Abstract
The cellular complexity of the brain is a major issue in the planning, execution and interpretation of microarray studies. Recent technical advances allow for high-throughput study of specific cell populations and circuits. Here we review representative examples of currently available methods that allow high resolution and specificity in brain microarray studies, while maintaining the goal of comprehensive, high-throughput analysis.
The notion that tissue complexity and cellular heterogeneity pose major challenges to the progress of microarray-powered research in neuroscience is now widely accepted (Geschwind, 2000; Mirnics & Pevsner, 2004; Arlotta et al. 2005; Dougherty & Geschwind, 2005; Coppola & Geschwind, 2006). Microarray platforms are capable of querying the whole known transcriptome on a single slide but, in many cases, most of the genes on the array are not detected, or not detected at a level allowing assessment of differential expression. Not only is the number of cell types in many CNS samples large, but the complexity of their interactions with other cells is enormous. When different cell types are analysed together in the same sample with a microarray platform, only the common features may be detected by the assay, whereas the specific profiles may be diluted. For example, certain signalling molecules may be differentially expressed in opposite directions in adjacent inhibitory and excitatory neurons or pathways, cancelling each other out at the whole tissue level in a microarray experiment. In addition, low abundance mRNAs coding for receptors or transcription factors expressed in a small percentage of cells may not be detected at all. Depending on the goal of the experiment, this may pose significant problems for the subsequent analysis and interpretation of the data and, while whole tissue experimentation still can play a major role, one needs to be wary of basing conclusions on what is not observed, given the potential type II error (e.g. Geschwind, 2000).
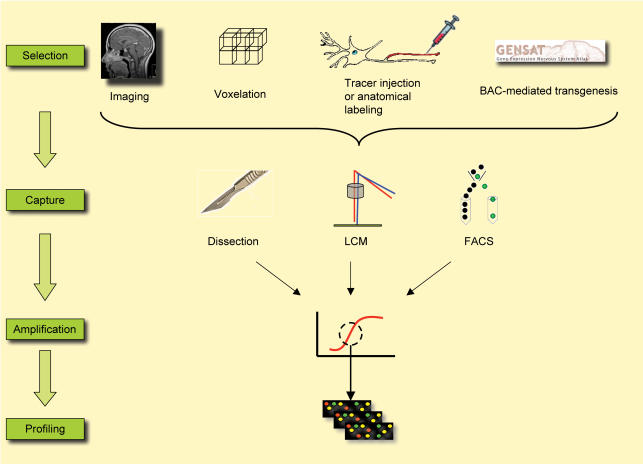
Technical advances now allow one to select experimental samples with complexity ranging from the regional to the cellular level. Still, RNA amplification is needed, adding potential sources of noise, and perhaps not providing a complete representation (Lobo et al. 2006; Nygaard & Hovig, 2006). Recent approaches to achieve increased sample specificity include microarray studies on specific regions or cells, using functional imaging methods to guide tissue selection, microdissection of specific nuclei or brain areas, and separation and sorting of individual cells based on fluorescent signals or other markers (recently reviewed in Nelson et al. 2006; Fig. 1). This review focuses on the currently available approaches to address molecular genomic profiling at the regional, circuit and cellular level.
Figure 1. Schematic illustrating the major methods currently available to address regional and cell specificity in human brain and experimental models.
In human brain, neuroanatomical methods can be used to guide dissection and laser-guided microdissection. Voxelation approaches allow visualization of 3D gene expression maps. In experimental models, in addition to image-based methods, cell specificity can be achieved with labelling methods, based on both neuroanatomical and genetic markers (e.g. http://www.ncbi.nlm.nih.gov/projects/gensat/). Such intrinsic labels can be used to guide dissection or FACS-based approaches. Most of these methods require an amplification step due to low RNA amounts, prior to gene expression profiling using microarrays. Many robust methods for such amplification are now widely available, making this a worthwhile approach.
