Abstract
Pancreatic carcinoma has an extremely bad prognosis due to lack of early diagnostic markers and lack of effective therapeutic strategies. Recently, we have established an in vitro model recapitulating the first steps in the carcinogenesis of the pancreas. SV40 large T antigen-immortalized bovine pancreatic duct cells formed intrapancreatic adenocarcinoma tumors on k-rasmut transfection after orthotopic injection in the nude mouse pancreas. Here we identified genes and proteins differentially expressed in the course of malignant transformation using reciprocal suppression subtractive hybridization and 2D gel electrophoresis and mass spectrometry, respectively. We identified 34 differentially expressed genes, expressed sequence tags, and 15 unique proteins. Differential expression was verified for some of the genes or proteins in samples from pancreatic carcinoma. Among these genes and proteins, the majority had already been described either to be influenced by a mutated ras or to be differentially expressed in pancreatic adenocarcinoma, thus proving the feasibility of our model. Other genes and proteins (e.g., BBC1, GLTSCR2, and rhoGDIα), up to now, have not been implicated in pancreatic tumor development. Thus, we were able to establish an in vitro model of pancreatic carcinogenesis, which enabled us to identify genes and proteins differentially expressed during the early steps of malignant transformation.
Keywords: Transcriptomics, proteomics, ki-ras, SV40 large T, pancreatic, carcinogenesis
Introduction
Pancreatic cancer is the fourth most common cause of cancer deaths [1]. Its incidence is increasing, affecting about 10 per 100,000 population per year in western countries. Because no screening markers are available up to now and due to lack of early symptoms, the majority of tumors are already in advanced nonresectable stages at the time of diagnosis, and no therapeutic strategy developed so far has resulted in a considerable increase in long-term survival. This is why the prognosis of this disease is still very dismal, with 5-year survival rates of < 5% [2]. An important approach to the understanding of the biology and molecular alterations involved in the carcinogenesis of this disease is the development of relevant in vitro and in vivo models. These models may provide new diagnostic tools that would allow diagnosis at earlier stages, and this in turn may result in an increase in long-term survival. Because most pancreatic tumors are of ductal phenotype [3] and may derive from ductal epithelial cells [4], the cultivation of isolated human pancreatic duct epithelial cells may play a crucial role in the development of an in vitro model for pancreatic carcinogenesis.
In recent years, numerous alterations associated with carcinogenesis have been identified, namely, mutation of the ki-ras oncogene. More than 90% of pancreatic adenocarcinomas harbor mutations of this gene; these mutations are detectable as well in early stages of tumor development (PanIN lesions) [5], as in metastatic tumors. Thus, mutation of the ki-ras oncogene seems to be an early and important event in the tumorigenesis of the pancreas.
Recently, we were able to establish an in vitro model by mimicking the first steps of the carcinogenesis of the pancreas. We immortalized bovine pancreatic duct cells through transfection with SV40 large T antigen. These immortal cells were then transfected additionally with a vector coding for a mutated ki-ras gene (codon 12: GGT→GTT). Both cell lines were characterized in detail and showed classic markers of differentiated pancreatic duct cells [carbonic anhydrase type II, cytokeratins (CK) 7 and 19, and others], but they exhibited marked differences in their tumorigenic potential. Although the only immortalized cell line was neither able to grow in soft agar nor able to induce tumor, mutated ki-ras-expressing cells grew in soft agar and formed tumors and liver metastases when inoculated orthotopically in the pancreas of nude mice [6]. In an attempt to characterize the transition from an almost benign cell lineage to an invasive ductal adenocarcinoma, we asked for changes in gene expression at both RNA and protein levels. Here we describe the analysis and identification of genes and proteins that are differentially expressed during this course of malignant transformation.
Materials and Methods
RNA Isolation
Total RNA was prepared from pancreatic tissue samples, using the ToTally RNA kit (Ambion, Inc., Austin, TX), according to the protocol supplied by the manufacturer. RNA from pancreatic cell lines VA and VArasmut was isolated with RNeasy mini kit (Qiagen, Inc., Hilden, Germany) following the manufacturer's recommendations. For suppression subtractive hybridization (SSH) analysis, total RNA was treated with Dnase I (Ambion, Inc.) to remove traces of genomic DNA. Poly A+ RNA was isolated from total RNA preparations using oligo(dT)-conjugated magnetic Dynabeads (Dynal Biotech, Oslo, Norway).
SSH
SSH was performed using the PCR-Select cDNA Subtraction Kit (BD Clontech, Heidelberg, Germany). VArasmut and VA RNA isolations were reciprocally compared by forward and reverse subtractions. Driver and tester cDNA were produced, each from 2 µg of Poly A+ RNA, following the manufacturer's guidelines. Synthesized cDNA were digested with the restriction enzyme RsaI, and tester cDNA populations were divided into two tubes and ligated to adaptor 1 or adaptor 2R. Subtractive hybridization was performed by adding 1.5 µl of driver cDNA to each tube, one containing 1.5 µl of adaptor 1 and the other containing 1.5 µl of adapter 2R-ligated diluted tester cDNA in 1 µl of 4x hybridization buffer. After denaturation, samples were allowed to anneal at 68°C for 8 hours. Following the first hybridization, the two samples were combined simultaneously with the addition of 1 µl of freshly denatured driver cDNA, and hybridization was continued overnight at 68°C. Products from the second hybridization were diluted in 200 µl of dilution buffer (20 mM HEPES pH 8.3, 50 mM NaCl, and 0.2 mM EDTA), heated at 68°C for an additional 7 minutes, and stored at -20°C.
Polymerase Chain Reaction (PCR) Amplification of Subtracted Products
PTC-200 Thermal Cycler (MJ Research, Waltham, MA) was used to perform PCR amplification of subtracted tester products. Primary PCR amplifications were conducted for each tester using diluted subtracted products following the second hybridization. One microliter of sample was added to 24 µl of PCR master mix prepared using the reagents supplied in the kit, and cycling conditions commenced as follows: 75°C for 5 minutes to extend the adaptors; 94°C for 25 seconds; and 27 cycles at 94°C for 10 seconds, 66°C for 30 seconds, and 72°C for 1.5 minutes. Amplified products were diluted 10-fold in sterile water, and 1 µl of diluted primary PCR products was added to 24 µl of secondary PCR master mix containing nested primers (1 and 2R) to ensure the specific amplification of double-stranded templates containing both adaptors. Secondary PCR was performed at 94°C for 10 seconds, 68°C for 30 seconds, and 72°C for 1.5 minutes (cycle numbers were 18 and 19 for VArasmut and VA, respectively). PCR products were analyzed on a 2% agarose gel.
Cloning of Subtracted cDNA Templates
Following secondary PCR amplification, subtracted products from each tester cDNA population were purified using the QIAquick PCR Purification Kit (Qiagen, Inc.). Thus, short DNA fragments below 100 bp were removed before cloning. Purified products were ligated with the pGEM T-Easy vector (Promega Corp., Madison, WI) and used for the electrotransformation of competent DH5α Escherichia coli cells. Colonies were grown in a selective LB agar medium containing X-gal and isopropyl β-d-1-thiogalactopyranoside (IPTG) for the screening of blue/white colony. For each tester, 384 colonies were randomly picked up in microtiter plates and grown overnight in a liquid LB medium.
Differential Screening By Microarray Hybridization
To confirm the unique expression of subtracted products, all cDNA clones were subjected to differential screening by microarray hybridization. First, inserts from 768 selected clones (384 clones for each tester) were PCR-amplified with T7 and SP6 primers. PCR was performed by adding 0.5 ml of saturated growth liquid culture to 100-µl PCRs containing 10 mM Tris (pH 9.0), 50 mM KCl, 150 µM dNTP, 3 M betaine, 30 mM cresol red, and 2.0 U of Taq DNA polymerase (MBI Fermentas, St. Leon-Rot, Germany) in 96-well plates. Thermal cycling conditions consisted of initial denaturation at 95°C for 3 minutes, followed by 35 cycles of denaturation at 95°C for 30 seconds, annealing at 53°C for 30 seconds, and elongation at 72°C for 3 minutes, with a final 10-minute extension at 72°C in PTC-200 Thermal Cycler (MJ Research). Five microliters of each PCR was examined on a 2% agarose gel. PCR fragments were purified by isopropanol precipitation.
