Abstract
The lipid peroxidation product 4-hydroxynon-2-enal (4-HNE) forms as a consequence of oxidative stress, and acts as a signaling molecule or, at superphysiological levels, as a toxicant. The steady-state concentration of the compound reflects the balance between its generation and its metabolism, primarily through glutathione conjugation. Using an RNAi-based screen, we identified in Caenorhabditis elegans five glutathione transferases (GSTs) capable of catalyzing 4-HNE conjugation. RNAi knock-down of these GSTs (products of the gst-5, gst-6, gst-8, gst-10, and gst-24 genes) sensitized the nematode to electrophilic stress elicited by exposure to 4-HNE. However, interference with the expression of only two of these genes (gst-5 and gst-10) significantly shortened the life span of the organism. RNAi knock-down of the other GSTs resulted in at least as much 4-HNE adducts, suggesting tissue-specificity of effects on longevity. Our results are consistent with the oxidative stress theory of organismal aging, broadened by considering electrophilic stress as a contributing factor. According to this extended hypothesis, peroxidation of lipids leads to the formation of 4-HNE in a chain reaction which amplifies the original damage. 4-HNE then acts as an "aging effector" via the formation of 4-HNE-protein adducts, and a resulting change in protein function.
Keywords: Longevity, Aging, 4-Hydroxynonenal, 4-HNE, Caenorhabditis elegans, Glutathione transferase, GST
1. Introduction
A role for the accumulation of oxidative and free radical damage in aging was proposed many decades ago (Pearl, 1928; Harman, 1956). This theory went through numerous modifications and refinements, but has withstood the test of time remarkably well, and remains a major paradigm of research into the aging process. Recent work suggests that other types of molecular damage, brought about by reactive xenobiotics and/or products of endogenous metabolism, also play a key role in organismal senescence (McElwee et al., 2004; Gems and McElwee, 2005). Integrating the two lines of reasoning, we proposed an extension to the oxidative damage theory of aging. This hypothesis (Ayyadevara et al., 2005a; Ayyadevara et al., 2005b; McEwen et al., 2005) postulates that the lipid peroxidation chain reaction, initiated by a reaction of ROS (reactive oxygen species) with lipids, amplifies an original oxidative insult. The end products of lipid peroxidation, in particular electrophilic aldehydes exemplified by 4-HNE (4-hydroxynon-2-enal) are the effectors which act in parallel with ROS to cause molecular damage, and ultimately aging. We have previously provided evidence for the above hypothesis by experimentally modulating the capacity of Caenorhabditis elegans to metabolize 4-HNE. To this end, we transgenically overexpressed glutathione transferases (GSTs) with high catalytic activity for 4-HNE, including the murine enzyme mGSTA4-4, and the endogenous C. elegans CeGSTP2-2 (the gst-10 gene product), or knocked down the latter using RNAi, and observed the predicted effects on life span of the nematode (Ayyadevara et al., 2005a; Ayyadevara et al., 2005b).
Forty-four annotated gst genes, and a number of additional GST-like proteins, are listed in release 156 of WormBase (www.wormbase.org). A bioinformatics analysis of the C. elegans genome identified 57 genes encoding proteins that match the C-terminal portion of GSTs (supplemental Table S4 in Holt et al., 2002). Therefore, we posed the question whether C. elegans GSTs other than the already characterized gst-10 gene product have the ability to modulate life span by affecting 4-HNE levels in critical cells of the organism. To answer this question, in the present work we carried out an RNAi-based screen of GSTs. The results indicate that, in addition to the gst-10 gene product, at least 4 other C. elegans GSTs have the ability to metabolize 4-HNE to a significant extent. Each of these 5 enzymes protects the organism against electrophilic stress caused by exposure to 4-HNE, but only two of these significantly affect life span.
2. Materials and Methods
2.1. C. elegans culture conditions
C. elegans strain Bristol-N2 was used in all experiments except for the life span reported in Fig. 9 which was carried out on strain NL2099, obtained from the Caenorhabditis Genetics Center (St. Paul, MN). The animals were cultured at 20°C in nematode growth medium (NGM: 25 mM potassium phosphate, pH 6.0, 50 mM NaCl, 0.25% (w/v) peptone, 0.5% (w/v) cholesterol, 1 mM MgCl2, 1 mM CaCl2) and fed with Escherichia coli strain OP50 (2 × 109 cells/ml) grown in 3XD medium (10.5 g/l of Na2HPO4, 4.5 g/l of KH2PO4, 0.6 g/l of NH4Cl, 15 g/l of casein hydrolysate, 24 g/l of glycerol and 3 ml/l of 1 M MgSO4). In RNAi studies, worms were fed E. coli strain HT115(DE3) transformed with an insert-free L4440 feeding vector (control) or with the appropriate gst clone (in the same vector) from the RNAi library (Kamath and Ahringer, 2003; Kamath et al., 2003) purchased from Geneservice Ltd, Cambridge, U.K. The bacteria were treated with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) to induce the expression of double-stranded RNA. The identity of selected clones from the library, including gst-5, gst-6, gst-8, and gst-24, was confirmed by partial sequencing. RNAi targeted to gst-10, a gene not represented in the RNAi library, was carried out as described previously (Ayyadevara et al., 2005a).
Fig. 9.
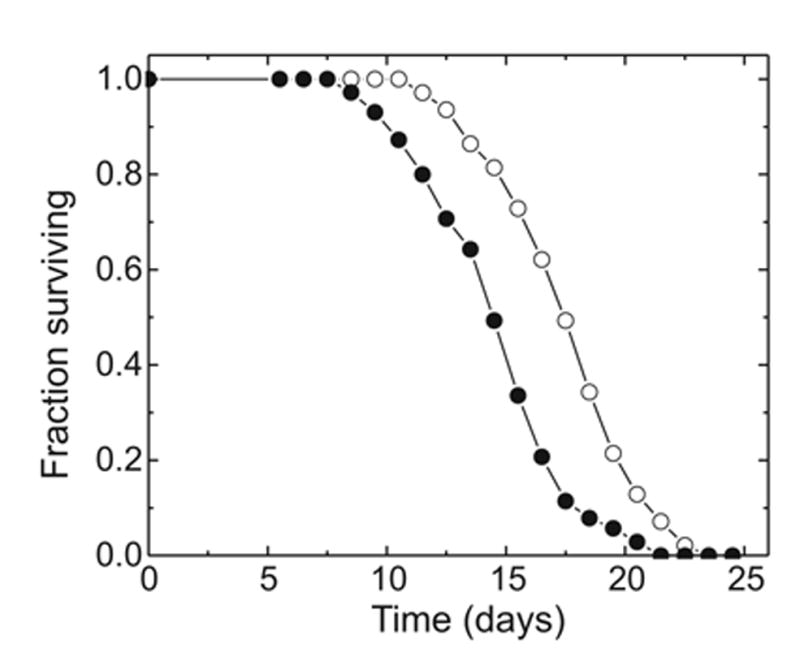
Reduction of life span in C. elegans strain NL2099 by RNAi targeted to gst-10. Open symbols: control worms fed bacteria transformed with insert-free vector; closed symbols: RNAi against gst-10. Each group consisted of 150 worms. The median life span is shortened by gst-10 knock-down by 17.3%, as calculated from approximating the survival curves by fitting the Gompertz function.
2.2. Resistance of C. elegans to electrophilic stress caused by 4-HNE
4-HNE was synthesized according to Gree et al. (1986) and Chandra and Srivastava (1997). Worms were transferred two days after hatching onto plates containing E. coli expressing the appropriate double-stranded RNA (see section 2.1), and were maintained under these conditions (with daily transfers to fresh plates) for 3 days. The animals were then rinsed off the plates using S buffer (0.1 M NaCl, 0.05 M potassium phosphate, pH 6.0) (Brenner, 1974) supplemented with 0.5% cholesterol, and were suspended in the same solution containing 10 mM 4-HNE. Worms were placed in 24-well plates at 50 worms in 0.3 ml buffer per well. Plates were kept at 20°C, and the worms were scored every hour for survival. Fifty animals were used per experimental group.
2.3. Life span determinations of C. elegans
Longevity of C. elegans in which individual GSTs were knocked down by RNAi was measured as described previously (Ayyadevara et al., 2005a).
