Abstract
Objective: To describe the development and evaluation of computational tools to identify concepts within medical curricular documents, using information derived from the National Library of Medicine's Unified Medical Language System (UMLS). The long-term goal of the KnowledgeMap (KM) project is to provide faculty and students with an improved ability to develop, review, and integrate components of the medical school curriculum.
Design: The KM concept identifier uses lexical resources partially derived from the UMLS (SPECIALIST lexicon and Metathesaurus), heuristic language processing techniques, and an empirical scoring algorithm. KM differentiates among potentially matching Metathesaurus concepts within a source document. The authors manually identified important “gold standard” biomedical concepts within selected medical school full-content lecture documents and used these documents to compare KM concept recognition with that of a known state-of-the-art “standard”—the National Library of Medicine's MetaMap program.
Measurements: The number of “gold standard” concepts in each lecture document identified by either KM or MetaMap, and the cause of each failure or relative success in a random subset of documents.
Results: For 4,281 “gold standard” concepts, MetaMap matched 78% and KM 82%. Precision for “gold standard” concepts was 85% for MetaMap and 89% for KM. The heuristics of KM accurately matched acronyms, concepts underspecified in the document, and ambiguous matches. The most frequent cause of matching failures was absence of target concepts from the UMLS Metathesaurus.
Conclusion: The prototypic KM system provided an encouraging rate of concept extraction for representative medical curricular texts. Future versions of KM should be evaluated for their ability to allow administrators, lecturers, and students to navigate through the medical curriculum to locate redundancies, find interrelated information, and identify omissions. In addition, the ability of KM to meet specific, personal information needs should be assessed.
With the advent of the Internet and widespread availability of tools for computer-based instruction, medical schools often provide course content materials1,2,3 and methods for curricular content evaluation online.4 Such systems typically require labor-intensive data entry and substantial manual organization.5,6 Because lecturers and course directors have limited time and resources, automated concept “mapping” and display of “relevant” segments of curricular content are highly desirable. Mapping tools could facilitate efficient concept-level integration of curricular components, affording lecturers and students easy access to relevant course documents and correlated biomedical literature.7 Concept mapping also would allow administrators and lecturers to revise curricula by highlighting redundancies and omissions.8 For these reasons, we have developed and evaluated the KnowledgeMap (KM) system, a prototypic biomedical concept identifier designed to improve access over time to curricular content at Vanderbilt University School of Medicine.
Background
For decades, informatics researchers have applied Natural Language Processing (NLP) techniques and heuristic concept matching tools based on standard lexicons (such as those found within the Unified Medical Language System—UMLS9—and the Galen Knowledge representation scheme10) to identify and extract “key” concepts from a number of biomedical sources.11,12,13,14,15,16 Older reports describe automated mapping of medical curricula using Medical Subject Headings (MeSH)5,11 or the UMLS.12 In the latter, a lack of concept representation in the older versions of the standard lexicon and, possibly, imperfect algorithms led to unacceptable recall results.12 Using more recent UMLS versions and newer approaches, investigators have mapped clinical free text more effectively.13,14,15,16 We developed the KM concept identifier system to address the format and content structure of medical curricular documents, including document organization, sentence structure, and concept clustering.
Curricular Document Organization
Medical curricular documents often use outline formats and include ad hoc abbreviations. Mapping tools must remove outline headings, preventing errors such as misidentification of “V. Cranial Nerves” as “fifth cranial nerve” while avoiding the removal of “E.” from a line starting with “E. coli.” Lecturers often use parenthetically defined abbreviations for word efficiency. Mapping tools must recognize document defined abbreviations (as does (the AbbRE [abbreviation recognition and extraction] program)17) and subsequently expand them for unambiguous concept identification.
Curricular Sentence Structure
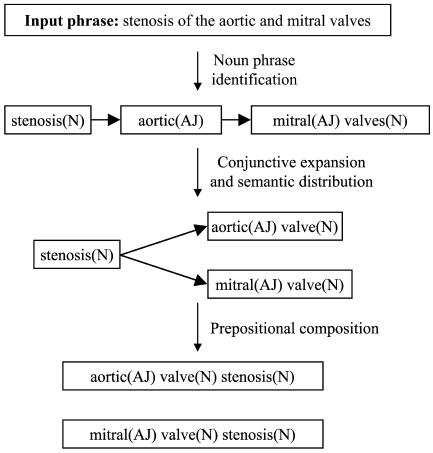
“Complex” noun phrases, which we define as noun phrases connected by prepositions, coordinating conjunctions, and linking verbs, frequently are used in medical documents. Mapping tools must appropriately recognize across prepositional connectors18 (for example, “carcinoma of the lung” is the same as “lung carcinoma”) while remaining sensitive to irreducible concepts, such as “activities of daily living” and “range of motion.”
Complex noun phrases may involve conjunctions that link two nouns sharing a common modifier (e.g., “dissection or aneurysm of the aorta”), or conjunctions linking two modifiers that share the same head noun (e.g., “abdominal or inguinal hernia”). Mapping tools should match the semantic types of adjectives or nouns before distributing them in conjunctional noun phrases.19
Many medical school documents also contain large noun phrases that are not represented singly in the 2001 Edition of the Metathesaurus. The term “small cell bronchogenic carcinoma” is not listed as a unique concept in the Metathesaurus, even though it contains the two overlapping concepts “small cell carcinoma” and “bronchogenic carcinoma.” Mapping tools should accurately recognize overlapping concepts by utilizing specific head noun–modifier pair heuristics.20
Curricular Concept Clustering
Each medical school curricular document, designed as a teaching tool, represents a circumscribed area of medical knowledge that permits context-dependent ambiguity resolution. “Envelope” indicates “viral envelope” in a lecture about herpes viruses and “nuclear envelope” in a lecture about eukaryote DNA. Mapping tools can use heuristic methods to deduce the meaning of ambiguous acronyms or abbreviations through reference to the frequency of concepts seen with them in Medline-indexed articles. In a clinical lecture on chest pain, Medline co-occurrence data for candidate concepts12,21 can prioritize “coronary artery disease” over “chronic actinic dermatitis” as an expansion of the acronym “CAD.”
