Abstract
It is a challenging question to understand how different neuronal types are organized into a complex architecture in the cortex, an architecture which is also adapted in different regions to subserve very different functions. Recent developments in genetic and molecular techniques have opened up the possibility of using gene expression profiling for neuronal cell typing, with the aim of uncovering novel cell types and the underlying mechanisms which generate and maintain neuronal heterogeneity in the cortex. This review introduces some current ideas about neuronal cell types in the cortex and describes recent approaches to expression profiling for defining cortical neuronal cell types.
Basic concept of neuronal cell type
In a strict sense, a neuronal cell type means a group of neurons that have a specific localization, protein expression pattern, excitability, and connectivity with other types of neurons. Potentially, a cell type population might also be expected to have a common function. A point of possible confusion is that, for example, ‘pyramidal cells’ could mean neurons in cortex with a ‘pyramidal’ morphology (triangular-shaped soma perpendicular to the layers, long prominent apical dendrite, etc.) or a group of neurons which, in addition to having pyramidal morphology, use glutamate as a neurotransmitter and exhibit a ‘regular-spiking’ pattern (Connors & Gutnick, 1990). A public debate on this issue is in progress at the moment (Yuste, 2005) but here we refer to a grouping of the first kind as a ‘neuronal class’ and refer to, for example, Cajal-Retzius cells or Martinotti cells as distinct neuronal cell types. In the future, classification of neuronal cell types should be standardized by an international organization as done for gene and protein names.
Classification of cortical neurons in the postgenomic era
Many neuronal classes in cortex have been discovered by morphological or neurochemical techniques (Peters & Jones, 1984). Such approaches remain important, not just because of the amount of knowledge accumulated over the long history of research, but also because they are well correlated with their functions. For this reason, it is becoming more common to use a combination of protein markers for classification of cortical neurons. The discriminatory power of these protein markers has been demonstrated by the development of reliable antibodies and confocal microscopy to visualize them, transgenic mice expressing markers with fluorescent proteins or knockout mice lacking transcription factors essential for development of particular cell types (Xu et al. 2004; Cheng et al. 2005b).
In theory, all the proteins expressed in cortical neurons could be candidate markers, and the GENSAT project (see this issue of The Journal of Physiology) is now examining the specific distributions of a large number of proteins in the brain, some of which seem to be localized to subsets of cortical neurons. The current list of commonly used markers includes metabolic enzymes and transporter proteins for neurotransmitters (e.g. the GABA synthesizing enzyme glutamate decarboxylase (GAD) and VGLUT) (Esclapez et al. 1994; Hisano, 2003), calcium binding proteins (e.g. parvalbumin, calbindin and calretinin) (Hof et al. 1999) and neuropeptides (e.g. VIP, cholesystokinin, somatostatin, neuropeptide Y and substance P) (Baraban & Tallent, 2004). Coexpression of calcium-binding proteins and neuropeptides has been examined at the single-cell level (Cauli et al. 2000; Wang et al. 2002). These markers have typically been studied, owing to their functional importance as neurotransmitter systems or cell-signalling components, or owing to the availability of good quality antibodies for immunohistochemistry.
Unfortunately, it is still unclear for many markers why they are specifically expressed in certain classes of neurons and how their expression is regulated. Addressing these questions is potentially important for understanding the complex spatial and temporal patterns of their expression during development, when marker expression changes in different ways in different groups of neurons (Hof et al. 1999; Lee et al. 1998; Schuske & Jorgensen, 2004). Moreover, the subcellular distribution of markers can also change during development (Guo et al. 1997; Sheikh et al. 1999), which sometimes gives the false impression that the density and localization of cell types have changed. Overall, understanding the cell-specific control of expression of genes and their protein products that confer distinct functions on neuronal types is part of the broader question of neuronal cell fate determination in the developing nervous system, and of the nature of the genetic networks that operate to control terminal neuronal differentiation (Livesey & Cepko, 2001).
