Abstract
This study investigated the molecular alterations underlying the physiological adaptations to starvation and refeeding in human skeletal muscle. Forty-eight hours' starvation reduced whole-body insulin sensitivity by 42% and produced marked changes in expression of key carbohydrate (CHO) regulatory genes and proteins: SREBP1c and hexokinase II (HKII) were downregulated 2.5- and 5-fold, respectively, whereas the pyruvate dexydrogenase kinase 4 (PDK4) was upregulated 4-fold. These responses were not dependent on the phosphorylation status of Akt and FOXO1. On the other hand, starvation and the concomitant increase in circulating free fatty acids did not upregulate the expression of transcription factors and genes involved in fat metabolism. Twenty-four hours' refeeding with a CHO-rich diet completely reversed the changes in PDK4, HKII and SREBP1c expression in human skeletal muscle but failed to fully restore whole-body insulin sensitivity. Thus, during starvation in healthy humans, unlike rodents, regulation of fat metabolism does not require an adaptive response at transcriptional level, but adaptive changes in gene expression are required to switch off oxidative glucose disposal. Lack of effect on key proteins in the insulin-signalling pathway may indicate that changes in intracellular substrate availability/flux may be responsible for these adaptive changes in glucose metabolism. This may represent an important aspect of the molecular basis of the development of insulin resistance in metabolic conditions characterized by energy restriction.
In healthy humans, starvation is associated with a shift in substrate utilization from carbohydrate (CHO) to fat, and a significant reduction in insulin sensitivity (Mansell & Macdonald, 1990; Webber et al. 1994; Fery et al. 1999). Skeletal muscle rather than the splanchnic bed appears to be responsible for the development of insulin resistance in starvation (Bjorkman & Eriksson, 1985; Mansell & Macdonald, 1990; Webber et al. 1994). This decrease in muscle glucose uptake is accompanied by a reduction in oxidative glucose disposal (Mansell & Macdonald, 1990; Webber et al. 1994; Fery et al. 1999), with either no change (Mansell & Macdonald, 1990; Webber et al. 1994) or a modest increase in non-oxidative glucose disposal (Fery et al. 1999). The underlying mechanisms controlling the starvation-induced switch in skeletal muscle substrate utilization and reduction in glucose uptake are unclear.
Experiments primarily carried out in rodents suggest that transcriptional regulation of a number of key genes involved in lipid and CHO metabolism is responsible for the metabolic adaptation in response to starvation and refeeding. Short-term starvation in rats and mice has been previously shown to increase the expression of genes involved in fatty acid transport and beta oxidation such as LPL, CPT1, LCAD, CD36 and UCP3 (Holst et al. 2003; de Lange et al. 2004), indicating that increased fatty acid influx into muscle during starvation is accompanied by an adaptive increase in gene expression. Regulation of oxidative glucose disposal appears to be controlled at the transcriptional and post-transcriptional level. Suppression of skeletal muscle pyruvate dehydrogenase complex (PDC) activity plays a major role in the downregulation of CHO oxidation in response to starvation (Sugden et al. 1993). Regulation of the activity of PDC in skeletal muscle is of critical importance in the integration of CHO and fat metabolism (Randle et al. 1963). Skeletal muscle PDC activity is inhibited by phosphorylation of the E1 component of the complex by pyruvate dehydrogenase kinase (PDK). Of the four PDK isoenzymes that have been identified in skeletal muscle, the PDK4 mRNA and protein expression was observed to increase in starvation and diabetes in animal models (Wu et al. 1998, 1999). In humans, starvation was shown to increase PDK4 transcription (Pilegaard et al. 2003) and mRNA content in skeletal muscle (Pilegaard et al. 2003; Spriet et al. 2004). However, neither study examined the effects of starvation and refeeding on PDK4 protein expression and the potential role of transcription factors (which animal studies have implicated in the regulation of PDK4 gene expression) such as peroxisome proliferator-activated receptors (PPARα and PPARδ), peroxisome proliferator-activated receptor gamma coactivator-1α (PGC1α)) and forkhead transcription factor (FOXO1).
Indeed, the integration of lipid and CHO metabolism is thought to be at least partly under the control of PPARs and/or PGC1, although their precise role in human skeletal muscle is not well understood (Lee et al. 2003). Evidence from rat studies also suggests that starvation represses the expression of sterol regulatory element-binding protein 1c (SREBP1c) and carbohydrate response element-binding protein (ChREBP); transcription factors shown to be integral to the control of genes in the glycolytic and lipogenic pathways (Yamashita et al. 2001; Guillet-Deniau et al. 2002; Bizeau et al. 2003; Commerford et al. 2004; Gosmain et al. 2004; He et al. 2004). However, their role in the adaptive response to fasting in humans has not been examined. A number of differences exist between rodent and human skeletal muscle metabolism. For example, the relatively high basal metabolic rate of rodents means that they have to rely more on CHO metabolism to sustain their daily energy expenditure (Suarez et al. 2004). Therefore, starvation and other conditions known to elevate lipid metabolism are expected to have a greater impact on CHO metabolism in rodents than in humans, which may have implications for the regulation of substrate integration in different species. Hence, extrapolations from studies on rodents to humans may not always be relevant.
The aim of this study was to (a) examine the effect of starvation and refeeding with a high CHO diet on changes in expression of key metabolic genes associated with CHO and fat oxidation pathways in human skeletal muscle, and relate those measurements to changes in insulin action; and (b) elucidate some of the signalling pathways responsible for the adaptive response to fasting in humans.
Methods
Subjects
Ten healthy males (age 26 ± 1 years, body mass 81 ± 4 kg, BMI 25.5 ± 1.2 kg m−2) gave written informed consent and volunteered to participate in this study. All procedures used in this study were performed according to the Declaration of Helsinki and approved by the University of Nottingham Medical School Ethics Committee. Subjects were asked to weigh and record their normal food intake for 3 days. The dietary information was analysed using Microdiet software (Downlee Systems Limited, UK).
Experimental design and protocol
All subjects underwent a 48-h period of starvation followed by a 24-h period of refeeding with a high CHO diet (total energy 3086 ± 19 kcal, of which 75% energy was CHO, 10% fat and 15% protein). On three occasions, before and after 48 h of starvation and after 24 h of refeeding, subjects underwent a 16 min insulin tolerance test (ITT) to quantify insulin sensitivity (Bonora et al. 1989). Muscle biopsies were obtained from the vastus lateralis at the start of the study and after 24 h and 48 h of starvation, and after 24 h of refeeding.
On day 1, subjects arrived at the laboratory 4 h after consumption of a standardized breakfast (providing 1 g CHO (kg body mass)−1, having abstained from caffeine on the day of the study, and from alcohol and heavy exercise for the previous 72 h. A muscle sample from the vastus lateralis of one leg was then obtained and an indwelling cannula (venflon, 21G, Ohmeda, Hatfield, Herts, UK) was inserted into an antecubital vein for glucose and insulin infusion and into a dorsal vein on the non-dominant hand for sampling, with the hand placed in a hot-air box maintained at 50–55°C to arterialize the blood. Samples were taken for baseline determination of blood glucose, lactate, β-hydroxybutyrate, insulin, glycerol, free fatty acids (FFA) and cytokines (IL1β, IL6, IL8, IL10, IL12 and TNFα) concentrations. An ITT was commenced by administering an intravenous bolus ((0.1 U (kg body mass)−1) of human soluble insulin (Actrapid, Novo, Copenhagen, Denmark). Blood samples were taken every 2 min, and the test was terminated after 16 min by infusing a 20% dextrose solution to prevent symptoms of hypoglycaemia. Following this, a high-CHO meal was provided, after which subjects were asked to fast for the next 48 h. During the period of starvation, subjects refrained absolutely from food but were allowed water, electrolytes, non-sugared carbonated drinks and, except on the day of the study, black non-sugared decaffeinated coffee and tea. While starving, subjects consumed 80 mmol sodium and 40 mmol potassium daily, as slow-release tablets to minimize the potentially confounding effects of fluid deprivation and intravascular volume depletion on cardiovascular reflexes and sympathetic nervous system activity. Subjects were required to continue with their normal daily activities but to refrain from formal heavy exercise sessions during the period of starvation.