SSH products from both subtraction experiments were combined on a slide. Quadruplicate spotting onto a custommade poly-l-lysine surface was carried out with SDDC-2DNA Microarrayer (Engineering Services, Inc., Toronto, Canada), using betaine spotting solution, according to the protocol described previously [7]. Poly A+ RNA isolated from VArasmut and VA cells (0.5 mg each) was labeled by direct incorporation of either Cy3 or Cy5 fluorescent dye using SuperScript II reverse transcriptase (Invitrogen, Inc., Karlsruhe, Germany) and oligo-d(T)20 as a primer. Labeled probes were purified through QIAquick PCR purification columns (Qiagen, Inc.), dried in a SpeedVac (Qiagen, Inc.), and resuspended in a total volume of 16 µl of hybridization buffer [3x SSC (0.45 M NaCl, 0.045 M sodium citrate), 1% sodium dodecyl sulfate (SDS), 5x Denhardt's solution, 100 mg/ml sheared salmon sperm DNA, 50% formamide, and 10% dextran sulfate]. The probes were denatured at 80°C for 10 minutes and applied to arrayed/denatured slides at 45°C for 16 hours in a humidified chamber (Telechem, Inc., Sunnyvale, CA). Hybridized slides were washed in 2x SSC and 0.1% SDS for 5 minutes at room temperature and in 0.2x SSC for 5 minutes before scanning by the ScanArray 5000 Microarray Analysis System (GSI Lumonics, Inc., Watertown, MA).
Separate images with 10-µm resolution were captured for each of two fluorophores used. GenePix v. 4.0 (Axon Instruments, Inc., Union City, CA) software was used to quantitate signals at each spot. Signal intensity filtering and background signal correction for spots were performed using GP3 perl script (http://www.bch.msu.edu/∼zacharet/microarray/gp3.html) [8].
Feature spot signals were normalized relative to signals from external controls: Arabidopsis thaliana genes spotted on arrays together with SSH PCR products (the corresponding Arabidopsis mRNA was spiked into labeling reactions in known concentrations).
The mean ratios of Cy3 and Cy5 signal intensities for individual spots were calculated by averaging the data obtained in four independent hybridizations with “dye flip.” Genes were considered differentially expressed when all hybridizations showed a > 2-fold change. The sequencing of PCR products was performed at Genotype, Inc. (Hirschhorn, Germany), a company providing commercial DNA analysis service.
Homology searches were performed using the Basic Local Alignment Search Tool (BLAST) on the combined GenBank/EMBL nonredundant (nr), expressed sequence tag (EST), and SwissProt databases (National Center for Biotechnology Information, which can be accessed online at www.ncbi.nlm.nih.gov).
Two-Dimensional Gel Electrophoresis
Protein extraction from cells obtained from different passages was performed with 1 ml of lysis buffer (9.5 M urea, 2% 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 0.8% Pharmalyte 3-10, 1% dithiothreitol (DTT), and 5 mM Pefabloc SC PLUS; Roche Diagnostics, Inc., Mannheim, Germany). After centrifugation at 13,000g at 15°C for 60 minutes, the protein solution was collected and stored at -80°C until use. Protein concentration was determined by the Bio-Rad Protein Assay (Bio-Rad, Munich, Germany), using a UV/Visible spectrophotometer (Utrospec 2000; Pharmacia Biotech, Uppsala, Sweden).
Samples (protein concentration, ∼ 1–3 µg/µl) were loaded into linear immobilized gradient (IPG) strips (Immobiline DryStrips, pH 4–7; Amersham Biosciences, Freiburg, Germany) using 50 µg of total protein per strip. The IPG strips were rehydrated (10 hours, 30 V, and 20°C) in a solution (350 µl) of 8 M urea, 2% CHAPS, 0.5% IPG buffer pH 4–7, 16 mM DTT, and a calculated amount of protein sample. Separation in the first dimension was performed on an IPGphor unit (Amersham Biosciences). The IPG strips were focused for a total of 125 kV hour. Before the second dimension separation, the IPG strips were equilibrated in equilibration buffer twice for 20 minutes (50 mM Tris-HCl pH 8.8, 6 M urea, 30% glycerol, and 2% SDS) containing DTT (10 mg/ml) and iodoacetamide (40 mg/ml), respectively.
The second dimension [SDS polyacrylamide gel electrophoresis (PAGE)] was performed on laboratory-made polyacrylamide gels (12.5% T; 1.5 x 200 x 250 mm) running on a vertical Hoefer DALT Electrophoresis Tank (Amersham Biosciences). Electrophoresis was carried out for about 22 hours in TGS electrode buffer (25 mM Tris, 192 mM glycine, and 0.1% SDS pH 8.3) at 10°C, applying a constant voltage of 85 V.
Gel Analysis
2-DE separated protein spots were visualized with a modified silver-staining method compatible with matrixassisted laser desorption/ionisation-time of flight-mass spectrometry (MALDI-TOF-MS) [9]. Silver-stained gels were digitized using a GS-800 densitometer (Bio-Rad), and images were imported in a 2D gel image analysis program on a set of six gels per sample. PDQuest 7.1 (Bio-Rad) was used to locate and quantify protein spots and to match spots through the gels. The quantity of each protein spot was normalized against the total quantity arising from all valid protein spots on the gel. Statistical comparison between individual protein abundances was conducted by the calculation of Student's t test within the PDQuest analysis, with a significance level of at least 90%.
Protein Identification By MALDI-MS
Gel pieces containing proteins of interest were manually excised and subjected to in-gel digestion. Proteins were reduced, alkylated, and digested with sequencing-grade modified trypsin (Promega Corp.) using 10 µl of trypsin (10 ng/µl in 40 mM NH4HCO3). Tryptic digested MALDI samples were prepared by cocrystallization of a saturated solution of the matrix (α-cyano-4-hydroxycinnamic acid in 50/50 vol/wt.% acetonitrile/0.1% trifluoroacetic acid in water) with ZipTipC18 (Millipore, Inc., Schwalbach, Germany) concentrated samples.
MALDI mass fingerprint spectra were recorded in positive ion mode with delayed extraction with a Reflex II TOF instrument (Bruker Daltonics, Inc., Bremen, Germany). Calibration was performed internally by a two-point linear fit using the autolysis products of trypsin at m/z = 1045.56 and m/z = 2211.10. Tryptic monoisotopic peptide masses were searched against NCBInr and SwissProt databases using the Mascot (Matrix Science, Ltd., London, Great Britain) and ProFound (Rockefeller University, New York, NY) programs.
Verification of Differential Gene and Protein Expression in Chronic Pancreatitis and Pancreatic Adenocarcinoma
To verify that the differential gene expression identified in our model system holds true in vivo, the expression of selected genes was analyzed in samples from normal pancreas, chronic pancreatitis, and pancreatic adenocarcinoma, using semiquantitative reverse transcription (RT) PCR.
Intron spanning primers for the respective genes (Table 1) were designed, using the online software Primer3 (www.broad.mit.edu/cgi-bin/primer/primer3_www.cgi), to identify possible genomic DNA contamination. To ensure that the PCR was in its logarithmic phase, the appropriate cycle number for each single gene was determined in prior experiments. PCR conditions were as follows: 94°C for 15 minutes, cycling at 94°C for 45 seconds, 60°C for 45 seconds, and 72°C for 1 minute, followed at the end by an extension step at 72°C for 10 minutes.
Table 1.
Primers Used for the RT-PCR of Human Tissue Samples.
| Gene Name | Primer Sequence | Product Size (bp) |
| Annexin 1 | Forward: 5′-ATGTCGCTGCCTTGCATAA | 432 |
| Reverse: 5′-CCTCAGATCGGTCACCCTTA | ||
| BBC1 | Forward: 5′-GTTCGGTACCACACGAAGG | 480 |
| Reverse: 5′-ACTGCCGACTGATTCCAAGT | ||
| rhoGDIα | Forward: 5′-TTTCCGCAGACCCCAAC | 406 |
| Reverse: 5′-GAGATTCCACTCCCAGGACA | ||
| SCART1 | Forward: 5′-TACAGCAGCTGCGAGACAGT | 222 |
| Reverse: 5′-TCCTCATCCCGTTCAAAGTC |
The differential protein expression of annexin 1 was analyzed by immunohistochemistry in paraffin-embedded tissue samples obtained from resections due to chronic pancreatitis and pancreatic carcinoma using standard protocols. In brief, tissue sections were deparaffinized in xylene (2 x 5 minutes), rehydrated in a graded series of ethanol, blocked with 10% fetal calf serum, and incubated for 45 minutes with anti-annexin 1 antibody [1:100 in phosphate-buffered saline (PBS)-bovine serum albumin (BSA); Santa Cruz Biotechnology, Santa Cruz, CA]. After incubation with the second antibody (rabbit-anti-mouseHRP, 1:100, inPBS-BSA; Dako, Inc., Hamburg, Germany), slides were developed using AEC substrate (Chemicon, Hampshire, Great Britain).