All of the techniques outlined above typically study the coexpression of one or two genes or gene products. Thus they are ill-suited for a post-genomic approach which uses genomic data and high-throughput techniques to study neuronal gene expression at the whole genome level. A recent development is the combined use of flow cytometry and transgenic mice expressing markers tagged with green fluorescent protein to isolate a population of neurons and obtain expression profiles (Arlotta et al. 2005; Sugino et al. 2006). It has also been demonstrated in lymphocytes that multiparameter flow cytometry can be used to detect phosphorylation of signalling proteins and thereafter infer causal networks using Bayesian analysis (Sachs et al. 2005), and this may be applicable to neuronal cell subpopulations after improvement in cell yields or protein detection sensitivity. With the increasing application of genomics methods to studying development and function of the CNS, there are now opportunities to apply these methods to classifying neocortical cell types, particularly in combination with electrophysiology.
Single cell gene expression profiling of neurons with patch clamping and microarray
Expression profiling by microarray has been used for characterizing cell types in various systems, such as cancer cells (Bucca et al. 2004), blood cells (Klein et al. 2003) and stem cells (Perez-Iratxeta et al. 2005). One of the advantages of using microarrays is that they can provide a large amount of expression data for genes associated with a wide variety of biological processes. This means that each cell type can be characterized not just by a few selected markers but also by a group of genes, which may be in the same gene family or ontology (Ashburner & Lewis, 2002) or have the same temporal expression patterns. Moreover, microarrays can be used to uncover new genes expressed in known cell types, and to infer their functions by comparing their spatial and temporal expression patterns with those of well-characterized genes.
Microarray analysis can be applied to a group of pooled neurons expressing a fluorescent-tagged protein (Arlotta et al. 2005; Sugino et al. 2006) or to single neurons (Kamme et al. 2003; Seshi et al. 2003). Fluorescent tag-based cell pooling can significantly improve the yield of microarray-detectable transcripts (by 19% when using 30 or more cells instead of a single cell) (Sugino et al. 2006). Since the cells can be easily identified in brain tissues, the cell populations can be further characterized by morphological and immunohistochemical techniques. While this method is only applicable to well-established neuronal classes with known protein markers, single-cell expression profiling is potentially extremely powerful for discovering novel neuronal cell types, and offers great flexibility to characterize individual neurons by different combinations of classifications. In addition, single-cell expression profiling is potentially a powerful way to resolve the degree of heterogeneity within individual subtypes, and to test the very concept of a distinguishable subtype. The technique can be used when the expression patterns of proteins markers are uncharacterized, for examples in developing or genetically modified brains, or in stem cell-derived neurons.
Single-cell expression profiling can be combined with electrophysiological recordings which, unlike most other techniques, are obtainable from living single neurons before sampling mRNA (Monyer & Markram, 2004). Passive membrane properties, such as input resistance and membrane capacitance, can be used to infer the size and channel densities of neurons, and spiking patterns seen in cortical neurons during current injection are often correlated with specific neuronal cell types (Markram et al. 2004). For example, spikes in pyramidal neurons have a ‘regular spiking’ pattern which is characterized by low and smooth afterhyperpolarization, long spike duration and low maximum spike frequency (Tateno et al. 2004). As expected from their large morphological and neurochemical heterogeneity, spiking patterns in interneurons can be classified into several subtypes (Markram et al. 2004). One of the major subtypes is the ‘fast spiking’ (FS) neuron, which has fast and sharp afterhyperpolarization, short spike duration and high maximum spike frequency which is constant during stimulation (Tateno et al. 2004). Fast spiking neurons are associated with basket cells and chandelier cells in cerebral cortex and express parvalbumin (Kawaguchi & Kubota, 1997).