2.4. Biochemical methods
Pellets of freshly harvested (for 4-HNE-conjugating activity assays) or frozen (for all other biochemical assays) C. elegans were homogenized in 20 mM potassium phosphate, 1.4 mM 2-mercaptoethanol, pH 7.5, in 1.5-ml microcentrifuge tubes using a fitting pestle (Pellet Pestle, Kimble/Kontes, Vineland, NJ), followed by sonication for 10 s using a tip sonicator. Enzymatic activity of 4-HNE conjugation to glutathione was determined in worm homogenates as described by Alin et al. (1985). Western blots to detect CeGSTP2-2 were probed with an antibody generated by us previously (Ayyadevara et al., 2005b). 4-HNE-protein adducts were quantitated by competitive ELISA (Satoh et al., 1999) using a polyclonal antibody against 4-HNE-modified keyhole limpet hemocyanin generously provided by Dr. Dennis R. Petersen, University of Colorado, Denver. For determination of 4-HNE-protein adducts in worms subjected to RNAi, the animals were transferred two days after hatching onto plates containing E. coli expressing the appropriate double-stranded RNA (see section 2.1), and were maintained under these conditions (with daily transfers to fresh plates) for 2 days.
2.5. Statistics
The statistical tests used are mentioned in the description of the individual experiments. Statistical procedures were carried out using the NCSS software package (Number Cruncher Statistical Systems, Kaysville, UT).
3. Results
3.1. A subset of C. elegans GSTs is capable of conjugating 4-HNE
RNA interference targeted to the subset of twenty-six gst genes represented in the Ahringer RNAi library (Kamath and Ahringer, 2003; Kamath et al., 2003) was carried out in wild-type C. elegans (strain Bristol-N2). In addition, gst-10 was targeted as described previously (Ayyadevara et al., 2005a). Assays of worm homogenates demonstrated that knock-down of CeGSTP2-2 (gst-10 gene product) reduced 4-HNE-conjugating activity, as previously reported (Ayyadevara et al., 2005b). Furthermore, RNAi targeted to an additional four out of the twenty-seven GSTs tested (gst-5, gst-6, gst-8, and gst-24) decreased 4-HNE-conjugating activity to a level equal or lower of that seen for RNAi against gst-10 (Fig. 1). An independent biological replication of the experiment was carried out using RNAi targeted to the above five GSTs, plus several others which had a marginal effect in the first screen (a total of ten GSTs: gst-1, -3, -5, -6, -8, -10, -12, -16, -24, -38, and control). The second experiment yielded a profile similar to the first with respect to activity decreases relative to control (data not shown). The results of both experiments were analyzed together using two-factor ANOVA with experiment as one factor and RNAi as the other. The resulting estimate of the common standard deviation (ANOVA root MSE) was 7% of the activity of the control. This low coefficient of variation indicates reproducibility of the RNAi intervention. Overall, the screen indicates that at least five of the multiple C. elegans GSTs have the ability to conjugate 4-HNE with glutathione.
Fig. 1.
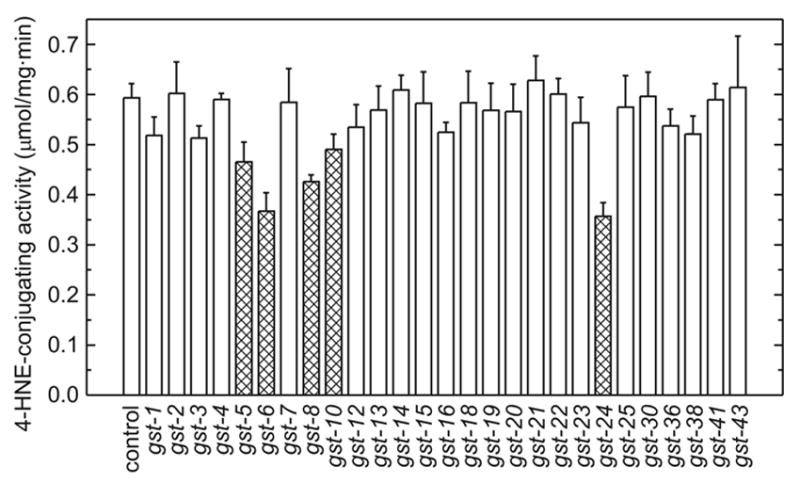
Effect of RNAi targeted to individual C. elegans GSTs on 4-HNE-conjugating enzyme activity in whole-worm homogenates. C. elegans strain Bristol-N2 was fed bacteria expressing double-stranded RNA as described in the Materials and Methods section. Animals were collected on day 5 after hatching, homogenized without freezing, and 4-HNE-conjugating activity was determined. The bars represent means ± S.D. of triplicate enzyme activity determinations carried out on the same homogenate. Cross-hatched bars represent RNAi treatments for which the resulting mean enzymatic activity is equal to or lower than that for RNAi targeted to gst-10. Control animals were fed bacteria transformed with the insert-free vector.
The specificity of RNAi is frequently of concern (Qiu et al., 2005; Birmingham et al., 2006), especially if the method is applied to members of a multi-gene family such as glutathione transferases. We have shown previously that RNAi targeted to gst-10 is likely to be specific (Ayyadevara et al., 2005a). There appears to be little or no cross-talk between gst-10 and gst-5, gst-6, gst-8, or gst-24-directed RNAi, since these treatments did not substantially decrease the expression of CeGSTP2-2, the product of the gst-10 gene (Fig. 2). While it is not practical to test possible RNAi mis-targeting for all gst pairs, the available evidence indicates considerable specificity, in agreement with an overall estimate of less than 1% false positives in the RNAi library used (Kamath et al., 2003). Therefore, the data presented in Fig. 1 suggest that gst-5, gst-6, gst-8, gst-10, and gst-24 together account for most of the 4-HNE-conjugating activity in C. elegans since the sum of activity decrements caused by these five RNAi interventions equals or exceeds the total activity in control worm homogenate.
Fig. 2.

Western blot of C. elegans in which individual GSTs (as labeled in the Figure) were knocked down by RNAi, probed with antibody against CeGSTP2-2 (gst-10 gene product). Control: worms fed bacteria transformed with insert-free vector. The blot was re-probed with anti-β-actin antibody to check for equal loading.
3.2. Knock-down of selected GSTs affects susceptibility of C. elegans to 4-HNE
To test whether RNAi-mediated knock-down of GSTs affects the susceptibility of C. elegans to electrophilic stress, worms were exposed to 10 mM 4-HNE, and survival was recorded. As shown in Fig. 3, RNAi targeted to gst-6 and gst-24 shortened survival more than two-fold. Knocking down gst-3, gst-5, gst-10, gst-13, and gst-15 had a lesser but still pronounced effect on electrophile susceptibility. Interference with several other GSTs, listed in Fig. 3C, had a statistically significant but relatively minor effect on stress resistance, while a large group of GSTs (Fig. 3D) had no effect. Median survival times for all knock-downs, calculated by a Gompertz approximation of the survival curves, are shown in Fig. 4.
Fig. 3.

Electrophile stress resistance of C. elegans in which individual GSTs were knocked down by RNAi. Worms fed bacteria expressing double-stranded RNA were exposed to 10 mM 4-HNE, and their survival time was recorded. Control: worms fed bacteria transformed with insert-free vector. For presentation clarity, the survival curves are divided into four groups: GSTs whose knock-down causes a lowering of resistance to 4-HNE versus control at P < 10−100 are shown in panel A; those with 10−100 < P ≤ 10−8, in panel B; those with 10−8 < P ≤ 0.05, in panel C; and those not significantly different from control (P > 0.05), in panel D. The control is common for the four panels. RNAi targeted to individual GSTs is denoted by distinct symbols listed in the respective panels. Each survival curve shown in the Figure represents a mean of 3 independent experiments with 50 worms per experiment. The P values listed above were calculated by Cox regression utilizing the original data from the 3 replicate experiments, adjusting for inter-experiments variability, and applying the Holm correction (Holm, 1979) for multiple comparisons versus control.
Fig. 4.
