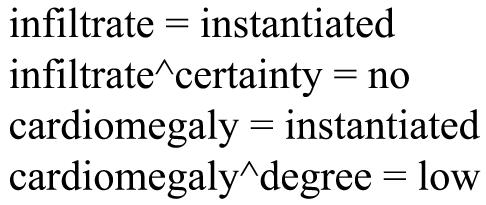Abstract
Objective: To analyze the effect of expert knowledge on the inductive learning process in creating classifiers for medical text reports.
Design: The authors converted medical text reports to a structured form through natural language processing. They then inductively created classifiers for medical text reports using varying degrees and types of expert knowledge and different inductive learning algorithms. The authors measured performance of the different classifiers as well as the costs to induce classifiers and acquire expert knowledge.
Measurements: The measurements used were classifier performance, training-set size efficiency, and classifier creation cost.
Results: Expert knowledge was shown to be the most significant factor affecting inductive learning performance, outweighing differences in learning algorithms. The use of expert knowledge can affect comparisons between learning algorithms. This expert knowledge may be obtained and represented separately as knowledge about the clinical task or about the data representation used. The benefit of the expert knowledge is more than that of inductive learning itself, with less cost to obtain.
Conclusion: For medical text report classification, expert knowledge acquisition is more significant to performance and more cost-effective to obtain than knowledge discovery. Building classifiers should therefore focus more on acquiring knowledge from experts than trying to learn this knowledge inductively.
Health care is an information-intensive industry, and health care delivery is dependent on accurate and detailed clinical data.1 An important goal of medical informatics is to facilitate access to and improve the quality of this information, thereby enhancing clinical outcomes. Data that are not available routinely in an easily accessible form represent a major challenge to this goal.
A prominent example of this challenge is accessing data contained in medical text reports. Medical text reports contain substantial and essential clinical data.2,3 For example, a recent study distinguishing between planned and unplanned readmissions found that information available in structured, coded format alone was not sufficient for classifying admissions and that information in text reports significantly improved this task.4 Although narrative text reports can be stored and retrieved electronically, clinical information represented in text reports often is not available in coded form and not easily used for automated decision support, analysis of patient outcomes, or clinical research. For computer analysis of patient data to effectively include clinical information from text reports, the data must be extracted from the reports and converted to a structured, coded form.5
One approach that may be used to convert this information to structured form is classification. Medical text reports can be classified according to the clinical conditions that are described in the reports (e.g., whether the report indicates the patient has pneumonia). Classifiers can be created to detect clinical conditions indicated in narrative text and to represent these indicated conditions as standardized codes or terms.5,6,7,8,9,10,11 However, manual creation of these classifiers (often represented as expert rules) is a difficult and expensive process, requiring the coordinated effort of both medical experts and knowledge engineers.8,12 Researchers therefore have investigated the use of inductive learning algorithms to automatically generate classifiers for medical documents.11,13,14
Studies of inductive learning and medical text reports typically have focused only on standard components of inductive learning, such as algorithm type or training set size.8,9,15,16 However, there is substantial variation in data preparation of medical text for inductive learning, dependent on the use of expert knowledge in the inductive learning process.14 Little is known about the effect of this expert knowledge. This study evaluated how expert knowledge affects the inductive learning process in creating classifiers for medical text reports.
Background
The preparation phase of inductive learning involves converting the original data, such as narrative text, to a form usable by inductive learning algorithms, such as a set of reports with attributes representing observations from the reports. Often, natural language processing (NLP) is used to convert unstructured text into a structured form that then is further modified for use with learning algorithms.8,9,15 NLP systems have been used to structure narrative clinical data by extracting observations and descriptive modifiers from free-text reports.
Expert knowledge can be used in the data preparation. An important task of data preparation is to determine the subset of attributes or features that are relevant to the classification task. This is done through feature selection or feature extraction. Domain experts can select specific attributes or features that are relevant to the classification task (feature selection). Using domain knowledge for feature selection has been suggested previously as a way to enhance the performance of machine learning algorithms.16 Gaines17 showed this effect of using expert knowledge to select relevant attributes. Clark and Matwin18 showed improved performance when using domain knowledge to restrict an algorithm's search space, but they also discussed the increased cost that can arise from using this knowledge. Domain knowledge can be used also to combine multiple features together, to create a new feature or variable (feature extraction). For example, variables assigned values indicating their presence or absence in a report could be extracted to new variables indicating their presence or absence as clinical conditions for a patient. Feature extraction not only changes the representation of the data, but may also reduce the number of variables used.
