Abstract
Protective immunity to Mycobacterium tuberculosis is poorly understood, but mounting evidence, at least in animal models, implicates major histocompatibility complex class I-restricted CD8+ T cells as an essential component. By using a highly sensitive assay for single cell interferon γ release, we screened an array of M. tuberculosis antigen-derived peptides congruent with HLA class I allele-specific motifs. We identified CD8+ T cells specific for epitopes in the early secretory antigenic target 6 during active tuberculosis, after clinical recovery and in healthy contacts. Unrestimulated cells exhibited peptide-specific interferon γ secretion, whereas lines or clones recognized endogenously processed antigen and showed cytolytic activity. These results provide direct evidence for the involvement of CD8+ cytotoxic T lymphocytes in host defense against M. tuberculosis in humans and support current attempts to generate protective cytotoxic T lymphocyte responses against M. tuberculosis by vaccination.
One-third of the world’s population is latently infected with Mycobacterium tuberculosis, which causes over 2 million deaths per year (1); more than any other single pathogen. The global resurgence of tuberculosis, together with the HIV pandemic and emerging multi-drug resistance, have heightened the need for an effective vaccine suitable for use in immunocompromised individuals. The current live attenuated vaccine, Bacille Calmette-Guerin (BCG), has variable and limited efficacy in tuberculosis-endemic regions (2). Knowledge of protective immune responses in tuberculosis is incomplete. Delayed type hypersensitivity, mediated by cytokine-secreting CD4+ T cells, contributes to both pathogenesis and protection. However, delayed type hypersensitivity, as measured by tuberculin skin testing, does not correlate with the partial protection induced by BCG vaccination, implicating another component of cell-mediated immunity (2). CD8+, as well as CD4+ T cells are a feature of the tuberculous granuloma. Recent evidence from murine models points to a protective role for CD8+ T cells in M. tuberculosis infection (3–6). However, major histocompatibility complex (MHC) class I-restricted CD8+ T cells specific for M. tuberculosis have not hitherto been found in humans, although BCG-specific CD8+ T cell responses of undefined antigen specificity have been reported (7). Low frequencies of MHC class I-restricted M. tuberculosis-specific CD8+ cytotoxic T lymphocytes (CTL) below the limits of detection of standard assays may have impeded their detection; we have adopted a novel, sensitive approach to identify such cells.
Two secreted antigens of M. tuberculosis, early secretory antigenic target 6 (ESAT-6) and the Antigen 85 complex, were studied because secreted antigens are implicated as targets of protective immune responses (8) and are more likely to access the host cell cytosol and hence the MHC class I antigen processing pathway. These antigens are the target of protective immune responses in mice (9, 10). ESAT-6 (6 kDa) is virtually specific for M. tuberculosis complex; the gene is present in all isolates of M. tuberculosis and virulent Mycobacterium bovis but is absent from all strains of BCG examined (11). The 30–32-kDa Antigen 85 complex consists of 3 homologous proteins, A, B, and C, which constitute 30% of M. tuberculosis culture filtrate (12) and elicit strong CD4+ T cell responses in tuberculin skin test-positive subjects (13).
The sequences of ESAT-6 and antigens 85A, 85B, and 85C were scanned with allele-specific peptide motifs for the HLA class I types HLA-A2, HLA-B7, HLA-B8, HLA-B35, HLA-B52, and HLA-B53, all of which were present in the study population. A total of 49 peptides congruent with these motifs were synthesized (Table 1). These candidate epitopes were then sorted into pools according to HLA class I alleles and used to stimulate peripheral blood mononuclear cells (PBMCs) from subjects with the corresponding HLA class I allele. In vitro-expanded populations of peptide-specific T cells were then detected with two different assays of effector function. In a preliminary study using the standard 51Cr release CTL assay we detected low levels of lytic activity against some peptide pools, but responses from the few remaining cells were too weak to define epitopes. We therefore adapted a more sensitive enzyme-linked immunospot (ELISPOT) technique for detecting single cell interferon γ (IFN-γ) secretion. We calibrated this assay against the 51Cr release CTL assay and found it an order of magnitude more sensitive for detecting low numbers of cultured human CD8+ peptide-specific T cells (14).
Table 1.
Peptides from ESAT-6
| HLA class I allele | Peptide motif | Peptide | Sequence | Position |
|---|---|---|---|---|
| HLA-A2 | -L/I/M------V/L/I | ES8 | GIEAAASAI | 10–18 |
| ES9 | AIQGNVTSI | 17–25 | ||
| ES10 | LLDEGKQSL | 28–36 | ||
| ES11 | ELNNALQNL | 64–72 | ||
| ES13 | AMASTEGNV | 82–90 | ||
| HLA-B8 | --K-K---L/I | ES7 | EGKQSLTKL | 31–39 |
| HLA-B52 | -Q-----I/V | ES12 | LQNLARTI | 69–76 |
We searched for 8-, 9-, or 10-residue sequences from ESAT-6, which were congruent with the peptide motifs of the HLA class I alleles -A2, -B7, -B8, -B35, -B52, and -B53. Sequences congruent with the peptide motifs for HLA-A2, HLA-B8, and HLA-B52 were identified; these peptides were synthesized and are displayed. No sequences congruent with HLA-B7, HLA-B35, and HLA-B53 were present in ESAT-6 and, thus, no peptides were synthesized for these HLA class I alleles. Peptides were sorted into pools that were used for in vitro restimulation of donor PBMCs. Peptides found to be CD8+ epitopes are shown in boldface. Similarly, 42 peptides were synthesized based on the sequences of antigens 85A and 85C. No CD8+ epitopes were identified among these and the peptides are not shown.