Resources
The 2001 UMLS is composed of three main components: the Metathesaurus, the SPECIALIST Lexicon, and the Semantic Network. The main component of the UMLS is the Metathesaurus, a composite of more than 50 separate source vocabularies containing nearly 1.5 million English strings organized into about 800,000 unique concepts. The Metathesaurus also contains files that provide metadata, relationships, and semantic information for each concept. The SPECIALIST Lexicon includes lexical information about a selected core group of biomedical terms, including their parts of speech, inflectional forms, common acronyms, and abbreviations. The Semantic Network is a classification system for the concepts in the Metathesaurus, identifying broader-than/narrower-than parent–child associations among different concepts and relationships represented within the Metathesaurus. For example, “disease or syndrome” is classified as a “pathologic function” in the semantic network, and itself has “child” concepts including “mental or behavioral dysfunction” and “neoplastic process.”
The 2001 UMLS Metathesaurus is composed of 19 main text files, each a table of concept and string-specific information. This information is accessible via concept unique identifiers (i.e., a CUI) and string unique identifiers (i.e., an SUI). All strings (i.e., SUIs) referring to one topic are assigned to the same CUI. For instance, the strings “hepatolenticular degeneration” and “Wilson's disease” both represent the same concept and are assigned the same CUI, although they have different SUIs. We used the following files from the Metathesaurus to create KM:
MRCON contains all strings for each concept, providing the CUI and SUI for each. For each term, MRCON identifies whether the term is a “preferred form” in its source vocabulary and whether it is a “suppressible synonym.” Examples of suppressible synonyms include the abbreviated term “fet heart rate variabil” for “variable fetal heart rate” and the term “bladder, unspecified” when used to represent “malignant neoplasm of the bladder.”
MRSO provides the vocabulary (terminology) source(s) for each string in MRCON. A particular string (e.g., “congestive heart failure”) may appear in multiple sources, such as the MeSH vocabulary used to index Medline and the International Classification of Diseases (ICD-9).
MRRANK contains a hierarchy of the vocabulary sources and string types used in composing MRCON. Certain vocabularies, such as MeSH, have higher precedence than others. From this information, one can determine which SUI in MRCON is the preferred term for a given concept.
MRSTY contains the semantic type (e.g., “disease or syndrome”) for each concept in MRCON. There are 134 different semantic types grouped into a hierarchy through the UMLS Semantic Network. For example, “Human” is in the “Vertebrate” subtree.
MRCOC provides the frequency of co-occurrence of two concepts in the same indexed articles from databases such as Medline (i.e., the number of articles discussing both concepts during a given time period). For example, MRCOC defines that the concept “myocardial infarction” co-occurred with the concept “electrocardiogram” in 1,012 Medline articles between 1992 and 1996.
Methods
We envision KM as the first in a series of concept-based tools for curricular analysis. The tools will allow students, faculty, and administration to view, manipulate, and improve the curriculum. To evaluate the current prototypic version of KM, we obtained 85% of the documents (handouts, presentations) used in the first two (preclinical) years of Vanderbilt Medical School lectures in 2000–2001. This resulted in a total of 571 documents. We tested the ability of KM to recognize manually identified “gold standard” concepts using a subset of the collected documents. We compared its performance to a state-of-the-art standard tool, the National Library of Medicine's MetaMap program.
KM System Resources
KM uses both UMLS-derived and author-developed resources for word and term normalization, language processing, and concept identification. KM currently uses the 2001 edition of the UMLS.9 We developed KM using Perl and Microsoft Visual C++. The evaluation tested KM running on a 1.0 GHz Pentium III Windows-based system with 512 MB of RAM.
Lexical Tools
We derived the KM lexicon from the UMLS SPECIALIST Lexicon,9 mapping each SPECIALIST word inflection and lexical variant to its unique base form (for example, “needs” maps to both “need” as a third-person singular noun form and to “need” as the infinitive form of a transitive verb). For many acronyms/abbreviations contained in SPECIALIST, we generated additional plural and period-containing variants, using each non–period-containing form as the base form. By this mechanism, we mapped “A.A.A.” to both “American Academy of Allergy” and “Abdominal Aortic Aneurysm,” although it maps only to the former in SPECIALIST. We created a list of “stopwords” (words considered not useful for information retrieval) containing all prepositions, pronouns, conjunctions, and determinants along with other nonmedical common words (such as “do,” “each,” and “other”).
To recognize additional word forms not in our base lexicon (but related to it), we utilized the SPECIALIST Neo-classical Combining Forms (prefixes, roots, and terminals). We also added 1,120 new prefixes, roots, and terminals using quasi-automated algorithms that analyzed word forms in the Metathesaurus for prefixes and suffixes that were not in the SPECIALIST list and that recurred with more than a threshold frequency across words in the Metathesaurus. We validated the new, automatically derived combining forms through manual review, using Webster's Third New International Dictionary, Unabridged.22
The authors manually created 156 suffix-based “form-rules” that allowed interconversion of lexical variants of “base” KM lexicon word entries. The form-rules provided mappings among common ending forms, based on parts of speech, for English, Latin, Greek, and inflections (▶). For example, to map noun form “appendix” to adjectival form “appendiceal,” apply form-rule “NO–ix→AJ–iceal.” KM does not apply form-rules or combining forms to “discover” matches for unmatched words unless the root or lexicon-matching stem is at least four letters long and not solely an abbreviation, determinant, conjunction, or preposition. To insure that we had created an adequate lexicon for normalization, we tested the normalization program on our corpus of lectures before further development. With the above heuristics, KM was able to match more than 97% of the non-stopwords in our corpus of lectures. KM does not normalize unrecognized words.
Table 1.
Example Form-rules Variant Generation
| Rule | Example |
|---|---|
| -ae NO ⇒ -a NO | fimbriae (NO) ⇒ fimbria (NO) |
| -as NO ⇒ -atic AJ | pancreas (NO) ⇒ pancreatic (AJ) |
| -nce NO ⇒ -nt AJ | absence (NO) ⇒ absent (AJ) |
| -oid AJ ⇒ oidy NO | diploid (AJ) ⇒ diploidy (NO) |
NO, noun; AJ, adjective.
To process documents to identify concepts, KM utilizes for its first pass a part-of-speech tagger developed by Cogilex R & D (Montreal, Canada).23 It is a versatile rule-based tagger that does not require frequency information and allowed incorporation of KM lexical information and combining forms to augment its base vocabulary.
UMLS Metathesaurus-based Resources
The Metathesaurus is a compendium of many controlled vocabularies developed for a variety of purposes. Consequently, it contains many strings that are not useful for concept identification in free text.13,24 The authors developed rules to filter out nonhelpful Metathesaurus terms, eliminated stopwords (including “NOS”), and normalized each string in the UMLS Metathesaurus (MRCON) by converting each word in each term (i.e., an SUI) to its base canonical form while leaving nonidentified words in their original form. From the MRCON terms, we removed unique terms (i.e., an SUI) fulfilling any of the following criteria:
Term was a suppressible synonym.