Regional analysis
A first level of specificity in expression studies in the nervous system is at the level of distinct brain regions or nuclei. Although studies have been reported analysing RNA from whole rodent brain, for example, the most intuitive and productive approach in recent years has involved the selection of specific brain regions for microarray studies. This makes sense at one level, since brain regions and nuclei are functionally relevant divisions of the CNS. Genes and pathways involved in human disorders in human brain and mouse models (Mirnics et al. 2000, 2005), and region-specific genes in rodent (Sandberg et al. 2000; Zhao et al. 2001; Zirlinger et al. 2001; Funatsu et al. 2004; Zapala et al. 2005) and human and primate brain (Bunney et al. 2003; Evans et al. 2003; Khaitovich et al. 2004), have been successfully identified using this approach. Although whole-tissue analysis still results in the inevitable inclusion of heterogeneous cell types, which may limit detection of rare transcripts, it still allows detection of many relevant gene expression changes. As such, it remains a reasonable approach, especially when coupled with extensive downstream verification. Progress towards specificity in regional brain analysis has been made combining microarrays with in situ hybridization (ISH; Zhao et al. 2001) and functional neuroimaging methods (Lewandowski & Small, 2005).
For example, Zhao et al. (2001) were able to microdissect distinct hippocampal regions, profile them with microarrays, and confirm the regional specificity of the identified genes through ISH. They report distinct anatomical and functional boundaries based on gene expression, providing evidence that gene expression studies from anatomical regions can be the first step on the path to the identification of molecularly based subregions in the hippocampus. Using a similar approach, a subset of amydgala-specific genes was identified with microarrays, followed by ISH confirmation which showed boundaries of expression of many of these genes as corresponding to cytoarchitectonically defined subnuclei (Zirlinger et al. 2001). The same group went a step further by microdissecting the amygdaloid subregions, hybridizing the extracted RNA in an additional microarray experiment, and identifying genes specifically enriched in amygdaloid subnuclei (Zirlinger, 2003; Zirlinger & Anderson, 2003).
Neuroimaging approaches using MRI allow one to visualize distinct hippocampal subregions in living subjects (Small et al. 2000). In an elegant study using neuroanatomical data, Small et al. were able to guide microdissection for microarray analysis identifying the retromer trafficking complex as possibly involved in the pathogenesis of Alzheimer's disease (AD) (Lewandowski & Small, 2005; Small et al. 2005). With this approach, coupling a regional, hypothesis-driven dissection with a microarray-based assay, Small and colleagues have begun to address the key issue of regional vulnerability in neurodegenerative disorders such as AD.
Two innovative methods, voxelation and gene expression tomography, are conceptually close to neuroimaging (Singh & Smith, 2003). Voxelation employs high-throughput gene expression analysis of discrete brain microareas (voxels), followed by 3D reconstruction (Brown et al. 2002a), whereas gene expression tomography involves the analysis of parallel tomographic sections (Brown et al. 2002b) and 3D image reconstruction from these sections, similar to tomographic imaging techniques. Proof-of-principle applications of these techniques have been reported on both human (Brown et al. 2002a) and mouse (Brown et al. 2002c) brain. At low-resolution, this technique suffers from the same issues of macro-regional dissection, i.e. cellular heterogeneity and dilution of rare signals. However, higher resolution maps and new RNA amplification procedures will probably improve these promising tools in the near future, allowing en masse regional analysis of gene expression across the brain.
These methods all essentially couple microarrays with neuroanatomical visualization and dissection methods, and have proven powerful in providing a proof of principle for future studies aimed at (1) identifying genes and pathways of interest in disease with known regional vulnerability, and (2) further defining the molecular anatomy of cortical and subcortical subregions with distinct functional identities.
Neuronal populations and circuits
The study of neuronal populations and brain circuitry components is key to understanding the molecular basis of neuronal diversity and circuit function. Cell types with a distinct ‘identity’ can be classified based on expressed markers, or on histological/structural features. Once tagged, the target cells can be isolated and studied using a variety of methods, including patch-clamp, fluorescence-activated cell sorting (FACS), or laser capture microdissection (LCM).