Thirty-nine adults were studied using the more sensitive ELISPOT assay. These included patients representing the full clinical spectrum of active tuberculosis (pulmonary, lymphadenitis, and extrapulmonary), as well as healthy contacts.
MATERIALS AND METHODS
Subjects.
Thirty-nine adult patients and contacts with suitable HLA haplotypes were studied. Most subjects originated from the Indian subcontinent, Africa, or northern Europe. The number of patients with different clinical types of disease, as well as the number of healthy contacts, are listed in Table 2. Most subjects were tuberculin skin test positive. All contacts, except one who was not tested, had a positive Heaf test of grade 2 or more. All patients had at least 5 mm of cutaneous induration in response to intradermal injection with 1 tuberculin unit of purified protein derivative, except four who were not tested and four who were negative (three with old, inactive pulmonary tuberculosis and one with active miliary disease). The distribution of each of the 6 HLA class I alleles for which we tested peptide pools was as follows: HLA-A2 was present in 26 subjects, HLA-B8 in 7 subjects, HLA-B7 in 6 subjects, HLA-B35 in 2 subjects, HLA-B52 in 2 subjects, and HLA-B53 in 2 subjects.
Table 2.
Numbers of healthy contacts and patients with different types of clinical disease
| Clinical phenotype | Subjects, n |
|---|---|
| Pulmonary (active) | 8 |
| Pulmonary (inactive) | 6 |
| Pleural | 2 |
| Lymphadenitis | 4 |
| Osteoarticular | 5 |
| Miliary | 1 |
| Gastrointestinal | 1 |
| Epididymo-orchitis | 1 |
| Healthy contacts | 11 |
Each subject was tested against the peptide pool (or pools if the subject had more than one suitable HLA class I allele) once only. Repeat blood samples were obtained and further testing carried out only if the donor had responded on the first occasion.
Diagnosis of Tuberculosis in Patients NPH54 and NPH97.
NPH54 was diagnosed with tuberculosis on the basis of clinical symptoms, acute erythema nodosum and mediastinal lymphadenopathy. Tuberculin skin test showed 25 mm induration 48 hr after intradermal injection of 1 tuberculin unit of purified protein derivative. In addition, a very high antibody titer of > 1/6,250 in response to the 16 kDa antigen of M. tuberculosis, helped to confirm active infection (15). For NPH97, the tuberculin skin test was positive and culture of a proximal phalangeal bone biopsy of the affected hand on Lowenstein–Jensen medium grew M. tuberculosis.
Tissue Typing.
Subjects were HLA typed serologically. NPH54, NPH97, WB, Akiba, and PG were additionally typed by amplification refractory mutation system (ARMS)–PCR using sequence-specific oligonucleotide primers (16).
Peptide Synthesis.
Peptides were synthesized on solid phase on a synthesizer (Zinsser, Frankfurt) using fluorenylmethoxycarbonyl chemistry. Purity was confirmed by HPLC.
ELISPOT Assay for IFN-γ.
Ninety-six-well polyvinylidene difluoride (PVDF)-backed plates (Millipore) precoated with the anti-IFN-γ mAb 1-D1K at 15 μg/ml (MABTECH, Stockholm) were washed with RPMI medium 1640 and blocked with R10 for 1 hr at room temperature. Short-term cell lines (STCLs), CTL lines, or clones were washed two times in RPMI medium 1640, resuspended in R10, and dispensed at known input cell numbers per well in duplicate wells. Peptide was added either directly to the supernatant at a final concentration of 2 μM (free peptide) or presented on a B cell line (BCL), prepulsed with 10 μM peptide for 1 hr at 37°C, and then washed three times in R10. Plates were incubated for 12 hr at 37°C, in 5% CO2/95% air. After washing six times with phosphate buffered saline (PBS)/0.05% Tween 20 to remove cells, plates were incubated for 3 hr with the second biotinylated anti-IFN-γ mAb 7-B6–1-biotin at 1 μg/ml (MABTECH). A further wash, as above, was followed by incubation with a 1:1,000 dilution of streptavidin–alkaline phosphatase conjugate for 2 hr. After another wash, chromogenic alkaline phosphatase substrate (Bio-Rad) was added to the wells, and 30 min later plates were washed with tap water. After drying, spot-forming cells (SFC) were counted under ×20 magnification. For ex vivo ELISPOT assays, 500,000 freshly isolated uncultured PBMCs were used per well. Responses were considered significant if a minimum of 10 SFCs were present per well and, additionally, this number was at least twice that in control wells.