On day 2, blood and muscle samples were obtained after exactly 24 h of starvation. On day 3, subjects arrived at the laboratory after exactly 48 h of starvation, and a muscle sample was obtained followed by an ITT (as described for day 1). A high-CHO meal was then provided as part of the high-CHO diet (described above) that subjects were asked to consume for the next 24 h. Subjects were also asked to abstain from alcohol and heavy exercise for the same period of time. On day 4, subjects arrived at the laboratory after exactly 24 h of refeeding, and a muscle sample was obtained followed by an ITT.
Blood analysis and calculation of insulin sensitivity
Whole-blood glucose and lactate concentrations were measured shortly after collection using glucose oxidase and l-lactate oxidase methods, respectively (Yellow Springs Instrument Analyser, YSI, 2300 STAT PLUS). Blood hydroxybutyrate concentrations were determined on whole-blood/10% PCA extracts (ratio 1: 1), using a colorimetric method by Roche (R-Biopharm GmbH, Germany). Plasma and serum were separated by low-speed centrifugation (15 min at 3000 g). Plasma was analysed for FFA (NEFA-C test, Wako, Germany) and glycerol (GPO-Trinder, Sigma Diagnostics, USA) concentrations. Serum was analysed for insulin concentrations by radioimmunoassay (Diagnostics Products Corporation, Llanberis, Wales, UK). Whole-body insulin sensitivity was calculated as the slope or first-order rate constant for blood glucose disappearance (log mmol min−1) from 4 to 16 min after insulin injection, since no changes in blood glucose concentration are observed within the first 4 min of the ITT (Bonora et al. 1989).
Cytokines were measured using a cytokine bead array (CBA; BD Biosciences) according to the manufacturer's instructions. Plasma samples were analysed on a Beckman Coulter ALTRA flow cytometer (Beckman Coulter, High Wycombe, UK), using 633 nm excitation to distinguish the bead populations, and 488 nm excitation of PE fluorescence for cytokine quantification. The PE fluorescence of each bead population was extracted, and cytokine concentration calculated against a 10-point standard curve using BD Cytometric Bead Array Software (BD Biosciences).
Muscle sampling and PDCa activity
All muscle samples were obtained from the vastus lateralis using the needle biopsy technique with suction being applied (Bergstrom, 1962). Each sample was taken through a separate skin incision (3–5 mm long). All incisions were made using a surgical blade under local anaesthetic (2–3 ml of 1% lidocaine (lignocaine)) while the subject was lying on an examination couch. After removing the biopsy needle from the leg, one piece of muscle tissue (20–40 mg) was used immediately for mitochondrial extraction (see Protein extraction), and the remaining part was kept in liquid nitrogen for subsequent enzyme, RNA and protein extraction. At a later date, 5–10 mg of frozen muscle was used to determine the active form of pyruvate dehydrogenase complex (PDCa) using a method exactly as described by Constantin-Teodosiu et al. (1991).
Total RNA extraction and reverse transcription
Total RNA was extracted from 10–20 mg of frozen muscle tissue by the method of Chomczynski & Sacchi (1987). Aliquots of RNA were quantified using a RiboGreen Quantification kit (Molecular Probes) and adjusted to the same total RNA concentration. Reverse transcription was carried out from 500 ng total RNA using Murine Moloney Leukaemia Virus (MMLV) reverse transcriptase (Promega) and random hexamer primers (Promega).
Real-time quantitative PCR
Taqman primers and probes (with the exception of FOXO1 which were obtained from Applied Biosystems, UK) were designed using Primer Express version 2.0 Software (Applied Biosystems, Warrington, UK) and are shown in Table 1. Real-time PCR was performed using PCR Universal Master Mix (Applied Biosystems) in a Micro-Amp 96-well plate using an ABI Prism 7000 Sequence Detection System (Applied Biosystems, UK). Primers and probes were used at a final concentration of 300 nm and 200 nm, respectively, in a reaction volume of 25 μl. Assays were performed in triplicate. The threshold (Ct) values for each triplicate were averaged and the quantification of expression of each gene relative to α-actin determined using the standard curve method. The Ct values for α-actin did not change significantly between time points and samples.
Table 1.
Sequences for primers and Taqman probes used for real-time PCR
| Gene | Forward primer | Probe | Reverse primer |
|---|---|---|---|
| HKII | AAGTTCTTGTCTCAGATTGAGAGTGACT | CTGCAACACTTAGGGCTTGAGAGCACCTG | CAGTGCACACCTCCTTAACAATG |
| PK | CACTAAAGGACCTGAGATCCGAACT | TCATCAAGGGCAGCGGCACTGCA | TGTTCTCGTCACACTTTTCCATGT |
| PDK1 | TTTACCCCCCTATTCAAGTTCATG | TCACAGTCAAATCCTCATTACCCAGCGTG | CCACCTCCTCGGTCACTCAT |
| PDK2 | CATCATGAAAGAGATCAACCTGCTT | CCGACCGAGTGCTGAGCACACCC | CAGGAGGCTCTGGACATACCA |
| PDK3 | CGCTACTCGCGCTTTTCG | CGTCGCCGCTCTCCATCAAACA | TGAAGTTTTCTCACATGCATTATCTCT |
| PDK4 | CAAGGATGCTCTGTGATCAGTATTATTT | CATCTCCAGAATTAAAGCTTACACAAGTGAATGGA | TGTGAATTGGTTGGTCTGGAAA |
| CD36/FAT | CTGGAGTCTGGAATTCAGAACGT | CCTGCAGGTTCAGTGCCCCC | GAAGTGAGGATGGGAGAGAAACA |
| CPT1 | TTTGGCCCTGTAGCAGATGAT | TCGTGTTCTCGCCTGCAATCATGT | ACTTGCTGGAGATGTGGAAGAAG |
| LCAD | GAGCATCTTGGTGGAATTGGA | TCCGCAGCTATTGTCTGGGAGGAGC | GGGCCTGAACAATTTGAATAAGC |
| PPARα | GCTTCCTGCTTCATAGATAAGAGCTT | AGCTCGGCGGCACAACCAGCA | CACCATCGCGACCAGATG |
| PPARδ | TGCGGCCATCATTCTGTGT | ACCGGCCAGGCCTCATGAACG | CAGGATGGTGTCCTGGATAGC |
| PGC1α | GGTGCAGTGACCAATCAGAAATAA | ATCCAATCAGTACAACAATGAGCCTTCAAACATAT | TTGCCTCATTCTCTTCATCTATCTTC |
| SREBP1c | GGAGGGGTAGGGCCAAC | CGCGGAGCCATGGATTGCAC | GTCAAATAGGCCAGGGAAGTC |
| ChREBP | AGTATCGACCCCACACTCACAC | GCCTGGCCTACAGTGGCAAGCTG | TTGTTCAGGCGGATCTTGTCT |
| α-ACTIN | GAGCCGAGAGTAGCAGTTGTA GCT | CCCGCCCAGAAACTAGACACAATG TGC | GCGGTGGTCTCGTCTTCGT |
Protein extraction
Total protein extracts were prepared from 20–30 mg of frozen biopsy tissue. Samples were first homogenized using a polytron homogenizer for 30 s on ice, in six volumes of buffer containing 20 mm Tris, 5 mm EDTA, 10 mm NaF, 2 mm DTT (pH 7.5) and the following protease inhibitors: 4-(2-aminoethyl)benzenesulphonyl fluoride (AEBSF; 10 mg ml−1), leupeptin (0.1 mg ml−1) and pepstatin A (10 μg ml−1). The homogenates were then centrifuged at 10 000 g for 20 min at 4°C, and the supernatants stored at −80°C until analysis. Mitochondria were extracted from 20–40 mg of fresh muscle tissue exactly as previously described (Wibom & Hultman, 1990) and used for the determination of PDK2 and PDK4 protein expression. Protein concentrations of mitochondrial suspensions and whole-tissue extracts were measured using the Bradford method (Bio-Rad Laboratories, UK).
