019 | 0.001 | A4 | G4 |
| CGCp | 0.009 | 0.001 | A4 | N/A |
| CGCAp | 0.008 | 0.001 | C5 | N/A |
| CGCACp | 0.018 | 0.003 | G6 | N/A |
| pACG | 0.009 | 0.001 | C3 | N/A |
| pCACG | 0.016 | 0.001 | G2 | N/A |
| pGCACG | 0.019 | 0.002 | C1 | N/A |
| CGTp | 0.009 | 0.001 | N/A | G4 |
| CGTGp+pTGCGa | 0.016 | 0.002 | N/A | C5, G2 |
| CGTGCp | 0.019 | 0.002 | N/A | G6 |
| pGCG | 0.009 | 0.001 | N/A | T3 |
| pGTGCG | 0.031 | 0.004 | N/A | C1 |
These two products are not resolved in ion-exchange chromatograms
The same yields are presented as bar graphs in Figure 3. The same procedure was applied to analyze the products formed in II. The major difficulty faced with this sequence, however, is that not all of the products originating from the CACGCG strand could be resolved so as to permit separate quantification. For this reason, our analysis of this system was limited to the CGCGTG strand only. Irradiation doses applied to this system did not exceed 400 kGy and data for the entire dose range were analyzed as linear dependencies as shown in Figure 4. The yields obtained from the slopes are reported in Table 2 and, in bar graph form, in Figure 5.
Fig 3.
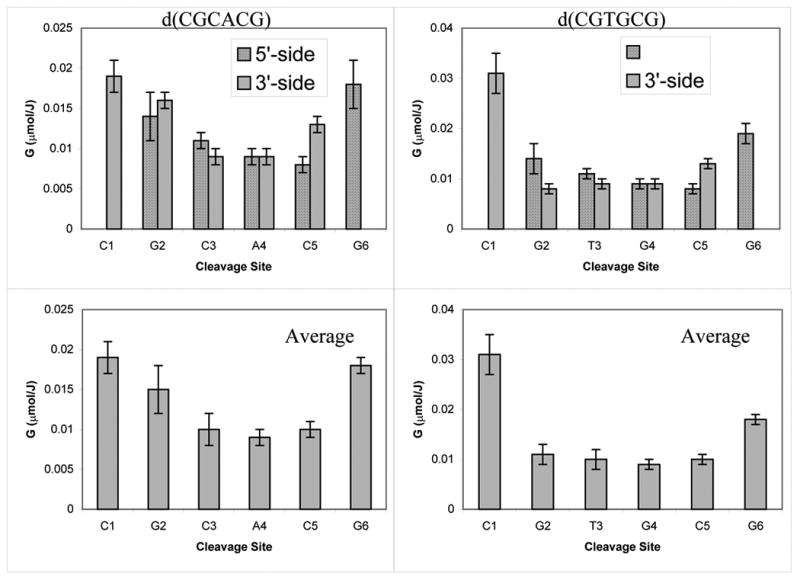
Product yields plotted as a function of cleavage site in I. The average yields are calculated as described in our earlier report.(9)
Fig 4.
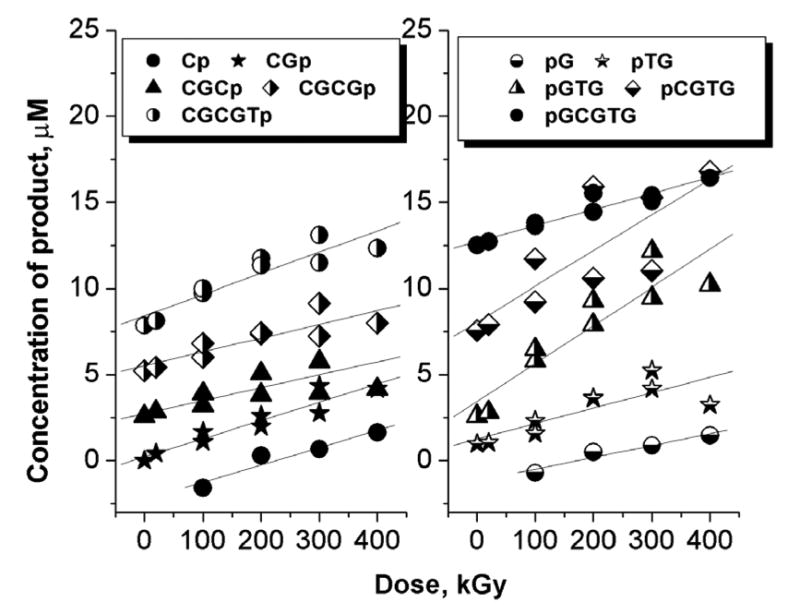
Dose response curves for the products generated in II. Only the products originating from the d(CGCGTG) strand are shown.