Results
Genes Differentially Expressed in ras-Transformed Pancreatic Duct Epithelial Cells
The secondary SSH PCR resulted in a distinct banding pattern and low background. Cloned subtraction SSH products were PCR-amplified and robotically spotted on microarrays along with normalization control DNA. The microarrays were hybridized with Cy5/Cy3-labeled RNA isolated from VArasmut and VA cells. After statistical analysis of hybridization results, 65 clones were considered differentially expressed and sequenced. Among these, 28 unique genes and 6 ESTs were identified; 16 of them were upregulated and 18 were downregulated in VArasmut cells. The results of the BLAST homology search are presented in Table 2. Of the 28 annotated genes, 14 already had been described to be either influenced by a mutated ras or differentially expressed in pancreatic adenocarcinoma (Table 2), thus proving the validity of our model. Other genes (e.g., epsin 1, apolipoprotein A1), up to now, have not been implicated in ras signaling. Most of the upregulated genes accounted for proteins of the cytoskeleton-like 40-kDa keratin, CK7, CK18, CK19, and myosin regulatory light chain. Moreover, several extracellular matrix proteins, as Col Iα1, Col Vα2, and osteonectin/SPARC, were upregulated. Additionally, two ribosomal proteins (mRPS34 and RPL26) and α enolase were upregulated. In contrast, the downregulated genes composed a rather heterogeneous group. Thus, three ribosomal proteins (RPS5, RPS19, and RPL39) and two putative tumorsuppressor genes, breast basic conserved 1 (BBC1) and GLTSCR2, were downregulated. Additionally, two genes involved in tumor immunology/tumor rejection, major histocompatibility complex (MHC) class 1 protein molecule D18.3 and SCART1, were downregulated. For the remaining downregulated genes (Table 2), no grouping concerning functional criteria was possible.
Table 2.
Putative Homology and Identities of cDNA Differentially Expressed in VArasmut Cells.
| Number | Expression Change | Homology | Base Pairs Submitted to BLAST | GenBank Accession Number | Identities | |
| Upregulated | ||||||
| 1 | * | Collagen, type I, α 1 | 534 | NM_000088 | Human 290/309 (93%) | |
| 2 | * | CK19 gene | 454 | X04198 | Bovine 149/151 (98%) | |
| 3 | * | Keratin 7 | 320 | BC002700 | Human 135/146 (92%) | |
| 4 | * | Osteonectin | 451 | J03233 | Bovine 194/195 (99%) | |
| 5 | † | 40-kDa keratin intermediate filament precursor gene | 429 | J03607 | Human 188/204 (92%) | |
| 6 | † | Keratin 18 | 365 | NM_000224 | Human 154/176 (87%) | |
| 7 | † | Myosin regulatory light chain | 377 | D82057 | Human 173/185 (93%) | |
| 8 | ‡ | α | Enolase mRNA | 433 | AF149256 | Bovine 193/201 (96%) |
| 9 | ‡ | Clathrin heavy chain | 549 | U31757 | Bovine 283/285 (99%) | |
| 10 | ‡ | EST | 336 | CB446148 | Bovine 207/237 (87%) | |
| 11 | ‡ | EST | 360 | CB464940 | Bovine 180/196 (91%) | |
| 12 | ‡ | Hypothetical protein MGC10731 | 540 | NM_030907 | Human 161/185 (87%) | |
| 13 | ‡ | Hypothetical protein MGC2963 | 700 | NM_031298 | Human 331/359 (92%) | |
| 14 | ‡ | Mitochondrial ribosomal protein S34 | 560 | NM_023936 | Human 312/365 (85%) | |
| 15 | ‡ | Procollagen, type V, α 2 | 458 | XM_193986 | Mouse 107/125 (85%) | |
| 16 | ‡ | Ribosomal protein L26 | 372 | AB093679 | Primate 112/137 (81%) | |
| Downregulated | ||||||
| 1 | * | Breast basic conserved 1 | 433 | AF192977 | Ovine 184/190 (96%) | |
| 2 | * | Ezrin | 576 | M98498 | Bovine 414/415 (99%) | |
| 3 | * | MHC class 1 protein molecule D18.3 | 463 | Y09207 | Bovine 266/266 (100%) | |
| 4 | * | Proliferating cell nuclear antigen | 550 | AF416380 | Ovine 304/318 (95%) | |
| 5 | * | Ribosomal protein S5 | 686 | BC018828 | Human 275/324 (84%) | |
| 6 | † | ADP/ATP translocase T2 | 367 | M24103 | Bovine 159/160 (99%) | |
| 7 | † | Apolipoprotein A-I | 520 | M35870 | Bovine 316/322 (98%) | |
| 8 | † | β Actin | 469 | AF129289 | Ovine 175/186 (94%) | |
| 9 | † | Chromosome 21 open reading frame 59 (C21orf59) | 664 | NM_021254 | Human 433/494 (87%) | |
| 10 | † | Epsin 1 | 577 | NM_057136 | Rat 461/515 (89%) | |
| 11 | † | EST | 272 | BM433009 | Bovine 134/142 (94%) | |
| 12 | † | Glioma-tumor-suppressor candidate region gene 2 | 413 | BC013307 | Human 199/235 (84%) | |
| 13 | † | Ribosomal protein L30 | 277 | AF063243 | Bovine 112/113 (99%) | |
| 14 | † | Squamous cell carcinoma antigen recognized by T cells 1 | 537 | BC028823 | Mouse 264/310 (85%) | |
| 15 | ‡ | EST | 443 | AV609800 | Bovine 226/228 (99%) | |
| 16 | ‡ | EST | 575 | BE751110 | Bovine 469/475 (98%) | |
| 17 | ‡ | EST | 523 | CB462154 | Bovine 309/324 (95%) | |
| 18 | ‡ | Ribosomal protein S19 | 324 | BC018616 | Human 145/153 (94%) | |
*Expression change, > 5-fold.
†Expression change, > 3-fold.
‡Expression change, > 2-fold.
Proteins Regulated By Activated ras in Pancreatic Duct Cells (PDCs)
Identification of proteins from 2D gels of VA and VArasmut cell lysates obtained from four independent experiments was conducted by MALDI mass fingerprinting. At the protein level, 36 protein spots were identified by mass spectrometry, representing 22 unique proteins (Table W1 and Figure 1). For several proteins [heat shock protein (HSP) 70, actin, vimentin, CK8, and CK19], different isoforms or fragmented proteins (CK8 and tubulin) were identified. Of the 36 analyzed protein spots, 11 were found to be upregulated and 9 were found to be downregulated in the majority of experiments by at least 1.5-fold in VArasmut cells (Table 3), whereas for the remainder, the expression was unchanged or expression changes were inconsistent. These 20 differentially expressed protein spots represented 15 unique proteins. Also at the protein level, several proteins (β actin, annexin 1, CK8, HSP70, myosin regulatory light chain, and vimentin) already had been demonstrated to be differentially expressed in pancreatic adenocarcinoma, again proving the validity of our model. Additionally, tropomyosin 1, fragmented α tubulin, γ actin, and proliferating cell nuclear antigen (PCNA) were found to be downregulated, whereas stressinduced phosphoprotein 1, HSP27, Grp58, rho guanine dinucleotide phosphate dissociation inhibitor α (rhoGDIα), and fragmented β tubulin were upregulated in VArasmut cells.
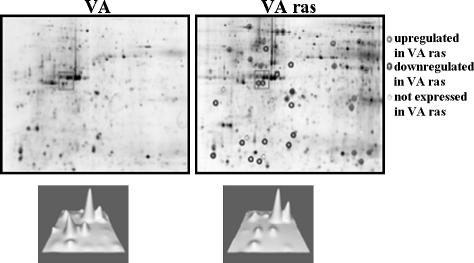
Figure 1.
Two-dimensional gel electrophoresis of VA and VArasmut cell lysates. Differentially expressed proteins are marked by circles. In the rectangles in the lower panel, the intensities of the respective spots are illustrated in 3D; the region shown corresponds to the rectangles in the upper panel.
Table 3.
Results of MALDI-TOF-MS and Database Search for Differentially Expressed Proteins in VArasmut Cells.