The types of domain knowledge used in data preparation also can vary between task-specific and representation-specific knowledge. Task-specific knowledge is conceptual knowledge about the general classification task. For the domain of classifying clinical reports, it is medical knowledge specific to the conditions being identified. In the context of feature selection, task-specific knowledge is the knowledge of which features or attributes are medically relevant to the clinical condition. Representation-specific knowledge is the knowledge about the specific data used. It involves understanding of the report representation, such as the different available values for features, or the meaning of those values. For example, representation-specific knowledge is the understanding of whether a value of “positive” for a feature indicates the feature is a word or phrase in the report, is a current condition for the patient, or was a previous diagnosis for the patient. In applications of machine learning, task-specific and representation-specific knowledge for feature selection is often used implicitly in the setup of the data, often in determining how data are represented. Manually selecting relevant parameters from original data sources, using predefined phrase lists, and modifying only relevant data are ways that researchers have used task-specific knowledge.8,9,11,19 Studies that used representation-specific knowledge have specifically designated negated phrases as separate concepts indicating the negation or only represented status information for observations.8,9,11 These studies used domain knowledge for feature selection or extraction but did not evaluate its impact on classifier performance.
In text classification, in which a large number of features is a major difficulty to machine learning, the potential of using domain knowledge is especially promising but somewhat unexplored. Whereas there are studies evaluating different automatic selection methods in text categorization,20 studies evaluating the effect of using domain knowledge for selection are limited. Therefore, the extent to which it can affect inductive learning performance is unknown.
Methods
We evaluated the effect of different methods for using expert knowledge in preparing medical text data for inductive learning. We evaluated the effect of expert knowledge in terms of learning algorithm performance, training set size efficiency, and creation costs. The classification tasks were to identify clinical observations indicated in chest radiograph reports and discharge summaries.
Data
The chest radiograph data were obtained from a data set used in an evaluation study of NLP.5 The set contained 200 randomly selected reports that had been classified by physicians for six clinical conditions: congestive heart failure, chronic obstructive pulmonary disease, acute bacterial pneumonia, neoplasm, pleural effusion, and pneumothorax. The reference standard for correct classification of a report for a condition was majority physician opinion. In that study, rules written by a physician and knowledge engineer were used to query the processor output and classify the reports.
Discharge summary data also came from a data set used to evaluate a natural language processor.21 The study automated a prediction rule for the prognosis of community-acquired pneumonia. Observations were extracted from sections of reports processed by a natural language processor, and these observations then were used by the prediction rule to assign patients with community-acquired pneumonia to one of five risk categories. The extracted observations included neoplastic disease, liver disease, congestive heart failure, cerebrovascular disease, renal disease, changes in mental status, cough, dyspnea, sputum production, fever, and pneumonia. The data set contained 79 discharge summaries, and a physician read the reports to establish a reference standard for that study.
Learning Algorithms
For this study, we used five algorithms from three different algorithm classes for comparison. These include rule-based (decision trees and rule induction), instance-based (nearest neighbor and decision tables), and probabilistic (naïve-Bayes) algorithms. The three types of algorithms were chosen because (1) the algorithm types are well known and studied both in machine learning and in applications of machine learning to medical data, and (2) the three algorithm types represent different approaches to learning.
The decision tree algorithm used was MC4, which is the same algorithm as C4.523 but with different default parameter settings. CN2, the rule induction algorithm, is a modification of the original AQ algorithm.24 The naïve-Bayes algorithm, also called “simple-Bayes,” is a common algorithm using predictive probabilities of attribute values.25 The nearest neighbor algorithm used was IB, developed by Albert and Aha,26 and we used the decision tables algorithm developed by Kohavi.27 These algorithms were used from the MLC++ machine learning library from Silicon Graphics.28 This library interfaces with many different machine learning algorithms, allowing different algorithms to be applied to the same data set for comparison using only one data format. To avoid biasing performance toward our expertise with particular algorithms,29 we used the default parameters already set for each algorithm.
Data Representations
We created report representations from the output of a natural language processor, specifically, the Medical Language Extraction and Encoding system (MedLEE) developed by Friedman at Columbia University.2 MedLEE is a semantic parser that takes narrative text as input and uses a clinical vocabulary to map words and phrases to standard terms. It has been trained to process many different types of clinical reports, including radiology reports and discharge summaries. MedLEE converts each report to a set of clinical observations, each of which is associated with descriptive modifiers. The processor attempts to encode all clinical information included in a report.