Western blotting
Proteins were separated using 5–20% gradient gels, and then transferred overnight to Hybond-C nitrocellulose membranes (Amersham Biosciences, UK). After blocking with 1% Western blocking reagent (Roche Diagnostics Co.) for 1 h at room temperature, the membranes were incubated for 1 h (except for SREBP1c which were incubated overnight at 4°C) with primary antibodies for PPARα (Cayman Chemical Co., USA), PPARδ, IRS1, IRS2, PDK2 and PDK4 (Santa Cruz Biotechnology Inc., USA), total Akt, phospho-Akt serine473, total FOXO1 and phospho-FOXO1 serine256 (Cell Signalling, USA), PGC1α (Calbiochem, USA), SREBP1c (mouse monoclonal IgG 2A4, American Type Culture Collection), prohibitin (Lab Vision Corp, USA) and α-actin (Sigma-Aldrich Co., USA). This was followed by incubation for 1 h at room temperature with either goat anti-rabbit HRP, rabbit anti-mouse HRP (Amersham Biosciences, UK) or rabbit anti-goat HRP (DakoCytomation, Denmark) secondary antibody as appropriate. All immunoreactive proteins were visualized using ECL plus (Amersham Biosciences, UK) and quantified by densitometry using the Quantity One 1-D Analysis Software version 4.5 (Bio-Rad Laboratories, Inc., USA). The mitochondria-specific protein prohibitin was used to normalize the mitochondrial protein expression of PDK2 and PDK4, whereas the content of all proteins determined from total muscle extracts was expressed relative to the content of α-actin. Neither prohibitin nor α-actin protein content was affected by starvation and refeeding.
The protein content of the slow- and fast-twitch fibres in the vastus lateralis of all subjects was determined using an immunochemical technique to quantify the slow (MHC-s) and fast (MHC-f) myosin heavy chain contents as previously described (Sazili et al. 2005). For each individual, the MHC-s and MHC-f contents were expressed relative to the content of the muscle-specific protein α-actin.
Statistical methods
Analysis of variance (ANOVA) for repeated measures across time was used to assess differences between physiological, metabolic and molecular responses to starvation and refeeding. Mauchly's test for sphericity was used; where asphericity was assumed, the Greenhouse-Geisser correction was used for epsilon < 0.75, if epsilon > 0.75 then the Huyn-Feldt correction was used. When a significant difference was obtained, the Holm-Bonferroni stepwise method was used to locate any differences. When there were only single comparisons Student's paired t test was used. Relationships between variables were determined by Pearson's product moment correlation coefficients. Statistical significance was accepted at a 5% level. Results are presented as means ± s.e.m.
Results
Whole-body metabolic responses and insulin sensitivity
Body mass decreased from 80.5 ± 4.3 kg to 79.0 ± 4.2 kg after 48 h of starvation (P < 0.001), but did not fully recover after 24 h refeeding with a high CHO diet (79.6 ± 4.1 kg; P < 0.05 from pre-starvation value). Starvation for 24 h increased plasma glycerol (P < 0.01) and blood hydroxybutyrate (P < 0.05) concentrations, and tended to increase plasma FFA (P = 0.06) concentrations. There was also a tendency for blood glucose concentration to fall (P = 0.078), whereas serum insulin concentration was unaffected by 24 h starvation. Starvation for 48 h decreased blood glucose (P < 0.001) and serum insulin (P < 0.05) concentrations, and increased plasma FFA (P < 0.01) and glycerol (P < 0.05) and blood hydroxybutyrate (P < 0.001) levels (Table 2). Refeeding restored metabolites to pre-starvation levels. There was no effect of starvation or refeeding on blood lactate concentrations (Table 2). Circulating levels of IL1β, IL6, IL8, IL10, IL12 and TNFα were not altered significantly by starvation or refeeding (data not shown).
Table 2.
Metabolic responses to 48 h starvation (fast) and 24 h refeeding (refed) with a high CHO diet
| Pre-fast | 24 h fast | 48 h fast | 24 h refed | |
|---|---|---|---|---|
| Plasma glucose (mm) | 4.34 ± 0.07 | 4.09 ± 0.09 | 3.51 ± 0.08**† | 4.44 ± 0.14 |
| Serum insulin (pm) | 29.5 ± 3.5 | 26.1 ± 3.0 | 16.4 ± 1.2** | 31.9 ± 5.3 |
| Plasma FFA (mm) | 0.46 ± 0.07 | 0.76 ± 0.12 | 0.97 ± 0.10** | 0.45 ± 0.08 |
| Plasma glycerol (μm) | 31.7 ± 4.7 | 46.9 ± 5.5** | 52.1 ± 6.3* | 31.8 ± 2.8 |
| Blood hydroxybutyrate (mm) | 0.20 ± 0.04 | 0.73 ± 0.15* | 1.51 ± 0.16**† | 0.19 ± 0.08 |
| Blood lactate (mm) | 0.90 ± 0.11 | 0.87 ± 0.07 | 0.93 ± 0.08 | 0.93 ± 0.07 |
Values are mean ± s.e.m.; n = 10 except for lactate (n = 7).
P < 0.05 from pre-fast
P < 0.01 from pre-fast
P < 0.05 from 24 h fast.
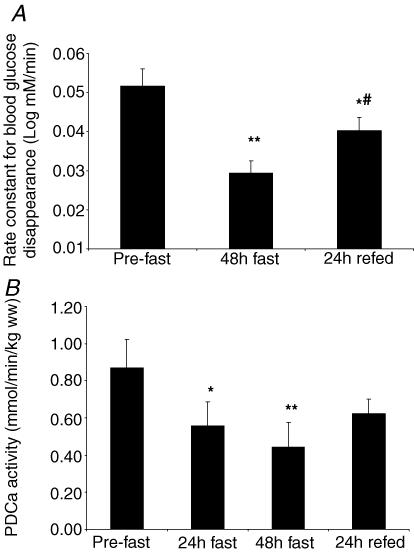
Whole-body insulin sensitivity (calculated from the first-order rate constant for blood glucose disappearance during ITT) decreased by 42 ± 5% after 48 h of starvation (from 0.052 ± 0.004 to 0.029 ± 0.003 log mmol min−1; P < 0.01) and recovered by 21 ± 7% (of initial value) upon refeeding (Fig. 1A). However, after 24 h refeeding, insulin sensitivity was lower (0.040 ± 0.003 log mmol min−1, P < 0.05) than the pre-starvation value.
Figure 1. . Whole-body insulin sensitivity (A) and skeletal muscle PDCa activity (B) responses to 48 h starvation (fast) and 24 h refeeding (refed) with a high-CHO diet.
Values are mean ± s.e.m.; n = 10 for insulin sensitivity and n = 9 for PDCa. *P < 0.05 from pre-fast; **P < 0.01 from pre-fast; #P < 0.05 from 48 h fast.
Skeletal muscle PDCa activity and PDK expression
Regulation of the activity of PDC in skeletal muscle is of critical importance in the integration of CHO and fat metabolism. Starvation decreased skeletal muscle PDCa activity by 37 ± 9% after 24 h (from 0.87 ± 0.15 to 0.56 ± 0.13 mmol min−1 (kg wet weight)−1; P < 0.05) and 51 ± 7% after 48 h (to 0.44 ± 0.13 mmol min−1 (kg wet weight)−1; P < 0.01) (Fig. 1B). Although refeeding increased PDCa activity, there was a tendency for PDCa to be lower after 24 h refeeding (0.62 ± 0.10 mmol min−1 (kg wet weight)−1,P = 0.098) when compared with the pre-starvation value.