A critical step in microarray analysis at the single-cell level is million-fold amplification of the starting picogram amounts of mRNA. Either linear T7-based amplification methods or exponential PCR-based amplification techniques can be used to amplify the initial single-cell mRNA. While the exponential PCR-based amplification technique has been shown to outperform linear amplification methods in precision, time and cost-efficiency, single-cell PCR-based amplification still has some limitations and problems. The most important problem is the mRNA sampling effect, which has been shown to introduce a high level of noise in microarray data, particularly for low abundant transcripts. A lower limit of 80 copies of mRNA of a particular gene per cell is reported to be necessary to register a two-fold change in input RNA (Theilgaard-Monch et al. 2001). Estimates of these limits by Nygaard and co-workers (Nygaard et al. 2005) are even higher. For a sample of 1000 pooled cells, differences in transcripts expressed with 121 gene mRNA copies per cell can be detected with statistical reliability, while for 250 cells, the limit was 1806 transcripts per cell. However, the sampling effect depends not only on transcript abundance, but also is strongly affected by the efficiency of RNA preparation and the first amplification steps, including an initial reverse transcription. Previously we have demonstrated that the global polyadenylated PCR-based amplification technique generates reliable data from picogram amounts of RNA (Subkhankulova & Livesey, 2006). More recently we estimated that the most crucial steps of amplification of original mRNA (reverse transcription, polyadenylation and the subsequent first cycles of PCR) reproduce the original mRNA profile with approximately 90% efficiency (unpublished observations; TS & FJL). We found that such a high efficiency produces random noise which is comparable with technical noise from microarray hybridizations, and allows us to estimate the expression ratios for the majority of tested genes.
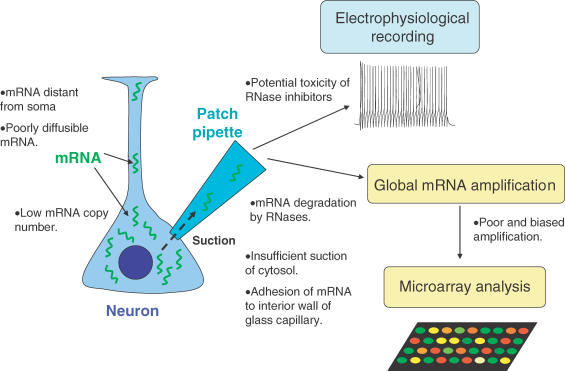
Although it is an exciting technology, the data from single-cell gene expression analysis of neuronal cell require careful scrutiny, even with the best possible mRNA amplification techniques (Fig. 1). It is possible that transcripts which are crucial for distinguishing the subtypes are missed during cytoplasmic harvesting or amplification. It has been observed in previous single-cell expression studies that even highly abundant transcripts can frequently be lost (Nygaard & Hovig, 2006). Some of the cytoplasmic mRNA can be located away from the soma from which the cytoplasm is collected, and they can be poorly mobile because of cytosolic RNA binding proteins such as Hu antigen D and Fragile X mental retardation protein (FMRP) (Bolognani et al. 2004; Ule & Darnell, 2006). Therefore, gene expression profiles from single cells should be interpreted as a reflection of somatic mRNA distributions which may still be used to classify neuronal cell types, without naively assuming that observed mRNA levels can predict total mRNA or protein expression levels. Expression profiles of some well-studied genes, particularly calcium binding proteins and ion channels have been confirmed by single-cell RT-PCR and immunohistochemistry, and expanding the list of genes which show consistent expression profiles in both techniques will significantly improve the use of single-cell microarray data.
Figure 1. Potential problems with single-cell gene expression profiling by cytoplasmic harvesting via patch-pipette.
Collecting extremely low amounts of mRNA from single cells is the biggest challenge of this technique. Only a small proportion of cytosol can be obtained by suction via patch-pipette, and poorly diffusible mRNA or dendritic mRNA are particularly hard to collect. The yield of mRNA can be improved by including inhibitors of RNases in the pipette, but these are often cytotoxic and can be detrimental to electrophysiological recording. It is also possible that some mRNA adheres to the interior wall of the glass capillary and evades expulsion from the patch-pipette.
Future perspectives
With new technological developments, it now seems possible to generate a comprehensive classification of neuronal cell types in the cortex. Armed with a reliable cell type list, we can start asking, for example, if specific cell types are related to certain higher cognitive functions or neurological disorders. Detailed phenotypical data on neuronal cell types can also be used to characterize neurons derived from stem cells, using a variety of parameters. For this purpose, it would be highly desirable to have a public repository for information on each cell type obtained by a series of standardized techniques, so that neuroscientists can deposit information on their cells and compare it against known cell types, similarly to BLAST for nucleotide and protein sequences. We have already witnessed how comprehensive knowledge of genomes can reshape the understanding of complex cellular systems, and, over the next few years, it should be possible to define the neuronal cell types of cerebral cortex and their functions, in much greater detail.