For example, the text “no evidence of infiltrate, but exam indicates slight cardiomegaly” occurring in a radiology report would produce the coded observations shown in ▶. To convert this hierarchical structure of MedLEE output to an attribute-value representation usable by learning algorithms, we first converted each occurring observation to a separate attribute with allowed values of “instantiated” or “not instantiated” (meaning whether the observation was instantiated in the report).8 Modifiers, or secondary attributes, were converted to individual primary attributes by combining them with their associated observations. Thus, the “certainty” modifier of “infiltrate” was represented by a new attribute, “infiltrate∧certainty.” Rather than the binary values of “instantiated” and “not instantiated,” these attributes had the values that were originally assigned to the modifiers by the processor. ▶ shows the hierarchical structure of ▶ in this attribute-value representation.
Figure 1.
Example of MedLEE output in hierarchical structure.
Figure 2.
Example of MedLEE output in flattened representation for use with inductive learning algorithms.
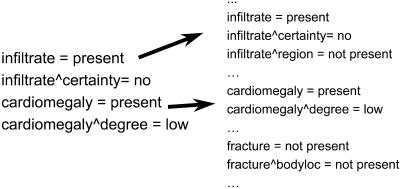
Each report was flattened to an attribute-value representation, creating a document vector representing the original report. The document vectors then were combined into a tabular structure representing each vector in terms of all attributes available in the data set. For each document vector, those attributes that did not occur in the original vector were assigned values of “not present,” indicating they were not instantiated in the report. ▶ shows how the document vectors were expanded. It shows the original document vector from ▶, which contains only four attributes. Other attributes, such as “fracture” and “fracture & bodyloc” may be present in another report of the training set, although they are not present in this specific case. The resulting table of vectors can contain many attributes that are very sparsely populated for individual cases. For example, the flattened vectors created from raw text or NLP output of the 200 chest radiograph reports contained more than 1,000 attributes.
Figure 3.
Example of a document vector obtained from the flattened output of a parsed report. Variables not in the report, but in other reports in the data set, must be included in the document vector.
Attribute Limiting
To prevent overfitting of any classification model to the data,30 we limited attributes either by their predictive value within the training set or by their relevance to the classification task as determined by domain knowledge.
The first limiting method (predictive) calculated the predictive values for each attribute value, assuming the attribute was conditionally independent from other features. The predictive values were the proportions of positive (or negative) instances in the training set correctly classified by a test using only a specific attribute value. These predictive values, for both positive and negative classifications, were then added together to create a predictive score for each attribute value. Higher scores indicated values that more strongly predict a positive classification when present in a report and a negative classification when they do not occur. We ranked attributes by the highest predictive score of their associated values and selected the top attributes for use by the machine learning algorithms. This method was efficient, because it searched only one feature subset and scaled up easily for many attributes. In addition, it allowed limiting without requiring user interaction; i.e., it worked “outside of the box” without having to be tuned to the specific learning task.
The other limiting methods (medical, negated, med&neg) used task-specific and representation-specific domain knowledge for feature selection and extraction. We limited attributes using task-specific domain knowledge by selecting only relevant observations or concepts. To establish relevance, we analyzed the expert queries from the original studies evaluating MedLEE.5,21 These studies used the same chest radiograph and discharge summary reports used here. The queries were part of expert rules that classified the medical reports. We selected the observations and concepts used in those expert queries as relevant concepts. Because a medical expert originally determined which concepts to use in these queries, selecting them from the queries was similar to having a medical expert manually select the relevant concepts. We limited using representation-specific knowledge by first manually analyzing examples of MedLEE output and determining how the observation status was represented in MedLEE output and text. We then represented each observation only by its state in the report and ignored other modifiers. For processed reports, we selected, from all possible values for the “certainty” and “status” modifiers, those values that represented a negated observation state. For example, “certainty = negative” or “status = resolved” indicates a negated state for observations.