Genetic markers
Specific neuronal populations can be genetically labelled using reporters expressed from the promoter of a cell-specific gene, or using BAC-mediated transgenesis. Gustincich et al. (2004) used a transgenic mouse line in which catecolaminergic retinal neurons were genetically marked and, after cell selection using a monoclonal antibody and patch-clamp, performed single-cell global amplification and microarray analysis, identifying a number of transcripts specific for this rare retinal cell type (Gustincich et al. 2004). Using genetically and retrogradely labelled mouse samples, Sugino et al. (2006) studied with microarrays the gene expression profile of ∼100 hand-sorted neurons from each of 12 neuronal populations in a tour-de-force study. Microarray data mostly confirmed the known classification based on expression markers and provided new markers for neuronal characterization, but also suggested that some neuronal populations identified by one marker were actually heterogeneous, supporting the role of microarray studies in characterizing neuronal types (Diaz, 2006; Sugino et al. 2006). This method – while high-resolution – may be hard to adapt to very high-throughput analysis of multiple samples and models, since it relies on manual dissociation and sorting. Further, these studies rely on small numbers of neurons, and it is likely that sample sizes of several hundred cells or more provide more comprehensive detection of low-abundance messages (Lobo et al. 2006).
BAC-mediated transgenesis permits region- and cell-specific reporter expression without altering the promoter region of the gene of interest (Heintz, 2000). In particular, the gene expression nervous system atlas (GENSAT) (Gong et al. 2003) is a repository of BAC-transgenic mice with hundreds of genetically labelled neuronal types. A conceptually simple way to obtain expression profiling of distinct neuronal populations involves the sorting of Green fluorescent protein (GFP)-labelled cell types prior to RNA extraction and microarray hybridization. Overcoming the technical issues related to tissue dissociation and RNA amplification, Lobo et al. (2006) were able to dissociate juvenile and adult striatonigral and striatopallidal neurons, do FACS-sort them based on the expression of known markers and, after RNA extraction and amplification, run microarrays. This approach allowed identification of numerous genes – with a significant overlap between postnatal day 20 (P20) and adult neurons – whose expression clearly distinguishes these two cell populations, including a lineage-specific transcription factor. Lobo et al. showed the functional significance of their work by showing specific disruption of striatonigral pathways in an Ebf1 knockout mouse. This study demonstrated the feasibility of such an approach in young and adult mice, and provided a clear proof-of-principle supporting the use of this technology (FACS-array) in studies addressing other neuronal populations. The GENSAT resource makes this approach scalable across a wide range of cell types. The main advantage of FACS-based sorting methods is the number of cells obtained: thousands of neurons can be quickly obtained per sort, so one is not limited to a few dozens or a hundred cells, which may affect the number of genes detected (Lobo et al. 2006). Furthermore, this method works in adult neurons, without induction of injury- or apoptosis-related genes, providing a higher-throughput alternative to current methods (Nelson et al. 2006).
In simpler organisms, labelling and FACS sorting can be coupled to cell culture to obtain homogeneous cell populations. In C. elegans, it is possible to dissociate embryos genetically expressing markers for specific cell types, culture dissociated embryonic cells (which undergo morphological differentiation in vitro), collect GFP-marked differentiated neurons in sufficient quantity to extract RNA, and perform gene expression studies (Christensen et al. 2002; Zhang et al. 2002). Using this technique, touch (Zhang et al. 2002), sensory (Colosimo et al. 2004), and motor (Cinar et al. 2005; Fox et al. 2005) neurons have been successfully labelled, FACS-sorted and studied using microarrays.
Histological/structural markers
Injection of neural tracers (such as fluorescent microspheres) in axonal terminals results in retrograde transport and cell body labelling. This approach selects neuronal populations based on their functionally relevant projections, in the absence of antigenic markers. In the first study based on sorting in mammals, Arlotta et al. (2005) injected fluorescent microspheres in the axonal projection fields of developing corticospinal motor neurons. After labelling of the cell bodies in the sensorimotor cortex by retrograde transport, they were able to dissociate and FACS-purify this specific cellular type in enough quantity to perform highly reproducible RNA amplification and expression profiling with microarrays (Arlotta et al. 2005). These investigations identified a number of transcripts involved in differentiation programs related to specific motor neuron subsets, based on their projections (Molyneaux et al. 2005) – a first step in molecularly defining functionally relevant circuiting.