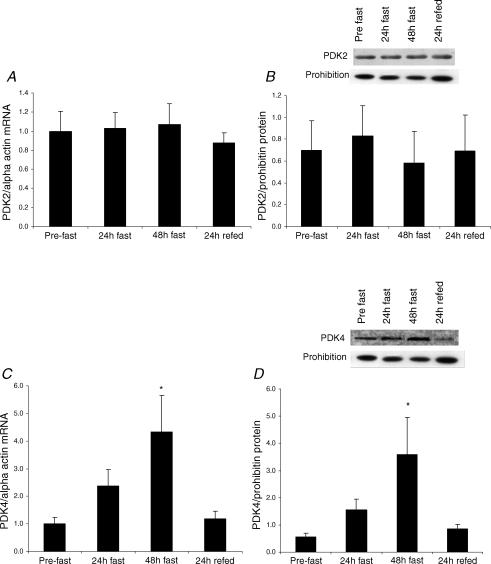
As PDK, the kinase that inactivates PDC, has been shown to be a major contributor to the long-term control of the complex, we measured the expression of all four isoforms of PDK identified in human skeletal muscle. Starvation increased muscle PDK4 mRNA content by 2.9-fold after 24 h and 4.3-fold after 48 h (P < 0.05) (Fig. 2C). Similarly, PDK4 protein expression increased by 3.5-fold after 48 h (P < 0.05) (Fig. 2D). Refeeding completely reversed these responses. In contrast, there was no effect of starvation or refeeding on mRNA or protein expression of PDK2, the other major isoform in skeletal muscle (Fig. 2A and B). PDK1 and 3 were found to be expressed at very low levels and not affected by starvation or refeeding (data not shown).
Figure 2. PDK2 and PDK4 mRNA (A and C) and protein (B and D) responses to 48 h starvation (fast) and 24 h refeeding (refed) with a high-CHO diet.
Values are mean ± s.e.m.; n = 10 for mRNA and n = 9 for protein data. *P < 0.05 from pre-fast.
Skeletal muscle SREBP1c, HKII, ChREBP and PK
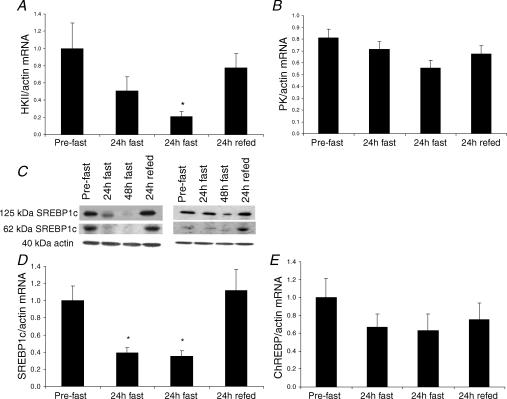
In order to understand the molecular basis for the reduction in muscle glucose uptake and utilization with starvation, we measured the expression of key transcription factors and genes associated with those processes. Starvation decreased muscle HKII mRNA content by 2-fold after 24 h and 5-fold after 48 h (P < 0.05) (Fig. 3A). Similarly, SREBP1c mRNA expression decreased by 2.5-fold after both 24 h and 48 h starvation (P < 0.05) (Fig. 3D). Refeeding completely reversed these responses. Due to limited amount of muscle tissue we could only perform SREBP1c protein analysis on two subjects, and the representative Western blots are shown in Fig. 3C. As expected, the changes in SREBP-1c mRNA were accompanied by a corresponding change in the amount of both precursor and mature forms of SREBP-1c protein (Fig. 3C). There was a trend for both ChREBP (P = 0.071) and PK (P = 0.087) mRNA content to decrease after 48 h starvation, but it failed to reach significance in either case (Fig. 3E and B, respectively).
Figure 3. HKII (A), PK (B), SREBP1c (D) and ChREBP (E) mRNA responses to 48 h starvation (fast) and 24 h refeeding (refed) with a high-CHO diet.
Two representative Western blots of SREBP1c protein are shown in Fig. 3C. Values are mean ± s.e.m.; n = 10 for all mRNA data. *P < 0.05 from pre-fast.
Skeletal muscle PPARs, PGC1α, CD36, CPT1 and LCAD
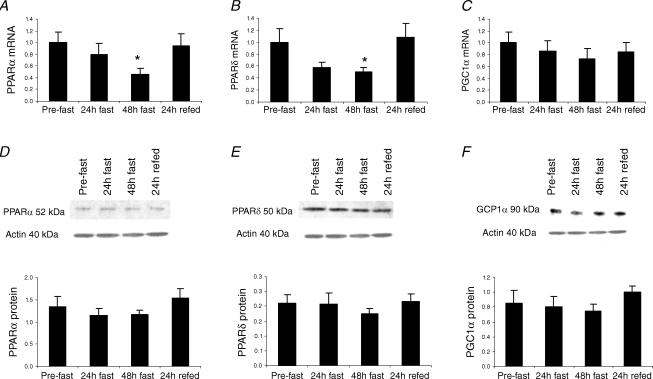
Since the starvation-induced shift in substrate utilization from CHO to fat might be a PPAR-dependent process, we measured the expression of PPARα and PPARδ, the most abundant isoforms in human skeletal muscle, and their coactivator PGC1α. Figure 4 shows the PPARα, PPARδ and PGC1α mRNA (Fig. 4A–C) and protein (Fig. 4D–F) responses to 48 h starvation and 24 h refeeding with a high CHO diet. There was no effect of starvation or refeeding on gene and protein expression of PGC1α (Fig. 4C and F), although it should be noted that seven out of the ten subjects showed a decrease in PGC1α mRNA expression with starvation. Interestingly, PPARα and PPARδ mRNA content decreased by ∼2-fold after 48 h starvation (P < 0.05) and returned to basal levels upon refeeding (Fig. 4A and B). In contrast, PPARα and PPARδ protein levels were unaffected by starvation or refeeding (Fig. 4D and E).
Figure 4. PPARα, PPARδ and PGC1α mRNA (A–C) and protein (D–F) responses to 48 h starvation (fast) and 24 h refeeding (refed) with a high-CHO diet.
Values are mean ± s.e.m.; n = 10 for all mRNA data, n = 8 for PPARα protein and n = 9 for PPARδ and PGC1α protein data. *P < 0.05 from pre-fast.
As fatty acid translocase (CD36, a transmembrane putative transporter of long-chain fatty acids), carnitine palmitoyltransferase 1 (CPT1, which catalyses the rate-limiting step in the transport of long-chain fatty acids into mitochondria) and long-chain acyl-CoA dehydrogenase (LCAD, which catalyses the first reaction in β-oxidation) are known transcriptional targets for both PPARα and PPARδ, we measured their mRNA expression as an index of PPAR activity in response to starvation. There was no effect of starvation or refeeding on mRNA expression of CD36 (pre-fast: 1.00 ± 0.14 arbitrary units (AU), 24 h fast: 0.99 ± 0.18 AU, 48 h fast: 0.95 ± 0.18 AU; 24 h refed: 1.04 ± 0.21 AU), CPT1 (pre-fast: 1.00 ± 0.08 AU, 24 h fast: 1.04 ± 0.10 AU, 48 h fast: 1.06 ± 0.09 AU; 24 h refed: 0.86 ± 0.10 AU) and LCAD (pre-fast: 1.00 ± 0.15 AU, 24 h fast: 1.03 ± 0.14 AU, 48 h fast: 0.99 ± 0.16 AU; 24 h refed: 1.10 ± 0.17 AU) in skeletal muscle.