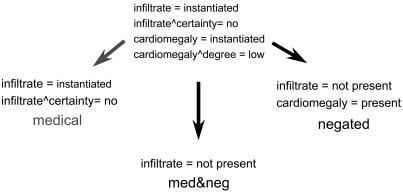
▶ gives an example showing how task-specific and representation-specific attribute limiting was performed. The attributes from ▶, which were created by flattening the NLP output, are based on two observations: “infiltrate” and “cardiomegaly.” The medical representation uses task-specific knowledge to limit attributes. If the classification task was to identify reports that indicated pneumonia, only the “infiltrate” observation is relevant, and attributes based on nonrelevant observations would be removed. The negated representation uses representation-specific information to limit attributes. Attributes based on modifiers are removed. However, the modifier-value pair “certainty = no” indicates that “infiltrate,” although instantiated in the report, is not present in the patient. Therefore, the value of the “infiltrate” attribute is changed to “not present.” (Values of “instantiated” are also changed to “present,” indicating the observation is present as a condition, rather than just instantiated in the report.) The med&neg representation combines both task-specific and representation-specific knowledge to further limit the data.
Figure 4.
Feature selection and extraction methods (limiting) using different types of expert knowledge to detect pneumonia.
The negated representation was also limited by the predictive values, as used in the predictive method. This was done because even with the representation-specific limiting, there were still many more features than cases in the data set. We limited the number of attributes for the predictive and negated data set to one tenth of the number of reports in the data set, consistent with a standard recommendation for feature selection.31,32,33 With the task-specific limiting methods (medical and med&neg), no secondary limiting was done. The task-specific limiting already had reduced the number of features to nearly a tenth of the data set size. In addition, feature selection methods are intended to discover which variables may be relevant to the classification task. Limiting task-specific features further would delete features that already have been determined to be important to classification.
Algorithm Performance
To evaluate the effect of expert knowledge on inductive learning performance, we compared the performance of five machine-learning algorithms, using different degrees of domain knowledge for feature selection/extraction (predictive, medical, negated, med&neg). These algorithms, as described above, were MC4, CN2, naïve-Bayes (NB), IB, and decision tables (DT). We measured performance in classifying six clinical conditions for radiology reports and 11 clinical conditions for discharge summaries. We processed each report using MedLEE, flattened the MedLEE output of these reports, and applied the four feature selection methods. We used 200 chest radiograph reports and 79 discharge summaries that had been classified previously by experts and used leave-one-out cross-validation to maximize the training set sizes while avoiding bias from testing on the training set. Each algorithm also was trained separately for each of the clinical conditions and representations and thus generated different classifiers for each disease and representation to determine whether the condition was present or absent. Performance was measured in terms of sensitivity and specificity, from which A' an estimate of receiver operating characteristic curve (ROC) area, was computed.34 Finally, we used bootstrapping to compute estimates of variance.35
Training Set Size Efficiency
In addition to testing classifier performance, we evaluated the effect of expert knowledge on classifiers at different training set sizes. We used learning curves, which show how the performance of an algorithm improves as the number of training examples increases. We computed learning curves of the four different feature selection/extraction methods using a separate set of 300 radiograph reports, classified according to the six clinical conditions by a single physician. For the test set, we used the same 200 cases used in the evaluation of algorithm performance described above. A learning curve was generated by building separate classifiers created by training on different subsets of the training set. We computed classifier performance when using training set sizes ranging from 30 to 300 cases, in increments of 30. Five training sets were built at each size for each disease and each algorithm, and the performance of A' was averaged among the five classifiers.
Classifier Creation Cost
We also evaluated expert knowledge and inductive learning in terms of costs for creating classifiers for chest radiographs. We measured this cost in terms of the human time component necessary to perform a task. Using the learning curves, we extrapolated the number of cases necessary for each method to reach expert-level performance or the performance of an expert manually classifying reports. We also used estimates of the time necessary to collect different types of expert knowledge based on experience in writing rules and creating training sets. We estimated costs for four components of building a classifier for medical text reports: manually classifying one report for use in a training set, writing manual rules, specifying relevant observations, and determining negation criteria.
We used MC4 as the machine learning algorithm for this cost analysis, because it is implemented easily and its performance was consistently high in all the studies. For each method, we measured the cost to obtain equivalent performance with expert rules, which were not different from manual physician review. To determine this cost when methods previously never reached the rules performance level, we either extrapolated the number of training cases that would be needed from the learning curves or inferred from existing data the costs of creating domain knowledge.
The cost of classifying chest x-ray reports was determined from the original evaluation of MedLEE.5 That study reported that it took a physician about two hours to analyze 100 reports. These reports were analyzed to detect six clinical conditions, although the bulk of the time probably was spent reading the report. Therefore, we estimated the cost of manually classifying one report by one physician for one condition (represented by CASE) to be about 1 minute. The time required to write rules for seven clinical conditions has been reported as one week.12 The average cost of writing rules for one condition (RULES) is between six and 20 hours. This includes the time to specify task-specific and representation-specific knowledge as rules, and to debug/test the rules.