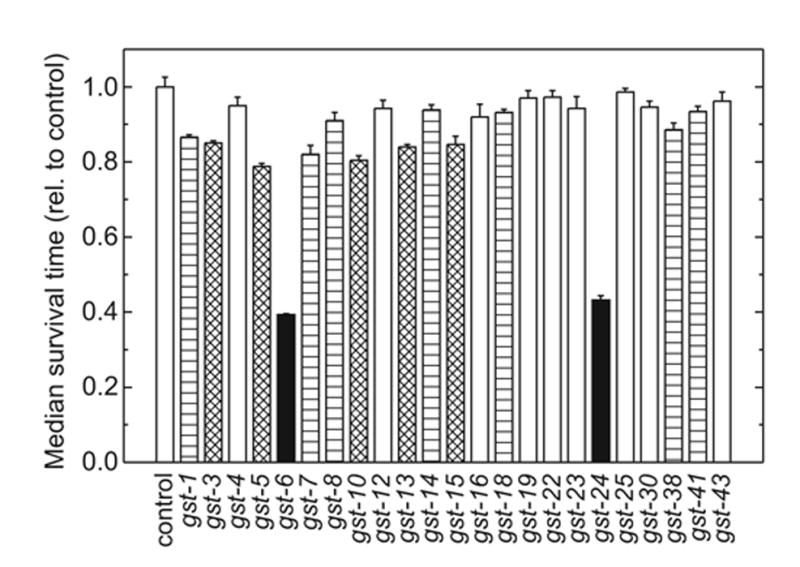
Median survival time in the presence of 10 mM 4-HNE of C. elegans in which individual GSTs were knocked down by RNAi. The Gompertz function was fitted to each survival curve shown in Fig. 3, and the resulting calculated median survival times ± asymptotic standard errors, normalized to those of control animals (5.00 ± 0.13 hr), are plotted. GSTs whose knock-down causes a lowering of resistance to 4-HNE versus control at P < 10−100 are shown as filled bars; those with 10−100 < P ≤ 10−8, as cross-hatched bars; those with 10−8 < P ≤ 0.05, as horizontally hatched bars; and those not significantly different from control (P > 0.05), as open bars. P values were calculated as described in the legend to Fig. 3.
Millimolar concentrations of 4-HNE were required to cause acute mortality in C. elegans. The range of effective 4-HNE concentrations was fairly narrow. For example, a preliminary experiment (not shown) revealed no mortality within 16 hr after exposure to 2.5 mM 4-HNE, while 10 mM (as used in the present work) caused 100% mortality in approximately 8 hr (Fig. 3). Millimolar 4-HNE concentrations significantly exceed those measured physiologically, at least in mammals; for example, 4-HNE was variously reported to be 0.03 ± 0.01 μM (Spies-Martin et al., 2002) or 0.7 ± 0.4 μM (Selley et al., 1989; Strohmaier et al., 1995) in normal human plasma, and perhaps ten times that during oxidative stress (Kimura et al., 2005; Zhang et al., 2005). However, we have now shown experimentally that effects of 4-HNE (as measured by the biologically relevant adduct formation on proteins) are equivalent for C. elegans treated with 10 mM 4-HNE, and cultured mammalian cells treated with 10 μM 4-HNE (Fig. 5).
Fig. 5.

Total 4-HNE-protein adduct formation in C. elegans treated with millimolar 4-HNE (open circles and upper abscissa), and in cultured mammalian cells (mouse embryonic fibroblasts) treated with micromolar 4-HNE (open squares and lower abscissa). Five-day-old worms in S-buffer (see Materials and Methods) supplemented with 0.5% cholesterol, and mouse embryonic fibroblasts in serum-free medium were treated with the indicated concentration of 4-HNE for 6 hr, homogenized, and assayed for 4-HNE adducts as described in Materials and Methods. Means ± S.D. of triplicate measurements on the same homogenate are shown; error bars smaller than the plotting symbol are drawn inside the symbol.
3.3. 4-HNE-protein adduct levels increase in C. elegans with age
The level of 4-HNE adducts increased with age in C. elegans (Fig. 6). The steady-state concentration of 4-HNE-protein adducts could reflect the rate of formation, which depends on the average tissue concentration of free 4-HNE, the rate of degradation of the modified proteins, or both. Regardless of the mechanism, the increase in 4-HNE adducts could be relevant to aging since it is thought that the biological effects of 4-HNE are mediated by adduct formation, predominantly on proteins.
Fig. 6.
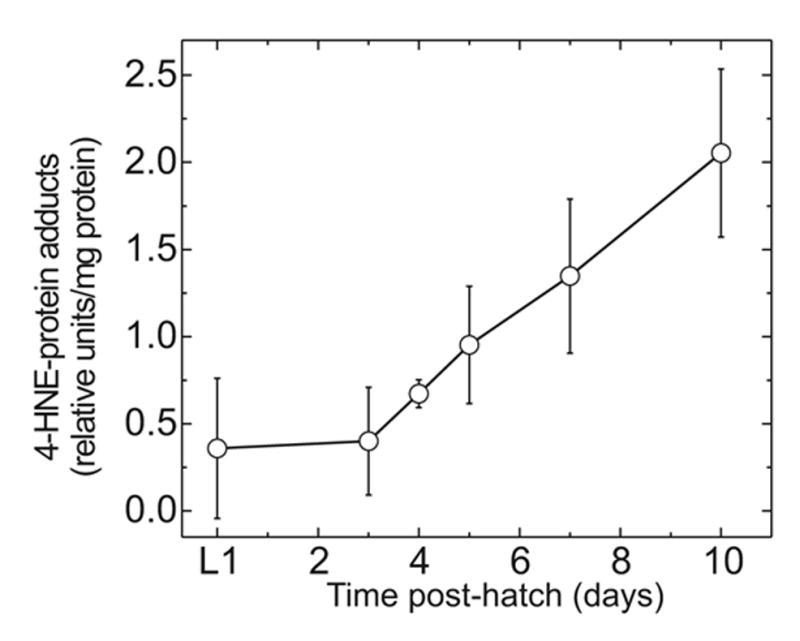
Level of 4-HNE-protein adducts in C. elegans Bristol-N2 as a function of age. Adducts were measured by ELISA as described in Materials and Methods. The values shown are means ± S.D. of three independent experiments, each normalized to its average level of adducts over the time period measured (0 to 10 days). Analysis of the data by General Linear Model ANOVA indicates that the adduct levels are not equal at the different ages at which they were measured (P = 0.0015).
3.4. Effect of RNAi-mediated GST knockdown on C. elegans life span
The increase of 4-HNE-protein adducts with age (Fig. 6) could indicate a cause-effect relationship. To test this hypothesis, the subset of C. elegans GSTs shown by RNAi knock-down to have 4-HNE-conjugating activity, i.e., gst-5, gst-6, gst-8, gst-10, and gst-24 (Fig. 1), was selected for the determination of a possible effect on life span. As shown in Fig. 7G and H, interference with the expression of the gst-10 gene reduced life span, in agreement with our previous results (Ayyadevara et al., 2005a). RNAi against gst-5 had a similar effect (Fig. 7A and B). In contrast, RNAi against gst-6 (Fig. 7C and D), gst-8 (Fig. 7E and F) and gst-24 (Fig. 7I and J) did not significantly affect life span under the conditions used.
Fig. 7.