Laser capture microdissection (LCM) (Emmert-Buck et al. 1996; Bohm et al. 2005) allows the specific and selective isolation of single cells, without disruption of the surrounding tissue. Fluoro-gold-labelled neurons can be used to visually guide LCM. For example, this technique has been used to mark dopaminergic (Yao et al. 2005) and motor (Cui et al. 2006) neurons. Using a similar approach, Lombardino et al. (2005) differentially marked (injecting dye-marked tracers) replaceable and non-replaceable projection neurons in the vocal centres of songbirds, isolated them through LCM, and compared their expression profile. Their results provided a possible link between cell renewal and neurodegeneration (Lombardino et al. 2005).
In a multifaceted approach, Diaz et al. (2002) achieved specificity by combining data from multiple conditions (whole cerebella, purified and cultured granule cells), multiple time points, and multiple models (wild-type and genetic mutant analysis) to dissect the genetic program of ponto-cerebellar system development. The data integration at the bioinformatics level – and a solid confirmation strategy with ISH – allowed them to draw a map of gene expression of this relatively simple developing system (Diaz et al. 2002). This work demonstrates how careful experimental design and detailed experimentation can lead to many functionally relevant insights.
Single-cell profiling
Intuitively, one could consider the single-cell level as the gold standard of gene expression studies: looking at a few or even one cell should yield the maximum specificity, and should allow one to classify cell types based on a molecular expression profile. Still, this apparent simplicity belies a number of technical issues, including increasing the number of experiments, cost, analytic burden, etc., which paradoxically increases the complexity of the experiments, as we discuss below.
In the past, single cells have been obtained with microdissection using a needle or a micromanipulator (O'Dell et al. 1998). Since the earliest studies in the field, the aim of many single-neuron analyses was to characterize the expression abnormalities associated with the neuropathological hallmarks of Alzheimer's disease, after single neuron microaspiration and RNA amplification (Chow et al. 1998; Ginsberg et al. 2000, 2006; Mufson et al. 2002). In a landmark paper, Luo et al. (1999) coupled LCM with T7 RNA amplification and microarray technology to study gene expression in single neuronal types. The composite strategy combining LCM, RNA amplification, microarray analysis, and ISH has been used in studies addressing a variety of conditions and cell types, including diseases such as amyotrophic lateral sclerosis (Jiang et al. 2005), brain nuclei (Bonaventure et al. 2002), hippocampal neurons (Kamme et al. 2003; Torres-Munoz et al. 2004), and the highly heterogeneous mammalian olfactory system (Tietjen et al. 2003). Paradoxically, the study of cells considered to be homogeneous has in some cases allowed identification of distinct groups based on gene expression profile (Kamme et al. 2003).
LCM allows collection of a higher number of cells compared with aspiration and other manual methods, but less than with FACS-based methods. RNA quality is dependent on fixation, staining and general handling of the tissue prior to LCM, and is generally better preserved in fresh-frozen samples. How FACS sorting of live cells compares with LCM-derived samples is not known, since no side-by-side comparisons of the two methods have been published; such studies would be valuable contributions to the field.
The idea of capturing single cells for molecular profiling has been tightly linked to the available options to achieve reliable and accurate RNA amplification. A typical yield from LCM is 10–30 pg of RNA per cell, and technologies for single cell mRNA analysis have been developed (Eberwine et al. 1992; Eberwine, 2001). Two issues inherent in single-cell analysis are worth mentioning: the first is somewhat of a paradox, in that microarray experimentation requires replication, yet some single-cell analyses strive to profile unique features of individual cells. The more replicates performed, the less individual variability may be detected, depending on the analytic strategy (e.g. Zirlinger, 2003), especially since we do not know how much heterogeneity to expect in gene expression studies within a defined cellular population. In this case, application of analytic methods (such as ANOVA) suited to detect variable genes, rather than simple differential expression across groups, may be more powerful. The second technical limitation is inherent to RNA amplification: since there can be a loss of linearity for very diluted transcripts (Sugino et al. 2006), transcripts with lower levels of expression may be too rare to be detected with microarrays, even after two rounds of amplification, leading to the possibility of obtaining different transcription profiles from the same cell types, depending on the starting amount of RNA. Therefore, a critical mass of cells is needed to detect rarer transcripts with current high-throughput technologies relying on RNA amplification. Attempts to estimate this number have been recently reported, and it may lie between 30 (Sugino et al. 2006) and 300 (Lobo et al. 2006) cells, possibly depending on the cell type. It is becoming clear that once a threshold of several hundred cells is reached, as in the case of FACS-array in this issue is less of a concern (Lobo et al. 2006, e.g. supplementary Fig. 5).