To determine the cost of specifying task-specific observations (TASK), we measured the time it took a physician to select relevant observations for one disease from a list. Initially, we limited the list of all possible observations to those that were more likely to be relevant to a disease using automated methods. First, a physician selected ICD-9 codes that were relevant to congestive heart failure (CHF). Using these codes, we compiled a set of 10,000 chest radiographs from New York-Presbyterian Hospital where the discharge diagnosis code of the inpatient visit associated with a report was relevant to CHF. We used a large set of reports here, rather than the 200 chest radiograph reports used above, to ensure a more comprehensive list of possible relevant observations. We processed these reports using MedLEE and compiled a list of all observations occurring in these reports. We then selected those observations occurring in at least 1% of all the reports, resulting in a list of about 200 observations. Finally, we measured the time a physician took to manually select from this list those findings that would be strongly relevant to identifying CHF in a chest x-ray report. It took between 5 and 15 minutes to determine discharge diagnoses of CHF and 5 to 15 minutes to select relevant observations from this list. Thus, we estimated that it took between 10 and 30 physician minutes to select relevant observations from NLP output.
Determining negation criteria was done by a nonphysician medical informatics researcher (ABW), with the assistance of a physician (not a coauthor). The researcher examined the list of possible MedLEE modifiers, selected those relevant to negation, and reviewed the list of possible values for these modifiers. There were 103 modifier values considered, which took between 10 and 30 minutes, with less than 15 minutes of physician time to answer questions. Thus, we estimated the time to determine negation criteria (NEG) to be about 45 minutes.
Results
▶ shows the performance of machine learning algorithms classifying chest radiograph reports when using limiting methods that use different types of domain knowledge (predictive, medical, negated, med&neg) for NLP output. The predictive method, which limits by the characteristics of the training set data without domain knowledge, performed significantly worse than the other methods (p < 0.001). The med&neg method, however, performed significantly better than all other methods (p < 0.001). There was no difference between the methods using one type of domain knowledge exclusively (i.e., medical and negated). These findings were consistent for individual algorithms, as well as for averaged performance across algorithms.
Figure 5.
Comparison of machine learning algorithms and feature selection methods using natural language processing (NLP) output from radiology reports. ROC = receiver operating characteristic [curve]; MC4, CN2, NB, IB, and DT are algorithms.
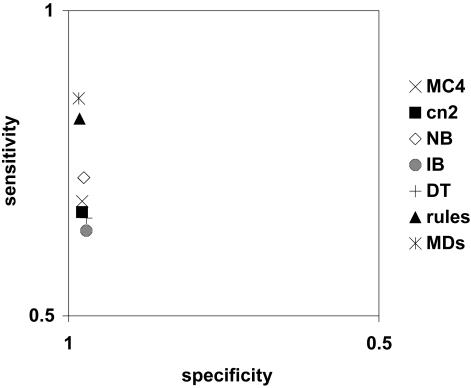
▶ shows the sensitivity and specificity of the various learning algorithms using the best-performing limiting method (med&neg) in receiver operating characteristic (ROC) space. In addition, it shows the average performance of expert rules and physicians in classifying the reports, as reported by Hripcsak et al.5 All algorithms performed worse than these expert rules or physicians.
Figure 6.
Receiver operating characteristic (ROC) plot of algorithm performance using med&neg feature selection. MC4, CN2, NB, IB, and DT are algorithms; rules is a classifier; and MDs refers to physicians.
▶ shows machine learning algorithm and limiting method performance when applied to NLP output from discharge summaries. No algorithm was superior to any other algorithm for all limiting methods, although decision tables were better than all other algorithms when using task-specific limiting. There was no difference in average performance between medical and med&neg, although there were significant differences between the average performances of the other limiting methods.
Figure 7.
Comparison of machine learning algorithms and feature selection methods using natural language processing (NLP) output from discharge summaries. ROC = receiver operating characteristic [curve]; MC4, CN2, NB, IB, and DT are algorithms.