| Protein Name | Taxonomy* | Accession Number | Mascot Score† | Peptides Matched | Peptide Coverage (%) | Molecular Weight Mw (Theoretical/Experimental; kDa) | pI (Theoretical/Experimental) | VArasmut/VA‡ | ± SD |
| Keratin type II, cytoskeletal 8 | B | P05786 | 239 | 17 | 55 | 42.2/53.3 | 5.13/5.92 | +3.3 | ±0.3 |
| Stress-induced phosphoprotein 1 | M | AAH03794 | 102 | 10 | 20 | 63.2/64.6 | 6.40/6.27 | +3.0 | ±1.2 |
| Heat shock 70-kDa protein 8 | B | NP_776770 | 133 | 12 | 24 | 71.4/71.5 | 5.49/5.50 | +2.8 | ±0.9 |
| Heat shock 27-kDa protein | C | P42929 | 92 | 7 | 30 | 22.9/25.0 | 6.23/6.11 | +2.6 | ±1.0 |
| Heat shock 70-kDa protein 8 isoform | B | NP_776770 | 66 | 7 | 12 | 71.4/71.5 | 5.49/5.45 | +2.3 | ±0.2 |
| Glucose-regulated protein 58 kDa | B | NP_776758 | 280 | 20 | 42 | 57.3/58.6 | 6.23/6.16 | +2.1 | ±0.4 |
| Annexin 1 | B | NP_786978 | 190 | 13 | 48 | 39.1/35.8 | 6.44/6.39 | +1.9 | ±1.4 |
| Rho GDP dissociation inhibitor α | B | NP_788823 | 129 | 8 | 37 | 23.5/26.5 | 5.12/5.24 | +1.8 | ±0.5 |
| β/γ Actin | M | CAA31455 | 95 | 8 | 26 | 41.3/44.1 | 5.56/5.43 | +1.8 | ±1.0 |
| Heat shock 70-kDa protein 5 | H | NP_005338 | 147 | 12 | 24 | 72.4/75.2 | 5.07/5.12 | +1.6 | ±0.2 |
| β Tubulin [fragmented protein] | M | NP_076205 | 197 | 14 | 27 | 50.4/36.0 | 4.78/5.59 | +1.6 | ±0.6 |
| γ Actin | H | AAA51580 | 81 | 5 | 21 | 26.1/26.1 | 5.65/5.65 | -1.6 | ±0.7 |
| α Tubulin [fragmented protein] | H | NP_116093 | 93 | 8 | 26 | 50.5/38.3 | 4.96/5.67 | -1.8 | ±0.7 |
| Tropomyosin 1 | H | NP_000357 | 105 | 9 | 25 | 32.8/31.2 | 4.81/4.69 | -1.9 | ±0.8 |
| Keratin, type II cytoskeletal 8 [fragmented protein] | B | P05786 | 115 | 8 | 25 | 42.4/26.4 | 5.13/5.01 | -2.0 | ±1.1 |
| Vimentin | B | NP_776394 | 187 | 15 | 42 | 53.7/47.7 | 5.20/4.89 | -2.1 | ±0.7 |
| β/γ Actin | B | ATBOB | 86 | 6 | 20 | 41.9/41.9 | 5.31/5.29 | -2.1 | ±1.2 |
| Vimentin | B | NP_776394 | 130 | 11 | 31 | 53.7/44.9 | 5.20/4.68 | -2.5 | ±0.9 |
| Myosin regulatory light chain | H | P_006462 | 60 | 4 | 29 | 19.8/21.2 | 4.67/4.60 | -3.0 | ±0.2 |
| Proliferating cell nuclear antigen | M | NP_035175 | 89 | 6 | 26 | 29.1/32.7 | 4.66/4.60 | -5.0 | ±0.9 |
pI = isoelectrical point; SD = standard deviation.
C = Canis familaris; B = Bos taurus; H = Homo sapiens; M = Mus musculus.
The mascot score is measured as -10*log(P), where P is the absolute probability that the observed match is a random event.
Fold change: (+) an increase in protein expression in VArasmut cells; (-) a decrease in protein expression in VArasmut cells.
Verification of Differential Gene and Protein Expression in Chronic Pancreatitis and Pancreatic Adenocarcinoma
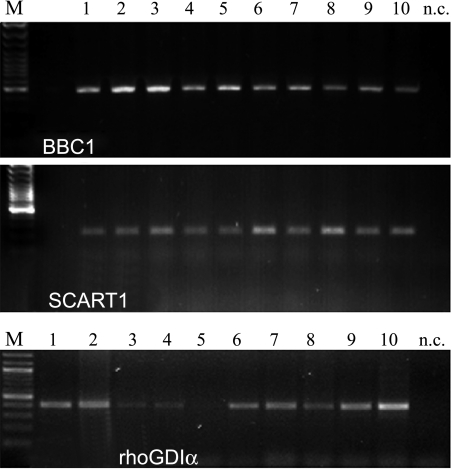
We attempted to verify the differential gene expression identified in our model system in vivo; thus, the expression of selected genes and proteins was analyzed in samples from normal pancreas, chronic pancreatitis, and pancreatic adenocarcinoma using semiquantitative RT-PCR or immunohistochemistry. We concentrated our expression analysis on genes, which up to now have not been reported to be differentially expressed in pancreatic adenocarcinoma. However, annexin 1, known for being overexpressed in pancreatic cancer, was included. For the putative tumor-suppressor BBC1, we were able to verify the downregulation observed in VArasmut cells in samples of pancreatic cancer (Figure 2, upper panel). Moreover, also for the gene rhoGDIα, which is overexpressed in VArasmut cells, a trend for an overexpression in samples of pancreatic adenocarcinoma was detectable (Figure 2, lower panel). In contrast, the expression of the gene squamous cell carcinoma antigen recognized by T cells (SCART1), which was downregulated in VArasmut cells, was not changed in pancreatic adenocarcinoma (Figure 2, middle panel).
Figure 2.
Expression of the genes BBC1, SCART1, and rhoGDIα in normal pancreas (lanes 1–5) and pancreatic cancer (lanes 6–10), analyzed using semiquantitative RT-PCR. BBC1 was moderately downregulated and rhoGDIα was moderately upregulated, whereas for SCART1, no obvious change in expression level was detectable. The housekeeping gene RPL13A (lanes 1–5 and 11–15 in Figure 3 correspond to lanes 1–5 and 6–10 in this figure) was used for normalization. nc = negative control.
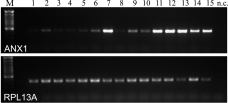
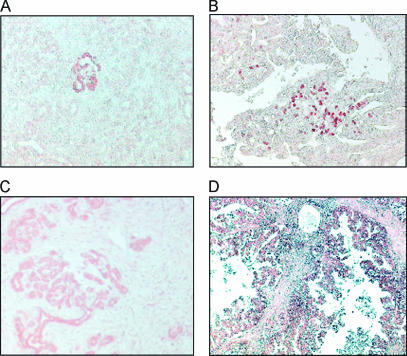
The expression of annexin 1 was analyzed in more detail. At the RNA level, ANX1 expression was barely detectable in normal pancreatic tissues, and most of the cases of chronic pancreatitis showed only a slight increase in ANX1 expression, whereas in one case, the expression was markedly upregulated to levels comparable to the expression level detectable in pancreatic carcinoma (Figure 3). Comparable results were obtained at the protein level using immunohistochemistry. The staining pattern for ANX1 in CP ranged from samples with no obvious staining to samples with single nests of ANX1-positive ductal structures (Figure 4A) and samples in which the majority of the remaining exocrine tissues were ANX1-positive (Figure 4C); in one case, nuclear staining was also observable (Figure 4B). In ductal pancreatic adenocarcinoma, tumor cells stained positive (Figure 4D).
Figure 3.
Marked upregulation of annexin 1 expression in pancreatic carcinoma (lanes 11–15) compared to normal pancreas (lanes 1–5) and chronic pancreatitis (lanes 6–10), analyzed using semiquantitative RT-PCR. The housekeeping gene RPL13A was used for normalization. nc = negative control.
Figure 4.
Annexin 1 (ANX1) staining in chronic pancreatitis (A–C) and pancreatic adenocarcinoma (D). The staining pattern for ANX1 in CP ranged from single nests of ANX1-positive ductal structures (A) to samples in which the majority of the remaining exocrine tissues were ANX1-positive (C); in one case, nuclear staining was also observable (B). In pancreatic carcinoma, tumor cells stained positive (D).
Discussion
In the last decades, knowledge concerning the molecular pathology of pancreatic ductal adenocarcinoma (PDAC) has increased markedly. However, this disease still remains an unresolved problem due to late diagnosis, low resectability, and the almost complete resistance to conventional radiotherapy/chemotherapy [10]. Several studies attempted to identify new diagnostic markers or new therapeutic targets for PDAC by gene expression microarray analysis of either bulk tissue [11,12] or microdissected pancreatic cancer tissue [13,14]. These studies have identified a multitude of differentially expressed genes in PDAC and have contributed to a better understanding of the aggressive behavior of PDAC. However, there is little overlap of identified genes among various gene expression studies. Thus, comparison of the results of several expression studies in PDAC revealed that 148 of 978 [15] and 64 of 568 [16] differentially expressed genes were identified in at least 2 of 10 studies analyzed. As discussed in the latter two publications, this low concordance may be due to type, histology, and number of samples used, or to different platforms and analysis procedures applied. Moreover, changes at the RNA level do not always correlate with protein expression [17]; therefore, in the last years, proteomic approaches have been used to identify differentially expressed proteins in PDAC [18,19]. Taking the results of these proteomic approaches, only about 20% to 30% of differentially expressed proteins were reflected by concomitant changes at the mRNA level [20]. However, these studies, too, showed only little concordance of the results, probably due to the same factors already discussed for transcriptomic approaches.