Term had fewer than 50% non-stopwords recognized by KM (in a multiword string).
Term had more than six non-stopwords (excluding semantic type “chemical”).
Term began with “other,” or contained variants of “without mention” or “not elsewhere classified.”
Term semantic type (such as “Clinical Drug” from UMLS MRSTY file) was on list of types authors chose to exclude from concept matching.
We used heuristics, similar to previously described methods, to extract acronyms and their expansions from both MRCON25 and lecture documents.17 To facilitate concept identification, we augmented the processed MRCON file (creating MRCON-PS) by adding MRCON-related strings with abbreviations and/or their expansions. For instance, “LV failure” (not in MRCON) was appended to the set of strings representing the concept “left ventricular failure” (present in MRCON). We created an inverted word index, mapping normalized words to their corresponding SUI in the MRCON-PS file. We chose a preferred name (CUI-PN) for each concept using MRRANK and MRSO. This processing resulted in more than 1,000,000 English strings and nearly 600,000 concepts in MRCON-PS.
To process complex noun phrases, we created a linkage precedence hierarchy (analogous to the order of operations in mathematical formulae) with coordinating conjunctions first, followed by linking verbs, and then by prepositions. We ranked prepositions by their frequency in MRCON strings, using only those occurring more than 70 times in MRCON. As such, “of,” “to,” and “with,” respectively, received the highest linkage priorities for prepositions.
We used the MRSTY semantic type information to assign “derived semantic types” (DSTY) to all one- and two-word normalized strings (similar to Campbell and Johnson26). For example, “concentration” only has semantic type “mental process” in the Metathesaurus, but the algorithm extracted “laboratory or test result” and “quantitative concept” as additional DSTYs from the strings “hemoglobin concentration” and “drug concentration,” respectively. Using patterns to extract one- or two-word terms from multiword phrases, we created DSTYs for about 90,000 additional entries and for approximately 12,000 terms not listed separately in MRCON. In addition, we manually added other DSTYs not in MRSTY.
Document Processor
To process documents, KM first removes outline headings by distinguishing, from position and context, among ambiguous headers such as “IV” meaning “intravenous” versus the Roman numeral outline marker. KM next eliminates carriage returns, tabs, and multiple spaces from within identified sentences. KM then tags words with their part of speech using the Cogilex part-of-speech tagger. Using the part-of-speech information, KM identifies noun phrases (defined as containing, as a minimum, either a solitary adjective or a solitary noun). KM retains adverbs, verb particles, and numbers only when they are contained within or linked to a noun phrase. Thus, the verb participle “enlarged” is not further processed unless it links to a noun phrase, acting as an adjective (▶). Noun phrases can span parentheses and dashes, with higher precedence given to the parentheses in the case of overlap. As seen in ▶, the KM algorithm considers possessive nouns as separate noun phrases with an implied linkage using the preposition “of.” Finally, KM normalizes each word.
Figure 1.
KM noun phrase identification for a simple sentence. NP, noun phrase. Initially, NP1 is linked only to NP2, but after conjunctive expansion, NP1 would be linked to both NP2 and NP3 because “heart” and “liver” are both DSTY “Body Part, Organ, or Organ Component.” Likewise, NP2's link_to field world also be changed to NP4.
KM Concept Identification
Before concept identification, KM searches the document for parenthetically defined acronyms. KM then identifies concepts in all simple noun phrases, considering all linked simple noun phrases for combinatory matching. Finally, KM attempts score-based ambiguity resolution for noun phrases with multiple MRCON-PS string candidates.
KM Methodology for Simple Noun Phrase Concept Identification
Similar to previous efforts,7,27 KM sequentially intersects, for each normalized word in each identified noun phrase, the list of “candidate MRCON canonical terms” (i.e., terms from MRCON-PS) matching the canonical word. Unless KM finds a common MRCON-PS candidate string for a document-defined acronym and its expansion, KM will use the acronym expansion to find candidate strings. KM stops simple noun phrase processing when (1) an exact canonical string match for a noun phrase has been identified or (2) linked noun phrases with non-null intersections are identified. Otherwise, KM performs the following:
Semantic and derivational variant generation: Similar to MetaMap, KM dynamically generates word variants for each noun phrase word, using both KM form-rules and UMLS derivational and semantically related terms (from the SPECIALIST database files DM.DB and SM.DB, respectively).13
Lexical/part-of-speech filtering: KM excludes adjectival verb participles, provided the remaining noun phrase matches (“chronic recurring arrhythmia”→“chronic arrhythmia”). KM also eliminates words not contained in any form in MRCON-PS.
Matched simple noun phrases are eligible for further matching across noun phrase linkages (“combinatory matching”). However, for simple noun phrases still yielding a null set, KM evaluates MRCON-PS strings containing any combination of noun phrase component words or their variants, allowing exact-matching candidates to overlap. Thus, for “glomerular endothelial cell” (not present in MRCON-PS), KM considers “glomerular endothelium,” “glomerular cell,” and “endothelial cell” as well as the words “glomerulus,” “endothelium,” and “cell.” If no exact string matches exist, KM considers overmatching candidates (candidates with additional words, numbers, or letters).
KM Methodology for Combinatory Matching
KM processes noun phrase linkages via the assigned priority of the linking word (see “UMLS Metathesaurus-based Resources” above). The algorithm first processes noun phrases joined by coordinating conjunctions, performing conjunctive expansion and modifier distribution. Conjunctive expansion occurs if the noun phrases share a DSTY and are linked to another noun phrase, as in the example “heart and liver are enlarged” in ▶. Modifier distribution allows distribution of adjectives or adjectival nouns with the same DSTY to a shared head noun across a conjunction as seen in ▶. As explained above, KM then constructs “candidate MRCON canonical terms” by intersecting candidate MRCON-PS strings associated with each word in the noun phrase (generally proceeding from left to right through the text). If the intersection process yields a null set, KM expands possibilities by including strings with derivational and semantic variants of the noun phrase words.
Figure 2.
Example of KM semantic-based conjunctive expansion and modifier distribution.