The learning curves for the feature selection methods, averaged over all five learning algorithms, are shown in ▶. The solid line indicates the best performance level reached by the predictive method on the learning curve. The point at which a learning curve for a limiting method intercepts this line indicates the number of training cases needed, along with the expert knowledge of the limiting method, to get equivalent performance to the predictive method on the curve. The med&neg method outperforms the 300-case predictive method at all training set sizes above 30, whereas medical and negated outperforms predictive at 60 cases and 210 cases, respectively.
Figure 8.
Comparison of machine learning algorithms and feature selection methods with different training set sizes. The solid horizontal line indicates the best performance level reached by the predictive method on the learning curve. ROC = receiver operating characteristic [curve].
▶ lists the estimated costs in terms of physician hours for building classifiers using different approaches, including manually writing rules. We determined an optimistically small number of training cases needed for the inductive learning algorithms, assuming linear increase in performance. We also determined the number of training cases assuming a logarithmic curve, which is similar in curvature to standard learning curves. The rules classifier was 50% less in cost than the most optimistic estimates for inductive learning-based classifiers.
Table 1.
Costs and Cost Components for Different Classifiers
| Classifier | Cost Components | Cost |
|---|---|---|
| rules | Rules | 6 hours |
| negated | Neg + (510 to 2,200) Case | 9 hours to 1 week |
| predictive | (670 to 7,700) Case | 11 hours to 3 weeks |
| medical | Task + (800 to 12,300) Case | 14 hours to 5 weeks |
| med&neg | Task + Neg + (800 to 16,500) Case | 15 hours to 2 months |
Discussion
The most significant factor in improving performance and decreasing costs of classifiers, or the performance of learning algorithms that create the classifiers, is the use of expert knowledge. Improved performance because of expert knowledge was much more significant than improvement because of differences in learning algorithms, as shown in ▶. The use of expert knowledge also affected comparisons between machine learning algorithms. For example, the predictive and negated limiting methods had significantly better performance with MC4 than decision trees. However, there was no performance difference between the algorithms using the med&neg method. This result was similar to that in previous work, which found that the actual data representation and the knowledge used to build it could even reverse conclusions drawn from comparisons between algorithms.14 Conclusions from comparisons between inductive learning algorithms must therefore take into account the domain knowledge and data representation used in preparing data for the learning task. In machine learning research, it has been shown that comparisons between learning algorithms are not inherently generalizable to other tasks. This research shows that the comparisons may not even be generalized to other approaches of the same task.
The analysis of learning curves (▶) found that the characteristics of the curves were sensitive to the use of expert knowledge for feature selection. There are noticeable differences in the slopes of the learning curves for different limiting methods, indicating that the addition of training cases affects some methods more than others. A conclusion would seem to be that the effect of additional training cases is less for the med&neg method than for others. However, the differences in slope may also be caused by the asymptotic quality of the learning curves. As performance approaches that of the reference standard (the physicians), we would expect the curves to flatten. Differing slopes seen here may be caused more by the actual performance level than by the type of limiting method.
A characteristic of a curve that may be more interesting than the slope is where the performance of another method surpasses the best performance of the predictive method. This point indicates the real value of the domain knowledge in terms of the training set size. When one type of knowledge (task-specific or representation-specific) is used, the number of training cases needed for equivalent performance decreases by about half. When both types of knowledge are used, the number of cases is less than one tenth.
Expert knowledge needed for constructing classifiers can be obtained separately as task-specific and representation-specific components. We represented expert knowledge in two separate forms: task-specific knowledge and representation-specific knowledge. Task-specific knowledge consisted of clinically relevant observations or features for a specific classification task. Representation-specific knowledge included state information (presence or absence in the patient) of observations. Both methods were used specifically in the feature selection stage of inductive learning. Task-specific and representation-specific knowledge significantly improved learning algorithm performance and could also be combined to further improve performance.
An important observation is that no inductive learning performed as well as physicians or expert-written rules (▶). Although we were able to show significant improvement over other inductive learning methods, the performance was not sufficient to justify using inductive learning instead of expert rules to create classifiers. Part of this problem could be due to the small training set size used, and good performance might be obtained if larger training sets were used. Still, creating such training sets is expensive and requires a domain expert to classify the training cases. In fact, the cost–benefit analysis of expert knowledge also showed that with a limited training set, it is more efficient to have experts write rules than to create a new training set (▶).