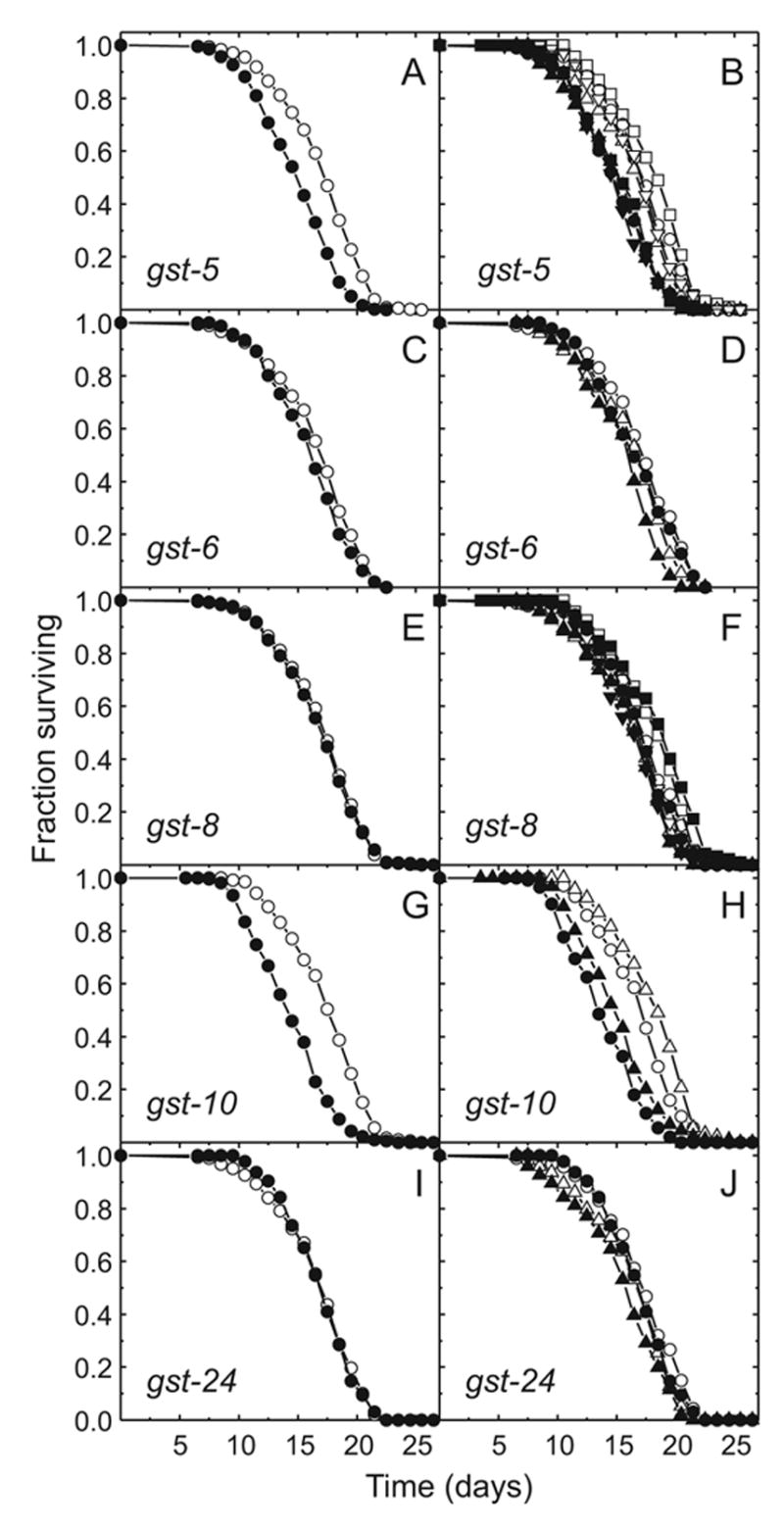
Effect of GST knock-down by RNAi on life span of C. elegans Bristol-N2. The life span of control animals fed bacteria transformed with insert-free vector (open symbols) was recorded simultaneously with that of animals in which individual GSTs (listed in the Figure) were knocked down by RNAi (closed symbols). Panels A, C, E, G, and I show means of independent life span determinations plotted separately and denoted by distinct symbols in panels B (4 independent experiments), D (2 experiments), F (4 experiments), H (2 experiments), and J (2 experiments), respectively. Each survival curve shown in panels B, D, F, H, and J represents a population of 100 or 150 worms. For each GST, data from the individual experiments were analyzed by Cox regression. After adjusting for inter-experiment variability and applying the Holm correction for multiple comparisons (Holm, 1979), RNAi targeted to gst-5 was found to be different from control at P = 4.8 × 10−21, gst-6 at P = 0.09, gst-8 at P = 0.27, gst-10 at P = 4.8 × 10−20, and gst-24 at P = 0.14.
3.5. Effect of RNAi-mediated GST knockdown on 4-HNE-protein adduct formation
We have previously shown that RNAi directed against gst-10 leads to an increased level of 4-HNE-protein adducts (Ayyadevara et al., 2005a; Ayyadevara et al., 2005b). We now extend this observation to additional GSTs which were identified (Fig. 1) as having 4-HNE-conjugating activity. A knock-down of these GSTs caused an up to two-fold increase in the levels of 4-HNE-protein adducts which correlated well (R2 = 0.5) with the decrement of 4-HNE-conjugating activity resulting from RNAi (Fig. 8A). For the two GSTs (gst-5 and gst-10 gene products) whose knock-down has been shown to affect longevity (Fig. 7), there was also a good inverse correlation (R2 = 0.7) between the amount of 4-HNE-protein adducts formed and median life span (Fig. 8B). In contrast, an equal or even greater increase of 4-HNE adducts due to interference with gst-6, gst-8, or gst-24 expression had little effect of life span (Fig. 8B). This indicates that accumulation of excessive 4-HNE-protein adducts is by itself not sufficient to affect longevity. To influence aging, 4-HNE adducts may need to accumulate in particular tissues, such as these to which the gst-5 and gst-10 gene products are localized.
Fig. 8.
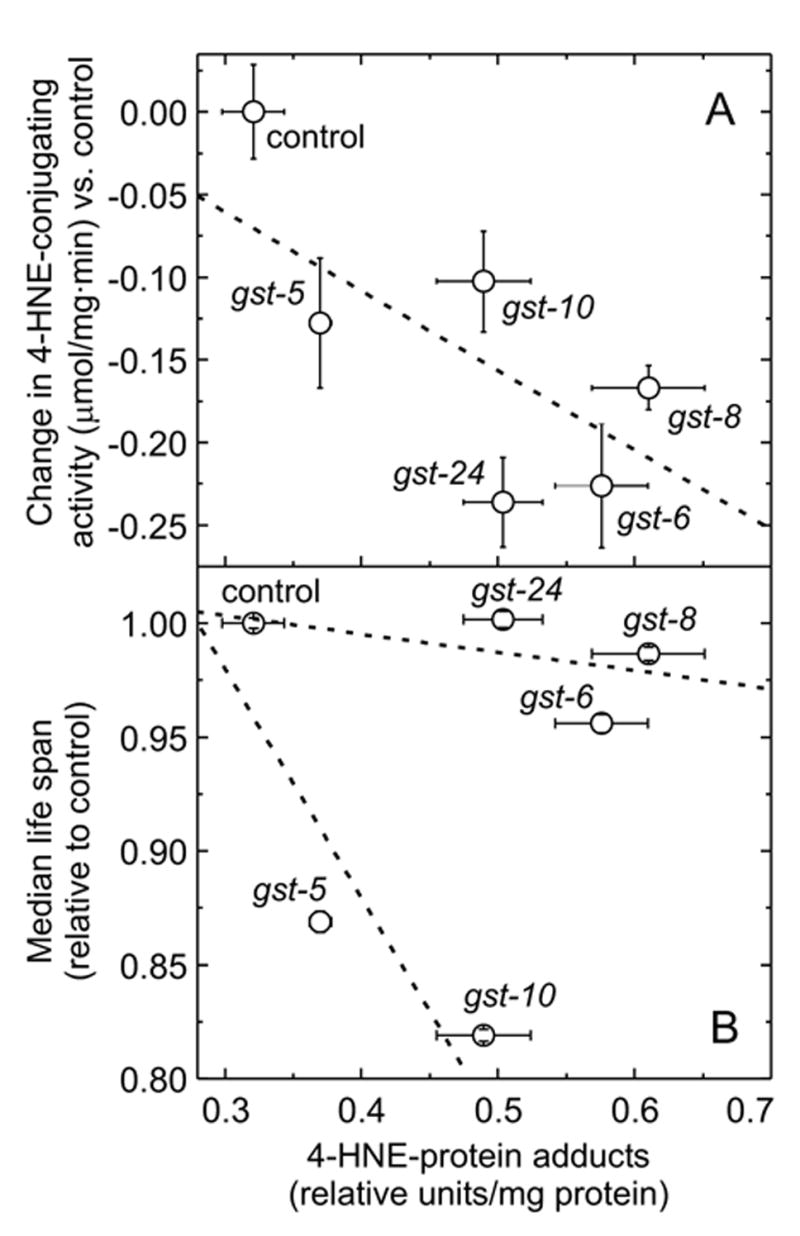
Effect of GST knock-down by RNAi on formation of 4-HNE-protein adducts. The amount of 4-HNE-protein adducts in whole-body homogenates of worms subjected to RNAi against gst-5, gst-6, gst-8, gst-10, and gst-24 is shown on the abscissa; control: insert-free feeding vector. Panel A: 4-HNE-protein adducts correlate (R2 = 0.50) with the decrement of 4-HNE-conjugating activity (taken from Fig. 1) in worms in which expression of individual GSTs was knocked down by RNAi. Panel B: for control animals and worms subjected to RNAi against gst-5 and gst-10, 4-HNE-protein adducts correlate well (R2 = 0.70) with the decrease in median life span (taken from Fig. 7). For control animals and worms subjected to RNAi against gst-6, gst-8, and gst-24, the correlation of 4-HNE-protein adducts with change in median life span is poor (R2 = 0.37). Regression lines were calculated using as the weight for each point the square root of the product of the standard deviations in the x-dimension and y-dimension. Error bars are shown inside the plotting symbol when they are smaller than the symbol diameter.