KM Methodology for Ambiguity Resolution
The KM scoring algorithm operates on phrase, context, and document levels. Similar to the MetaMap algorithm, KM gives phrase-level precedence to candidate strings based on cohesiveness, head-noun matching, derivational distance, and number of words spanned.13 KM calculates derivational distance with exact matches receiving the highest priority, then UMLS derivationally related terms (from DM.DB), followed by UMLS semantically related terms (from SM.DB), and finally form-rules. Because we found that the word with lowest frequency in MRCON-PS often represents the most meaningful word in the phrase (e.g., “end-systolic” in “high end-systolic volume”), KM favors multiword candidates that include the lowest-frequency word. KM also scores each candidate string based on its similarity to its CUI-PN (before or after normalization). Because the CUI-PN for an acronym usually is its expansion, KM augments scoring of document acronyms based on number, frequency, and clustering (words often appearing together) of CUI-PN words in the document.
For context-level scoring, KM scores candidate strings using semantic type and the extra words in overmatches. KM favors candidate strings based on previously described28,29 and author-derived semantic type rules. The program applies these rules based on the words in the noun phrase, their part of speech, and the surrounding words. For example, if a number follows the word “protein,” the KM scores the “laboratory procedure” higher than the “biologically active substance.” KM scores overmatching strings based on the document frequency and proximity of its extra word(s). For example, KM would prefer “central liver hemorrhagic necrosis” for “central hemorrhagic necrosis” if “liver” was in the sentence or appeared commonly in the document. Candidate overmatching concepts also are favored if the extra word(s) tends to cluster with the other words in the string. For example, “blood donor screening” would be favored as an overmatch for “donor screening” if “blood donor” occurred often in the document.
For document-level disambiguation, KM constructs, during document phrase processing, a list of concept numbers and associated semantic types for concepts exactly mapped (unambiguous “exact-matched” concepts) to UMLS concepts. KM subsequently favors those candidate concepts in “ambiguous matches” that are found in this set of previously seen exact-matched concepts. For example, after KM recognizes “beta adrenergic receptor” in a document, the ambiguous “beta receptor” will be interpreted as the former and not as “beta C receptor.” Likewise, candidate concepts with semantic types seen with high frequency in the document also are favored, thereby favoring “gentamycin” as semantic type “antibiotic” instead of “carbohydrate” or “laboratory procedure” in a lecture about antibiotics. KM also favors candidate concepts based on their Medline (literature-based) frequency of co-occurrence with exact-matched concepts from the UMLS MRCOC file.7 For instance, KM would expand “MAO” as “monoamine oxidase” in a lecture about major depression and as “maximum acid output” in a lecture about Helicobacter pylori infection.
KM Evaluation
We selected MetaMap as the standard state-of-the-art comparison metric for evaluation of the KM concept-matching algorithm. Appealing features of MetaMap include its long-standing development, accessibility by Web-based submission, and robust, score-based concept-matching algorithm for biomedical text. MetaMap also performs intensive variant generation and does not rely on word order, factors we believe are important to high recall in medical curricular documents.
We compared the abilities of KM and MetaMap to identify “important” concepts in selected subsets of medical school curriculum documents. In the first phase, we compared KM with MetaMap on an initial set of five “pilot” curricular documents; based on the results of this comparison, several adjustments were made to the KM algorithm. After then “freezing” KM, we compared it with MetaMap on a final “definitive” set of ten curricular documents.
Evaluation: Identification of Study Documents and “Gold Standard” Concepts
We asked lecturers and their course directors to help us manually identify the “important” concepts in their own course documents. The first four “lecturer–course director” pairs who agreed to participate were included in the study (two pairs taught in the first-year curriculum and two in the second year). We provided each pair with general instructions, a sample highlighted document, and their respective lecture notes (See ). We suggested that they highlight all “medically relevant and important terms” in their curricular document in a manner similar to the example from the training document. Participants highlighted terms using the Microsoft Word highlight function or on paper using a marker. Two authors (JDS, AS) independently highlighted the same four documents. We generated a “consensus” highlighted document for each lecturer–course director pair and for the author pair by merging those terms highlighted by either member of the pair.
We determined interrater reliability on “pilot” documents by comparing the consensus documents for each of the four lectures. Given that the interrater agreement was high (kappa 0.75) for the “pilot” documents, we elected to use just the author pair to subsequently identify “gold standard” concepts from the “definitive” set of ten documents. For purposes of the initial pilot study, the author pair highlighted one additional lecture, making a total of five pilot documents. The “definitive” study was comprised of five author-pair consensus documents from the first-year curriculum and five from the second year, each from a different course. These documents were selected randomly from each of the major courses from which we had received lecture materials. The authors did not view them until the evaluation.
Evaluation: Categorization of Concepts
We next categorized each highlighted concept as either a “meaningful term” or a “composite phrase.” We defined a meaningful term as either a meaningful word that describes a medical concept or a meaningful phrase that, when reduced to the word level, loses its meaning. Examples include the term “heart” and the phrases “Wilson's disease” and “volume of distribution.” A composite phrase was defined as a phrase that is composed of meaningful terms. For example, the composite phrase “elevation of blood pressure” retains meaning as “elevation” of “blood pressure.” We categorized all highlighted concepts prior to running KM and MetaMap on the documents.
Evaluation: Comparison of Documents
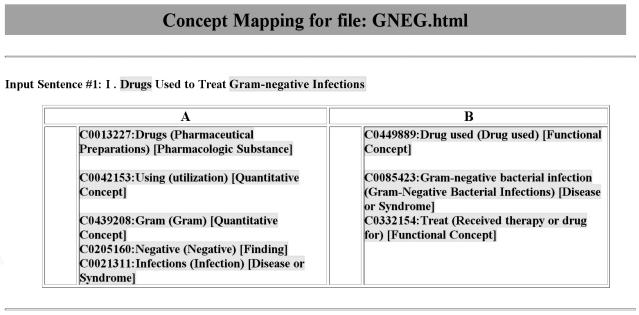
The KM document parser processed the study documents into sentences. KM and MetaMap then performed concept indexing on each document. MetaMap includes a rich variety of settings allowing the user to control its concept-identification behavior. Through experimentation prior to this analysis and consultation of earlier work,30 we found the optimal settings for MetaMap to be “default options” plus “quick composites” and “ignore stop phrases” using the “strict model”13 of the UMLS. Allowing “concept gaps” and “overmatches” produced worse recall and precision. A Perl script standardized the output of KM and MetaMap into an identical format that masked the source (KM vs. MetaMap). This script selected only the top-scoring candidate concept from each algorithm. If more than two candidates for the same concept had an equal score, the script chose the first candidate concept. ▶ shows an example of this output. The author pair previously had identified the “gold standard” meaningful terms and composite phrases in the study documents. Blinded to the identity of the concept indexers, the authors then determined the number of “gold standard” meaningful terms and composite phrases “correctly matched” by MetaMap and by KM and the false-positive rates for both, allowing recategorization if both authors were in agreement.