Recently, we have demonstrated that the expression of a mutated ki-ras oncogene in immortalized pancreatic duct epithelial cells yielded a tumorigenic transformed phenotype [6]. Using this in vitro model of pancreatic carcinogenesis, we sought to overcome some of the problems described above. First, by the use of two cell lines differing only in the expression of a mutated ki-ras, a comparison of different cell types found in pancreatic cancer tissues was avoided. Moreover, our survey of ras transformation targets should augment the chance to identify marker molecules with expression changes early in the course of pancreatic carcinogenesis, as ki-ras mutations are detectable already in about 40% of PanIN-1 lesions associated with PDAC [5]. Comparable strategies were applied in two studies either by the inhibition of the expression of a mutated ras using antisense k-ras-transduced AsPC-1 cells [21] or by the expression of a mutated k-ras in HPV16-E6E7-immortalized human pancreatic duct epithelial cells [22]. Both studies analyzed differentially expressed genes at the RNA level. In the former study using the differential display technique, 20 differentially expressed genes were identified, of which > 50% were mitochondrial genes. In the latter study using Affymetrix gene chip arrays, about 1050 differentially expressed genes induced by the expression of the mutated k-ras gene were identified. However, only 5% of these genes have been reported previously as differentially expressed in PDAC or pancreatic tumor cell lines. To overcome these limitations, we combined transcriptomic and proteomic techniques in one study to identify targets with differential expressions both at the mRNA level and at the protein level. Using this approach, we identified 28 unique genes and 6 ESTs differentially expressed at the mRNA level using SSH analysis, and 15 unique differentially expressed proteins using 2D PAGE and MS analysis. In our study, as in others [17,20], concordance between mRNA and protein data was only marginal. Thus, only highly abundant proteins such as actin and CKs were identified by both techniques. This may be due to the lower dynamic range of the 2D PAGE protocol, which allows less abundant proteins to escape detection [23]. However, many of the differentially expressed molecules identified with either method (14 of 28 genes and 6 of 15 proteins; Tables 2 and 3) already had been demonstrated to be differentially expressed in pancreatic adenocarcinoma or to be influenced by a mutated ras, thus proving the validity of our model. To further validate our model, the expression of several genes was analyzed by RT-PCR and immunohistochemistry in samples from normal human pancreas, chronic pancreatitis, and PDAC. For three of four genes analyzed (BBC1, rhoGDIα, and ANXA1), the trend observed in our model system was also detectable in PDAC samples, again proving the validity of our model.
Due to the high number of differentially expressed genes and proteins, only a selected set of these molecules will be discussed in more detail. The majority of the expression changes identified affected cytoskeletal proteins. The cytoskeleton is involved in numerous cellular functions such as cell motility, mitogenesis, morphology, muscle contraction, cytokinesis, and establishment of cell polarity. Deregulated expression and reorganization of cytoskeletal proteins are associated with the development of various forms of cancer [24]. In our study, we found an overexpression at the RNA level of CKs such as CK7, CK8, and CK19, as well as of myosin regulatory light chain. Overexpression of these CKs already has been reported in pancreatic carcinoma in several other gene expression studies [11,12], and myosin regulatory light chain has been shown to be involved in the invasion and adhesion of pancreatic cancer cells [25]. At the protein level, the results concerning CK expression were quite different. Here only a translationally modified isoform of CK8 was detectable, showing an increased apparent molecular weight compared to the theoretical value, and this isoform was upregulated in VArasmut cells.
A fragmented form of CK8 and additionally fragmented forms of the cytoskeletal proteins α tubulin, β tubulin, and vimentin were identified, which, except for β tubulin, were all downregulated in VArasmut cells. Although we cannot rule out that the fragmentation of these proteins happened during the isolation of the proteins, the other proteins identified in this study were not fragmented, which argues against this possibility. Proteolytic degradation of several proteins, including nuclear lamins, CKs, and vimentin, is a hallmark of the dramatic cytoskeletal reorganization that occurs during apoptosis [26–28]. Caspase-mediated fragmentation of CK18 and CK19 during apoptosis generated stable fragments with a defined molecular weight (CK18: 29 and 23 kDa; CK19: 28 and 20 kDa), whereas type II CKs (e.g., CK8) are virtually resistant to this degradation [27]. Because CK8 fragments have been detected, caspase-mediated fragmentation appears not to be the cause of the observed CK degradation. However, the release of proteolytically processed CKs (CK8, CK18, and CK19) into the culture medium was reported for the mammary tumor cell line MCF-7 [29]. This release of fragmented CKs resulted in the development of serum tumor markers such as TPS (CK18 fragment) and CYFRA 21-1 (CK19 fragment) [30]. Although these markers traditionally have been considered as markers of tumor proliferation, more recently, it has been shown that these markers are also released during the apoptosis of epithelial tumor cells [31]. Therefore, the reduced level of CK8 fragmentation may be an indicator of a reduced basal apoptotic rate in VArasmut cells.
The α and β tubulin fragments were slightly underrepresented and overrepresented in VArasmut cells, respectively. In contrast, vimentin fragments were markedly decreased in VArasmut cells. Like CKs, tubulin [32] and vimentin [33] are also cleaved during apoptosis. Caspase-mediated proteolysis of vimentin results in the generation of several fragments, including fragments of about 48 and 45 kDa [28], which were also identified in our study. Thus, the decreased fragmentation of α tubulin and vimentin again indicates a reduced basal rate of apoptosis in VArasmut cells.
Another large group of proteins with altered expression consisted of HSPs and cochaperones. All of these proteins (HSP70, HSP27, Grp58, and stress-induced phosphoprotein 1, also called HSP70/HSP90 organizing protein/HOP) were upregulated in VArasmut cells. HSPs were discovered as a group of proteins that are induced by various kinds of stress [34]; they facilitate the correct folding of other proteins under physiological and stress conditions with the help of cochaperones such as HOP [35]. Overexpressed HSP27 and HSP70 prevent the apoptosis induced by various stimuli, including hyperthermia, oxidative stress, CD95 ligation, or chemotherapy, by interfering with the action of key apoptotic proteins, such as Bid, Bax, apoptosis-inducing factor, and apoptosis protease-activating factor 1. Moreover, the signaling of survival factors through the PI3K/Akt pathway is promoted by the binding of HSP27 to Akt (reviewed in Garrido et al. [36]). HSPs are overexpressed in various forms of cancer, such as breast cancer [37], gastric cancer [38], colorectal carcinoma [39], and pancreatic carcinoma [40]. HSP27 and HSP70 expressions are associated with the invasion and metastasis of xenotransplanted human breast cancer cells [41] and with an increase in the tumorigenic potential of a mouse fibrosarcoma model [42], respectively. Increased HSP27 and HSP70 expression in acute myeloid leukemia, esophageal carcinomas, and colonic cancers has been associated with poor prognosis [43], resistance to radiotherapy and chemotherapy [44], and metastasis [45]. In contrast, pancreatic carcinoma patients with extensive HSP70 staining of tumor cells had a better prognosis [46]. In normal cells, the chaperone Grp58, a member of the protein disulfide isomerase family, is an integral part of the peptide-loading complex of the MHC class I molecules [47] and also is implicated in signaling by Stat3 [48]. Expression of Grp58 was induced in v-onc-transformed and v-src-transformed kidney cells and fibroblasts, respectively [49]. Additionally, overexpression of Grp58 was associated with chemoresistance in oral squamous cell carcinoma [50] and serous epithelial ovarian carcinoma [51]. However, inhibition of Grp58 expression by siRNA decreased mitomycin C-induced cytotoxicity in human colon carcinoma cells [52]. Thus, the increased expression of HSPs, HOP, and Grp58 observed in VArasmut cells may confer an increased resistance to apoptosis in cells. The resulting reduced apoptotic rate could be reflected by the observed reduced fragmentation of cytoskeletal proteins, as discussed above.
Two putative tumor-suppressor genes, GLTSCR2 and BBC1, were downregulated in ki-ras-transfected cells. Recently, the stabilization of the tumor-suppressor PTEN by the protein PICT-1, encoded by GLTSCR2, was demonstrated in human breast cancer MCF-7 cells [53]. Thus, the reduced expression of GLTSCR2 may explain the loss of PTEN observable in pancreatic cancer cells without concomitant mutation or promoter methylation of the PTEN gene [54,55]. The gene BBC1 located on chromosome 16 was originally described as a putative breast-tumor-suppressor gene [56]. Allelic loss of the chromosome region of BBC1 (16q22–q24) was associated with sporadic breast cancer [57]. However, in a later study, no tumor-specific mutations in the BBC1 gene were detected in a selected set of breast tumors that showed loss of heterozygosity (LOH) at 16q24, thus excluding BBC1 as a candidate breast-tumor-suppressor gene [58]. Also in prostate cancer, several studies demonstrated LOH of the BBC1 region in prostatic cancer [59,60], which increased in higher-grade tumors and metastases. Up to now, only one publication has reported a loss of the chromosomal region of BBC1 (16q22–q24) in pancreatic cancer [61], and in our small series of five pancreatic adenocarcinoma samples, a clear trend toward a reduced BBC1 expression was detectable, encouraging further studies on BBC1 as a tumorsuppressor gene in PDAC.
rhoGDIα is a cellular regulatory protein that acts primarily by controlling the cellular distribution and activity of rho GTPases [62], which transduce external signals to multiple downstream targets to elicit a variety of cellular responses such as organization of the actin cytoskeleton, cell cycle progression, cell polarity, and morphology [63]. GDIs typically act as negative regulators of rho GTPases through inhibition of GDP-GTP exchange [64]. Nevertheless, it has been demonstrated that rhoGDI, despite being a negative regulator of Cdc42 activation, is required for Cdc42-mediated cellular transformation [65]. Moreover, rhoGDI is overexpressed in a variety of cancers such as ovarian cancer [66], breast cancer [67], and lung cancer [68]. To our knowledge, except for the present study, there is only one additional publication concerning the overexpression of rhoGDIα in PDAC [69]. Additionally, rhoGDI overexpression is associated with chemoresistance in ovarian cancer [70], breast cancer [71], and melanoma [72]. Given the almost complete resistance of PDAC to conventional chemotherapy, the role of rhoGDIα overexpresson in pancreatic cancer warrants further analysis.