The next frontier of specificity is at the subcellular level. Zhong et al. (2006) took advantage of the anatomy of the rodent hippocampus, which permits the dissection of dendritic projections of hippocampal neurons, and compared the dendritic transcripts to the mRNA pool present in the cell body (Zhong et al. 2006). Finally, coimmunoprecipitation techniques allow one to extract and study transcripts included in a macromolecular complex. In one of the earliest studies in the field, Brown et al. (2001) used this method to identify transcripts binding the fragile X mental retardation protein (Brown et al. 2001). A similar approach, involving cell-specific expression of a FLAG-tagged polyA binding protein, followed by cross-linking and purification of cell-type-specific mRNA, has been successfully applied in worm and fly models (Kunitomo et al. 2005; Yang et al. 2005). PolyA- or other tagging/IP-based approaches will not allow detection of micro RNAs or other non-coding RNA species (Cao et al. 2006; Furuno et al. 2006).
How gene expression studies can contribute to system level knowledge: new analytical approaches
The knowledge of behaviour at the level of a single cell is a necessary component of understanding the system in which it acts. Given the high number of variables and confounding factors involved, bioinformatics methods naturally have been involved in dealing with such complexity. This field represents a meeting point between ‘wet’ molecular biology and computational approaches. The availability of datasets from distinct cell types and brain regions allows comprehensive approaches, aimed at detecting general mechanisms of cellular behaviour, as well as cell-type specific features. A considerable number of datasets and computationally intensive approaches are needed to obtain a clear signal-to-noise ratio. From analysis of coexpression across different species (Stuart et al. 2003) to evolutionary analysis of coexpression networks (Oldham et al. 2005), this largely unexplored approach is beginning to show its power, a trend that will continue as the number of available microarray datasets grows. Coexpression networks have modular architecture that closely follows neuroanatomy and other key functional elements of the CNS (Oldham et al. 2005). It is likely that this strategy will be more able (compared with conventional analysis) to detect genes involved in maintaining (and disrupting) the wiring patterns of the brain, and is expected to provide significant advances at every level of complexity (Zhang & Horvath, 2005; Pocklington et al. 2006), as hundreds of datasets on specific cell populations are expected to be generated and made available to the scientific community in the near future. This will facilitate more rapid translation of this high-dimensional data to provide new functional insights.