Generation of CD8+ T Cell Lines.
STCLs were generated as described (17). Briefly, PBMCs separated from whole blood were prepulsed with 30–50 μM peptide for 1 hr in a cell pellet and then diluted up to 1 × 106 cells/ml in RPMI medium 1640 supplemented with 10% pooled heat-inactivated human AB serum (R10) and 25 ng/ml rhIL-7 (R & D Systems), and seeded at 200 μl per well in a U-bottomed 96-well plate. Lymphocult-T (Biotest, Dreiech, Germany) was added to 10% of each well at regular intervals. STCLs were assayed at day 14. Lines 3–9, 3–10, and 3–20 were generated from individual STCLs by two rounds of restimulation (on days 14 and 21) with autologous peptide-pulsed irradiated BCL at a 1:3 feeder:responder ratio. Line 4–1 was generated by pooling four ES12-specific STCLs (originally established with CD4-depleted PBMCs), followed by restimulation with peptide-pulsed autologous BCL.
Generation of CD8+ T Cell Clones.
Enumeration of IFN-γ SFCs after CD4 and CD8 depletions indicated that the frequency of CD8+ ES12-specific T cells in NPH54-derived monospecific lines 3–9 and 3–20 was 1:44 and 1:67 CD8+ T cells, respectively. This indicated that for T cells seeded at one cell per well, 1.5–2.3 specific clones should be generated per 100 wells seeded. This quantitative data guided our cloning procedure: for each line, 240 wells were seeded at one cell per well (following CD4 depletion). Cloning mix consisted of three-way mixed lymphocyte reaction, 10% Lymphocult-T, phytohemagglutinin at 1 μg/ml, and ES12-pulsed autologous irradiated washed BCL, with a total of 100,000 feeders per well. Clones were screened in ELISPOT assays and three ES12-specific clones were subsequently recovered from line 3–9 and two from line 3–20. Clones were maintained by peptide-pulsed autologous irradiated BCL restimulation and supplementation with 10% Lymphocult-T and rhIL-7 at 25 ng/ml.
51Cr Release Cytotoxicity Assays.
Standard chromium release assays were performed as described (18). In brief, BCLs were labeled with 100 mCi 51Cr (Amersham), washed in RPMI medium 1640, and then pulsed with peptide as above and plated out at 5,000 cells per well. CTL, R10, or 5% Triton X-100 were added. Test wells were in duplicate, other wells in quadruplicate. Plates were incubated for 5 hr at 37°C, in 5% CO2/95% air and harvested supernatant read on filtermats in an LKB 1205 beta-plate scintillation counter (Wallac, Gaithersburg, MD). Background 51Cr release was less than 20%. Percent lysis was calculated from the formula 100 × (E − M)/(T − M), where E is the experimental release, M is the spontaneous release, and T is the maximal release.
Specific Cell Depletions.
CD4+ and CD8+ T cells were depleted by 30-min incubation with anti-CD4 or anti-CD8 mAbs conjugated to ferrous beads, DYNABEADS M-450, (Dynal, Oslo) in 500 μl of R10 on ice. Following dilution of up to 5 ml in R10, the conjugate-coated cells were removed by a magnetic field. CD8+ T cell depletions were highly effective and were not toxic, because there was no detectable loss of viability in the depleted population and responses of antigen 85-specific CD4+ T cell lines were unaffected by depletion.
Recombinant Vaccinia Virus Construction.
Construction of the rVV is described in detail elsewhere (ref. 19; and A.S.M., unpublished work). In brief, the amplification product of the ESAT-6 gene obtained by PCR using the plasmid template pAA249 (P. Andersen, Staatens Seruminstitut, Copenhagen-S, Denmark) was cloned into plasmid p1108-tPA to create p1108-tPA–ESAT-6. Homologous recombination into the thymidine kinase locus of vaccinia strain WR with cationic lipid transfection was followed by selection of rVV using mycophenolic acid. A second negative control rVV was also constructed by using p1108 minus the coding sequence. Recombinants were verified by sequencing and expression confirmed by PCR and capture ELISA (M. Harboe, unpublished work).
RESULTS
Identification of ESAT-6-Specific Effector T Cells Direct from Peripheral Blood.