The protein annexin 1 was found to be overexpressed in VArasmut cells. Annexin 1, also called lipocortin 1, belongs to a large family of Ca2+-dependent phospholipid-binding proteins [73]. It is a glucocorticoid-regulated protein and shares many anti-inflammatory effects with these drugs, such as inhibition of cell proliferation, regulation of cell migration, and apoptosis [74,75]. Overexpression of annexin 1 has been reported for various cancers such as breast cancer [76] and hepatocellular carcinoma [77]. However, its downregulation was found, for example, in esophageal and prostate carcinoma [78]. In pancreatic carcinoma, overexpression of annexin 1 was demonstrated at the RNA level [12,14] and at the protein level [19] and correlated with a poorly differentiated phenotype of tumor cells [79]. Increased annexin 1 expression was demonstrated in drug-resistant tumor cells of the stomach [80], prostate [81], and breast [82]. Thus, the overexpression of annexin 1 may be responsible, at least in part, for the chemoresistance found in virtually all pancreatic carcinomas.
In conclusion, we have demonstrated that the transfection of a mutated ki-ras was accompanied by a specific expression pattern of several genes and proteins, which may result in an increase in malignant potential and an increased increased resistance to the apoptosis of transfected VArasmut cells. Thus, our in vitro model is a valuable tool that may be used to analyze the role of genetic alterations implicated in the early stages of tumor development in PDAC. The combination of two different approaches studying gene expression, namely, transcriptomics and proteomics, allowed for the identification of potential targets for the early diagnosis and/or therapy of pancreatic carcinoma, whereas these targets would have been missed by using only one of these two techniques.
Supplementary Material
Acknowledgements
We would like to thank Swelana Sander-Naderi, Sarina Lö ffler, and Anette Funk for technical assistance, and Wolfgang Hagmann for critical review of the manuscript.
Footnotes
This article refers to supplementary material, which is designated by “W” (i.e., Table W1) and is available online at www.bcdecker.com.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Freelove R, Walling AD. Pancreatic cancer: diagnosis and management. Am Fam Phys. 2006;73:485–492. [PubMed] [Google Scholar]
- 3.Morohoshi T, Held G, Klöppel G. Exocrine pancreatic tumours and their histological classification. A study based on 167 autopsy and 97 surgical cases. Histopathology. 1983;7:645–661. doi: 10.1111/j.1365-2559.1983.tb02277.x. [DOI] [PubMed] [Google Scholar]
- 4.Grapin-Botton A. Ductal cells of the pancreas. Int J Biochem Cell Biol. 2005;37:504–510. doi: 10.1016/j.biocel.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Löhr M, Klöppel G, Maisonneuve P, Lowenfels AB, Lüttges J. Frequency of K-ras mutations in pancreatic intraductal neoplasias associated with pancreatic ductal adenocarcinoma and chronic pancreatitis: a meta-analysis. Neoplasia. 2005;7:17–23. doi: 10.1593/neo.04445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Löhr M, Müller P, Zauner I, Schmidt C, Trautmann B, Thevenod F, Capella G, Farre A, Liebe S, Jesnowski R. Immortalized bovine pancreatic duct cells become tumorigenic after transfection with mutant k-ras. Virchows Arch. 2001;438:581–590. doi: 10.1007/s004280100397. [DOI] [PubMed] [Google Scholar]
- 7.Diehl F, Grahlmann S, Beier M, Hoheisel JD. Manufacturing DNA microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res. 2001;29:E38. doi: 10.1093/nar/29.7.e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fielden MR, Halgren RG, Dere E, Zacharewski TR. GP3: GenePix post-processing program for automated analysis of raw micro-array data. Bioinformatics. 2002;18:771–773. doi: 10.1093/bioinformatics/18.5.771. [DOI] [PubMed] [Google Scholar]
- 9.Mortz E, Krogh TN, Vorum H, Gorg A. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics. 2001;1:1359–1363. doi: 10.1002/1615-9861(200111)1:11<1359::AID-PROT1359>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 10.Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- 11.Friess H, Ding J, Kleeff J, Fenkell L, Rosinski JA, Guweidhi A, Reidhaar-Olson JF, Korc M, Hammer J, Büchler MW. Microarraybased identification of differentially expressed growth- and metastasisassociated genes in pancreatic cancer. Cell Mol Life Sci. 2003;60:1180–1199. doi: 10.1007/s00018-003-3036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iacobuzio-Donahue CA, Hruban RH. Gene expression in neoplasms of the pancreas: applications to diagnostic pathology. Adv Anat Pathol. 2003;10:125–134. doi: 10.1097/00125480-200305000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, Loader J, Terris B, Stamp G, Baron A, Scarpa A, Lemoine NR. Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene. 2002;21:4587–4594. doi: 10.1038/sj.onc.1205570. [DOI] [PubMed] [Google Scholar]
- 14.Grützmann R, Foerder M, Alldinger I, Staub E, Brummendorf T, Ropcke S, Li X, Kristiansen G, Jesnowski R, Sipos B, et al. Gene expression profiles of microdissected pancreatic ductal adenocarcinoma. Virchows Arch. 2003;443:508–517. doi: 10.1007/s00428-003-0884-1. [DOI] [PubMed] [Google Scholar]
- 15.Brandt R, Grützmann R, Bauer A, Jesnowski R, Ringel J, Löhr M, Pilarsky C, Hoheisel JD. DNA microarray analysis of pancreatic malignancies. Pancreatology. 2004;4:587–597. doi: 10.1159/000082241. [DOI] [PubMed] [Google Scholar]
- 16.Grützmann R, Boriss H, Ammerpohl O, Lüttges J, Kalthoff H, Schackert HK, Klöppel G, Saeger HD, Pilarsky C. Meta-analysis of microarray data on pancreatic cancer defines a set of commonly dysregulated genes. Oncogene. 2005;24:5079–5088. doi: 10.1038/sj.onc.1208696. [DOI] [PubMed] [Google Scholar]
- 17.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19: 1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen R, Yi EC, Donohoe S, Pan S, Eng J, Cooke K, Crispin DA, Lane Z, Goodlett DR, Bronner MP, et al. Pancreatic cancer proteome: the proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology. 2005;129:1187–1197. doi: 10.1053/j.gastro.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Crnogorac-Jurcevic T, Gangeswaran R, Bhakta V, Capurso G, Lattimore S, Akada M, Sunamura M, Prime W, Campbell F, Brentnall TA, et al. Proteomic analysis of chronic pancreatitis and pancreatic adenocarcinoma. Gastroenterology. 2005;129:1454–1463. doi: 10.1053/j.gastro.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Löhr JM, Faissner R, Findeisen P, Neumaier M. Proteome analysis—basis for individualized pancreatic carcinoma therapy? Internist (Berlin) 2006;47(1):S40–S48. doi: 10.1007/s00108-006-1634-7. [DOI] [PubMed] [Google Scholar]
- 21.Ohnami S, Matsumoto N, Nakano M, Aoki K, Nagasaki K, Sugimura T, Terada M, Yoshida T. Identification of genes showing differential expression in antisense K-ras-transduced pancreatic cancer cells with suppressed tumorigenicity. Cancer Res. 1999;59:5565–5571. [PubMed] [Google Scholar]
- 22.Qian J, Niu J, Li M, Chiao PJ, Tsao MS. In vitro modeling of human pancreatic duct epithelial cell transformation defines gene expression changes induced by K-ras oncogenic activation in pancreatic carcinogenesis. Cancer Res. 2005;65:5045–5053. doi: 10.1158/0008-5472.CAN-04-3208. [DOI] [PubMed] [Google Scholar]
- 23.Shen J, Person MD, Zhu J, Abbruzzese JL, Li D. Protein expression profiles in pancreatic adenocarcinoma compared with normal pancreatic tissue and tissue affected by pancreatitis as detected by two-dimensional gel electrophoresis and mass spectrometry. Cancer Res. 2004;64:9018–9026. doi: 10.1158/0008-5472.CAN-04-3262. [DOI] [PubMed] [Google Scholar]