3.6. Efficacy of RNAi in Bristol-N2 and in the sensitized NL2099 strain of C. elegans
With the exception of four proteins, no information is currently available on the tissue localization of C. elegans GSTs. The four GSTs whose tissue distribution has been at least partially characterized include the gst-10 gene product which has a limited tissue distribution consistent with expression in sensory neurons (Ayyadevara et al., 2005b, and the C. elegans Gene Expression Consortium), and additional three GSTs (gst-4, gst-33, and gst-38 gene products) characterized by the C. elegans Gene Expression Consortium (http://elegans.bcgsc.ca/perl/eprofile/index). At least some neuronally expressed genes have been shown to be relatively or completely refractory to RNAi-mediated knock-down in wild-type C. elegans (Tavernarakis et al., 2000) but not in the sensitized NL2099 strain (Simmer et al., 2002). To test whether the observed reduction of life span caused by RNAi targeted to gst-10 (Ayyadevara et al., 2005a) (Fig. 7) could be blunted by an incomplete knock-down, RNAi against gst-10 was used in the NL2099 strain. A comparison of the resulting decrease in life span (Fig. 9) with that caused by gst-10-directed RNAi in the Bristol-N2 strain (Fig. 7G and H) indicates that the effect of RNAi against gst-10 is similar in both strains. This is reflected in a similar reduction of median life span (by 17.3% in NL2099 versus 18.1% in Bristol-N2, as calculated from fitting of the Gompertz function to the survival curves). The equal susceptibility of gst-10 to RNAi in the wild-type and the RNAi-hypersensitive strain does not rule out a possible localization of gst-10 expression in sensory neurons since amphids (a subset of sensory neurons, Lanjuin and Sengupta, 2004) have been previously shown to be sensitive to RNAi (Bianchi et al., 2003).
4. Discussion
We have previously reported that 4-HNE, and/or a chemically related product of lipid peroxidation, affects life span of C. elegans (Ayyadevara et al., 2005a; Ayyadevara et al., 2005b). A part of the experimental evidence supporting this conclusion was the finding that overexpression of CeGSTP2-2, the gst-10 gene product, extends life span of C. elegans, while RNAi knock-down of gst-10 has the opposite effect. However, both immunodepletion of CeGSTP2-2 and the yield of its biochemical purification indicated that the enzyme accounts for only a quarter to a third of the total 4-HNE-conjugating activity of the worm (Ayyadevara et al., 2005b). We have now used an RNAi screen to identify additional GST(s) with 4-HNE-conjugating activity. Of the 44 gst genes currently listed in WormBase, 26 are represented in the Ahringer RNAi library (Kamath and Ahringer, 2003; Kamath et al., 2003). In addition, we have previously developed RNAi for gst-10 (Ayyadevara et al., 2005a). A screen of the 27 genes, using 4-HNE-conjugating enzymatic activity as the endpoint, identified a subset of 5 GSTs (gst-5, gst-6, gst-8, gst-10, and gst-24) whose knock-down decreased the ability of the worm homogenate to conjugate 4-HNE with glutathione (Fig. 1). As expected, this subset contained the previously found gst-10 (Engle et al., 2001; Ayyadevara et al., 2005b). RNAi targeted to gst-10 led to the loss of 17% of total activity (Fig. 1), in reasonable agreement with the 20 – 25% estimated from immunodepletion studies (Ayyadevara et al., 2005b).
Although the screen covered only 27 out of 44, or approximately two-thirds, of the known gst genes, the sum of activity decrements observed upon RNAi knock-down of the 5 enzymes readily accounts for the total 4-HNE-conjugating activity of control worms, in spite of the fact that RNAi is rarely quantitative. It should be pointed out, however, that errors associated with RNAi-triggered activity decrements are compounded upon summation. Moreover, off-target RNAi, if present, could also skew the results by overestimating the contributions of individual GSTs to the total activity. In light of these uncertainties, the possibility cannot be ruled out that some of the GSTs not represented in the RNAi library (and thus not tested) may also have 4-HNE-conjugating activity. It appears likely, however, that many or most C. elegans GSTs with that activity have been identified, with the possible exception of enzymes expressed at low levels. Low-abundance GSTs could be missed, even if they had a high catalytic efficiency for 4-HNE, since the screen summarized in Fig. 1 relies on total 4-HNE-conjugating activity in C. elegans lysates.
A second RNAi screen of gst genes utilized susceptibility to 4-HNE-elicited electrophilic stress as the endpoint (Fig. 3 and Fig. 4). Not surprisingly, the 5 enzymes found to have 4-HNE-conjugating activity were also protective against acute 4-HNE toxicity. Enzyme activity and survival time in the presence of 4-HNE correlated highly for C. elegans knock-downs of these 5 enzymes (Fig. 10A; R2 = 0.76, P = 0.02). In addition, loss of GSTs other than the 5 enzymes characterized by 4-HNE-conjugating activity sensitized C. elegans to 4-HNE stress. In these cases, the effect was statistically significant but generally smaller than that observed for some of the 4-HNE-metabolizing enzymes, most notably for gst-6 and gst-24 (Fig. 3 and Fig. 4). It is possible that exposure to 4-HNE leads to secondary events such as a generalized oxidative stress (Uchida, 2003) against which GSTs can be protective, even if they lack the ability to conjugate 4-HNE which was the primary trigger of the stress.
Fig. 10.
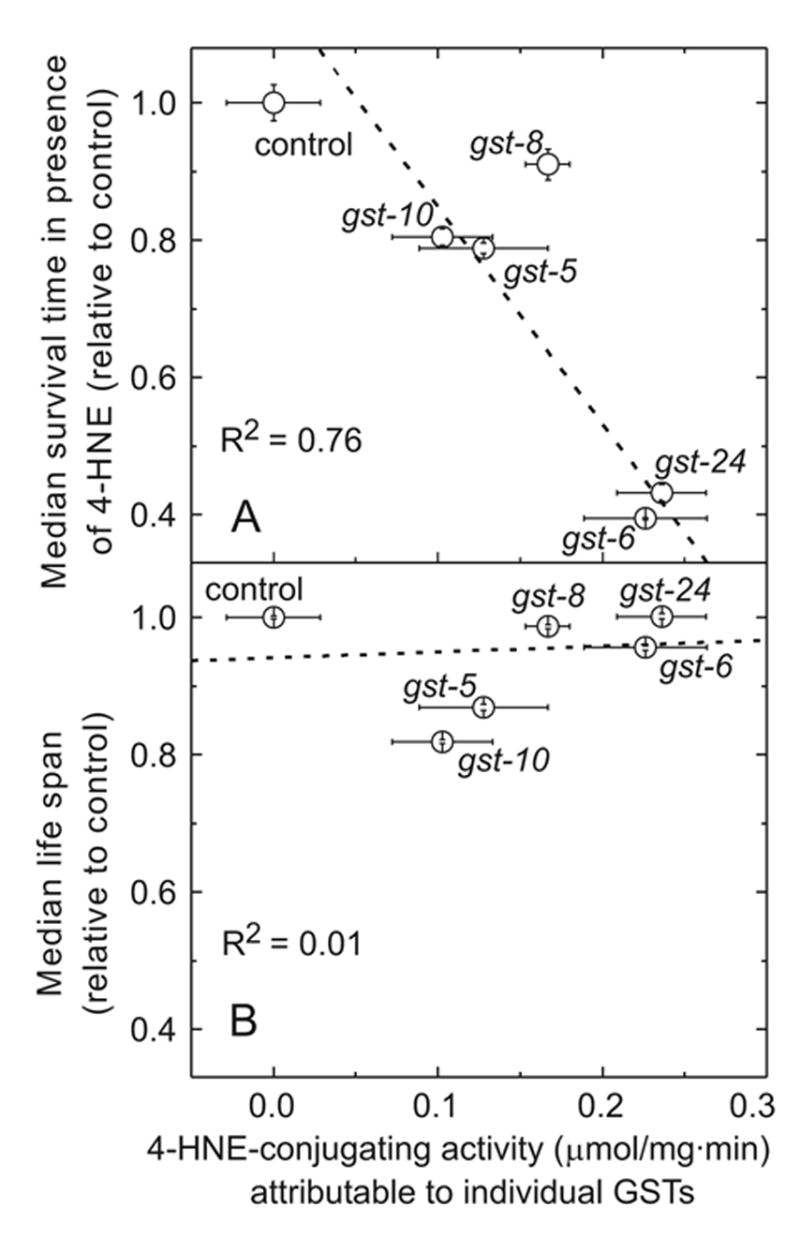
Correlation between 4-HNE-conjugating activity of selected C. elegans GSTs and (panel A) resistance against stress evoked by exposure to 4-HNE, or (panel B) life span. Shown are only those GSTs which have been identified as able to conjugate 4-HNE. 4-HNE-conjugating activity (abscissa) is equal to the decrease in activity caused by RNAi knock-down of each GST (from Fig. 1). Resistance to 4-HNE (ordinate, panel A) is expressed as the median survival time in the presence of 10 mM 4-HNE (from Fig. 4). Life span (ordinate, panel B) is the median survival time under unstressed conditions (from Fig. 7). Regression lines were calculated using as the weight for each point the square root of the product of the standard deviation of enzyme activity (x-dimension error) and the asymptotic standard error of the median survival time (y-dimension error). Error bars are shown inside the plotting symbol when they are smaller than the symbol diameter.