Another conclusion that could be drawn from these results is that creating rules may be more a process of knowledge acquisition than knowledge discovery. Inductive learning, a knowledge discovery process, must incorporate expert knowledge to be even reasonably efficient. Otherwise, a lot of effort is wasted relearning what is already known. Therefore, efforts should focus on obtaining and representing that expert knowledge in the rule-generation process. Here we show that expert knowledge could be separated into components effectively. Other approaches could focus on efficiently collecting the different types of knowledge from experts, or incorporating information from other knowledge sources. Such efforts would be more effective at improving classifier performance than more in-depth analysis of inductive learning.
This research was performed on tasks in which expert-written rules had already been shown to be effective. Thus, the expert knowledge already existed in some form. There may be other tasks in which such information is not available and must be discovered. The cost analysis showed how expensive it could be to create a large-enough training set to discover knowledge. In such cases, efforts should be made to maximize the efficiency of collecting a training set. For example, some cases in a training set would add more information than others, especially when many examples are redundant. Maximizing the value added by each case classified by an expert for a training set would improve the efficiency of the inductive learning process.
Conclusion
We analyzed the effect of expert knowledge on the inductive learning process in terms of data representation, classifier performance, and costs. This analysis showed expert knowledge to be the most significant factor affecting inductive learning performance, outweighing differences in learning algorithms. In addition, we found that the use of expert knowledge can affect comparisons between learning algorithms. This expert knowledge may be obtained and represented separately as knowledge about the clinical task or about the data representation used. The benefit of the expert knowledge is more than that of inductive learning, with less cost to obtain. For this task, building classifiers should focus, therefore, more on acquiring knowledge from experts than trying to learn this knowledge inductively.
This work was supported by National Library of Medicine Grants R01 LM06910 “Discovering and Applying Knowledge in Clinical Databases,” R01 LM06274 “Unlocking Data from Medical Records with Text Processing,” and Pfizer, Inc. grant “Using Information Systems to Advance Clinical Research and Clinical Care.”
References
- 1.Chute CG, Cohn SP, Campbell JR. A framework for comprehensive health terminology systems in the United States: development guidelines, criteria for selection, and public policy implications. ANSI Healthcare Informatics Standards Board Vocabulary Working Group and the Computer-Based Patient Records Institute Working Group on Codes and Structures. J Am Med Inform Assoc. 1998;5:503–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1:161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spyns P. Natural language processing in medicine: an overview. Methods Inf Med. 1996;35:285–301. [PubMed] [Google Scholar]
- 4.Kossovsky MP, Sarasin FP, Bolla F, Gaspoz JM, Borst F. Distinction between planned and unplanned readmissions following discharge from a department of internal medicine. Methods Inf Med. 1999;38:140–3. [PubMed] [Google Scholar]
- 5.Hripcsak G, Friedman C, Alderson PO, DuMouchel W, Johnson SB, Clayton PD. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med. 1995;122:681–8. [DOI] [PubMed] [Google Scholar]
- 6.Jain NL, Friedman C. Identification of findings suspicious for breast cancer based on natural language processing of mammogram reports. Proc AMIA Annu Fall Symp. 1997:829–33. [PMC free article] [PubMed]
- 7.Jain NL, Knirsch CA, Friedman C, Hripcsak G. Identification of suspected tuberculosis patients based on natural language processing of chest radiograph reports. Proc AMIA Annu Fall Symp. 1996:542–46. [PMC free article] [PubMed]
- 8.Zingmond D, Lenert LA. Monitoring free-text data using medical language processing. Comput Biomed Res. 1993;26:467–81. [DOI] [PubMed] [Google Scholar]
- 9.Chapman WW, Fizman M, Chapman BE, Haug PJ. A comparison of classification algorithms to automatically identify chest x-ray reports that support pneumonia. J Biomed Inform. 2001;34:4–14. [DOI] [PubMed] [Google Scholar]
- 10.Chapman WW, Haug PJ. Comparing expert systems for identifying chest x-ray reports that support pneumonia. Proc AMIA Symp. 1999:216–20. [PMC free article] [PubMed]
- 11.Aronow DB, Fangfang F, Croft WB. Ad hoc classification of radiology reports. J Am Med Inform Assoc. 1999;6:393–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hripcsak G, Kuperman GJ, Friedman C. Extracting findings from narrative reports: software transferability and sources of physician disagreement. Methods Inf Med. 1998;37:1–7. [PubMed] [Google Scholar]
- 13.Hersh WR, Leen TK, Rehfuss PS, Malveau S. Automatic prediction of trauma registry procedure codes from emergency room dictations. Medinfo. 1998;9(pt 1):665–9. [PubMed] [Google Scholar]
- 14.Wilcox A, Hripcsak G. Medical text representations for inductive learning. Proc AMIA Symp. 2000:923–27. [PMC free article] [PubMed]
- 15.Wilcox A, Hripcsak G. Classification algorithms applied to narrative reports. Proc AMIA Symp. 1999:455–59. [PMC free article] [PubMed]
- 16.Wilcox A, Hripcsak G. Knowledge discovery and data mining to assist natural language understanding. Proc AMIA Annu Fall Symp. 1998:835–39. [PMC free article] [PubMed]
- 17.Gaines BR. An ounce of knowledge is worth a ton of data: quantitative studies of the trade-off between expertise and data based on statistically well-founded empirical induction. Proceedings of the Sixth International Workshop on Machine Learning. San Mateo, CA: Morgan Kaufmann, 1989, pp 156–9.