Skeletal muscle insulin-signalling proteins
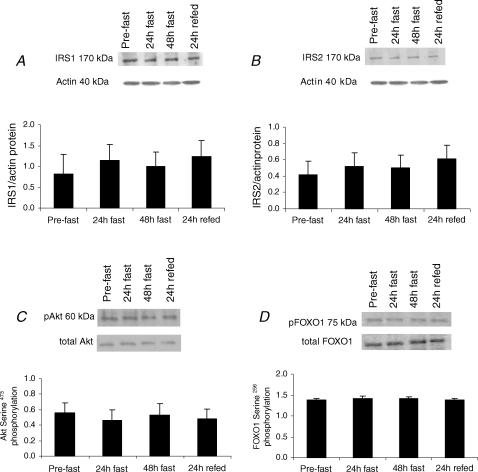
Starvation is known to decrease circulating levels of insulin, which is an important regulator of SREBP1c, HKII and PDK4 expression. In order to elucidate the role of insulin signalling in the transcriptional modulations in these key regulators of glucose utilization in human skeletal muscle, we measured the expression and phosphorylation states of Akt and the forkhead transcription factor FOXO1, a direct substrate for Akt and a known transcriptional activator of PDK4. The expression of IRS1 and IRS2, upstream molecules in the insulin-signalling pathway, were also measured. There was no effect of starvation or refeeding on skeletal muscle total protein Akt expression and serine473 phosphorylation (Fig. 5C), FOXO1 mRNA (data not shown), total protein expression and serine256 phosphorylation (Fig. 5D), and protein levels of IRS1 (Fig. 5A) and IRS2 (Fig. 5B).
Figure 5. Skeletal muscle IRS1 (A) and IRS2 (B) protein, and Akt serine473 phosphorylation (C) and FOXO1 serine256 phosphorylation (D) responses to 48 h starvation (fast) and 24 h refeeding (refed) with a high-CHO diet.
Akt and FOXO1 serine phosphorylations were determined using specific anti-phospho-antibodies and are expressed relative to total Akt and FOXO1 protein content (determined after stripping and reprobing the immunoblots for protein expression). Values are mean ± s.e.m.; n = 9.
Muscle fibre type composition
In order to evaluate the effect of interindividual differences in muscle fibre type composition on the extent of the metabolic and molecular responses to starvation in our subjects, we determined the content of the slow and fast MHC in their vastus lateralis. These results cannot be used to determine the percentage of slow and fast fibres in the muscle biopsy, but do indicate the range of the slow- and fast-twitch fibres between subjects. There was low interindividual variability in the MHC-s content (mean ± s.d. of 0.21 ± 0.06 AU, range 0.12–0.28) and modest variability in the MHC-f content (0.47 ± 0.20 AU, range 0.17–0.75) in the vastus lateralis. No correlation was observed between the MHC-s or MHC-f contents and the starvation-induced changes in whole-body insulin sensitivity, muscle PDCa activity and gene and protein expression of key metabolic enzymes and transcription factors.
Discussion
Previous studies on humans from our laboratory using insulin clamps and stable isotopes demonstrated a reduction in insulin sensitivity and a shift in basal and insulin-stimulated substrate utilization from CHO to fat after 36–72 h of starvation (Mansell & Macdonald, 1990; Webber et al. 1994). In the basal state after a 48 h fast, forearm muscle demonstrated a reduced glucose uptake and markedly increased glycerol output, indicating increased fat utilization. During insulin infusion there was marked reduction in glucose uptake by the forearm muscle, indicating profound insulin resistance (Mansell & Macdonald, 1990). However, the molecular mechanisms underlying the effects of starvation on insulin resistance were not examined and remain unclear. The findings from the present study confirmed the starvation-induced development of insulin resistance and indicated that in healthy humans there is sufficient capacity in skeletal muscle to utilize the substantial increase in fatty acid influx that occurs during starvation, but adaptive changes in gene expression are required to switch off oxidative glucose disposal. The increased expression of PDK4 in skeletal muscle appears to be an important adaptation to starvation, and other metabolic conditions characterized by elevated lipid metabolism, which contributes to the long-term control of PDC activity and thus downregulation of CHO oxidation under those conditions (Bowker-Kinley et al. 1998; Peters et al. 2001). Of the four isoenzymes that have been identified in humans, PDK4 has been shown to exhibit low responsiveness to short-term metabolic regulators (e.g. pyruvate, acetyl-CoA and NADH) whereas an increase in PDK4 mRNA and protein expression was observed in starvation and diabetes in animal models (Wu et al. 1998, 1999). In humans, starvation was also shown to upregulate PDK4 mRNA expression in skeletal muscle after 20 h (Pilegaard et al. 2003) and 40 h (Spriet et al. 2004), but no measurements of PDK4 protein expression were made in those studies. The present study confirmed the starvation-induced upregulation in PDK4 mRNA expression, and additionally showed a decrease in PDCa activity and a marked increase in PDK4 protein expression in human skeletal muscle, a response which was reversed by refeeding with a CHO-rich diet. Since PDC is the rate-limiting step in glucose oxidation, these responses provide the adaptive mechanisms within skeletal muscle underlying the starvation-induced reduction in CHO oxidation demonstrated in our previous studies (Mansell & Macdonald, 1990; Webber et al. 1994).
Increased circulating FFAs have been proposed as the mediator of starvation-induced upregulation of PDK4 expression, and it has been suggested that this might be a PPAR-dependent process (Wu et al. 1999; Huang et al. 2002). The PPARs are nuclear hormone receptors which regulate transcription of genes involved in fat oxidation and storage, and certain long-chain fatty acids, in particular polyunsaturated fatty acids, are potent activators for both PPARα and PPARδ (Forman et al. 1997). Administration of selective PPARδ agonists to mice fed a high-fat diet ameliorated weight gain and insulin resistance and increased fat oxidation in skeletal muscle, whereas in genetically obese mice it improved glucose tolerance (Tanaka et al. 2003). There is also evidence that feeding rats with WY-14, 643, a selective agonist for PPARα, induces a large increase in PDK4 expression (Wu et al. 1999). However, the starvation-induced increase in PDK4 expression remains intact in PPARα-null mice suggesting that upregulation of PDK4 expression in skeletal muscle does not require the activation of PPARα (Holness et al. 2002). In support of this, expression of PPARα in skeletal muscle is not affected by starvation in humans (Spriet et al. 2004).
The present study extends those findings by showing that the response of skeletal muscle PDK4 expression to starvation and subsequent refeeding in healthy humans is not mediated by an increase in PPARα or PPARδ protein expression. Interestingly, PPARα and PPARδ mRNA content actually decreased after 48 h of starvation and returned to basal levels upon refeeding, whereas protein levels were unaffected. This is in agreement with some animal studies that have observed a decrease in the mRNA levels of both PPARα and PPARδ during starvation in rat gastocnemius muscle (de Lange et al. 2004) and rat heart (Van der Lee et al. 2001), but not other studies which showed no effect of starvation on PPARα in rats (Escher et al. 2001) and mice (Holst et al. 2003). Post-translational regulation of PPAR activity, such as phosphorylation and ligand activation (Shalev et al. 1996; Forman et al. 1997), can mediate the increase in fat oxidation during starvation. However, given that there were no changes in gene expression of key transcriptional targets for PPARs that mediate fat utilization, namely CD36, CPT1 and LCAD, it would appear that changes in PPARα or PPARδ activity are not likely to mediate the adaptive response to starvation in human skeletal muscle. Similarly, no effect of starvation or refeeding on gene and protein expression of PGC1α in human skeletal muscle was observed. PGC1α is a transcription factor involved in a number of important biological processes including mitochondrial biogenesis and respiration, fatty acid oxidation, hepatic gluconeogenesis and glucose transport (Puigserver & Spiegelman, 2003), and is a potent coactivator for PPARs in skeletal muscle (Tanaka et al. 2003; Wang et al. 2003). PGC1α expression has previously been reported to be reduced in the skeletal muscle of insulin-resistant and type 2 diabetic patients (Mootha et al. 2003; Patti et al. 2003). However, PGC1α does not appear to be involved in the development of insulin resistance in fasted humans.