Two CD8+ epitopes in ESAT-6 were identified. The T cells from donor NPH54, who had tuberculous mediastinal lymphadenitis, recognized peptides corresponding to both of these epitopes. Uncultured PBMCs isolated at the time of the diagnosis from NPH54, who has HLA-B52 and HLA-A2.01, secreted IFN-γ in response to an ESAT-6-derived peptide pool for these class I alleles in an ex vivo ELISPOT assay. The mean number of IFN-γ SFCs enumerated from 5 × 105 PBMCs in duplicate wells was 19 for the ESAT-6 peptides compared with 2 in the control wells with no peptide. A subsequent assay tested freshly isolated PBMCs against each of the individual peptides within the responding pools; IFN-γ SFCs were detected in response to peptides ES12 and ES13, whose sequences are congruent with the HLA-B52 and HLA-A2.01 peptide motifs, respectively. The frequency of ES12- and ES13-specific IFN-γ SFCs is of the same order of magnitude as SFCs for HLA-A2.01-restricted influenza matrix epitope M1 58–66 (Fig. 1A). Unrestimulated PBMCs from a second donor, NPH97, with tuberculous osteomyelitis, also recognized the ES12 peptide. This patient also has HLA-B52 and HLA-A2.01, and the magnitude of the ES12-specific response was similar to the response to the HLA-A2-restricted influenza matrix epitope. Single cell IFN-γ release by freshly isolated T cells in these short 12-hr ex vivo assays, employing no stimulus other than cognate peptide, indicates that these cells are highly likely to be circulating activated effector T cells (14).
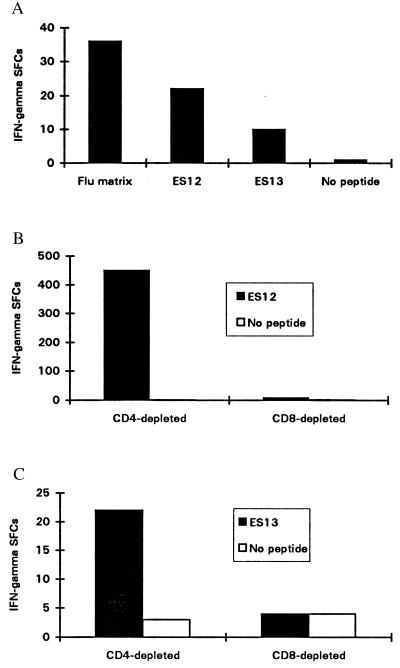
Figure 1.
(A) Enumeration of circulating peptide-specific effectors from the peripheral blood of NPH54 (HLA class I haplotype: HLA-A2.01, HLA-A68; HLA-B52, HLA-B38). The sequences of peptides ES12 and ES13 are congruent with the peptide motifs of HLA-B52 and HLA-A2.01, respectively. The frequency of ES12-specific effectors during active tuberculosis (1:23,000) is slightly less than the frequency of effectors specific for the HLA-A2-restricted epitope in influenza virus M1 58–66 (1:14,000). Activated effectors specific for ES13, an HLA-A2-restricted epitope, are also evident, but at a lower frequency (1:50,000). Freshly isolated PBMCs (500,000) were seeded in duplicate wells and peptide added to a final concentration of 2 μM in a 12-hr ex vivo ELISPOT assay for IFN-γ. (B) ES12-specific IFN-γ secreting T cells are CD8+. Enumeration of IFN-γ SFCs in a 12-hr ELISPOT assay for IFN-γ with cell line 3–20 from donor NPH54 was performed and is shown. After depletion of CD4+ or CD8+ cells, 20,000 cells were added to each of a pair of duplicate wells and peptide ES12 was added to a final concentration of 2 μM: the mean number of SFCs is shown. No SFCs were observed in the absence of peptide. CD8+ cell depletion completely abrogates the response. Similar results were obtained with cell line 3–9. (C) ES13-specific IFN-γ release by an STCL raised against ES13 with PBMCs from donor WB; a healthy contact with HLA haplotype HLA-A2, HLA-A1; HLA-B7, HLA-B8. After magnetic depletion of CD4+ or CD8+ cells, 20,000 cells were added to each of a pair of duplicate wells in a 12-hr ELISPOT assay and peptide added at 2 μM to the supernatant. The mean number of IFN-γ SFCs for each pair of wells is shown.
ESAT-6 Epitope-Specific T Cells Are CD8+.
ES12-specific T cell lines were generated from NPH54 and NPH97 PBMCs. Depletion experiments demonstrated that the ES12-specific T cells are CD8+ (Fig. 1B). Similar depletion studies on ES13-specific STCLs from donor WB, a healthy contact with HLA-A2.01, confirmed that these ES13-specific T cells are also CD8+ (Fig. 1C). CD4-depleted ES13-specific STCLs from donor NPH43, a patient with lymphadenitis (class I HLA haplotype: HLA-A2.01, HLA-A29; HLA-B7, and HLA-B51), recognized peptide presented through HLA-A2.01 on ES13-prepulsed HLA-A2.01-matched heterologous BCL. Ninety-eight IFN-γ SFCs were enumerated in response to the ES13-pulsed BCL, compared with 48 IFN-γ SFCs for the unpulsed control BCL; the high backgrounds are probably due to alloresponse (13). Responses to ES13 were transient and often undetectable in PBMCs from subsequent blood samples drawn later in the course of therapy, thus precluding comprehensive analysis of this epitope.
No CD8+ epitopes were identified among any of the 42 peptides from antigens 85A, 85B, or 85C. Despite stimulation in vitro with nonamer peptides, the resultant STCLs were all CD4+. Interestingly, certain peptides elicited IFN-γ secretion by freshly isolated uncultured CD4+ cells in 12-hr ex vivo ELISPOT assays (A.L., R.B., R.J.W., G.P., and A.V.S.H., unpublished data.)