- 24.Yamazaki D, Kurisu S, Takenawa T. Regulation of cancer cell motility through actin reorganization. Cancer Sci. 2005;96:379–386. doi: 10.1111/j.1349-7006.2005.00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaneko K, Satoh K, Masamune A, Satoh A, Shimosegawa T. Myosin light chain kinase inhibitors can block invasion and adhesion of human pancreatic cancer cell lines. Pancreas. 2002;24:34–41. doi: 10.1097/00006676-200201000-00005. [DOI] [PubMed] [Google Scholar]
- 26.Rao L, Perez D, White E. Lamin proteolysis facilitates nuclear events during apoptosis. J Cell Biol. 1996;135:1441–1455. doi: 10.1083/jcb.135.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ku NO, Liao J, Omary MB. Apoptosis generates stable fragments of human type I keratins. J Biol Chem. 1997;272:33197–33203. doi: 10.1074/jbc.272.52.33197. [DOI] [PubMed] [Google Scholar]
- 28.Byun Y, Chen F, Chang R, Trivedi M, Green KJ, Cryns VL. Caspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosis. Cell Death Differ. 2001;8:443–450. doi: 10.1038/sj.cdd.4400840. [DOI] [PubMed] [Google Scholar]
- 29.Chan R, Rossitto PV, Edwards BF, Cardiff RD. Presence of proteolytically processed keratins in the culture medium of MCF-7 cells. Cancer Res. 1986;46:6353–6359. [PubMed] [Google Scholar]
- 30.Pujol JL, Grenier J, Daures JP, Daver A, Pujol H, Michel FB. Serum fragment of cytokeratin subunit 19 measured by CYFRA 21-1 immunoradiometric assay as a marker of lung cancer. Cancer Res. 1993;53:61–66. [PubMed] [Google Scholar]
- 31.Sheard MA, Vojtesek B, Simickova M, Valik D. Release of cytokeratin-18 and -19 fragments (TPS and CYFRA 21-1) into the extracellular space during apoptosis. J Cell Biochem. 2002;85:670–677. doi: 10.1002/jcb.10173. [DOI] [PubMed] [Google Scholar]
- 32.Asumendi A, Andollo N, Boyano MD, Hilario E, Perez-Yarza G, Atencia R, Arechaga J, Garcia-Sanz M. The role of cleavage of cell structures during apoptosis. Cell Mol Biol (Noisy-le-Grand) 2000;46:1–11. [PubMed] [Google Scholar]
- 33.Lavastre V, Chiasson S, Cavalli H, Girard D. Viscum album agglutinin-I induces apoptosis and degradation of cytoskeletal proteins via caspases in human leukaemia eosinophil AML14. 3D10 cells: differences with purified human eosinophils. Br J Haematol. 2005;130:527–535. doi: 10.1111/j.1365-2141.2005.05633.x. [DOI] [PubMed] [Google Scholar]
- 34.Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 35.Odunuga OO, Longshaw VM, Blatch GL. Hop: more than an Hsp70/Hsp90 adaptor protein. Bioessays. 2004;26:1058–1068. doi: 10.1002/bies.20107. [DOI] [PubMed] [Google Scholar]
- 36.Garrido C, Brunet M, Didelot C, Zermati Y, Schmitt E, Kroemer G. Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell Cycle. 2006;5:2592–2601. doi: 10.4161/cc.5.22.3448. [DOI] [PubMed] [Google Scholar]
- 37.Tauchi K, Tsutsumi Y, Hori S, Yoshimura S, Osamura RY, Watanabe K. Expression of heat shock protein 70 and c-myc protein in human breast cancer: an immunohistochemical study. Jpn J Clin Oncol. 1991;21:256–263. [PubMed] [Google Scholar]
- 38.Liu X, Ye L, Wang J, Fan D. Expression of heat shock protein 90 beta in human gastric cancer tissue and SGC7901/VCR of MDR-type gastric cancer cell line. Chin Med J (Engl) 1999;112:1133–1137. [PubMed] [Google Scholar]
- 39.Kanazawa Y, Isomoto H, Oka M, Yano Y, Soda H, Shikuwa S, Takeshima F, Omagari K, Mizuta Y, Murase K, et al. Expression of heat shock protein (Hsp) 70 and Hsp 40 in colorectal cancer. Med Oncol. 2003;20:157–164. doi: 10.1385/MO:20:2:157. [DOI] [PubMed] [Google Scholar]
- 40.Ogata M, Naito Z, Tanaka S, Moriyama Y, Asano G. Overexpression and localization of heat shock proteins mRNA in pancreatic carcinoma. J Nippon Med Sch. 2000;67:177–185. doi: 10.1272/jnms.67.177. [DOI] [PubMed] [Google Scholar]
- 41.Lemieux P, Oesterreich S, Lawrence JA, Steeg PS, Hilsenbeck SG, Harvey JM, Fuqua SA. The small heat shock protein hsp27 increases invasiveness but decreases motility of breast cancer cells. Invasion Metastasis. 1997;17:113–123. [PubMed] [Google Scholar]
- 42.Jaattela M. Over-expression of hsp70 confers tumorigenicity to mouse fibrosarcoma cells. Int J Cancer. 1995;60:689–693. doi: 10.1002/ijc.2910600520. [DOI] [PubMed] [Google Scholar]
- 43.Steiner K, Graf M, Hecht K, Reif S, Rossbacher L, Pfister K, Kolb HJ, Schmetzer HM, Multhoff G. High HSP70-membrane expression on leukemic cells from patients with acute myeloid leukemia is associated with a worse prognosis. Leukemia. 2006;20:2076–2079. doi: 10.1038/sj.leu.2404391. [DOI] [PubMed] [Google Scholar]
- 44.Miyazaki T, Kato H, Faried A, Sohda M, Nakajima M, Fukai Y, Masuda N, Manda R, Fukuchi M, Ojima H, et al. Predictors of response to chemo-radiotherapy and radiotherapy for esophageal squamous cell carcinoma. Anticancer Res. 2005;25:2749–2755. [PubMed] [Google Scholar]
- 45.Wang XP, Qiu FR, Liu GZ, Chen RF. Correlation between clinicopathology and expression of heat shock protein 70 and glucoseregulated protein 94 in human colonic adenocarcinoma. World J Gastroenterol. 2005;11:1056–1059. doi: 10.3748/wjg.v11.i7.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sagol O, Tuna B, Coker A, Karademir S, Obuz F, Astarcioglu H, Kupelioglu A, Astarcioglu I, Topalak O. Immunohistochemical detection of pS2 protein and heat shock protein-70 in pancreatic adenocarcinomas. Relationship with disease extent and patient survival. Pathol Res Pract. 2002;198:77–84. doi: 10.1078/0344-0338-00190. [DOI] [PubMed] [Google Scholar]
- 47.Antoniou AN, Ford S, Alphey M, Osborne A, Elliott T, Powis SJ. The oxidoreductase ERp57 efficiently reduces partially folded in preference to fully folded MHC class I molecules. EMBO J. 2002;21:2655–2663. doi: 10.1093/emboj/21.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu X, Guo ZS, Marcu MG, Neckers L, Nguyen DM, Chen GA, Schrump DS. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst. 2002;94:504–513. doi: 10.1093/jnci/94.7.504. (April 3) [DOI] [PubMed] [Google Scholar]
- 49.Hirano N, Shibasaki F, Sakai R, Tanaka T, Nishida J, Yazaki Y, Takenawa T, Hirai H. Molecular cloning of the human glucose-regulated protein ERp57/GRP58, a thiol-dependent reductase. Identification of its secretory form and inducible expression by the oncogenic transformation. Eur J Biochem. 1995;234:336–342. doi: 10.1111/j.1432-1033.1995.336_c.x. [DOI] [PubMed] [Google Scholar]
- 50.Nakatani K, Nakamura M, Uzawa K, Wada T, Seki N, Tanzawa H, Fujita S. Establishment and gene analysis of a cisplatin-resistant cell line, Sa-3R, derived from oral squamous cell carcinoma. Oncol Rep. 2005;13:709–714. [PubMed] [Google Scholar]
- 51.Bernardini M, Lee CH, Beheshti B, Prasad M, Albert M, Marrano P, Begley H, Shaw P, Covens A, Murphy J, et al. High-resolution mapping of genomic imbalance and identification of gene expression profiles associated with differential chemotherapy response in serous epithelial ovarian cancer. Neoplasia. 2005;7:603–613. doi: 10.1593/neo.04760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su S, Adikesavan AK, Jaiswal AK. si RNA inhibition of GRP58 associated with decrease in mitomycin C-induced DNA cross-linking and cytotoxicity. Chem Biol Interact. 2006;162:81–87. doi: 10.1016/j.cbi.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 53.Okahara F, Ikawa H, Kanaho Y, Maehama T. Regulation of PTEN phosphorylation and stability by a tumor suppressor candidate protein. J Biol Chem. 2004;279:45300–45303. doi: 10.1074/jbc.C400377200. [DOI] [PubMed] [Google Scholar]