PDK4 expression has been shown to be regulated by insulin (Huang et al. 2002; Kwon et al. 2004). In rat hepatoma cells, insulin suppresses PDK4 through Akt-mediated inactivation of FOXO1 and FOXO3 transcription factors (Kwon et al. 2004). Both FOXOs can bind directly to the promoter region of the PDK4 gene, and in fasting mice induction of PDK4 mRNA expression is associated with an increase in the mRNA expression of both FOXO1 and FOXO3, and an increase in FOXO1 protein expression and phosphorylation at serine256 (Furuyama et al. 2003). In the present study, serum insulin concentrations were reduced after 48 h of starvation and returned to pre-starvation levels following refeeding. In contrast to findings from previous animal and in vitro studies, the mRNA and protein expression of FOXO1 in human skeletal muscle was unaffected by starvation or refeeding. Furthermore, the basal level of phosphorylation of FOXO1 at serine256 was also unaffected by starvation or refeeding. FOXO1 phosphorylation is increased by insulin, which acts via Akt. When Akt total and phosphorylated at serine473 protein levels were measured, no significant changes were observed with fasting and refeeding. Also we did not observe any changes in protein expression of IRS1 and IRS2. Interestingly, in a recent human study low-rate lipid infusion for 24 h in the absence of insulin infusion induced insulin resistance without impairment of either basal or insulin-stimulated phosphorylation of IRS1 and Akt (Storgaard et al. 2004). Thus, in contrast to results from animal and in vitro studies, the findings from the present study indicate that upregulation of skeletal muscle PDK4 expression in fasted humans may occur in a novel manner distinct, at least in part, from the PPARs and Akt/FOXO1 pathways.
Our finding that during starvation in healthy humans, unlike rodents, regulation of fat metabolism does not require an adaptive response at the level of gene expression might not be totally unexpected if one considers the number of differences that exist between rodent and human skeletal muscle metabolism. For example, humans have relatively lower basal metabolic rate and rely more than rodents on fat metabolism to sustain their daily energy expenditure (Suarez et al. 2004). This, combined with a very high maximum rate for energy utilization from fatty acids when compared with the energy requirement of resting skeletal muscle (0.40 versus 0.07 mol ATP min−1, respectively) (van der Vusse & Reneman, 1996), may explain its capacity to substantially increase fatty acid influx and oxidation without requiring an adaptive response at the level of gene expression in healthy humans.
In this study, we sought to examine in healthy humans the effect of nutrient availability on expression of SREBP-1c and ChREBP, two transcription factors that mediate the effects of insulin and glucose on the regulation of key genes (HKII and PK) associated with glucose metabolism in skeletal muscle. A few studies to date have investigated the functional role of SREBP-1c in skeletal muscle (Ducluzeau et al. 2001; Guillet-Deniau et al. 2002; Ikeda et al. 2002; Muscat et al. 2002; Sewter et al. 2002), but only two have used human subjects (Ducluzeau et al. 2001; Sewter et al. 2002). However, the role and regulation of ChREBP in human skeletal muscle have not been studied. Animal studies have shown that high-CHO diets activate ChREBP, and high-fat diets inhibit its activity in liver (Yamashita et al. 2001). Interestingly, there was no effect of starvation on ChREBP gene expression in adipose tissue, whereas refeeding with a high CHO diet resulted in its upregulation (He et al. 2004). In the present study, starvation tended to decrease the expression of ChREBP and PK, suggesting that changes in nutrient availability may have the same effect on human skeletal muscle ChREBP gene expression as that reported in liver and adipose tissue in animals. Starvation was also accompanied by a decrease in expression of SREBP-1c and HKII in human skeletal muscle, and expression of both genes was normalized by 24 h refeeding with a CHO-rich diet. Since insulin sensitivity had not returned to normal after refeeding, it is possible that nutrient availability and/or flux rather than returning insulin sensitivity may at least in part be responsible for the restoration of SREBP-1c and HKII expression following refeeding. It is likely that the latter responses, in light of the tendency for PDCa to be lower after 24 h refeeding when compared with the pre-starvation value, may facilitate an increase in non-oxidative glucose disposal during the refeeding period. Although muscle glycogen levels were not measured in the present study, previous studies in humans have shown that, unlike liver glycogen, which is rapidly depleted during fasting, skeletal muscle glycogen is quite resistant to changes, with only ∼20% reduction observed after 48 h of starvation (Hultman & Bergstrom, 1967). If a similar reduction in muscle glycogen content occurred in the present study, it is likely that 24 h of refeeding (which provided ∼600 g of CHO intake) would have led to complete restoration of muscle glycogen content within that period, and thus dissociation of the non-oxidative glucose disposal from the incomplete reversal of insulin sensitivity.
Circulating insulin concentrations did not decline until after the first 24 h of starvation when SREBP1c and HKII expression had already decreased. However, there was a tendency for plasma FFA to increase and blood glucose to decrease at 24 h of starvation, and these changes became significant after 48 h. Thus, it is possible that in the in vivo setting of our experiment, changes in circulating FFAs (Xu et al. 1999, 2002) and glucose (Dentin et al. 2005) may be important negative and positive regulators, respectively, of SREBP1c expression in human skeletal muscle during starvation. During refeeding with a CHO-rich diet, the increased glucose availability would have been accompanied by increased serum insulin and decreased circulating FFA levels. In agreement with a previous animal study (Commerford et al. 2004), these changes during refeeding were accompanied by an upregulation in SREBP-1c expression in human skeletal muscle. However, the independent, if any, effects of insulin, glucose and FFA on SREBP-1c expression in skeletal muscle are not clear. Insulin-clamp studies performed in humans (Ducluzeau et al. 2001; Sewter et al. 2002) have shown that hyperinsulinaemia increases skeletal muscle SREBP1c mRNA expression. However, as plasma FFA levels are suppressed during insulin clamps it is not possible to delineate the separate effects of elevated insulin and decreased FFA on SREBP1c gene expression from those studies. Interestingly, intralipid infusion for 3 h, designed to elevate plasma FFA concentrations to levels seen after starvation, tended to increase rather than decrease SREBP1c expression in human skeletal muscle (authors' unpublished observation). However, during both insulin clamps and intralipid infusions there is an increased supply of FFA and/or glucose (energy surplus), whereas starvation is a state of energy restriction. Thus, the difference in nutritional status between these in vivo settings may act to mask the effect of plasma FFA and glucose availability on SREBP1c gene expression.
The relative muscle fibre type composition may influence the extent of skeletal muscle adaptive responses to starvation. Indeed, animal studies have shown that the reduction in whole-body glucose disposal during starvation is largely due to a decline in glucose uptake and utilization in oxidative slow-twitch skeletal muscles, which also happen to exhibit high rates of glucose utilization in the fed state (Holness & Sugden, 1990). More recently, starvation was shown to increase PDK4 protein expression in both slow-oxidative and fast-oxidative-glycolytic rat skeletal muscles, with a greater response observed in the latter type of muscle (Holness et al. 2002). The results from the present study showed no association between the MHC-s and MHC-f contents in the vastus lateralis (which were used as indices of the abundance of slow- and fast-twitch fibres, respectively) and the starvation-induced changes in whole-body insulin sensitivity, muscle PDCa activity and gene and protein expression of key metabolic enzymes and transcription factors, possibly due to the low/modest variability in relative muscle fibre type composition of the vastus lateralis of our subjects. Thus, it appears that under the conditions of this study the relative muscle fibre type composition did not appear to influence the metabolic and molecular responses to starvation in healthy humans.