ES12-Specific CD8+ T Cells Are MHC Class I-Restricted and Recognize Endogenously Processed Antigen.
The epitope ES12 was further characterized with ES12-specific lines and clones. T cell recognition of peptide had until now relied on presentation of ES12 on autologous cells by adding free peptide to the ELISPOT assay supernatant. To demonstrate HLA class I restriction, ES12 peptide was presented to clones with ES12-prepulsed BCL matched or mismatched at B52; only HLA-B52-matched ES12-prepulsed BCL were recognized by cells pooled from clones 3–1, 3–15, and 3–98 (Fig. 2A). ES12-specific lines raised from NPH97’s PBMCs were similarly confirmed to be HLA-B52 restricted (data not shown).
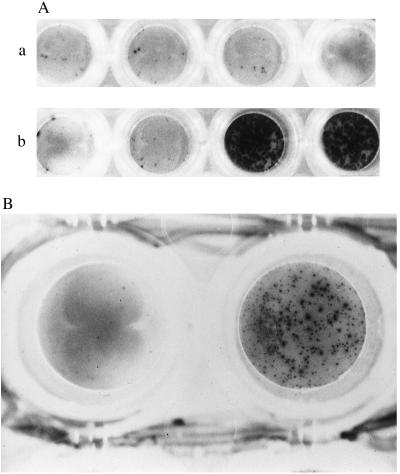
Figure 2.
(A) Photomicrograph showing peptide-specific HLA-B52-restricted IFN-γ release by ES12-specific CD8+ clones derived from donor NPH54. Cells from clones 3–1, 3–15, and 3–98 were pooled and assayed against unpulsed (first pair of wells in a and b), and ES12-pulsed (second pair of wells in a and b). HLA-B52-mismatched (A) and matched (B) target BCLs. The mismatched BCL is PG: HLA-A2.01, HLA-A3; HLA-B7, HLA-B51 and is shown in A. The matched BCL, a homozygous typing line, is Akiba: HLA-A24; HLA-B52, and is shown in B. Assays were performed in duplicate wells with 5,000 T cells and 50,000 B cells per well. Only the pair of duplicate wells with ES12-pulsed HLA-B52-matched targets are positive; the spots are so numerous that they appear confluent. (B) Photomicrograph showing IFN-γ release by ES12-specific CD8+ clone 3–15 of donor NPH54, in response to autologous BCL infected with vaccinia virus recombinant for the gene encoding ESAT-6 (rVV-ESAT-6). The negative control is the left-hand well, using autologous BCL infected with rVV control. BCL were infected the night before with the respective recombinant viruses at a multiplicity of infection (m.o.i) of 7 plaque-forming units per cell in serum-free medium; after 90 min, cells were diluted up to 1 million/ml in R10 and incubated overnight. Infected BCL (100,000) were then added to each well along with 5,000 cloned T cells. The photomicrograph shows the result with clone 3–15, giving in excess of 450 SFCs. The results with the other two clones, 3–1 and 3–98, were so strongly positive that the spots were confluent. (×20.)
To show that the ES12-specific CD8+ T cell clones were capable of recognizing endogenously processed antigen, autologous BCL, infected with vaccinia virus recombinant for ESAT-6 (rVV-ESAT-6) or a control lacking the ESAT-6 sequence, were used to stimulate cytokine release. Only clones 3–1, 3–15, and 3–98 incubated with the ESAT-6 recombinant vaccinia-infected BCL-secreted IFN-γ in the ELISPOT assay (Fig. 2B).
M. tuberculosis Antigen-Specific CD8+ T Cells Are Cytolytic.
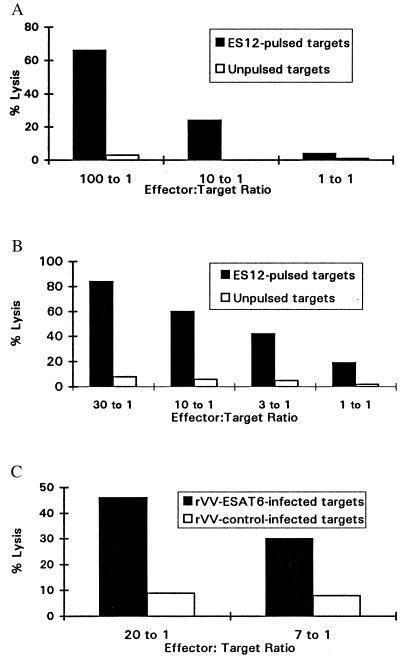
By using a sensitive measurement of cytokine release, we characterized CD8+ T cells specific for ES12. We now returned to conventional 51Cr release assays to test whether these cells were also capable of lytic activity. This was confirmed over a broad range of effector: target ratios for CD8+ T cell lines from both NPH54 and NPH97 using ES12-pulsed heterologous HLA-B52-matched BCL as targets (Fig. 3 A and B). Peptide-specific lysis was detectable even when the peptide prepulse concentration was titrated as low as 1 nM (data not shown). Finally, to establish whether ES12-specific CTL could also kill targets expressing endogenously processed antigen, we demonstrated HLA-B52-restricted lysis of heterologous HLA-B52-matched targets infected with rVV-ESAT-6 (Fig. 3C).
