Abstract
The authors describe a research group that supports the needs of investigators seeking data from an electronic medical record system. Since its creation in 1972, the Regenstrief Medical Records System has captured and stored more than 350 million discrete coded observations on two million patients. This repository has become a central data source for prospective and retrospective research. It is accessed by six data analysts—working closely with the institutional review board—who provide investigators with timely and accurate data while protecting patient and provider privacy and confidentiality. From January 1, 1999, to July 31, 2002, data analysts tracked their activities involving 47,559 hours of work predominantly for physicians (54%). While data retrieval (36%) and analysis (25%) were primary activities, data analysts also actively collaborated with researchers. Primary objectives of data provided to investigators were to address disease-specific (35.4%) and drug-related (12.2%) questions, support guideline implementation (13.1%), and probe various aspects of clinical epidemiology (5.7%). Outcomes of these endeavors included 117 grants (including $300,000 per year salary support for data analysts) and 139 papers in peer-reviewed journals by investigators who rated the support provided by data analysts as extremely valuable.
Providing accurate data from electronic medical record systems for the purposes of research requires a supportive infrastructure. Such an infrastructure has evolved at Regenstrief Institute over the 30 years of existence of the Regenstrief Medical Records System (RMRS). This electronic medical record system was created as a modular system to provide service functions for clinics, laboratory, radiology, and pharmacy. Each module is linked to a central database that captures and stores data from each modular source. These data then may be linked by using a unique patient identifier. The types and quantity of data within these modular sources have been previously defined.1 From its modest beginning in 1972 as a pilot project within a diabetes care clinic, the RMRS currently contains more than 350 million discrete coded observations on two million patients encompassing much of the city of Indianapolis. Researchers sometimes are overwhelmed envisioning possible hypotheses to test using the enormity of data contained within this system. However, going from a worthwhile hypothesis to analysis and interpretation of data using such a large warehouse requires fast computers, specialized programs, protocols to protect personal data, and many professional and technical personnel to guide researchers to the answers they seek.
Background
At Regenstrief Institute, a core group of data analysts and a variety of processes have evolved to fulfill the research needs of scholars on the campus of the Indiana University Medical Center who wish to obtain data from the RMRS. Much of the background and history of the RMRS have been described in previous reports.1,2,3,4,5 Further, Tierney and McDonald6,7 have published several reports describing the research applications of clinical data repositories including the RMRS. Yet, there has been little description of the structural components, processes, and supportive activities necessary to provide data for research from such repositories. We felt that the increasing numbers of centers capturing and analyzing data from their electronic record systems would benefit from an available description of our current efforts to provide and analyze data for research projects involving data from the RMRS.
The purpose of this report is to describe the structural components, functions, and activities of the Health Care Data & Epidemiology Section at Regenstrief Institute. The report aims to provide ideas to those considering the implementation of similar research support systems and to others considering changes to existing systems. We identify the types of requesters of secondary data, describe the activities of data analysts in providing these data, and categorize the types of projects for which data were requested during 3.5 years. Finally, we report on the outcomes of providing these data to 14 active Regenstrief Institute investigators who responded to an informal survey about funding, research papers, and their perceived value of this research support.
System Description
Setting and Structural Components
Setting
The RMRS captures patient data from three hospitals on the Indiana University Medical Center campus and from 30 clinics around the inner city of Indianapolis.1 The site of longest tenure and greatest development as it relates to the RMRS is Wishard Health Services. This city–county hospital uses the RMRS at its 250-bed acute care hospital (21,000 annual admissions), its primary care and specialty outpatient clinics (1.2 million visits by 185,500 patients), and its emergency department (110,000 annual emergent and urgent visits) located adjacent to the hospital, as well as a network of 30 neighborhood clinics throughout the city. Pharmacies affiliated with these clinics fill and refill more than 900,000 prescriptions per year, data for which are contained within the RMRS.
Other health care systems contribute data to RMRS data repositories. These include two tertiary care hospitals, Indiana University Hospital (330 beds) and Riley Hospital (195 beds), that in 1998 merged with Methodist Hospital (775 beds) to form Clarian Health Partners, Inc. (1,300 total beds, >57,000 annual admissions, and >900,000 outpatient visits). Furthermore, two large grants from the National Library of Medicine and the National Cancer Institute (Clement J. McDonald, MD, principal investigator) have tremendously facilitated integration of clinical data from the emergency departments of all hospitals throughout the greater Indianapolis area as well as limited inpatient data from these hospitals.