It is worth noting that, for evaluation of 4-HNE stress resistance, approximately 10 mM 4-HNE was necessary to trigger mortality within a time consistent with acute toxicity (8 hr in the present work). This is in contrast to cultured cells in which similar mortality is elicited by micromolar concentrations of 4-HNE. Since biological effects of 4-HNE are thought to be mediated mostly by reaction of the compound with targets on proteins within the cell, we compared the concentration dependence of 4-HNE-protein adduct formation in C. elegans and in cultured mammalian cells. The results (Fig. 5) show a pronounced lag phase in worms. A similar increment of adducts (over the basal level which was similar in both systems) was reached at 10 mM 4-HNE in C. elegans and at 10 μM 4-HNE in cultured cells. This indicates that access of external 4-HNE to internal tissues is restricted, perhaps due to a shielding effect of the cuticle, and/or reduced ingestion of a noxious compound which would limit exposure through the gut (Kaletta and Hengartner, 2006). At 10 mM 4-HNE, the effective concentrations of the compound reaching C. elegans tissues was reduced by three orders of magnitude; at 2.5 mM external 4-HNE, apparently no 4-HNE reached internal tissues since no additional adduct formation (Fig. 5) and no short-term toxicity was observed. A relatively high resistance of C. elegans has been also observed for other compounds. For example, 100 mM paraquat was used to evaluate oxidative stress in C. elegans (Leiers et al., 2003), exceeding by three orders of magnitude the concentration necessary to achieve a comparable toxicity in cultured cells (Bagley et al., 1986).
If, as we postulate, modifications of proteins caused by 4-HNE contribute to organismal aging, the level of 4-HNE-protein adducts would be expected to increase with age. This was indeed the case (Fig. 6). The steady-state level of 4-HNE-protein adducts is determined by the formation rate on one hand, and by the turnover rate of adducted proteins on the other. The overall turnover rate of proteins decreases with age in many organisms, including nematodes (Ryazanov and Nefsky, 2002; Tavernarakis and Driscoll, 2002; Samara and Tavernarakis, 2003), but the turnover of at least some moderately 4-HNE-modified proteins may be in fact higher than that of native proteins (Carbone et al., 2004; Tsuchiya et al., 2005). This complex behavior makes it difficult to predict the age-dependence of the half-life of 4-HNE-protein adducts. In any case, however, conjugation of 4-HNE to glutathione would spare proteins from 4-HNE adduction by decreasing free 4-HNE levels. This interpretation is consistent with our finding that, in worms in which the expression of the five GSTs identified to conjugate 4-HNE (Fig. 1) was knocked down by RNAi, the level of 4-HNE-protein conjugates in whole-body homogenates inversely correlated with the remaining 4-HNE-conjugating activity (Fig. 8A).
As we have previously demonstrated for gst-10 by overexpressing the enzyme (Ayyadevara et al., 2005b), a GST able to conjugate 4-HNE may lower the concentration of the electrophile, reduce the steady-state 4-HNE-protein adduct level, and increase life span. Conversely, the loss of expression of gst-10 led to a reduction of life span (Ayyadevara et al., 2005a). In the present study, in addition to the already characterized gst-10, only RNAi targeted to gst-5 caused life span reduction in C. elegans (Fig. 7). Strikingly, the knock-down of gst-8, gst-6, or gst-24 did not affect life span, even though the individual contributions of these three GSTs to the 4-HNE metabolizing capacity (Fig. 1) and to the maintenance of low levels of 4-HNE-protein adducts (Fig. 8) equal or exceed those of gst-5 or gst-10. Consequently, there was no significant correlation between the 4-HNE-conjugating activity and median longevity of worms in which the five GSTs which possess 4-HNE-conjugating activity were individually knocked down by RNAi (Fig. 10B; R2 = 0.01, P = 0.85). Similarly, 4-HNE-protein adducts inversely correlated with median life span only for gst-5 and for gst-10, but not for gst-8, gst-6, or gst-24 (Fig. 8B). The existence of two classes of 4-HNE-conjugating GSTs, with and without effect of life span, can be explained in at least two ways. One possibility is that the properties of gst-10 and gst-5 gene products which contribute to longevity assurance are not linked to the 4-HNE-conjugating activity of these enzymes. Although at present this possibility cannot be ruled out conclusively, we consider it unlikely for the following reason. Tissue-specific expression of either the ectopic murine mGSTA4-4 or the endogenous CeGSTP2-2 (gst-10 gene product) led to an extension of life span which was proportional to the increment of 4-HNE-conjugating activity, and inversely proportional to the level of 4-HNE-protein adducts (Ayyadevara et al., 2005b). No physiological substrates other than 4-HNE (or related electrophilic aldehydes derived from lipid peroxidation) are presently known to be common to the murine and nematode enzymes. Although this does not preclude the identification of such common substrates in the future, we propose that the most parsimonious interpretation of the data is that 4-HNE is an aging effector in C. elegans (Ayyadevara et al., 2005a; Ayyadevara et al., 2005b). For this reason, we favor a hypothesis which provides a second possible explanation of the lack of correlation between 4-HNE-conjugating activity and life span (Fig. 10B). According to this hypothesis, a 4-HNE-conjugating enzyme has to be expressed in a critical tissue or cell type to affect longevity.
We have previously demonstrated that CeGSTP2-2 is expressed at a high level in only a small number of cells (Ayyadevara et al., 2005b), probably including amphids and phasmids. The tissue specificity was determined using experimental expression of several proteins (green fluorescent protein, mGSTA4-4, and CeGSTP2-2 itself) under the control of a gst-10 promoter fragment, but also by indirect immunofluorescence to detect endogenous CeGSTP2-2 (Ayyadevara et al., 2005b). Agreement of the latter approach with the transgenic experiments strongly indicates that the observed tissue specificity pattern reflects the native localization of CeGSTP2-2. It could be argued that CeGSTP2-2 is also present at low levels (below detection threshold of the methods used) in other, or even all, C. elegans tissues, and that the effects of modulating CeGSTP2-2 expression on life span (Ayyadevara et al., 2005a; Ayyadevara et al., 2005b) are mediated by generalized detoxification in all tissues. Alternatively, the effect of CeGSTP2-2 on longevity could be amphid-specific. Since ablation of individual amphid neurons is known to modulate life span (Alcedo and Kenyon, 2004), CeGSTP2-2 could affect longevity by affecting 4-HNE-protein adduct formation and thus altering the physiological state of amphids. The gst-5 gene product could be expressed in the same or another tissue whose function is involved in longevity assurance.
Of the two models that could account for life span modulation by GSTs (i.e., generalized low-level expression versus high abundance in a limited number of cells), we favor the latter because 4-HNE-metabolizing GSTs other than the gst-10 or gst-5 gene products affect the level of 4-HNE-protein adducts without modulating life span. This demonstrates that detoxification of 4-HNE, while important under certain conditions, is by itself not sufficient for longevity assurance. Tissue localization is likely to be another requirement that must be met; in the case of CeGSTP2-2, the relevant tissue may be sensory neurons. No information is currently available on the tissue distribution of either the gst-5 gene product or the 4-HNE-conjugating GSTs which do not affect life span. Further work will be needed to test the tissue localization hypothesis.