Figure 3.
(A) ES12-specific cytolytic activity of CTL line 4–1 from donor NPH54 in a 5 hr 51Cr release cytotoxicity assay. Peptide-specific lysis titrated downwards with diminishing effector-to-target cell ratio, and nonspecific lysis of unpulsed targets was less than 5%. Target cells were heterologous HLA-B52-matched BCL (homozygous typing line, Akiba: HLA-A24; HLA-B52, as in Fig. 2A), prepulsed with 10 μM ES12. (B) ES12-specific cytolytic activity of a CTL line from donor NPH97 in a 5 hr 51Cr release cytotoxicity assay. The CTL line was generated by restimulation of 14-day STCLs (cultured as described in Materials and Methods) with ES12-pulsed, washed, irradiated autologous BCL. Peptide-specific lysis titrated downwards with each 3-fold diminution in effector-to-target cell ratio, and nonspecific lysis of unpulsed targets was less than 5%. Target cells were heterologous HLA-B52-matched BCL (Akiba) as in Fig. 2A, prepulsed with 10 μM ES12. (C) Lysis of HLA-B52-matched heterologous targets (Akiba BCL) expressing endogenously processed ESAT-6. Targets were infected with rVV-ESAT-6 and rVV-control as in Fig. 2B and were labeled with 51Cr the following day. CTL line 4–1 raised against ES12 specifically lysed the rVV-ESAT-6-infected targets; lysis of rVV-control-infected targets was below 10%.
DISCUSSION
We have identified CD8+ HLA class I-restricted T cells specific for epitopes in the M. tuberculosis protein ESAT-6 in 4 of 39 infected individuals. This is almost certainly an underestimate of the actual prevalence of M. tuberculosis-specific CD8+ CTL in infected individuals for the following reasons. First, we limited the search to epitopes restricted through six HLA class I alleles. Second, of the very large number of antigens secreted by M. tuberculosis, we studied only two. And third, M. tuberculosis-specific CD8+ CTL are more likely to be found in draining lymph nodes or at the site of infection rather than in the peripheral circulation. Nevertheless, the frequencies of circulating ESAT-6-specific effectors in the peripheral blood of NPH54 approximates to that for an influenza virus epitope (Fig. 1A). This relatively high frequency of CTL effectors specific for a single bacterial epitope is comparable to that found in malaria, a protozoal disease, where there is substantial indirect evidence for a protective role for Plasmodium falciparum-specific CTL (18, 20, 21). ESAT-6-specific effectors among uncultured PBMCs were detectable in the ex vivo ELISPOT assay in NPH54 with tuberculous lymphadenitis and in NPH97 with tuberculous osteomyelitis. For the common HLA class I allele, HLA-A2, the number of subjects studied is sufficient to permit a preliminary comparison of the prevalence of responders between the different clinical subgroups. For donors with HLA-A2, ES13-specific IFN-γ secretion in freshly isolated PBMCs or STCLs was observed in 1 of 7 healthy contacts and 2 of 3 patients with lymphadenitis, but in none of 12 patients with other more disseminated forms of disease (pulmonary, pleural, and gastrointestinal). This distribution of responses suggests that responses to ESAT-6 may be associated with an immune response capable of containing M. tuberculosis, but larger numbers of subjects will be required to establish this.
The ELISPOT assay for IFN-γ release may measure an effector function of more protective relevance than the 51Cr release cytotoxicity assay. IFN-γ, a potent activator of macrophages, is essential for resistance to M. tuberculosis infection in mice (22, 23), whereas recent studies in perforin gene and Fas receptor gene knockout mice indicate that the lytic activity of CD8+ T cells is not required to control virulent M. tuberculosis infection (24). Moreover, humans homozygous for a point mutation in the IFN-γ receptor 1 gene, in whom cell surface expression of this receptor is absent, are highly susceptible to fatal disseminated mycobacterial infection (25), and human T cell-derived IFN-γ has recently been reported to inhibit the intracellular growth of M. tuberculosis.**
CD8+ CTL-derived IFN-γ may be especially important both for cells lacking MHC class II molecules, e.g., in the lung (26) and for macrophages where mycobacteria can evade recognition during chronic infection by sequestering their antigens away from sensitized CD4+ T cells (27). Moreover, infection of murine macrophage cell lines with live M. tuberculosis has recently been shown to down-regulate MHC class II expression while simultaneously enhancing the presentation of exogenous soluble antigen through the MHC class I antigen processing pathway (28).