Acknowledgments
This work is supported by European Commission Framework Programme 6.
References
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Lewis S. On ontologies for biologists: the gene ontology – untangling the web. Novartis Found Symp. 2002;247:66–80. [PubMed] [Google Scholar]
- Baraban SC, Tallent MK. Interneuron diversity series: Interneuronal neuropeptides – endogenous regulators of neuronal excitability. Trends Neurosci. 2004;27:135–142. doi: 10.1016/j.tins.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Merhege MA, Twiss J, Perrone-Bizzozero NI. Dendritic localization of the RNA-binding protein HuD in hippocampal neurons: association with polysomes and upregulation during contextual learning. Neurosci Lett. 2004;371:152–157. doi: 10.1016/j.neulet.2004.08.074. [DOI] [PubMed] [Google Scholar]
- Bucca G, Carruba G, Saetta A, Muti P, Castagnetta L, Smith CP. Gene expression profiling of human cancers. Ann N Y Acad Sci. 2004;1028:28–37. doi: 10.1196/annals.1322.003. [DOI] [PubMed] [Google Scholar]
- Cauli B, Porter JT, Tsuzuki K, Lambolez B, Rossier J, Quenet B, Audinat E. Classification of fusiform neocortical interneurons based on unsupervised clustering. Proc Natl Acad Sci U S A. 2000;97:6144–6149. doi: 10.1073/pnas.97.11.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Samad OA, Xu Y, Mizuguchi R, Luo P, Shirasawa S, Goulding M, Ma Q. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci. 2005b;8:1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- Connors BW, Gutnick MJ. Intrinsic firing patterns of diverse neocortical neurons. Trends Neurosci. 1990;13:99–104. doi: 10.1016/0166-2236(90)90185-d. [DOI] [PubMed] [Google Scholar]
- Esclapez M, Tillakaratne NJ, Kaufman DL, Tobin AJ, Houser CR. Comparative localization of two forms of glutamic acid decarboxylase and their mRNAs in rat brain supports the concept of functional differences between the forms. J Neurosci. 1994;14:1834–1855. doi: 10.1523/JNEUROSCI.14-03-01834.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Kaplan IV, Cooper NG, Mower GD. Expression of two forms of glutamic acid decarboxylase (GAD67 and GAD65) during postnatal development of the cat visual cortex. Brain Res Dev Brain Res. 1997;103:127–141. doi: 10.1016/s0165-3806(97)81789-0. [DOI] [PubMed] [Google Scholar]
- Hisano S. Vesicular glutamate transporters in the brain. Anat Sci Int. 2003;78:191–204. doi: 10.1046/j.0022-7722.2003.00059.x. [DOI] [PubMed] [Google Scholar]
- Hof PR, Glezer II, Conde F, Flagg RA, Rubin MB, Nimchinsky EA, Vogt Weisenhorn DM. Cellular distribution of the calcium-binding proteins parvalbumin, calbindin, and calretinin in the neocortex of mammals: phylogenetic and developmental patterns. J Chem Neuroanat. 1999;16:77–116. doi: 10.1016/s0891-0618(98)00065-9. [DOI] [PubMed] [Google Scholar]
- Kamme F, Salunga R, Yu J, Tran DT, Zhu J, Luo L, Bittner A, Guo HQ, Miller N, Wan J, Erlander M. Single-cell microarray analysis in hippocampus CA1: demonstration and validation of cellular heterogeneity. J Neurosci. 2003;23:3607–3615. doi: 10.1523/JNEUROSCI.23-09-03607.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kubota Y. GABAergic cell subtypes and their synaptic connections in rat frontal cortex. Cereb Cortex. 1997;7:476–486. doi: 10.1093/cercor/7.6.476. [DOI] [PubMed] [Google Scholar]
- Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Jr, Miljkovic V, Cattoretti G, Califano A, Dalla-Favera R. Gene expression dynamics during germinal center transit in B cells. Ann N Y Acad Sci. 2003;987:166–172. doi: 10.1111/j.1749-6632.2003.tb06045.x. [DOI] [PubMed] [Google Scholar]