- 18.Clark P, Matwin S. Using qualitative models to guide inductive learning. Proceeding of the Tenth International Machine Learning Conference (ML-93). San Mateo, CA: Morgan Kaufmann, 1993, pp 49–56.
- 19.Ohmann C, Moustakis V, Yang Q, Lang K. Evaluation of automatic knowledge acquisition techniques in the diagnosis of acute abdominal pain. Acute Abdominal Pain Study Group. Artif Intell Med. 1996;8:23–36. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Pederson JP. A comparative study on feature selection in text categorization. Proceedings of the Fourteenth International Conference on Machine Learning (ICML'97). San Mateo, CA: Morgan Kaufmann, 1997, pp 412–20.
- 21.Friedman C, Knirsch C, Shagina L, Hripcsak G. Automating a severity score guideline for community-acquired pneumonia employing medical language processing of discharge summaries. Proc AMIA Symp. 1999:256–60. [PMC free article] [PubMed]
- 22.Salzberg S. On comparing classifiers: a critique of current research and methods. Data Mining and Knowledge Discovery. 1997;1:317–27. [Google Scholar]
- 23.Quinlan J. AnonymousC4.5: Programs for Machine Learning. Redwood City, CA: Morgan Kaufmann, 1993.
- 24.Clark P. Machine learning: techniques and recent developments. In Mirzai AR (ed). Artificial Intelligence: Concepts and Applications in Engineering. London: Chapman and Hall, 1990, pp 65–93.
- 25.Langley P, Iba W, Thompson K. An analysis of Bayesian classifiers. Proceedings of the Tenth National Conference on Artificial Intelligence. San Jose, CA: AAAI Press, 1992, pp 223–8.
- 26.Albert MK, Aha DW. Analyses of instance-based learning algorithms. Proceedings of the Ninth National Conference on Artificial Intelligence. Anaheim, CA: AAAI Press, 1991, pp 553–8.
- 27.Kohavi R. The power of decision tables. Proceedings of the 8th European Conference on Machine Learning. Berlin: Springer, 1995, pp 174–89.
- 28.Kohavi R, Sommerfield D, Dougherty J. Data mining using MLC++: a machine learning library in C++. Toulouse, France: IEEE Computer Society Press. Tools with Artificial Intelligence, 1996, pp 234–45.
- 29.Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognition. 1997;30:1145–59. [Google Scholar]
- 30.Almuallim H, Dietterich TG. Learning with many irrelevant features. Proceedings of the Ninth National Conference on Artificial Intelligence (AAAI-91). Anaheim, CA: AAAI Press, 1991, pp 547–52.
- 31.Peduzzi P, Concato J, Feinstein AR, Holford TR. Importance of events per independent variable in proportional hazards regression analysis.II. Accuracy and precision of regression estimates. J Clin Epidemiol. 1995;48:1503–10. [DOI] [PubMed] [Google Scholar]
- 32.Peduzzi P, Concato J, Kemper E, Holford TR, Feinstein AR. A simulation study of the number of events per variable in logistic regression analysis. J Clin Epidemiol. 1996;49:1373–9. [DOI] [PubMed] [Google Scholar]
- 33.Tierney WM, Murray MD, Gaskins DL, Zhou XH. Using computer-based medical records to predict mortality risk for inner-city patients with reactive airways disease. J Am Med Inform Assoc. 1997;4:313–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grier JB. Nonparametric indexes for sensitivity and bias: computing formulas. Psychol Bull. 1971;75:424–9. [DOI] [PubMed] [Google Scholar]
- 35.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. New York: Chapman and Hall, 1993.