Data Elements
At Wishard Health Services, where the RMRS has been in operation since 1972, the database has captured, in coded form, all diagnostic studies (chemistry, hematology, cytology, surgical pathology, bone marrow biopsy, obstetric ultrasounds, electrocardiograms, echocardiography, electromyelograms, electroencephalograms, radiology studies) and all orders (including prescriptions). It also captures clinical encounter information and the full text of all dictated reports (operative notes, discharge summaries, visit notes, radiology). The RMRS carries every electrocardiogram tracing produced at Wishard for the last ten years and every digital radiology image produced at Indiana University/Riley and Wishard Hospitals since August 1999. The data now contained within the RMRS are used heavily for research and are complemented by external data. For example, on a yearly basis, patient records in this database are matched against the Indiana State death tapes to identify patients who have died outside of the hospital.
Because RMRS data are archived and retrievable, investigators may use these secondary data to perform retrospective research (retrospective cohort or case–control studies) or prospective clinical trials. As described below, carefully conceived guidelines are followed before data are provided to investigators for research; these include protocols for communicating with administrators from participating health care institutions that contain vaults of data within the RMRS and following federal guidelines aimed at protecting patient privacy and confidentiality. By doing so, the RMRS is protected as a data source, relieving many anxieties of laypersons and professionals about the use of patient-specific data for research.
Personnel
Data analysts are at the center of this research support aimed at providing data for research. Regenstrief Institute provides space and resources for six data analysts who provide the core functional capacity of this research support section. However, their salaries often are supported from grants and contracts. From July 1, 2000, to June 30, 2002, the Institute provided salary support for three full-time equivalent (FTE) positions, but income from grants and contracts supported all but one FTE position. For this period, mean direct income to the research section from grants and contracts was approximately $300,000 per year.
The type of formal training data analysts have varies considerably. Two data analysts have master's level statistics degrees, two have bachelor degrees in the sciences, one has a master's degree in economics, and one has a PhD degree in child and family health sciences. We have found that it takes six to eight months of on-site training and several formal training sessions learning SAS before data analysts can perform the majority of their work independently. Three data analysts have formal training in Microsoft Access and Visual Basic programming, and several are well versed in advanced applications of commonly used software such as Microsoft Excel. These commonly available software packages are used to create databases and analysis tools that provide investigators the ability to monitor patients in their clinical trials. One Microsoft Access application was created to enable drug therapy monitoring of 447 patients with reactive airways disease for a randomized controlled trial.8,9 Requests involving charge data or cost conversions are assigned to the data analyst with economics training, those involving statistical analysis go to data analysts with advanced statistical training, and requests from Riley Children's Hospital investigators are assigned to the data analyst with training in child and family health sciences. Each analyst then manages all aspects of a particular data request including communication with the investigator. This includes meeting with the investigator, maintaining records on each request, assuring that patient and provider privacy and confidentiality are maintained, and tracking the time spent on various key activities on each data request. Input from a physician or other clinician is uniformly required for the data analyst to understand the full scope of the data being extracted and the relationship of these data to the clinical question or problem being addressed by the investigator. This is particularly important because the investigators often do not fully comprehend the vast data contained within the RMRS, whereas the data analyst may not fully comprehend the clinical relevance of the data to the research question. A seemingly simple task is the identification of patients with a certain disease, which is usually not so straightforward. For example, identifying patients with heart failure could be done using the physician's diagnosis, echocardiogram results, chest x-rays, prescription medications, or all of these variables. Data analysts and investigators would work closely together on such a project to find the most appropriate case definition.
A particularly important function of the data analyst is to help the clinician-investigator translate his or her research idea into a question that the data source can answer. However, this often requires input from a medical informatics researcher who understands both clinical medicine and the data structure of RMRS to serve as a liaison between the clinician-investigator (who is intimately familiar with the various dimensions of clinical medicine but usually not informatics) and the data analyst (who is intimately familiar with the data dictionary and the structure and content of the RMRS database but not clinical medicine). Hence, the collaboration among the investigator, data analyst, and medical informatics researcher often is necessary to reconcile the data source (structure and types of data) and the research question being addressed.
Because extracted data may be complex, the services of programmers and biostatisticians often are important. Data analysts frequently visit Regenstrief Institute programmers for assistance with their software programs, data extractions, and adding new data streams. In turn, the data analysts provide programmers feedback with their careful vigilance in calling attention to important issues involving RMRS data before these issues can become concerns. For example, a change in a file name by a programmer can easily disrupt a batched program, which the data analyst might find and fix. In a recent example, data analysts made the observation that patient procedures were being recorded without corresponding visit data on patients undergoing these procedures. This problem was reported to the programmers who determined that the recently installed clinic software was overwriting patient visit data. Similarly, biostatisticians are frequently called on for sophisticated statistical procedures necessary for addressing questions or testing hypotheses using secondary data sources. Regenstrief Institute houses three PhD-level biostatistician research scientists and several others are trained at the master's level.