- Lee EY, Lee TS, Baik SH, Cha CI. Postnatal development of somatostatin- and neuropeptide Y-immunoreactive neurons in rat cerebral cortex: a double-labeling immunohistochemical study. Int J Dev Neurosci. 1998;16:63–72. doi: 10.1016/s0736-5748(97)00040-3. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- Monyer H, Markram H. Interneuron diversity series: Molecular and genetic tools to study GABAergic interneuron diversity and function. Trends Neurosci. 2004;27:90–97. doi: 10.1016/j.tins.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Nygaard V, Holden M, Loland A, Langaas M, Myklebost O, Hovig E. Limitations of mRNA amplification from small-size cell samples. BMC Genomics. 2005;6:147. doi: 10.1186/1471-2164-6-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard V, Hovig E. Options available for profiling small samples: a review of sample amplification technology when combined with microarray profiling. Nucl Acids Res. 2006;34:996–1014. doi: 10.1093/nar/gkj499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Iratxeta C, Palidwor G, Porter CJ, Sanche NA, Huska MR, Suomela BP, Muro EM, Krzyzanowski PM, Hughes E, Campbell PA, Rudnicki MA, Andrade MA. Study of stem cell function using microarray experiments. FEBS Lett. 2005;579:1795–1801. doi: 10.1016/j.febslet.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Peters A, Jones EG. Classification of cortical neurons. In: Peters A, Jones EG, editors. Cellular Components of the Cerebral Cortex. Vol. 1. New York: Plenum Press; 1984. pp. 107–121. In Cerebral Cortex. [Google Scholar]
- Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- Schuske K, Jorgensen EM. Vesicular glutamate transporter – shooting blanks. Science. 2004;304:1750–1752. doi: 10.1126/science.1100475. [DOI] [PubMed] [Google Scholar]
- Seshi B, Kumar S, King D. Multilineage gene expression in human bone marrow stromal cells as evidenced by single-cell microarray analysis. Blood Cells Mol Dis. 2003;31:268–285. doi: 10.1016/s1079-9796(03)00150-5. [DOI] [PubMed] [Google Scholar]
- Sheikh SN, Martin SB, Martin DL. Regional distribution and relative amounts of glutamate decarboxylase isoforms in rat and mouse brain. Neurochem Int. 1999;35:73–80. doi: 10.1016/s0197-0186(99)00063-7. [DOI] [PubMed] [Google Scholar]
- Subkhankulova T, Livesey FJ. Comparative evaluation of linear and exponential amplification techniques for expression profiling at the single-cell level. Genome Biol. 2006;7:R18. doi: 10.1186/gb-2006-7-3-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Tateno T, Harsch A, Robinson HP. Threshold firing frequency-current relationships of neurons in rat somatosensory cortex: type 1 and type 2 dynamics. J Neurophysiol. 2004;92:2283–2294. doi: 10.1152/jn.00109.2004. [DOI] [PubMed] [Google Scholar]
- Theilgaard-Monch K, Cowland J, Borregaard N. Profiling of gene expression in individual hematopoietic cells by global mRNA amplification and slot blot analysis. J Immunol Meth. 2001;252:175–189. doi: 10.1016/s0022-1759(01)00340-4. [DOI] [PubMed] [Google Scholar]
- Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gupta A, Toledo-Rodriguez M, Wu CZ, Markram H. Anatomical, physiological, molecular and circuit properties of nest basket cells in the developing somatosensory cortex. Cereb Cortex. 2002;12:395–410. doi: 10.1093/cercor/12.4.395. [DOI] [PubMed] [Google Scholar]
- Xu Q, De Cobos I, La CE, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J Neurosci. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R. Origin and classification of neocortical interneurons. Neuron. 2005;48:524–527. doi: 10.1016/j.neuron.2005.11.012. [DOI] [PubMed] [Google Scholar]