References
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Bohm C, Newrzella D, Sorgenfrei O. Laser microdissection in CNS research. Drug Discov Today. 2005;10:1167–1174. doi: 10.1016/S1359-6446(05)03555-5. [DOI] [PubMed] [Google Scholar]
- Bonaventure P, Guo H, Tian B, Liu X, Bittner A, Roland B, et al. Nuclei and subnuclei gene expression profiling in mammalian brain. Brain Res. 2002;943:38–47. doi: 10.1016/s0006-8993(02)02504-0. [DOI] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Brown VM, Ossadtchi A, Khan AH, Cherry SR, Leahy RM, Smith DJ. High-throughput imaging of brain gene expression. Genome Res. 2002a;12:244–254. doi: 10.1101/gr.204102. [DOI] [PubMed] [Google Scholar]
- Brown VM, Ossadtchi A, Khan AH, Gambhir SS, Cherry SR, Leahy RM, Smith DJ. Gene expression tomography. Physiol Genomics. 2002b;8:159–167. doi: 10.1152/physiolgenomics.00090.2001. [DOI] [PubMed] [Google Scholar]
- Brown VM, Ossadtchi A, Khan AH, Yee S, Lacan G, Melega WP, et al. Multiplex three-dimensional brain gene expression mapping in a mouse model of Parkinson's disease. Genome Res. 2002c;12:868–884. doi: 10.1101/gr.229002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunney WE, Bunney BG, Vawter MP, Tomita H, Li J, Evans SJ, et al. Microarray technology: a review of new strategies to discover candidate vulnerability genes in psychiatric disorders. Am J Psychiatry. 2003;160:657–666. doi: 10.1176/appi.ajp.160.4.657. [DOI] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Chow N, Cox C, Callahan LM, Weimer JM, Guo L, Coleman PD. Expression profiles of multiple genes in single neurons of Alzheimer's disease. Proc Natl Acad Sci U S A. 1998;95:9620–9625. doi: 10.1073/pnas.95.16.9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M, Estevez A, Yin X, Fox R, Morrison R, McDonnell M, et al. A primary culture system for functional analysis of C. elegans neurons and muscle cells. Neuron. 2002;33:503–514. doi: 10.1016/s0896-6273(02)00591-3. [DOI] [PubMed] [Google Scholar]
- Cinar H, Keles S, Jin Y. Expression profiling of GABAergic motor neurons in Caenorhabditis elegans. Curr Biol. 2005;15:340–346. doi: 10.1016/j.cub.2005.02.025. [DOI] [PubMed] [Google Scholar]
- Colosimo ME, Brown A, Mukhopadhyay S, Gabel C, Lanjuin AE, Samuel AD, Sengupta P. Identification of thermosensory and olfactory neuron-specific genes via expression profiling of single neuron types. Curr Biol. 2004;14:2245–2251. doi: 10.1016/j.cub.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Coppola G, Geschwind DH. Querying the genome with microarrays: progress and hope for neurological disease. Nat Clin Pract Neurol. 2006;2:147–158. doi: 10.1038/ncpneuro0133. [DOI] [PubMed] [Google Scholar]
- Cui D, Dougherty KJ, Machacek DW, Sawchuk M, Hochman S, Baro DJ. Divergence between motoneurons: gene expression profiling provides a molecular characterization of functionally discrete somatic and autonomic motoneurons. Physiol Genomics. 2006;24:276–289. doi: 10.1152/physiolgenomics.00109.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz E. A functional genomics guide to the galaxy of neuronal cell types. Nat Neurosci. 2006;9:10–12. doi: 10.1038/nn0106-10. [DOI] [PubMed] [Google Scholar]
- Diaz E, Ge Y, Yang YH, Loh KC, Serafini TA, Okazaki Y, et al. Molecular analysis of gene expression in the developing pontocerebellar projection system. Neuron. 2002;36:417–434. doi: 10.1016/s0896-6273(02)01016-4. [DOI] [PubMed] [Google Scholar]
- Dougherty JD, Geschwind DH. Progress in realizing the promise of microarrays in systems neurobiology. Neuron. 2005;45:183–185. doi: 10.1016/j.neuron.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Eberwine J. Single-cell molecular biology. Nat Neurosci. 2001;4(Suppl):1155–1156. doi: 10.1038/nn1101-1155. [DOI] [PubMed] [Google Scholar]
- Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, et al. Analysis of gene expression in single live neurons. Proc Natl Acad Sci U S A. 1992;89:3010–3014. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, et al. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- Evans SJ, Choudary PV, Vawter MP, Li J, Meador-Woodruff JH, Lopez JF, et al. DNA microarray analysis of functionally discrete human brain regions reveals divergent transcriptional profiles. Neurobiol Dis. 2003;14:240–250. doi: 10.1016/s0969-9961(03)00126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox RM, Von Stetina SE, Barlow SJ, Shaffer C, Olszewski KL, Moore JH, et al. A gene expression fingerprint of C. elegans embryonic motor neurons. BMC Genomics. 2005;6:42. doi: 10.1186/1471-2164-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funatsu N, Inoue T, Nakamura S. Gene expression analysis of the late embryonic mouse cerebral cortex using DNA microarray: identification of several region- and layer-specific genes. Cereb Cortex. 2004;14:1031–1044. doi: 10.1093/cercor/bhh063. [DOI] [PubMed] [Google Scholar]
- Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, et al. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH. Mice, microarrays, and the genetic diversity of the brain. Proc Natl Acad Sci U S A. 2000;97:10676–10678. doi: 10.1073/pnas.97.20.10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Che S, Counts SE, Mufson EJ. Shift in the ratio of three-repeat tau and four-repeat tau mRNAs in individual cholinergic basal forebrain neurons in mild cognitive impairment and Alzheimer's disease. J Neurochem. 2006;96:1401–1408. doi: 10.1111/j.1471-4159.2005.03641.x. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Hemby SE, Lee VM, Eberwine JH, Trojanowski JQ. Expression profile of transcripts in Alzheimer's disease tangle-bearing CA1 neurons. Ann Neurol. 2000;48:77–87. [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gustincich S, Contini M, Gariboldi M, Puopolo M, Kadota K, Bono H, et al. Gene discovery in genetically labeled single dopaminergic neurons of the retina. Proc Natl Acad Sci U S A. 2004;101:5069–5074. doi: 10.1073/pnas.0400913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. Analysis of mammalian central nervous system gene expression and function using bacterial artificial chromosome-mediated transgenesis. Hum Mol Genet. 2000;9:937–943. doi: 10.1093/hmg/9.6.937. [DOI] [PubMed] [Google Scholar]
- Jiang YM, Yamamoto M, Kobayashi Y, Yoshihara T, Liang Y, Terao S, et al. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann Neurol. 2005;57:236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- Kamme F, Salunga R, Yu J, Tran DT, Zhu J, Luo L, et al. Single-cell microarray analysis in hippocampus CA1: demonstration and validation of cellular heterogeneity. J Neurosci. 2003;23:3607–3615. doi: 10.1523/JNEUROSCI.23-09-03607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitomo H, Uesugi H, Kohara Y, Iino Y. Identification of ciliated sensory neuron-expressed genes in Caenorhabditis elegans using targeted pull-down of poly(A) tails. Genome Biol. 2005;6:R17. doi: 10.1186/gb-2005-6-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski NM, Small SA. Brain microarray: finding needles in molecular haystacks. J Neurosci. 2005;25:10341–10346. doi: 10.1523/JNEUROSCI.4006-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Lombardino AJ, Li XC, Hertel M, Nottebohm F. Replaceable neurons and neurodegenerative disease share depressed UCHL1 levels. Proc Natl Acad Sci U S A. 2005;102:8036–8041. doi: 10.1073/pnas.0503239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, et al. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Korade Z, Arion D, Lazarov O, Unger T, Macioce M, et al. Presenilin-1-dependent transcriptome changes. J Neurosci. 2005;25:1571–1578. doi: 10.1523/JNEUROSCI.4145-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- Mirnics K, Pevsner J. Progress in the use of microarray technology to study the neurobiology of disease. Nat Neurosci. 2004;7:434–439. doi: 10.1038/nn1230. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Mufson EJ, Counts SE, Ginsberg SD. Gene expression profiles of cholinergic nucleus basalis neurons in Alzheimer's disease. Neurochem Res. 2002;27:1035–1048. doi: 10.1023/a:1020952704398. [DOI] [PubMed] [Google Scholar]