Figure 3.
Standardized study document output for indexing algorithms identifying MRCON concepts. The highlighted terms of the input sentence are the “gold standard” concepts. Our script also highlighted differences between the outputs of the two algorithms (i.e., the “A” and “B” columns). In this case, A, MetaMap output; B, KnowledgeMap output.
Study definitions included: (1) MEDICAL TRUE-POSITIVE (MTP): a correctly identified concept that is a gold standard meaningful term (e.g., “DNA”); (2) NONMEDICAL TRUE-POSITIVE (NMTP): a correctly identified concept that is not a gold standard meaningful term (e.g., adverbs such as “likely”); (3) MEDICAL FALSE-POSITIVE (MFP): a misidentified concept that is a gold standard meaningful term (e.g., selecting the element “lead” for the document concept “leading strand”); (4) NONMEDICAL FALSE-POSITIVE (NMFP): a misidentified concept that is not a gold standard meaningful term (e.g., selecting “pocket mouse” for the document concept “[pants] pocket”); (5) RECALL for meaningful terms: the number of MTPs divided by the total number of meaningful terms; (6) MEDICAL PRECISION: the number of MTPs divided by total meaningful term attempts (MTP/[MTP + MFP]); (7) OVERALL PRECISION: for an indexing algorithm, the ratio of the number of correctly matched concepts to the total number of identified (i.e., proposed) concepts ([MTP + NMTP]/[MTP + MFP + NMTP + NMFP]).
We randomly selected two study documents, one from each curricular year, to categorize KM algorithm successes (relative to MetaMap) and failures for matching of meaningful terms. A KM “success” occurred when KM correctly matched a “gold standard” concept not matched by MetaMap; a KM “failure” occurred when KM did not match a “gold standard” concept in the study document (regardless of MetaMap's performance on the concept). We then determined the component of KM's algorithm (e.g., MRCOC co-occurrences, CUI-PN scoring) that resulted in each success and failure. When two components contributed equally, we assigned each component half a success or failure.
We calculated statistical significance between the indexers with a paired, two-tailed t-test using SigmaStat (Chicago, IL) for Microsoft Windows.
Results
Recall and Precision for KM and MetaMap
The five documents in the pilot study contained an average (±SD) of 391 ± 54 meaningful terms and 90 ± 10 composite phrases. The ten documents in the definitive study contained an average of 427 ± 109 meaningful terms and 110 ± 27 composite phrases. ▶ contains overall recall and medical precision results for both studies; ▶ shows results for each document in the definitive study. The overall precision was 81% for MetaMap and 86% for KM (p < 0.01). There were no differences in recall between the pilot and definitive studies.
Table 2.
Recall and Precision of Meaningful Terms by Both Concept-indexing Algorithms
| Concept Identifier | Pilot Study (n = 5) | Definitive Study (n = 10) |
|---|---|---|
| Gold standard | ||
| Meaningful terms† | 1,955 | 4,281 |
| Composite phrases‡ | 448 | 1,105 |
| MetaMap | ||
| Meaningful terms | 1,580 (81%) | 3,325 (78%) |
| Medical precision | 85% | |
| Composite phrases | 156 (35%) | 308 (28%) |
| KM | ||
| Meaningful terms | 1,677 (86%)* | 3,510 (82%)* |
| Medical precision | 89%* | |
| Composite phrases | 154 (34%) | 382 (35%)* |
KM, KnowledgeMap; n, number of documents evaluated.
p < 0.01.
Meaningful terms are irreducible medically significant terms, such as “heart” or “Wilson's disease.”
Composite phrases include multiple meaningful terms that together represent a medical concept, such as “stenosis of the aortic valve” or “elevated thyroid stimulating hormone.”
Table 3.
Recall and Precision for Meaningful Terms for Each Document in the Definitive Study*
| MetaMap |
KM |
||||
|---|---|---|---|---|---|
| Year and Lecture Topic (by Course Title) | Gold Standard Concepts | Recall | Precision | Recall | Precision |
| First-year lectures | |||||
| Biochemistry | 484 | 297 (61%) | 75% | 343 (71%) | 77% |
| Histology | 357 | 296 (83%) | 89% | 313 (88%) | 92% |
| Embryology | 263 | 204 (78%) | 85% | 211 (80%) | 84% |
| Physiology | 531 | 425 (80%) | 84% | 423 (80%) | 88% |
| Immunology | 411 | 292 (71%) | 79% | 324 (79%) | 87% |
| Total | 2,046 | 1,514 (74%) | 82% | 1,614 (79%) | 86% |
| Second-year lectures | |||||
| Nutrition | 324 | 276 (85%) | 93% | 282 (87%) | 94% |
| Lab diagnosis | 377 | 301 (80%) | 83% | 330 (88%) | 91% |
| Physical diagnosis | 644 | 530 (82%) | 88% | 547 (85%) | 90% |
| Pathology | 435 | 381 (88%) | 94% | 387 (89%) | 95% |
| Pharmacology | 455 | 323 (71%) | 80% | 350 (77%) | 86% |
| Total | 2,235 | 1,811 (81%) | 88% | 1,896 (85%)† | 91% |
| All documents | 4,281 | 3,325 (78%) | 85% | 3,510 (82%)** | 89%** |
“Precision” in this table is calculated as “medical precision” as defined in “Methods.”
p < 0.05.
p < 0.01.
Detailed KM Algorithm Analysis
We further characterized in detail KM performance relative to MetaMap (▶) on a first-year biochemistry lecture on DNA replication and a second-year pharmacology lecture on antibiotics using the following categories and the definition of relative success and failure given above:
Heuristic disambiguation of competing candidate terms (18 successes, 5 failures): This occurs when KM identifies multiple canonical MRCON strings (including the correct match) that potentially match the document phrase. Heuristic CUI-PN scoring to select one term as the “match,” the most common cause for failure (2.5 failures*), was also the most important determinant of KM successes (8 successes). KM's heuristic use of MRCOC co-occurrences and document semantic type frequency to select “the best match” each caused 3.5 successes.* KM successfully identified “resistance” (semantic type “functional concept”), instead of “[psychotherapeutic] resistance,” selected by MetaMap. While all chemicals and proteins in these documents were more accurately represented as “biologically active substance,” Meta Map occasionally misidentified chemicals as their “laboratory procedure.”