Table 2.
Radiation-Chemical Yieldsa of Fragmentation Products Originating from the d(CGCGTG) strand in d(CACGCG:CGCGTG) (site numbering starts at the 5’-end)
| Site | 5'-side product
|
3'-side product
|
Average yield | ||
|---|---|---|---|---|---|
| Fragment | Yield (μmol/J) | Fragment | Yield (μmol/J) | (μmol/J) | |
| C1 | N/A | N/A | pGCGTG | 0.004 | 0.004 |
| G2 | Cp | 0.005b | pCGTG | 0.010c | 0.007 |
| C3 | CGp | 0.006 | pGTG | 0.010 | 0.008 |
| G4 | CGCp | 0.003 | pTG | 0.004 | 0.004 |
| T5 | CGCGp | 0.004 | pG | 0.004b | 0.004 |
| G6
|
CGCGTp
|
0.006
|
N/A
|
N/A
|
0.006
|
| Total: | 0.033 | ||||
|
|
|||||
Standard error at the 95% confidence level for all yields is about 0.001 μmol/J except 0.002 μmol/J for pGTG;
The reported yield is 1/2 of the actual yield since half of this product is presumed to originate from damage to the opposite strand;
This product is not resolved from pACGCG in ion-exchange chromatograms. The yield has been calculated under the assumption that both products contribute equally to the measured peak area.
Fig 5.
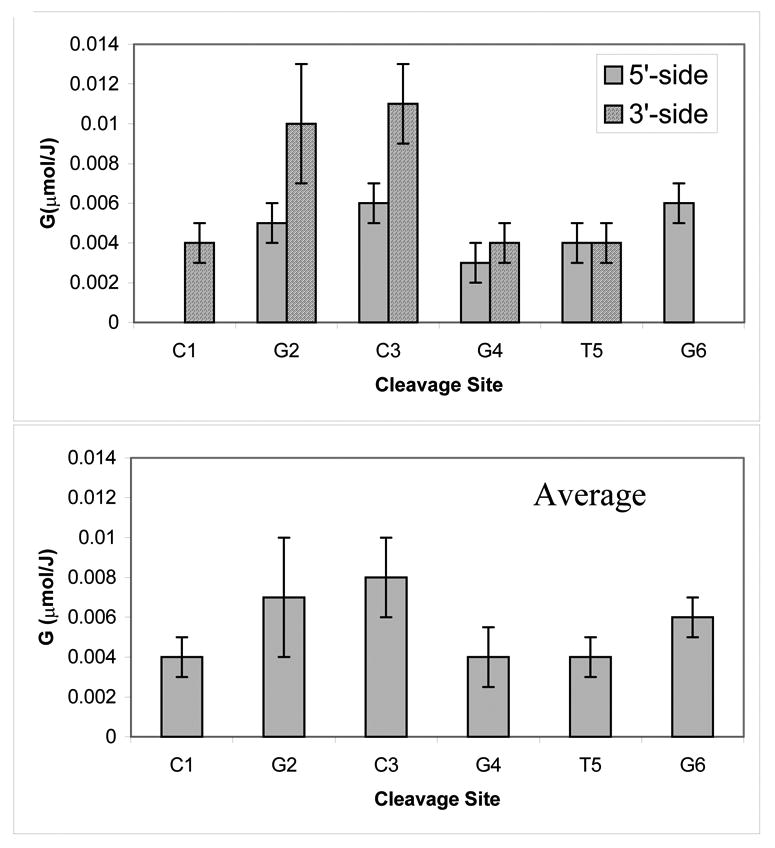
Product yields plotted as a function of cleavage site in II. The yields are reported for the d(CGCGTG) strand only.