- Nelson SB, Sugino K, Hempel CM. The problem of neuronal cell types: a physiological genomics approach. Trends Neurosci. 2006;29:339–345. doi: 10.1016/j.tins.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Nygaard V, Hovig E. Options available for profiling small samples: a review of sample amplification technology when combined with microarray profiling. Nucl Acids Res. 2006;34:996–1014. doi: 10.1093/nar/gkj499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dell DM, Raghupathi R, Crino PB, Morrison B, 3rd, Eberwine JH, McIntosh TK. Amplification of mRNAs from single, fixed, TUNEL-positive cells. Biotechniques. 1998;25:566–570. doi: 10.2144/98254bm04. [DOI] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. Network analysis of gene expression in human and chimpanzee brains identifies functional modules. Society for Neuroscience. 2005 2005 Online, Program No. 4098. [Google Scholar]
- Pocklington AJ, Cumiskey M, Armstrong JD, Grant SG. The proteomes of neurotransmitter receptor complexes form modular networks with distributed functionality underlying plasticity and behaviour. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100041. 10.1038/msb4100041/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Yasuda R, Pankratz DG, Carter TA, Del Rio JA, Wodicka L, et al. Regional and strain-specific gene expression mapping in the adult mouse brain. Proc Natl Acad Sci U S A. 2000;97:11038–11043. doi: 10.1073/pnas.97.20.11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RP, Smith DJ. Genome scale mapping of brain gene expression. Biol Psychiatry. 2003;53:1069–1074. doi: 10.1016/s0006-3223(03)00238-5. [DOI] [PubMed] [Google Scholar]
- Small SA, Kent K, Pierce A, Leung C, Kang MS, Okada H, et al. Model-guided microarray implicates the retromer complex in Alzheimer's disease. Ann Neurol. 2005;58:909–919. doi: 10.1002/ana.20667. [DOI] [PubMed] [Google Scholar]
- Small SA, Wu EX, Bartsch D, Perera GM, Lacefield CO, DeLaPaz R, et al. Imaging physiologic dysfunction of individual hippocampal subregions in humans and genetically modified mice. Neuron. 2000;28:653–664. doi: 10.1016/s0896-6273(00)00144-6. [DOI] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Tietjen I, Rihel JM, Cao Y, Koentges G, Zakhary L, Dulac C. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38:161–175. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- Torres-Munoz JE, Van Waveren C, Keegan MG, Bookman RJ, Petito CK. Gene expression profiles in microdissected neurons from human hippocampal subregions. Brain Res Mol Brain Res. 2004;127:105–114. doi: 10.1016/j.molbrainres.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Yang Z, Edenberg HJ, Davis RL. Isolation of mRNA from specific tissues of Drosophila by mRNA tagging. Nucleic Acids Res. 2005;33:e148. doi: 10.1093/nar/gni149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao FYuF, Gong L, Taube D, Rao DD, MacKenzie RG. Microarray analysis of fluoro-gold labeled rat dopamine neurons harvested by laser capture microdissection. J Neurosci Meth. 2005;143:95–106. doi: 10.1016/j.jneumeth.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Zapala MA, Hovatta I, Ellison JA, Wodicka L, Del Rio JA, Tennant R, et al. Adult mouse brain gene expression patterns bear an embryologic imprint. Proc Natl Acad Sci U S A. 2005;102:10357–10362. doi: 10.1073/pnas.0503357102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4(Issue 1) doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Delohery T, Nasipak B, Foat BC, Bounoutas A, et al. Identification of genes expressed in C. elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
- Zhao X, Lein ES, He A, Smith SC, Aston C, Gage FH. Transcriptional profiling reveals strict boundaries between hippocampal subregions. J Comp Neurol. 2001;441:187–196. doi: 10.1002/cne.1406. [DOI] [PubMed] [Google Scholar]
- Zhong J, Zhang T, Bloch LM. Dendritic mRNAs encode diversified functionalities in hippocampal pyramidal neurons. BMC Neurosci. 2006;7:17. doi: 10.1186/1471-2202-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirlinger M. Selection and validation of microarray candidate genes from subregions and subnuclei of the brain. Methods. 2003;31:290–300. doi: 10.1016/s1046-2023(03)00158-0. [DOI] [PubMed] [Google Scholar]
- Zirlinger M, Anderson D. Molecular dissection of the amygdala and its relevance to autism. Genes Brain Behav. 2003;2:282–294. doi: 10.1034/j.1601-183x.2003.00039.x. [DOI] [PubMed] [Google Scholar]
- Zirlinger M, Kreiman G, Anderson DJ. Amygdala-enriched genes identified by microarray technology are restricted to specific amygdaloid subnuclei. Proc Natl Acad Sci U S A. 2001;98:5270–5275. doi: 10.1073/pnas.091094698. [DOI] [PMC free article] [PubMed] [Google Scholar]