- 54.Okami K, Wu L, Riggins G, Cairns P, Goggins M, Evron E, Halachmi N, Ahrendt SA, Reed AL, Hilgers W, et al. Analysis of PTEN/MMAC1 alterations in aerodigestive tract tumors. Cancer Res. 1998;58:509–511. [PubMed] [Google Scholar]
- 55.Asano T, Yao Y, Zhu J, Li D, Abbruzzese JL, Reddy SA. The PI 3-kinase/Akt signaling pathway is activated due to aberrant Pten expression and targets transcription factors NF-kappaB and c-Myc in pancreatic cancer cells. Oncogene. 2004;23:8571–8580. doi: 10.1038/sj.onc.1207902. [DOI] [PubMed] [Google Scholar]
- 56.Adams SM, Helps NR, Sharp MG, Brammar WJ, Walker RA, Varley JM. Isolation and characterization of a novel gene with differential expression in benign and malignant human breast tumours. Hum Mol Genet. 1992;1:91–96. doi: 10.1093/hmg/1.2.91. [DOI] [PubMed] [Google Scholar]
- 57.Schmutzler RK, Fimmers R, Bierhoff E, Lohmar B, Homann A, Speiser P, Kubista E, Jaeger K, Krebs D, Zeillinger R, et al. Association of allelic losses on human chromosomal arms 11Q and 16Q in sporadic breast cancer. Int J Cancer. 1996;69:307–311. doi: 10.1002/(SICI)1097-0215(19960822)69:4<307::AID-IJC12>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 58.Moerland E, Breuning MH, Cornelisse CJ, Cleton-Jansen AM. Exclusion of BBC1 and CMAR as candidate breast tumoursuppressor genes. Br J Cancer. 1997;76:1550–1553. doi: 10.1038/bjc.1997.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Visakorpi T, Kallioniemi AH, Syvanen AC, Hyytinen ER, Karhu R, Tammela T, Isola JJ, Kallioniemi OP. Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res. 1995;55:342–347. [PubMed] [Google Scholar]
- 60.Hugel A, Wernert N. Loss of heterozygosity (LOH), malignancy grade and clonality in microdissected prostate cancer. Br J Cancer. 1999;79:551–557. doi: 10.1038/sj.bjc.6690087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang MC, Chang YT, Tien YW, Sun CT, Wu MS, Lin JT. Distinct chromosomal aberrations of ampulla of Vater and pancreatic head cancers detected by laser capture microdissection and comparative genomic hybridization. Oncol Rep. 2005;14:867–872. [PubMed] [Google Scholar]
- 62.Olofsson B. Rho guanine dissociation inhibitors: pivotal molecules in cellular signalling. Cell Signal. 1999;11:545–554. doi: 10.1016/s0898-6568(98)00063-1. [DOI] [PubMed] [Google Scholar]
- 63.Erickson JW, Cerione RA. Multiple roles for Cdc42 in cell regulation. Curr Opin Cell Biol. 2001;13:153–157. doi: 10.1016/s0955-0674(00)00192-7. [DOI] [PubMed] [Google Scholar]
- 64.Fukumoto Y, Kaibuchi K, Hori Y, Fujioka H, Araki S, Ueda T, Kikuchi A, Takai Y. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for the rho proteins, ras p21-like small GTP-binding proteins. Oncogene. 1990;5:1321–1328. [PubMed] [Google Scholar]
- 65.Lin Q, Fuji RN, Yang W, Cerione RA. RhoGDI is required for Cdc42-mediated cellular transformation. Curr Biol. 2003;13:1469–1479. doi: 10.1016/s0960-9822(03)00613-4. [DOI] [PubMed] [Google Scholar]
- 66.Jones MB, Krutzsch H, Shu H, Zhao Y, Liotta LA, Kohn EC, Petricoin EF., III Proteomic analysis and identification of new biomarkers and therapeutic targets for invasive ovarian cancer. Proteomics. 2002;2:76–84. [PubMed] [Google Scholar]
- 67.Fritz G, Brachetti C, Bahlmann F, Schmidt M, Kaina B. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Br J Cancer. 2002;87:635–644. doi: 10.1038/sj.bjc.6600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.MacKeigan JP, Clements CM, Lich JD, Pope RM, Hod Y, Ting JP. Proteomic profiling drug-induced apoptosis in non-small cell lung carcinoma: identification of RS/DJ-1 and RhoGDIalpha. Cancer Res. 2003;63:6928–6934. [PubMed] [Google Scholar]
- 69.Lu Z, Hu L, Evers S, Chen J, Shen Y. Differential expression profiling of human pancreatic adenocarcinoma and healthy pancreatic tissue. Proteomics. 2004;4:3975–3988. doi: 10.1002/pmic.200300863. [DOI] [PubMed] [Google Scholar]
- 70.Goto T, Takano M, Sakamoto M, Kondo A, Hirata J, Kita T, Tsuda H, Tenjin Y, Kikuchi Y. Gene expression profiles with cDNA microarray reveal RhoGDI as a predictive marker for paclitaxel resistance in ovarian cancers. Oncol Rep. 2006;15:1265–1271. [PubMed] [Google Scholar]
- 71.Zhang B, Zhang Y, Dagher MC, Shacter E. Rho GDP dissociation inhibitor protects cancer cells against drug-induced apoptosis. Cancer Res. 2005;65:6054–6062. doi: 10.1158/0008-5472.CAN-05-0175. [DOI] [PubMed] [Google Scholar]
- 72.Poland J, Sinha P, Siegert A, Schnolzer M, Korf U, Hauptmann S. Comparison of protein expression profiles between monolayer and spheroid cell culture of HT-29 cells revealed fragmentation of CK18 in three-dimensional cell culture. Electrophoresis. 2002;23:1174–1184. doi: 10.1002/1522-2683(200204)23:7/8<1174::AID-ELPS1174>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 73.Raynal P, Pollard HB. Annexins: the problem of assessing the biological role for a gene family of multifunctional calcium- and phospholipid-binding proteins. Biochim Biophys Acta. 1994;1197:63–93. doi: 10.1016/0304-4157(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 74.Kamal AM, Flower RJ, Perretti M. An overview of the effects of annexin 1 on cells involved in the inflammatory process. Mem Inst Oswaldo Cruz. 2005;100(1):39–47. doi: 10.1590/s0074-02762005000900008. [DOI] [PubMed] [Google Scholar]
- 75.Perretti M, D'Acquisto F. Novel aspects of annexin 1 and glucocorticoid biology: intersection with nitric oxide and the lipoxin receptor. Curr Drug Targets (Inflamm Allergy) 2006;5:107–114. doi: 10.2174/187152806776383170. [DOI] [PubMed] [Google Scholar]
- 76.Ahn SH, Sawada H, Ro JY, Nicolson GL. Differential expression of annexin I in human mammary ductal epithelial cells in normal and benign and malignant breast tissues. Clin Exp Metastasis. 1997;15:151–156. doi: 10.1023/a:1018452810915. [DOI] [PubMed] [Google Scholar]
- 77.Masaki T, Tokuda M, Ohnishi M, Watanabe S, Fujimura T, Miyamoto K, Itano T, Matsui H, Arima K, Shirai M, et al. Enhanced expression of the protein kinase substrate annexin in human hepatocellular carcinoma. Hepatology. 1996;24:72–81. doi: 10.1053/jhep.1996.v24.pm0008707286. [DOI] [PubMed] [Google Scholar]
- 78.Paweletz CP, Ornstein DK, Roth MJ, Bichsel VE, Gillespie JW, Calvert VS, Vocke CD, Hewitt SM, Duray PH, Herring J, et al. Loss of annexin 1 correlates with early onset of tumorigenesis in esophageal and prostate carcinoma. Cancer Res. 2000;60:6293–6297. [PubMed] [Google Scholar]
- 79.Bai XF, Ni XG, Zhao P, Liu SM, Wang HX, Guo B, Zhou LP, Liu F, Zhang JS, Wang K, et al. Overexpression of annexin 1 in pancreatic cancer and its clinical significance. World J Gastroenterol. 2004;10:1466–1470. doi: 10.3748/wjg.v10.i10.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sinha P, Hutter G, Kottgen E, Dietel M, Schadendorf D, Lage H. Increased expression of annexin I and thioredoxin detected by two-dimensional gel electrophoresis of drug resistant human stomach cancer cells. J Biochem Biophys Methods. 1998;37:105–116. doi: 10.1016/s0165-022x(98)00020-7. [DOI] [PubMed] [Google Scholar]
- 81.Carollo M, Parente L, D'Alessandro N. Dexamethasoneinduced cytotoxic activity and drug resistance effects in androgenindependent prostate tumor PC-3 cells are mediated by lipocortin 1. Oncol Res. 1998;10:245–254. [PubMed] [Google Scholar]
- 82.Wang Y, Serfass L, Roy MO, Wong J, Bonneau AM, Georges E. Annexin-I expression modulates drug resistance in tumor cells. Biochem Biophys Res Commun. 2004;314:565–570. doi: 10.1016/j.bbrc.2003.12.117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.