In contrast to the effect on longevity which is restricted to the gst-10 and gst-5 gene products, loss of any of the five 4-HNE-conjugating enzymes decreased the resistance of the organism to electrophilic stress caused by 4-HNE (Fig. 10A). This could be explained by the fact that, to affect longevity, 4-HNE needs to target the particular tissue that constitutes, or becomes, the "weak link" (Shmookler Reis, 2003) under the conditions used. However, knock-down of any 4-HNE-metabolizing enzyme may sensitize the organism to acute 4-HNE toxicity since damage to any tissue, if severe enough, would lead to death.
We have shown that at least 5 out of 44, or no less than one-tenth of the total number, of C. elegans GSTs are able to conjugate 4-HNE. A phylogenetic analysis (Fig. 11) indicates that enzymes which have this activity did not arise through a recent gene duplication. Instead, they are present in two distant branches of the phylogenetic tree. Even within each branch, the individual GSTs reactive toward 4-HNE – and in particular, the two that impact longevity (shown boxed in Fig. 11) – are separated by considerable evolutionary distance. This situation resembles that found by us in the fruitfly Drosophila melanogaster where 4-HNE-metabolizing GSTs belong to at least two distinct classes (Singh et al., 2001; Sawicki et al., 2003). The pattern depicted in Fig. 11 could have arisen if 4-HNE-conjugating activity of an early ancestral GST were retained in some but not all descendant GSTs. Alternatively, the ability to conjugate 4-HNE could have emerged in distantly related GSTs through convergent evolution. Regardless of the evolutionary mechanism, the presence of 4-HNE-conjugating activity in GSTs which diverged early indicates that this function is physiologically important. In fact, metabolism of 4-HNE is likely to be required in any aerobic organism which contains peroxidation-prone polyunsaturated fatty acids.
Fig. 11.
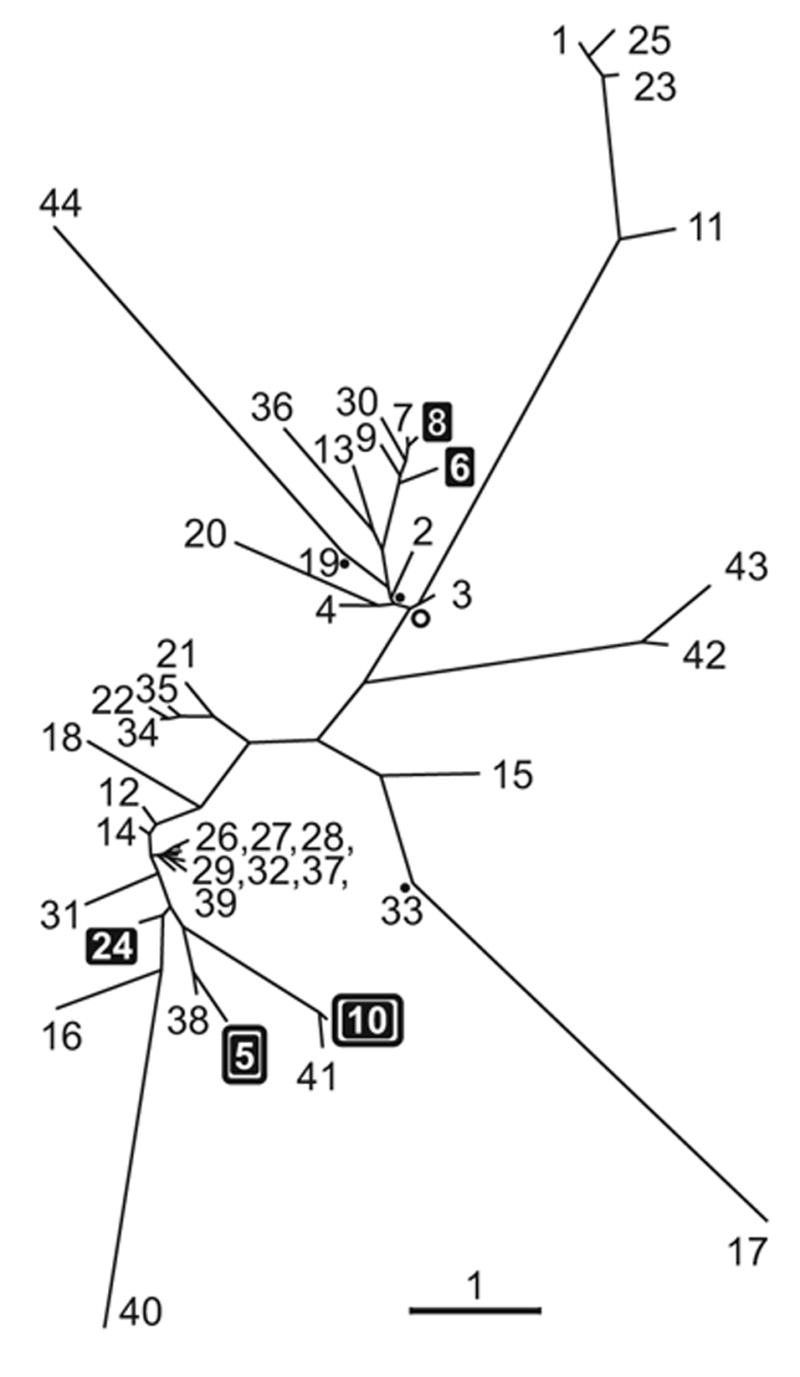
Unrooted radial phylogenetic tree of C. elegans gst coding sequences constructed by the maximum likelihood method, as implemented in the software fastDNAml (Olsen et al., 1994), and visualized using TreeView (Page, 1996). The numbers refer to gst gene designations. The 5 GSTs which we identified as having 4-HNE-conjugating activity are shown on black background; of these, the 2 enzymes (gst-5 and gst-10) whose knock-down by RNAi leads to life span shortening are boxed. The length of 3 branches in the tree (marked with filled circles) may not be significantly greater than zero (P ≥ 0.05), and the length of one branch (marked with open circle) is positive at 0.01 ≤ P < 0.05; all other branches are significant at P < 0.01. The scale bar represents a branch length corresponding to an evolutionary distance equivalent to one expected nucleotide substitution at any given site.
In summary, by an RNAi screen we have demonstrated that at least five C. elegans GSTs have catalytic activity for conjugation of 4-HNE with glutathione. Loss of any of these five enzymes leads to an increased level of 4-HNE-protein adducts and sensitizes the organism to electrophilic stress elicited by 4-HNE, but only the loss of the gst-5 and gst-10 gene products reduces the life span of the nematode. We propose that the latter effect requires expression in a tissue whose physiological state contributes to the determination of longevity.