KM handling of abbreviations, acronyms, and hyphens (13 successes, 22 failures): KM correctly identified “GPC” as “gram-positive cocci” by co-occurrence scoring. However, both indexing algorithms missed “SSB” (defined in the document as “single-stranded DNA binding protein”). Because the author defined “SSB” with an equal sign ( = ) rather than a parenthetical expression, KM could not link the document definition with the acronym. KM also did not normalize plural acronyms, misidentifying the document-defined concept “PBPs” (“penicillin-binding proteins”).
KM heuristic use of overmatches (34 successes, 7 failures): ▶ shows a successful KM overmatch: “gram-negative bacterial infection” for “gram-negative infection.” KM used sentence and document context to achieve 18 of its successes. Co-occurring concepts accounted for another nine successes. KM incorrectly overmatched “broad-spectrum” as “broad-spectrum penicillin” due to a high document frequency of “penicillin.”
KM heuristic use of form-rules (11 successes, 2 failures): Form-rules allowed KM to translate “dosing” into “dosage” and avoided some potential stemming errors by MetaMap, such as mapping “synergistic” to “SYN,” a gene. The two failures occurred when KM mapped “organism” in a nonmatching multiword noun phrase to “organ.”
KM use of multiword phrase pairing (10 successes, 6 failures): Failures resulted when the indexer combined the words of a composite phrase together in a way as to produce a different meaning than the intended concept(s). Because of sequential matching heuristic, KM misinterpreted the phrase “bacterial DNA replication fork” as “bacterial DNA,” “replication,” and “fork” instead of “bacterial” and “DNA replication fork.” However, KM correctly crossed parenthetical boundaries to match “omega protein” from “omega (ω) protein.”
MRCON-contained overmatches (44 failures): Both programs often misidentified “[DNA] replication fork” as “[genetic] replication” and “[cutlery] fork.” Furthermore, because only one Metathesaurus string exactly matches “replication,” the KM algorithm listed “[genetic] replication” as an exact-matched concept, thereby favoring it when evaluating ambiguous matches.
Failures due to concepts not present in MRCON (141 failures): For 62% of KM failures, we could not find a correct MRCON concept. These failures included the multiword terms “30s ribosomal subunit” and “positive predictive value,” the one-word concepts “concentration” (of a chemical) and “data,” along with compound words, such as “semidiscontinuous.” MRCON failures also affected mapping precision, causing KM to incorrectly match “concentration” (of a chemical) as “concentration” (the mental process) and “[DNA] helix” as “helix [of ear].”
Other reasons for success or failure (10 successes, 13 failures): Five failures were due to bacterial genus name abbreviations. For those bacteria listed with their genus abbreviated in the Metathesaurus, KM was able to correctly identify the organism. However, “E. faecium,” for example, is only listed in the Metathesaurus as “Enterococcus faecium,” leading to an invalid KM match. Further, because of our MRCON processing, KM misidentified “GI tract” as “US GI tract” (meaning “ultrasound of GI tract”) because we had considered “US” a stopword. The overlapping matches of KM did not cause any mismatches in these two documents. Successful overlaps included matching “history of penicillin allergy” as “history of allergy” and “penicillin allergy” along with matching “chromosomal DNA replication” as “chromosomal replication” and “DNA replication.”
Table 4.
Causes of Successful and Missed Matches by KM in Two Documents*
| KM Component/Method | Causes of Successful KM Matches | Causes of Failed KM Matches |
|---|---|---|
| Heuristic disambiguation of competing candidate terms | 18 (18%) | 5 (2%) |
| Abbreviation/acronym/hyphen handling | 13 (13%) | 22 (10%) |
| Heuristic overmatch utilization | 34 (35%) | 7 (3%) |
| Heuristic use of form-rules | 11 (11%) | 2 (1%) |
| Heuristic multiword phrase pairing | 10 (10%) | 6 (3%) |
| KM determination of part of speech | N/A | 4 (2%) |
| MRCON-contained overmatches | N/A | 44 (19%) |
| Failure to correct document misspelling | N/A | 2 (1%) |
| Concept not present in MRCON | N/A | 141 (62%) |
| Other reasons for success or failure | 12 (12%) | 13 (6%) |
| Total | 98 | 246 |
“Successful KM matches” were those gold standard meaningful terms matched correctly by KM but not MetaMap. The “Failed KM matches” were all gold standard meaningful terms not correctly identified by KM, irrespective of MetaMap's performance.
Discussion
The current study results (recall rates for meaningful terms of 78% and 82% for MetaMap and KM, respectively) are similar to those recently reported for mapping clinical text using the UMLS15,16 and better than previous results12 mapping medical curricular text. Recent UMLS expansion may explain these improvements as well as better mapping algorithms. Based on analysis of two study documents, we estimate that the 2001 Metathesaurus represents 89% of medically important curricular concepts. Lowe et al.31 found that the 1998 Metathesaurus represented 81% of the important concepts in radiology imaging reports. While UMLS provides good general coverage of concepts, some important concepts were absent, such as “double helix,” “positive predictive value,” “concentration” (of a chemical), and “loading dose” (although more specific forms such as “hemoglobin concentration” and “drug loading dose” were sometimes present). Many of these concepts could be added to the Metathesaurus without adding ambiguity.
The recall of KM for second-year, clinically oriented courses was higher than for first-year, basic science–oriented courses (▶). Because major components of the Metathesaurus, such as International Classification of Disease (ICD-9), Physicians' Current Procedural Terminology (CPT), and SNOMED, are clinical reporting tools, we expected that the indexing algorithms would perform better for clinically oriented documents. Although nonsignificant, KM appeared to have a higher recall in our pilot study than in the definitive study (86% vs. 82%). (Rerunning the current KM algorithm on the pilot documents yielded a recall of 88%.) The pilot study contained a higher proportion of clinically oriented documents, including three second-year documents and two first-year documents, one of which was a clinical microbiology lecture.
Because we defined composite phrases as being accurately represented by their component meaningful terms, our considerably lower composite phrase recall would not ultimately affect information retrieval. Thus, “disease of thyroid gland” would be represented equally by two concepts (“thyroid gland” and “disease”) or by one concept (“thyroid gland disease”). In fact, apart from further processing, an indexer would likely fail to retrieve “thyroid gland disease” for a query containing only “thyroid gland.” In general, KM identified more composite phrases in the definitive test because of conjunctive expansion (e.g., “renal or hepatic disease” expanded to “renal disease” and “hepatic disease”), successful overmatching (e.g., “decrease intake” to “decreased PO intake”), and improvements in crossing prepositions (e.g., “decrease in creatinine clearance” to “creatinine clearance decrease”).