Discussion
The products
It is well known that, for DNA in aqueous solution, hydrogen atom abstraction from any site of the deoxyribose moiety leads to the formation of free bases plus 3’- and 5’-phosphates (16). We show here that, for solid state DNA, the same relatively simple spectrum of major products is also generated by direct ionization (9). The similarity of the DNA fragmentation pattern produced by direct ionization with that produced by hydrogen abstraction in aqueous solution strongly supports the hypothesis that direct ionization of DNA gives rise to a similar set of sugar radicals. Such a set of neutral radicals is expected if sugar-phosphate radical cations deprotonate at rates that are comparable to, or faster than, competing reactions (such as hole transfer to the base stack). Direct ionization in the solid state also generates an equivalent number of electron adducts; these are located on the pyrimidine bases (17) and are known to be inconsequential with respect serving as precursors to strand breaks. This work, therefore, adds to the growing evidence that the primary precursor for direct-type strand breaks is ionization of the sugar-phosphate backbone.
A remarkable attribute of strand break accumulation in crystalline DNA is the resistance to dose saturation. In this study, we find no saturation in the production of any of the damage products in I or II up to doses of 500 kGy. It is known that electron-gain and electron-loss centers localized on DNA bases tend to saturate at much lower doses. On the other hand, the features attributed to the sugar radicals in EPR spectra are resistant to dose saturation (18). This resistance is almost certainly the consequence of two complementary factors. One is that the neutral sugar radical has a small cross section for radiation destruction (because the initial positive charge has been distanced from the unpaired electron by irreversible proton transfer). The other is that the base stack is a strong electron scavenger and, thereby, protects the sugar radical from electron return. The observation that both strand break products and sugar radicals share this remarkable attribute lends support to the conclusion that strand break products are derived from sugar radicals, and not from base centered radicals.
It is seen from Figure 2 that the dose saturation of strand break products occurs between 400 kGy and 650 kGy. This is almost certainly a consequence of parent compound depletion. Using a strand break yield of 0.1 μmole/J (see below), the fraction of oligomer strands that contain sugar damage after a dose of 500 kGy is ~10%. At this level of depletion, the slope of the dose response curve should decrease and, at higher doses, turn negative.
The yields
Figure 3 shows a bar graph of the product yields for I plotted as a function of the cleavage site. It is presumed that the sugar radical undergoes reactions that result in the release of two fragments, one with a 3’ and the other with a 5’ terminal phosphate. The average yields per damage site were calculated, as in our previous study, by averaging the yields of the 3’- and 5’-phosphates correlated with the damage site (9). While this treatment tends to underestimate strand break yields, it is the best we can do under the current conditions. The same approach was applied to the product analysis generated from the CGCGTG strand in II, and the results are shown in Figure 5.
There are two major conclusions that can be derived from these results. First, there is no obvious preference for the cleavage at A, T, C or G in either I or II. Therefore, the rate of hole transfer from the ionized sugar to the base is not controlled to any significant extent by the oxidation potential of the base. This can be explained by the high exothermicity of the process, independent of the base. Consistent with this, we observe that in duplex I the yield of ssb for the d(CGCACG) strand is approximately equal to the yield of ssb for the d(CGTGCG ) strand (0.08±0.02 μmol/J). The ssb yield found in II for the CTCGCG strand is somewhat lower, 0.033±0.01 μmol/J. This makes the total yield of single strand brakes in I and II around 0.16±0.04 μmol/J and 0.07±0.02 μmol/J, respectively. Given the accuracy of these measurements and the limited number of sequences studied, the significance of the difference between I and II is unclear.