In summary, the results from the present study demonstrated that during starvation in healthy humans, unlike rodents, utilization of the fatty acids after the substantial increase in their availability does not require an adaptive response at the level of gene expression. On the other hand, transcriptional regulation of key genes in glucose uptake (SREBP1c, HKII) and oxidation (PDK4) are required to switch off glucose disposal. In contrast to results from animal and in vitro studies, these molecular adaptations in response to fasting occurred in a manner distinct from the PPAR pathways and were not associated with changes in the phosphorylation and expression of key proteins in the insulin-signalling pathway, suggesting that changes in intracellular substrate metabolism may be involved in the molecular regulation of CHO utilization in skeletal muscle of fasted humans. Further studies to investigate the absolute relationship between muscle substrate utilization and regulatory gene expression would be valuable. The differential regulation of metabolic genes and transcription factors involved in CHO and fat utilization in human skeletal muscle during starvation may represent an important component of the molecular basis of the development of insulin resistance in other metabolic conditions and diseases characterized by elevated lipid metabolism and/or energy restriction.
Acknowledgments
This work was supported by a grant from the Biotechnology and Biological Sciences Research Council (BBSRC) of UK (grant 42/D1563 to K. Tsintzas) and a block grant to A. Bennett from FRAME.
References
- Bergstrom J. Muscle electrolytes in man. Scand J Clin Lab Invest. 1962;68:1–100. [Google Scholar]
- Bizeau ME, MacLean PS, Johnson GC, Wei Y. Skeletal muscle sterol regulatory element binding protein-1c decreases with food deprivation and increases with feeding in rats. J Nutr. 2003;133:1787–1792. doi: 10.1093/jn/133.6.1787. [DOI] [PubMed] [Google Scholar]
- Bjorkman O, Eriksson LS. Influence of a 60-hour fast on insulin-mediated splanchnic and peripheral glucose metabolism in humans. J Clin Invest. 1985;76:87–92. doi: 10.1172/JCI111982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonora E, Moghetti P, Zancanaro C, Cigolini M, Querena M, Cacciatori V, Corgnati A, Muggeo M. Estimates of in vivo insulin action in man: comparison of insulin tolerance tests with euglycemic and hyperglycemic glucose clamp studies. J Clin Endocrinol Metab. 1989;68:374–378. doi: 10.1210/jcem-68-2-374. [DOI] [PubMed] [Google Scholar]
- Bowker-Kinley MM, Davis WI, Wu P, Harris RA, Popov KM. Evidence for existence of tissue-specific regulation of the mammalian pyruvate dehydrogenase complex. Biochem J. 1998;329(1):191–196. doi: 10.1042/bj3290191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Commerford SR, Peng L, Dube JJ, O'Doherty RM. In vivo regulation of SREBP-1c in skeletal muscle: effects of nutritional status, glucose, insulin, and leptin. Am J Physiol Regul Integr Comp Physiol. 2004;287:R218–R227. doi: 10.1152/ajpregu.00377.2003. [DOI] [PubMed] [Google Scholar]
- Constantin-Teodosiu D, Carlin JI, Cederblad G, Harris RC, Hultman E. Acetyl group accumulation and pyruvate dehydrogenase activity in human muscle during incremental exercise. Acta Physiol Scand. 1991;143:367–372. doi: 10.1111/j.1748-1716.1991.tb09247.x. [DOI] [PubMed] [Google Scholar]
- de Lange P, Ragni M, Silvestri E, Moreno M, Schiavo L, Lombardi A, Farina P, Feola A, Goglia F, Lanni A. Combined cDNA array/RT-PCR analysis of gene expression profile in rat gastrocnemius muscle: relation to its adaptive function in energy metabolism during fasting. Faseb J. 2004;18:350–352. doi: 10.1096/fj.03-0342fje. [DOI] [PubMed] [Google Scholar]
- Dentin R, Girard J, Postic C. Carbohydrate responsive element binding protein (ChREBP) and sterol regulatory element binding protein-1c (SREBP-1c): two key regulators of glucose metabolism and lipid synthesis in liver. Biochimie. 2005;87:81–86. doi: 10.1016/j.biochi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Ducluzeau PH, Perretti N, Laville M, Andreelli F, Vega N, Riou JP, Vidal H. Regulation by insulin of gene expression in human skeletal muscle and adipose tissue. Evidence for specific defects in type 2 diabetes. Diabetes. 2001;50:1134–1142. doi: 10.2337/diabetes.50.5.1134. [DOI] [PubMed] [Google Scholar]
- Escher P, Braissant O, Basu-Modak S, Michalik L, Wahli W, Desvergne B. Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology. 2001;142:4195–4202. doi: 10.1210/endo.142.10.8458. [DOI] [PubMed] [Google Scholar]
- Fery F, Plat L, Balasse EO. Effect of fasting on the intracellular metabolic partition of intravenously infused glucose in humans. Am J Physiol. 1999;277:E815–E823. doi: 10.1152/ajpendo.1999.277.5.E815. [DOI] [PubMed] [Google Scholar]
- Forman BM, Chen J, Evans RM. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc Natl Acad Sci U S A. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Kitayama K, Yamashita H, Mori N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J. 2003;375:365–371. doi: 10.1042/BJ20030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosmain Y, Lefai E, Ryser S, Roques M, Vidal H. Sterol regulatory element-binding protein-1 mediates the effect of insulin on hexokinase II gene expression in human muscle cells. Diabetes. 2004;53:321–329. doi: 10.2337/diabetes.53.2.321. [DOI] [PubMed] [Google Scholar]
- Guillet-Deniau I, Mieulet V, Le Lay S, Achouri Y, Carre D, Girard J, Foufelle F, Ferre P. Sterol regulatory element binding protein-1c expression and action in rat muscles: insulin-like effects on the control of glycolytic and lipogenic enzymes and UCP3 gene expression. Diabetes. 2002;51:1722–1728. doi: 10.2337/diabetes.51.6.1722. [DOI] [PubMed] [Google Scholar]
- He Z, Jiang T, Wang Z, Levi M, Li J. Modulation of carbohydrate response element-binding protein gene expression in 3T3-L1 adipocytes and rat adipose tissue. Am J Physiol Endocrinol Metab. 2004;287:E424–E430. doi: 10.1152/ajpendo.00568.2003. [DOI] [PubMed] [Google Scholar]
- Holness MJ, Bulmer K, Gibbons GF, Sugden MC. Up-regulation of pyruvate dehydrogenase kinase isoform 4 (PDK4) protein expression in oxidative skeletal muscle does not require the obligatory participation of peroxisome-proliferator-activated receptor alpha (PPARalpha) Biochem J. 2002;366:839–846. doi: 10.1042/BJ20020754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holness MJ, Sugden MC. Glucose utilization in heart, diaphragm and skeletal muscle during the fed-to-starved transition. Biochem J. 1990;270:245–249. doi: 10.1042/bj2700245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst D, Luquet S, Nogueira V, Kristiansen K, Leverve X, Grimaldi PA. Nutritional regulation and role of peroxisome proliferator-activated receptor delta in fatty acid catabolism in skeletal muscle. Biochim Biophys Acta. 2003;1633:43–50. doi: 10.1016/s1388-1981(03)00071-4. [DOI] [PubMed] [Google Scholar]
- Huang B, Wu P, Bowker-Kinley MM, Harris RA. Regulation of pyruvate dehydrogenase kinase expression by peroxisome proliferator-activated receptor-alpha ligands, glucocorticoids, and insulin. Diabetes. 2002;51:276–283. doi: 10.2337/diabetes.51.2.276. [DOI] [PubMed] [Google Scholar]