Acknowledgments
This work was supported in part by National Institutes of Health grants R01 AG18845 and R01 ES07804 (to PZ), and Program Project grant P01 AG20641 (to RJSR). PZ and RJSR are recipients of VA Research Career Scientist Awards. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). We thank Dr. Dennis R. Petersen, University of Colorado, Denver, for the generous gift of an antibody against protein-4 HNE adducts.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41:45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- Alin P, Danielson UH, Mannervik B. 4-Hydroxyalk-2-enals are substrates for glutathione transferase. FEBS Lett. 1985;179:267–270. doi: 10.1016/0014-5793(85)80532-9. [DOI] [PubMed] [Google Scholar]
- Ayyadevara S, Dandapat A, Singh SP, Benes H, Zimniak L, Shmookler Reis RJ, Zimniak P. Lifespan extension in hypomorphic daf-2 mutants of Caenorhabditis elegans is partially mediated by glutathione transferase CeGSTP2-2. Aging Cell. 2005a;4:299–307. doi: 10.1111/j.1474-9726.2005.00172.x. [DOI] [PubMed] [Google Scholar]
- Ayyadevara S, Engle MR, Singh SP, Dandapat A, Lichti CF, Benes H, Shmookler Reis RJ, Liebau E, Zimniak P. Lifespan and stress resistance of Caenorhabditis elegans are increased by expression of glutathione transferases capable of metabolizing the lipid peroxidation product 4-hydroxynonenal. Aging Cell. 2005b;4:257–271. doi: 10.1111/j.1474-9726.2005.00168.x. [DOI] [PubMed] [Google Scholar]
- Bagley AC, Krall J, Lynch RE. Superoxide mediates the toxicity of paraquat for Chinese hamster ovary cells. Proc Natl Acad Sci USA. 1986;83:3189–3193. doi: 10.1073/pnas.83.10.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi L, Kwok SM, Driscoll M, Sesti F. A potassium channel-MiRP complex controls neurosensory function in Caenorhabditis elegans. J Biol Chem. 2003;278:12415–12424. doi: 10.1074/jbc.M212788200. [DOI] [PubMed] [Google Scholar]
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall WS, Khvorova A. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Meth. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone DL, Doorn JA, Petersen DR. 4-hydroxynonenal regulates 26S proteasomal degradation of alcohol dehydrogenase. Free Radic Biol Med. 2004;37:1430–1439. doi: 10.1016/j.freeradbiomed.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Chandra A, Srivastava SK. A synthesis of 4-hydroxy-2-trans-nonenal and 4-(3H) 4-hydroxy-2-trans-nonenal. Lipids. 1997;32:779–782. doi: 10.1007/s11745-997-0100-6. [DOI] [PubMed] [Google Scholar]
- Engle MR, Singh SP, Nanduri B, Ji X, Zimniak P. Invertebrate glutathione transferases conjugating 4-hydroxynonenal: CeGST 5.4 from Caenorhabditis elegans. Chem Biol Interact. 2001;133:244–248. [Google Scholar]
- Gems D, McElwee JJ. Broad spectrum detoxification: the major longevity assurance process regulated by insulin/IGF-1 signaling? Mech Ageing Dev. 2005;126:381–387. doi: 10.1016/j.mad.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Gree R, Tourbah H, Carrie R. Fumaraldehyde monodimethyl acetal: an easily accessible and versatile intermediate. Tetrahedron Lett. 1986;27:4983–4986. [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand J Statistics. 1979;6:65–70. [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai Z, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chaturverdi K, Christophides GK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu Z, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke Z, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin JJ, Lobo NF, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O'Brochta DA, Pfannkoch C, Qi R, Regier MA, Remington K, Shao H, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun J, Thomasova D, Ton LQ, Topalis P, Tu Z, Unger MF, Walenz B, Wang A, Wang J, Wang M, Wang X, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang H, Zhao Q, Zhao S, Zhu SC, Zhimulev I, Coluzzi M, della Torre A, Roth CW, Louis C, Kalush F, Mural RJ, Myers EW, Adams MD, Smith HO, Broder S, Gardner MJ, Fraser CM, Birney E, Bork P, Brey PT, Venter JC, Weissenbach J, Kafatos FC, Collins FH, Hoffman SL. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Kaletta T, Hengartner MO. Finding function in novel targets: C. elegans as a model organism. Nat Rev Drug Discov. 2006;5:387–399. doi: 10.1038/nrd2031. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kimura H, Liu S, Yamada S, Uchida K, Matsumoto K, Mukaida M, Yoshida K. Rapid increase in serum lipid peroxide 4-hydroxynonenal (HNE) through monocyte NADPH oxidase in early endo-toxemia. Free Radic Res. 2005;39:845–851. doi: 10.1080/10715760500161546. [DOI] [PubMed] [Google Scholar]
- Lanjuin A, Sengupta P. Specification of chemosensory neuron subtype identities in Caenorhabditis elegans. Curr Opin Neurobiol. 2004;14:22–30. doi: 10.1016/j.conb.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Leiers BR, Kampkotter A, Grevelding CG, Link CD, Johnson TE, Henkle-Duhrsen K. A stress-responsive glutathione S-transferase confers resistance to oxidative stress in Caenorhabditis elegans. Free Radic Biol Med. 2003;34:1405–1415. doi: 10.1016/s0891-5849(03)00102-3. [DOI] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- McEwen JE, Zimniak P, Mehta JL, Shmookler Reis RJ. Molecular pathology of aging and its implications for senescent coronary atherosclerosis. Curr Opin Cardiol. 2005;20:399–406. doi: 10.1097/01.hco.0000175517.50181.89. [DOI] [PubMed] [Google Scholar]
- Olsen GJ, Matsuda H, Hagstrom R, Overbeek R. fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- Page RDM. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pearl R. The rate of living. University of London Press; London: 1928. [Google Scholar]
- Qiu S, Adema CM, Lane T. A computational study of off-target effects of RNA interference. Nucl Acids Res. 2005;33:1834–1847. doi: 10.1093/nar/gki324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryazanov AG, Nefsky BS. Protein turnover plays a key role in aging. Mech Ageing Dev. 2002;123:207–213. doi: 10.1016/s0047-6374(01)00337-2. [DOI] [PubMed] [Google Scholar]
- Samara C, Tavernarakis N. Calcium-dependent and aspartyl proteases in neurodegeneration and ageing in C. elegans. Ageing Res Rev. 2003;2:451–471. doi: 10.1016/s1568-1637(03)00032-1. [DOI] [PubMed] [Google Scholar]
- Satoh K, Yamada S, Koike Y, Igarashi Y, Toyokuni S, Kumano T, Takahata T, Hayakari M, Tsuchida S, Uchida K. A 1-hour enzyme-linked immunosorbent assay for quantitation of acrolein- and hydroxynonenal-modified proteins by epitope-bound casein matrix method. Anal Biochem. 1999;270:323–328. doi: 10.1006/abio.1999.4073. [DOI] [PubMed] [Google Scholar]
- Sawicki R, Singh SP, Mondal AK, Benes H, Zimniak P. Cloning, expression, and biochemical characterization of one Epsilon-class (GST-3) and ten Delta-class (GST-1) glutathione S-transferases from Drosophila melanogaster, and identification of additional nine members of the Epsilon class. Biochem J. 2003;370:661–669. doi: 10.1042/BJ20021287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selley ML, Bartlett MR, McGuiness JA, Hapel AJ, Ardlie NG, Lacey MJ. Determination of the lipid peroxidation product trans-4-hydroxy-2-nonenal in biological samples by high-performance liquid chromatography and combined capillary column gas chromatography--negative-ion chemical ionisation mass spectrometry. J Chromatogr B. 1989;488:329–340. doi: 10.1016/s0378-4347(00)82957-6. [DOI] [PubMed] [Google Scholar]
- Shmookler Reis RJ. Toward a unified theory of aging – what mammals can learn from worms and other ephemeral creatures. In: Cutler RG, Rodriguez H, editors. Oxidative Stress and Aging: Advances in Basic Science, Diagnostics and Intervention. Vol. 2. World Scientific Publishing; Singapore: 2003. pp. 1263–1283. [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Singh SP, Coronella JA, Beneš H, Cochrane BJ, Zimniak P. Catalytic function of Drosophila melanogaster glutathione S-transferase DmGSTS1-1 (GST-2) in conjugation of lipid peroxidation end products. Eur J Bioch. 2001;268:2912–2923. doi: 10.1046/j.1432-1327.2001.02179.x. [DOI] [PubMed] [Google Scholar]
- Spies-Martin D, Sommerburg O, Langhans CD, Leichsenring M. Measurement of 4-hydroxynonenal in small volume blood plasma samples: modification of a gas chromatographic-mass spectrometric method for clinical settings. J Chromatogr B. 2002;774:231–239. doi: 10.1016/s1570-0232(02)00242-8. [DOI] [PubMed] [Google Scholar]
- Strohmaier H, Hinghofer-Szalkay H, Schaur RJ. Detection of 4-hydroxynonenal (HNE) as a physiological component in human plasma. J Lipid Mediators Cell Signal. 1995;11:51–61. doi: 10.1016/0929-7855(94)00027-a. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Driscoll M. Caloric restriction and lifespan: a role for protein turnover? Mech Ageing Dev. 2002;123:215–229. doi: 10.1016/s0047-6374(01)00341-4. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Wang SL, Dorovkov M, Ryazanov A, Driscoll M. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat Genetics. 2000;24:180–183. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Yamaguchi M, Chikuma T, Hojo H. Degradation of glyceraldehyde-3-phosphate dehydrogenase triggered by 4-hydroxy-2-nonenal and 4-hydroxy-2-hexenal. Arch Biochem Biophys. 2005;438:217–222. doi: 10.1016/j.abb.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Progr Lipid Res. 2003;42:318–343. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- Zhang H, Dickinson DA, Liu RM, Forman HJ. 4-Hydroxynonenal increases γ-glutamyl transpeptidase gene expression through mitogen-activated protein kinase pathways. Free Radic Biol Med. 2005;38:463–471. doi: 10.1016/j.freeradbiomed.2004.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]