The observation that ES12-specific T cell lines and clones recognize target cells infected with vaccinia virus recombinant for ESAT-6 indicates that this antigen can be endogenously processed through the MHC class I antigen processing pathway, resulting in the presentation of the epitope ES12 through HLA-B52. Because responses to the M. tuberculosis-specific peptide ES12 were elicited from freshly isolated, unrestimulated lymphocytes in an ex vivo assay, CD8+ T cells must have been primed through recognition of processed antigen in vivo. This study thus provides evidence that in humans an M. tuberculosis antigen is naturally processed in vivo through the MHC class I pathway leading to the induction of MHC class I-restricted effector T cells. Further support comes from preliminary data showing that human macrophages infected with M. tuberculosis in vitro are recognized by ES12-specific, HLA-B52-restricted CTL that suppress mycobacterial growth (R.B., et al., unpublished data).
Murine models, including studies with β2-microglobulin gene knockout mice (3), TAP-1 null mutant mice (28), and adoptive transfer experiments with HSP-65 immunized mice (4) show that CD8+ CTL are essential for protection against M. tuberculosis infection. CD8+ T cells also constitute a crucial effector mechanism in the protective immunity conferred by DNA vaccination against tuberculosis (9, 29). It has therefore been important to establish whether MHC class I-restricted CD8+ CTL play a role in M. tuberculosis infection in humans, for, if so, their induction could guide the rational design of subunit vaccines. However, it has proved very difficult to identify these cells in humans (7, 30). Indeed, the recent identification of human CD1-restricted CD8+ T cells specific for M. tuberculosis nonpeptide antigens has led to the suggestion that disruption of CD1-mediated antigen presentation may account for the enhanced susceptibility of β2-microglobulin gene knockout mice to M. tuberculosis infection (31). However, this study demonstrates the presence of classical MHC class I-restricted CD8+ CTL specific for an M. tuberculosis protein antigen in infected individuals. These cells circulate at a relatively high frequency in peripheral blood and freshly isolated, unrestimulated cells rapidly display effector function within 12 hr of antigen contact. Induction of MHC class I-restricted peptide-specific CD8+ CTL with new generation CTL-inducing vaccines is now feasible. The phenotype and specificity of the cells identified here not only endorses efforts to develop CTL-inducing vaccines against tuberculosis but also supports the candidacy of ESAT-6 as a component of such vaccines.
Acknowledgments
We thank the patients and doctors of the Infectious Diseases Unit and Osler Chest Unit, Churchill Hospital, Oxford, in particular C. Conlon, M. Benson, T. Peto, and R. Davies, and also the Lister Unit, Northwick Park Hospital, in particular R. N. Davidson and T. Whitehead. We also thank M. Mackett (Cancer Research Campaign, Manchester, U.K.) for assistance in producing the rVV and M. Bunce and T. Rostron (Churchill Hospital) for help with molecular tissue typing. We also acknowledge helpful discussions with A. J. McMichael and W. J. Britton. Informed consent was obtained from all subjects and the study was approved by the Harrow and Central Oxford Research Ethics Committees. A.L. and A.S.M. are Medical Research Council Clinical Training Fellows, R.B. and R.J.W. are supported by the Medical Research Council, A.A.P. is supported by the Wellcome Trust, and A.V.S.H. is a Wellcome Trust Principal Research Fellow.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: BCG, Bacille Calmette-Guerin; MHC, major histocompatibility complex; CTL, cytotoxic T lymphocyte; ESAT-6, early secretory antigenic target 6; PBMCs, peripheral blood mononuclear cells; IGN-γ, interferon-γ; SFC, spot-forming cells; STCL, short-term cell line; BCL, B cell line; ELISPOT, enzyme-linked immunospot.
Zhang, M., Gong, J., Lin, Y., Boylen, C. T. & Barnes, P. F. American Association of Immunologists Joint Meeting, June 2–6, 1996, New Orleans, LA.