- Hultman E, Bergstrom J. Muscle glycogen synthesis in relation to diet studied in normal subjects. Acta Med Scand. 1967;182:109–117. doi: 10.1111/j.0954-6820.1967.tb11504.x. [DOI] [PubMed] [Google Scholar]
- Ikeda S, Miyazaki H, Nakatani T, Kai Y, Kamei Y, Miura S, Tsuboyama-Kasaoka N, Ezaki O. Up-regulation of SREBP-1c and lipogenic genes in skeletal muscles after exercise training. Biochem Biophys Res Commun. 2002;296:395–400. doi: 10.1016/s0006-291x(02)00883-5. [DOI] [PubMed] [Google Scholar]
- Kwon HS, Huang B, Unterman TG, Harris RA. Protein kinase B-alpha inhibits human pyruvate dehydrogenase kinase-4 gene induction by dexamethasone through inactivation of FOXO transcription factors. Diabetes. 2004;53:899–910. doi: 10.2337/diabetes.53.4.899. [DOI] [PubMed] [Google Scholar]
- Lee CH, Olson P, Evans RM. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology. 2003;144:2201–2207. doi: 10.1210/en.2003-0288. [DOI] [PubMed] [Google Scholar]
- Mansell PI, Macdonald IA. The effect of starvation on insulin-induced glucose disposal and thermogenesis in humans. Metabolism. 1990;39:502–510. doi: 10.1016/0026-0495(90)90009-2. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Muscat GE, Wagner BL, Hou J, Tangirala RK, Bischoff ED, Rohde P, Petrowski M, Li J, Shao G, Macondray G, Schulman IG. Regulation of cholesterol homeostasis and lipid metabolism in skeletal muscle by liver X receptors. J Biol Chem. 2002;277:40722–40728. doi: 10.1074/jbc.M206681200. [DOI] [PubMed] [Google Scholar]
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters SJ, Harris RA, Wu P, Pehleman TL, Heigenhauser GJ, Spriet LL. Human skeletal muscle PDH kinase activity and isoform expression during a 3-day high-fat/low-carbohydrate diet. Am J Physiol Endocrinol Metab. 2001;281:E1151–E1158. doi: 10.1152/ajpendo.2001.281.6.E1151. [DOI] [PubMed] [Google Scholar]
- Pilegaard H, Saltin B, Neufer PD. Effect of short-term fasting and refeeding on transcriptional regulation of metabolic genes in human skeletal muscle. Diabetes. 2003;52:657–662. doi: 10.2337/diabetes.52.3.657. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- Randle PJ, Garland PB, Hales CN, Newsholme EA. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet. 1963;1:785–789. doi: 10.1016/s0140-6736(63)91500-9. [DOI] [PubMed] [Google Scholar]
- Sazili AQ, Parr T, Sensky PL, Jones SW, Bardsley RG, Buttery PJ. The relationship between slow and fast myosin heavy chain content, calpastatin and meat tenderness in different ovine skeletal muscles. Meat Sci. 2005;69:17–25. doi: 10.1016/j.meatsci.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Sewter C, Berger D, Considine RV, Medina G, Rochford J, Ciaraldi T, Henry R, Dohm L, Flier JS, O'Rahilly S, Vidal-Puig AJ. Human obesity and type 2 diabetes are associated with alterations in SREBP1 isoform expression that are reproduced ex vivo by tumor necrosis factor-alpha. Diabetes. 2002;51:1035–1041. doi: 10.2337/diabetes.51.4.1035. [DOI] [PubMed] [Google Scholar]
- Shalev A, Siegrist-Kaiser CA, Yen PM, Wahli W, Burger AG, Chin WW, Meier CA. The peroxisome proliferator-activated receptor alpha is a phosphoprotein: regulation by insulin. Endocrinology. 1996;137:4499–4502. doi: 10.1210/endo.137.10.8828512. [DOI] [PubMed] [Google Scholar]
- Spriet LL, Tunstall RJ, Watt MJ, Mehan KA, Hargreaves M, Cameron-Smith D. Pyruvate dehydrogenase activation and kinase expression in human skeletal muscle during fasting. J Appl Physiol. 2004;96:2082–2087. doi: 10.1152/japplphysiol.01318.2003. [DOI] [PubMed] [Google Scholar]
- Storgaard H, Jensen CB, Bjornholm M, Song XM, Madsbad S, Zierath JR, Vaag AA. Dissociation between fat-induced in vivo insulin resistance and proximal insulin signaling in skeletal muscle in men at risk for type 2 diabetes. J Clin Endocrinol Metab. 2004;89:1301–1311. doi: 10.1210/jc.2003-031243. [DOI] [PubMed] [Google Scholar]
- Suarez RK, Darveau CA, Childress JJ. Metabolic scaling: a many-splendoured thing. Comp Biochem Physiol B Biochem Mol Biol. 2004;139:531–541. doi: 10.1016/j.cbpc.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Sugden MC, Howard RM, Munday MR, Holness MJ. Mechanisms involved in the coordinate regulation of strategic enzymes of glucose metabolism. Adv Enzyme Regul. 1993;33:71–95. doi: 10.1016/0065-2571(93)90010-b. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Yamamoto J, Iwasaki S, Asaba H, Hamura H, Ikeda Y, Watanabe M, Magoori K, Ioka RX, Tachibana K, Watanabe Y, Uchiyama Y, Sumi K, Iguchi H, Ito S, Doi T, Hamakubo T, Naito M, Auwerx J, Yanagisawa M, Kodama T, Sakai J. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc Natl Acad Sci U S A. 2003;100:15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Lee KA, Willemsen PH, Samec S, Seydoux J, Dulloo AG, Pelsers MM, Glatz JF, Van der Vusse GJ, Van Bilsen M. Fasting-induced changes in the expression of genes controlling substrate metabolism in the rat heart. J Lipid Res. 2001;42:1752–1758. [PubMed] [Google Scholar]
- van der Vusse GJ, Reneman RS. Handbook of Physiology, section 12, Exercise: Regulation and Integration of Multiple Systems. New York: Oxford Press; 1996. Lipid metabolism in muscle. [Google Scholar]
- Wang YX, Lee CH, Tiep SYuRT, Ham J, Kang H, Evans RM. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell. 2003;113:159–170. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- Webber J, Taylor J, Greathead H, Dawson J, Buttery PJ, Macdonald IA. Effects of fasting on fatty acid kinetics and on the cardiovascular, thermogenic and metabolic responses to the glucose clamp. Clin Sci. 1994;87:697–706. doi: 10.1042/cs0870697. [DOI] [PubMed] [Google Scholar]
- Wibom R, Hultman E. ATP production rate in mitochondria isolated from microsamples of human muscle. Am J Physiol. 1990;259:E204–E209. doi: 10.1152/ajpendo.1990.259.2.E204. [DOI] [PubMed] [Google Scholar]
- Wu P, Inskeep K, Bowker-Kinley MM, Popov KM, Harris RA. Mechanism responsible for inactivation of skeletal muscle pyruvate dehydrogenase complex in starvation and diabetes. Diabetes. 1999;48:1593–1599. doi: 10.2337/diabetes.48.8.1593. [DOI] [PubMed] [Google Scholar]
- Wu P, Sato J, Zhao Y, Jaskiewicz J, Popov KM, Harris RA. Starvation and diabetes increase the amount of pyruvate dehydrogenase kinase isoenzyme 4 in rat heart. Biochem J. 1998;329:197–201. doi: 10.1042/bj3290197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Cho H, O'Malley S, Park JH, Clarke SD. Dietary polyunsaturated fats regulate rat liver sterol regulatory element binding proteins-1 and -2 in three distinct stages and by different mechanisms. J Nutr. 2002;132:3333–3339. doi: 10.1093/jn/132.11.3333. [DOI] [PubMed] [Google Scholar]
- Xu J, Nakamura MT, Cho HP, Clarke SD. Sterol regulatory element binding protein-1 expression is suppressed by dietary polyunsaturated fatty acids. A mechanism for the coordinate suppression of lipogenic genes by polyunsaturated fats. J Biol Chem. 1999;274:23577–23583. doi: 10.1074/jbc.274.33.23577. [DOI] [PubMed] [Google Scholar]
- Yamashita H, Takenoshita M, Sakurai M, Bruick RK, Henzel WJ, Shillinglaw W, Arnot D, Uyeda K. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc Natl Acad Sci U S A. 2001;98:9116–9121. doi: 10.1073/pnas.161284298. [DOI] [PMC free article] [PubMed] [Google Scholar]