Second, the earlier results obtained for the d(CGCG)2 and d(CGCGCG)2 crystals provided some evidence for the increased damage to the terminal sites in the strands (which do not carry terminal phosphates). This same effect appears to occur in I but the picture is not clear for II because of the poorer resolution of product in the chromatograms. The increased damage to the terminal sugars may result from the fact that the 3’- and 5’-termini in either oligonucleotide are not phosphorylated and, thereby, may participate in reactions not available to the internal sugars. Specifically, the presence of free OH groups in the terminal sugar may alter the chemistry of the sugar radical cation, providing an additional channel for its deprotonation. Deprotonation at the OH group by the radical cation would result in an alkoxy-radical (R-O•), which can be converted further into a carbon centered radical through hydrogen atom abstraction or transfer.
This study has demonstrated that the yield of strand breakage events is significant and consistent with the values obtained in the previous study using a different chromatographic technique (reverse phase as opposed to anion exchange). Also, it is known from previous work that there is also a significant fraction of free radical damage trapped at the bases (19, 20). Both of these direct-type lesions could play an important role in the formation of multiply damaged sites (MDS) (21, 22), and the neglect of this type of damage in calculations could cause significant under-estimation of the local severity of helical damage. For example, a direct-type strand break situated within 10 bases of an indirect-type strand break could result in a biologically more damaging double strand break (23) (24). In these types of calculations, it is important, therefore, to include the yields and distribution of direct-type damage.
Conclusions
The yield of direct-type strand breaks in crystalline I and II is not sequence-dependent but does show preferential breakage at the termini of the oligodeoxynucleotides. The total yield of strand breaks for these sequences ranges from 0.16±0.04 μmol/J to 0.07±0.02 μmol/J.
Acknowledgments
We thank Kermit R. Mercer for his invaluable technical assistance. The investigation was supported by PHS Grant 2-R01-CA32546, awarded by the National Cancer Institute, DHHS. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
References
- 1.von Sonntag C. The Chemical Basis of Radiation Biology. Taylor and Francis; New York: 1987. [Google Scholar]
- 2.Swarts SG, Sevilla MD, Becker D, Tokar CJ, Wheeler KT. Radiation-induced DNA damage as a function of hydration. I. Release of unaltered bases. Radiat Res. 1992;129:333–344. [PubMed] [Google Scholar]
- 3.Ito T, Baker SC, Stickley CD, Peak JG, Peak MJ. Dependence of the yields of strand breaks induced by gamma-rays in DNA on the physical conditions of exposure: water content and temperature. Int J Radiat Biol. 1993;63:289–296. doi: 10.1080/09553009314550391. [DOI] [PubMed] [Google Scholar]
- 4.Milligan JR, Limoli CL, Wu CCL, Ng JYY, Ward JF. In: Mechanisms of DNA strand breakage by ionizing radiation. Fuciarelli AF, Zimbrick JD, editors. Battelle Press; Columbus, Ohio: 1995. pp. 165–174. [Google Scholar]
- 5.LaVere T, Becker D, Sevilla MD. Yields of OH· in γ-irradaited DNA as a function of DNA hydration: Hole transfer vs. OH· formation. Radiation Research. 1996;145:673–680. [PubMed] [Google Scholar]
- 6.Mroczka NE, Mercer KR, Bernhard WA. The effects of lattice water on free radical yields in x-irradiated crystalline pyrimidines and purines: a low-temperature electron paramagnetic resonance investigation. Radiat Res. 1997;147:560–568. [PubMed] [Google Scholar]