References
- 1.Bloom B R, Murray C J. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 2.Bloom B R, Fine P E M. In: Tuberculosis: Pathogenesis, Protection and Control. Bloom B R, editor. Washington, DC: Am. Soc. Microbiol.; 1994. pp. 531–558. [Google Scholar]
- 3.Flynn J L, Goldstein M M, Triebold K J, Koller B, Bloom B R. Proc Natl Acad Sci USA. 1992;89:12013–12017. doi: 10.1073/pnas.89.24.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silva C L, Silva M F, Pietro R C L R, Lowrie D B. Immunology. 1994;83:341–346. [PMC free article] [PubMed] [Google Scholar]
- 5.Muller I, Cobbold S P, Waldmann H, Kaufmann S H. Infect Immun. 1987;55:2037–2041. doi: 10.1128/iai.55.9.2037-2041.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orme I M, Collins F M. Cell Immunol. 1984;84:113–120. doi: 10.1016/0008-8749(84)90082-0. [DOI] [PubMed] [Google Scholar]
- 7.Turner J, Dockrell H M. Immunology. 1996;87:339–342. doi: 10.1046/j.1365-2567.1996.512590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horwitz M A, Lee B-W E, Dillon B J, Harth G. Proc Natl Acad Sci USA. 1995;92:1530–1534. doi: 10.1073/pnas.92.5.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huygen K, Content J, Denis O, Montgomery D L, Yawman A M, Deck R R, DeWitt C M, Orme I M, Baldwin S, D’Souza C, Drowart A, Lozes E, Vandenbussche P, Van, Vooren J P, Liu M A, Ulmer J B. Nat Med. 1996;2:893–898. doi: 10.1038/nm0896-893. [DOI] [PubMed] [Google Scholar]
- 10.Andersen P, Andersen A B, Sorensen A L, Nagai S. J Immunol. 1995;154:3359–3372. [PubMed] [Google Scholar]
- 11.Harboe M, Oettinger T, Wiker H G, Rosenkrands I, Andersen P. Infect Immun. 1996;64:16–22. doi: 10.1128/iai.64.1.16-22.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiker H G, Harboe M. Microbiol Rev. 1992;56:648–661. doi: 10.1128/mr.56.4.648-661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roche P W, Peake P W, Billman-Jacobe H, Doran T, Britton W J. Infect Immun. 1994;62:5319–5326. doi: 10.1128/iai.62.12.5319-5326.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lalvani A, Brookes R, Hambleton S, Britton W J, Hill A V S, McMichael A J. J Exp Med. 1997;186:859–865. doi: 10.1084/jem.186.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkinson R J. Ph.D. thesis. London: London Univ.; 1997. [Google Scholar]
- 16.Krausa P M, Brywka M, III, Savage D, Hui K M, Bunce M, Ngai J L, Teo D L, Ong Y W, Barouch D, Allsopp C E, McMichael A J, Browning M. Tissue Antigens. 1995;45:223–231. doi: 10.1111/j.1399-0039.1995.tb02444.x. [DOI] [PubMed] [Google Scholar]
- 17.Ota K, Matsui M, Milford E L, Mackin G A, Weiner H L, Hafler D A. Nature (London) 1990;346:183–187. doi: 10.1038/346183a0. [DOI] [PubMed] [Google Scholar]
- 18.Hill A V S, Elvin J, Willis A C, Aidoo M, Allsopp C E, Gotch F M, Gao X M, Takiguchi M, Greenwood B M, Townsend A R, McMichael A J, Whittle H C. Nature (London) 1992;360:434–439. doi: 10.1038/360434a0. [DOI] [PubMed] [Google Scholar]
- 19.Mackett M. In: DNA Cloning 4: A Practical Approach. Glover D M, Hanes B D, editors. Oxford: IRL; 1995. p. 43. [Google Scholar]
- 20.Aidoo M, Lalvani A, Allsopp C E, Plebanski M, Meisner S J, Krausa P, Browning M, Morris-Jones S, Gotch F, Fidock D A, Takiguchi M, Robson K J H, Greenwood B M, Druilhe P, Whittle H C, Hill A V S. Lancet. 1995;345:1003–1007. doi: 10.1016/s0140-6736(95)90754-8. [DOI] [PubMed] [Google Scholar]
- 21.Wizel B, Houghten R A, Parker K C, Coligan J E, Church P, Gordon D M, Ballou W R, Hoffman S L. J Exp Med. 1995;182:1435–1445. doi: 10.1084/jem.182.5.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper A M, Dalton D K, Stewart T A, Griffin J P, Russell D G, Orme I M. J Exp Med. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flynn J L, Chan J, Triebold K J, Dalton D K, Stewart T A, Bloom B R. J Exp Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laochumroonvorapong P, Wang J, Liu C C, Ye W, Moreira A L, Elkon K B, Freedman V H, Kaplan G. Infect Immun. 1997;65:127–132. doi: 10.1128/iai.65.1.127-132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newport M J, Huxley C M, Huston S, Hawrylowicz C M, Oostra B A, Williamson R, Levin M. N Engl J Med. 1996;335:1941–1949. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 26.Orme I M. In: Host Response to Intracellular Pathogens. Kaufmann S E H, editor. Austin, TX: Landes; 1997. pp. 115–130. [Google Scholar]
- 27.Pancholi P, Mirza A, Bhardwaj N, Steinman R M. Science. 1993;260:984–986. doi: 10.1126/science.8098550. [DOI] [PubMed] [Google Scholar]
- 28.Mazzacaro R J, Gedde M, Jensen E R, van, Santen H M, Ploegh H L, Rock K L, Bloom B R. Proc Natl Acad Sci USA. 1996;93:11786–11791. doi: 10.1073/pnas.93.21.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tascon R E, Colston M J, Ragno S, Stavropoulos E, Gregory D, Lowrie D B. Nat Med. 1996;2:888–892. doi: 10.1038/nm0896-888. [DOI] [PubMed] [Google Scholar]
- 30.DeLibero G, Flesch I, Kaufmann S H E. Eur J Immunol. 1988;18:59–64. doi: 10.1002/eji.1830180110. [DOI] [PubMed] [Google Scholar]
- 31.Stenger S, Mazzacaro R J, Uyemura K, Cho S, Barnes P F, Rosat J P, Sette A, Brenner M B, Porcelli S A, Bloom B R, Modlin R L. Science. 1997;276:1684–1687. doi: 10.1126/science.276.5319.1684. [DOI] [PubMed] [Google Scholar]