The KM algorithm can improve in its disambiguation of acronyms and its identification of overmatches. A new scoring scheme for “previously seen concepts” that favors overmatches over partial matches could improve overmatch recall. Extraction of acronyms offset by dashes and equal signs may yield higher recall but could risk chance matching, because those characters do not clearly demarcate phrase end-points. The context-specific semantic type rules of KM could also improve, particularly for disambiguating chemical names from their corresponding “laboratory procedure,” a problem for both MetaMap and KM. We derived the current weighting schemes empirically; a decision-tree algorithm that selectively weights certain scoring elements in certain cases may prove superior.
Limitations caution the interpretation of these study results. We formally tested only 15 curricular documents from one institution. We chose documents that represent a wide range of topics and text formats. The use of a larger document set could confirm a similar performance when the system is scaled to the entire curriculum. Our concept indexing techniques have not been applied to other texts (i.e., journal articles, textbooks, or clinical free text). For example, the use of document-level disambiguation techniques in KM (such as Medline co-occurrences, concepts and semantic types exactly matched elsewhere in the document, and acronym discovery) may not be as effective for clinical free text. We used the 2001 edition of the UMLS. While we designed KM to easily accommodate newer editions of the UMLS, this has not been tested.
We chose to index all medically important concepts in the document; our goal was to create a system that would allow for exhaustive searching and comparison of documents. Students searching lectures, administrators seeking to identify areas of curricular overlap, and faculty preparing lectures are often interested in all instances in which a particular topic is taught. This task is simpler than that of finding the set of topics that most accurately and succinctly describes a document.
Acceptable “recall” rates for effective concept-based information retrieval in medical curricular documents have not been established. Nadkarni et al.15 concluded that higher recall rates than reported in the current study are needed for successful concept-based information retrieval of clinical text. This may not be true when identifying concepts in medical curriculum. In contrast to clinical documentation in which mention of a particular disease may only occur once as an item in the “past medical history,” educational documents typically mention important concepts many times. A quick review of the lecture on DNA replication, for instance, finds the concept “deoxyribonucleic acid” mentioned 29 times, “DNA helicase” 13 times, and “Okazaki fragments” seven times. Misidentification or omission of a concept does not carry the same significance in education as in the clinical context. KM should be modified to include text-based searching methods that can identify concepts not in the Metathesaurus.15,32
Finally, this study is one of the first to report recall data for the National Library of Medicine's MetaMap with respect to medical curricular documents. Overall, MetaMap performed well with preprocessed curricular documents. KM showed marginal advantages over MetaMap in successfully selecting overmatching concepts, correctly matching acronyms, heuristically disambiguating “best” candidates from sets of “tied” candidate concepts, processing distributed modifiers, and expanding conjunctive phrases.
Motivation and Future Directions
Given the rapid growth of medical knowledge, instructors must frequently revise lecture notes, and academic program committees must regularly review and change curricular content. Automated extraction of concepts represented in educational texts (curricular documents) is the first step toward developing tools to help educators locate, integrate, evaluate, and iteratively improve medical school curriculum content. The KM concept identifier's use of the UMLS represents important progress toward this end.
The authors plan to develop tools that build on KM concept indexing to identify similar documents from disparate sections of the curriculum, to create tools that can perform relevant Medline queries to supplement curricular content or clinical case descriptions, and to help faculty and students correlate clinical cases with available online educational materials.
Although effective concept recognition is an important foundation for a course management system, the “proof of the pudding is in the eating.”33 Only when faculty regularly use a designed system to accomplish their objectives can developers contemplate success.
Appendix A. Excerpt from the “Embryology” Document from the Definitive Study*
Embryology II:
Embryogenesis—Part 2
Fertilization to Gastrulation
[The First Two Weeks]
- I. Fertilization:
- Requirements:
- Mature male and female gametes having:
- * a haploid number of chromosomes (22 + X and 22 + Y chromosomes), and
- * half the amount of DNA of a normal somatic cell as a consequence of gametogenesis
- Ovum—a secondary oocyte which:
- * is arrested in metaphase of the 2nd maturation (meiotic) division
- * has ruptured from a mature tertiary or graafian follicle, leaving behind granulosa cells, which, together with cells from the theca interna, are vascularized and develop—under the influence of luteinizing hormone (LH)—a yellowish pigment to become a corpus luteum that secretes progesterone
- * is surrounded by zona pellucida and corona radiata (rearranged cumulus oophorus cells)
- * has been transported by fimbria and ciliary action into the ampulla of the uterine tube
- Spermatazoa:
- * have completed 2nd maturation division
- * have (via spermiogenesis) shed most cytoplasm and acquired:
- - acrosome
- - head (condensed nucleus w/ DNA)
- - neck, middle piece, and tail
- - ability to swim straight
- * have been ejaculated into the female reproductive tract and, arrival, have undergone capacitation:
- - ∼7-hour process
- - glycoprotein coat and seminal plasma proteins removed from plasma membrane overlying acrosome, enabling acrosome reaction
- * have—within 24 hours—passed from the vagina to the ampulla of the uterine tube
- Fertilization process
- Phase 1: Passage through corona radiata:
- * exposed acrosomes of capacitated sperm release hyaluronidase
- - causes separation and sloughing of cells of corona
- -thus capacitated sperm easily pass through corona
- * although only one sperm is required for fertilization, it is believed that teamwork by many facilitates in penetrating surrounding barriers.
- Phase 2: Penetration of zona pellucida
- The zona pellucida is an amorphous, glygoprotein shell that:
- * facilitates and maintains sperm binding (via ZP3 ligand)
- * induces acrosomal reaction when sperm cell binds to zona, causing acrosome to release acrosin and trypsinlike enzymes, allowing penetration. Once the fertilizing sperm penetrates the zona pellucida, a zona reaction occurs, and the zona and the oocyte plasma membrane become impermeable to other sperm.
- Phase 3: Formation of the zygote:
- Adhesion of oocyte (integrins) & sperm (disintegrins)/fusion of plasma membranes.
- * only sperm head and tail enter cytoplasm of oocyte
- Completion of oocyte's second meiotic division, formation of female pronucleus
- Formation of male pronucleus
- * head enlarges, tail degenerates
- As pronuclei form, they replicate their DNA
- Pronuclei fuse as their membranes break down, and their chromosomes intermingle and condense, becoming arranged for a mitotic cell division
The authors thank Mr. Michel Décary of Cogilex R&D, Inc., for providing the part-of-speech tagging software. The authors thank Alice Coogan, MD, David Wasserman, MD, Owen McGuiness, PhD, Terrence Dermody, MD, Luc Van-Kaer, PhD, Joseph Awad, MD, and Richard Shelton, MD, for their work toward establishing the gold standard terms in documents. Finally, the authors thank the National Library of Medicine for developing and making available the UMLS.