- 7.Debije MG, Strickler MD, Bernhard WA. On the efficiency of hole and electron transfer from the hydration layer to DNA: an EPR study of crystalline DNA X-irradiated at 4 K. Radiat Res. 2000;154:163–170. doi: 10.1667/0033-7587(2000)154[0163:oteoha]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baverstock KF, Will S. Evidence for the dominance of direct excitation of DNA in the formation of strand breaks in cells following irradiation. Int J Radiat Biol. 1989;55:563–568. doi: 10.1080/09553008914550611. [DOI] [PubMed] [Google Scholar]
- 9.Debije MG, Razskazovskiy Y, Bernhard WA. The Yield of Strand Breaks Resulting from Direct-Type Effects in Crystalline DNA X-Irradiated at 4 K and Room Temperature. J Am Chem Soc. 2001;123:2917–2918. doi: 10.1021/ja005790r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Razskazovskiy Y, Debije MG, Bernhard WA. Direct radiation damage to crystalline DNA: what is the source of unaltered base release? Radiat Res. 2000;153:436–441. doi: 10.1667/0033-7587(2000)153[0436:drdtcd]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meggers E, Dussy A, Schafer T, Giese B. Electron transfer in DNA from guanine and 8-oxoguanine to a radical cation of the carbohydrate backbone. Chem--Eur J. 2000;6:485–492. doi: 10.1002/(sici)1521-3765(20000204)6:3<485::aid-chem485>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 12.Marcus R, Sutin N. Electron transfer in chemistry and biology. Biochim Biophys Acta. 1985;811:265–322. [Google Scholar]
- 13.Sadasivan C, Gautham N. Sequence-dependent microheterogeneity of Z-DNA: the crystal and molecular structures of d(CACGCG)d(CGCGTG) and d(CGCACG)d(CGTGCG) J Mol Biol. 1995;248:918–930. doi: 10.1006/jmbi.1995.9894. [DOI] [PubMed] [Google Scholar]
- 14.Debije MG, Bernhard WA. Electron and Hole Transfer Induced by Thermal Annealing of Crystalline DNA X-Irradiated at 4 K. J Phys Chem B. 2000;104:7845–7851. [Google Scholar]
- 15.Mercer KR, Bernhard WA. Design and operation of a variable temperature accessory for Q-band ESR. J Magn Reson. 1987;74:66–71. [Google Scholar]
- 16.Pogozelski WK, Tullius TD. Oxidative Strand Scission of Nucleic Acids: Routes Initiated by Hydrogen Abstraction from the Sugar Moiety. Chem Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 17.Becker D, Sevilla MD. The chemical consequences of radiation damage to DNA. In: Sinclair JTL, Sinclair WK, editors. Advances in Radiation Biology. Vol. 17. Academic Press; San Diego: 1993. pp. 121–180. [Google Scholar]
- 18.Becker D, Razskazovskii Y, Sevilla MD. ESR investigation of heavy ion-beam (16O8+) irradiated DNA: Evidence for damage to the deoxyribose backbone. Radiation Research. 1996;146:361–368. [PubMed] [Google Scholar]
- 19.Debije MG, Bernhard WA. Free radical yields in crystalline DNA X-irradiated at 4 K. Radiat Res. 1999;152:583–589. [PMC free article] [PubMed] [Google Scholar]
- 20.Debije MG, Milano MT, Bernhard WA. DNA responds to ionizing radiation as an insulator, not as a "molecular wire". Angew Chem, Int Ed. 1999;38:2752–2756. [PMC free article] [PubMed] [Google Scholar]
- 21.Ward JF. Some biochemical consequences of the spatial distribution of ionizing radiation-produced free radicals. Radiat Res. 1981;86:185–195. [PubMed] [Google Scholar]
- 22.Ward JF, Milligan JR, Jones GDD, Fahey RC. Multiply damages sites. In: Fuciarelli AF, Zimbrick JD, editors. Damage DNA. Battelle Press; Columbus, Ohio: 1995. pp. 45–53. [Google Scholar]
- 23.Nikjoo H, O'Neill P, Goodhead DT, Terrissol M. Computational modelling of low-energy electron-induced DNA damage by early physical and chemical events. Int J Radiat Biol. 1997;71:467–483. doi: 10.1080/095530097143798. [DOI] [PubMed] [Google Scholar]
- 24.Moiseenko VV, Hamm RN, Waker AJ, Prestwich WV. Modelling DNA damage induced by different energy photons and tritium beta-particles. Int J Radiat Biol. 1998;74:533–550. doi: 10.1080/095530098141113. [DOI] [PubMed] [Google Scholar]