Footnotes
In these cases, there were two components of KM's algorithm that equally caused a success or a failure, so each was given a score of 0.5.
This appendix was written by H. Wayne Lambert, PhD, and is used here with his permission.
References
- 1.Candler C, Blair R. An analysis of Web-based instruction in a neurosciences course. Med Educ Online. 1998;3:3. [Google Scholar]
- 2.Zucker J, Chase H, Molholt P, Bean C, Kahn RM. A comprehensive strategy for designing a Web-based medical curriculum. Proc AMIA Symp. 1996:41–5. [PMC free article] [PubMed]
- 3.McNulty JA. Evaluation of Web-based computer-aided instruction in a basic science course. Acad Med. 2000;75:59–65. [DOI] [PubMed] [Google Scholar]
- 4.Nowacek G, Friedman CP. Issues and challenges in the design of curriculum information systems. Acad Med. 1995;70:1096–100. [PubMed] [Google Scholar]
- 5.Mattern WD, Anderson MB, Aune KC, et al. Computer databases of medical school curricula. Acad Med. 1992;67:12–6. [DOI] [PubMed] [Google Scholar]
- 6.Turchin A, Lehmann CU. Active Learning Centre: design and evaluation of an educational World Wide Web site. Med Inform Internet Med. 2000;25:195–206. [DOI] [PubMed] [Google Scholar]
- 7.Miller RA, Gieszczykiewicz FM, Vries JK, Cooper GF. CHARTLINE: providing bibliographic references relevant to patient charts using the UMLS Metathesaurus knowledge sources. Proc Annu Symp Comput Appl Med Care. 1992:86–90. [PMC free article] [PubMed]
- 8.Kanter SL. Using the UMLS to represent medical curriculum content. Proc Annu Symp Comput Appl Med Care. 1993:762–765. [PMC free article] [PubMed]
- 9.National Library of Medicine. UMLS Knowledge Sources (ed 12), 2001.
- 10.Rector AL, Nowlan WA, Glowinski A. Goals for concept representation in the GALEN project. Proc Annu Symp Comput Appl Med Care. 1993:414–8. [PMC free article] [PubMed]
- 11.Dimse SS, O'Connell MT. Cataloging a medical curriculum using MeSH Keywords. Proc Annu Symp Comput Appl Med Care. 1988:332–6.
- 12.Kanter SL, Miller RA, Tan M, Schwartz J. Using POSTDOC to recognize biomedical concepts in medical school curricular documents. Bull Med Libr Assoc. 1994;82:283–7. [PMC free article] [PubMed] [Google Scholar]
- 13.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: The MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed]
- 14.Lussier YA, Shagina L, Friedman C. Automating SNOMED coding using medical language understanding: A feasibility study. Proc AMIA Symp. 2001:418–22. [PMC free article] [PubMed]
- 15.Nadkarni P, Chen R, Brandt C. UMLS concept indexing for production databases: a feasibility study. J Am Med Inform Assoc. 2001;8:80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sneiderman CA, Rindflesch TC, Bean CA. Identification of anatomical terminology in medical text. Proc AMIA Symp. 1998:428–32. [PMC free article] [PubMed]
- 17.Yu H, Hripcsak G, Friedman C. Mapping abbreviations to full forms in biomedical articles. J Am Med Inform Assoc. 2002;9:262–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leroy G, Chen H. Filling preposition-based templates to capture information from medical abstracts. Pac Symp Biocomput. 2002:350–61. [PubMed]
- 19.Sager N, Friedman C, Lyman MS, Members of the Linguistic String Project Medical Language Processing: Computer Management of Narrative Data. Reading, MA: Addison-Wesley, 1987, pp 89–93.
- 20.Strzalkowski T, Carballo J. Recent developments in natural language text retrieval. In: Harman DK (ed). The Second Text Retrieval Conference (TREC-2). NIST Special Publication 500-215. Washington, DC: U.S. Government Printing Office; 1994, pp 123–36.
- 21.Zeng Q, Cimino JJ. Automated knowledge extraction from the UMLS. Proc AMIA Symp. 1998:568–72. [PMC free article] [PubMed]
- 22.Webster's Third New International Dictionary, Unabridged. ProQuest Information and Learning Company, 2001. <http://collections.chadwyck.com/mwd>. Accessed Jan 2002.
- 23.Cogilex R & D, Inc. <http://www.cogilex.com> Accessed June 2002.
- 24.McCray AT, Bodenreider O, Malley JD, et al. Evaluating UMLS strings for natural language processing. Proc AMIA Symp. 2001:448–52. [PMC free article] [PubMed]
- 25.Liu H, Lussier YA, Friedman C. A study of abbreviations in the UMLS. Proc AMIA Symp. 2001:393–7. [PMC free article] [PubMed]
- 26.Campbell DA, Johnson SB. A technique for semantic classification of unknown words using UMLS resources. Proc AMIA Symp. 1999:716–20. [PMC free article] [PubMed]
- 27.Cooper GF, Miller RA. Related articles, an experiment comparing lexical and statistical methods for extracting MeSH terms from clinical free text. J Am Med Inform Assoc. 1998;5:62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rindflesch TC, Aronson AR. Ambiguity resolution while mapping free text to the UMLS Metathesaurus. Proc Annu Symp Comput Appl Med Care. 1994:240–4. [PMC free article] [PubMed]
- 29.Johnson SB. Related articles, a semantic lexicon for medical language processing. J Am Med Inform Assoc. 1999;6:205–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein H, Weeber M, Jong-van den Berg LTW, et al. Evaluating MetaMap's text-to-concept mapping performance. Proc AMIA Symp. 1999:101.
- 31.Lowe HJ, Antipov I, Hersh W, Smith CA, Mailhot M. Automated semantic indexing of imaging reports to support retrieval of medical images in the multimedia electronic medical record. Methods Inf Med. 1999;38:303–7. [PubMed] [Google Scholar]
- 32.Aronson A, Rindflesch T, Browne A. Exploiting a large thesaurus for information retrieval. Proc RIAO '94. 1994:197–216.
- 33.Cervantes M. Don Quixote. <http://promo.net/pg/>. Accessed Jun 7, 2002.