Functions
Planning and Preliminary Process
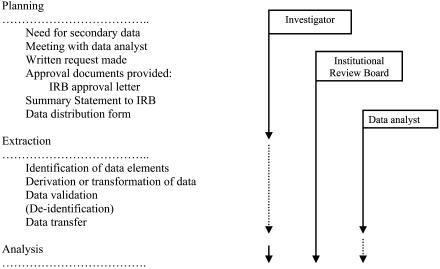
The overall processes for planning and data extraction for a given project are shown in ▶. We encourage investigators to inform the Regenstrief Institute of their data needs early in their project planning, preferably before the proposal has been written. By doing so, data analysts can advise investigators on available data and features of the system. When the project begins, investigators and data analysts communicate regularly by e-mail, telephone, and meetings. Data for the purposes of research must be requested in writing (letter, fax, or e-mail). There are three key reasons for written requests only. First, and the most obvious reason, is that a written request provides a permanent record of that request, which is a useful reference for both data analysts and investigators. Second, the written request provides a mechanism to compare the investigator's request with the documents submitted to the institutional review board (IRB). Third, our small experiments and anecdotal reports involving receipt of requests by various methods suggest that the act of writing is the best way to engage the investigator to consider the full scope of data needs for her or his project. Thus, the written request has been an important time saver for data analysts because it results in fewer iterations of data retrievals.
Figure 1.
Flow chart of planning and preliminary process functions performed by data analysts from January 1, 1999, to July 31, 2002, working closely with investigators and the institutional review board (IRB).
Investigators may actively engage themselves in the early planning process by using two particularly useful software applications, namely, Inquiry and Fast Retrieval. Inquiry allows the examination of all observations or records on a particular patient. When a patient's medical record number is entered, all of that patient's queried medical information is displayed temporally. This software is particularly useful in understanding the scope of care for a limited number of patients. Below is an example of a simple retrieval of hemoglobin A1c for an imaginary patient with a series of high values marked by *H.
INQUIRY DATA†
#000000-0 PATIENT'S NAME M W Age 69yr
Ad Hoc Flowsheet 31Aug99/08:37 04May99/08:38 2Feb99/11:00
HGB A1c | 10.6*H | 7.4*H | 9.0*H %
However, the majority of data requests involve the identification of a cohort or group of patients with pre-established characteristics followed by extraction of data on targeted variables for that cohort. Fast Retrieval is a program—driven by interactive menus—used to define a cohort of patients using specific inclusion and exclusion criteria and then report data distributions on that cohort. Unlike Inquiry, which searches for all data for individual patients, Fast Retrieval uses an inverted file structure to specify criteria pertaining to patients' observations, and the computer returns a list of patients who satisfy those criteria. Therefore, Fast Retrieval permits the investigator to do simple and fast queries to create cohorts meeting user-defined criteria, e.g., patients 65 years of age and older with a hemoglobin level less than 7 g/dL within the past year, men 50 years and older who have not had prostate-specific antigen testing, patients prescribed antihypertensive drugs from July 1, 2002, to December 1, 2002, whose last systolic blood pressure was greater than 140 mm Hg. Once the cohort is defined, unique patient identifiers can be used (with special access privileges and IRB approval) for more comprehensive retrievals using other software such as CARE (vide infra), or reports can be generated online without any direct access to the unique identifiers. Also, cohorts may be saved and run later to extract additional data, or the query may be saved and run at a later date to update the cohort.
Fast Retrieval can be used to quickly report patient counts and distributions of diagnoses and drugs for specific patient cohorts of interest. For example, a physician interested in studying patients with sarcoidosis found that at the health care center of interest there were 665 patients ever seen (a useful count for a retrospective study), 198 of whom were seen within the last year (a useful count for a prospective study or trial). Using Fast Retrieval, the total time of this query was less than 2 minutes, and reporting all prescriptions ever prescribed for these patients took another 25 seconds. In comparison, this query would require an enormous retrieval taking many hours to open and examine the records of the 2 million patients in the database. Importantly, all data are kept private and secure during Fast Retrieval processing. Thus, Fast Retrieval is a helpful tool for investigators in their study planning stage to determine whether sufficient patient records exist to conduct a study. Data analysts conduct hands-on training sessions on how to use Fast Retrieval for clinicians who wish to do more complicated queries by themselves. A manual containing examples also is provided to trainees. Although Fast Retrieval allows investigators to securely probe data contained within the RMRS, it has limitations. For example, extracting parameter data using complicated relative dates is difficult, and defining new variables is not supported.
More complicated querying requires a program called CARE.10 Compared with Fast Retrieval, CARE is accessible primarily to data analysts, programmers, and medical informaticians. The major reason for the limited access of CARE is that it takes several months to become versatile at CARE programming, and most investigators do not have the time for such training. To facilitate processing, data analysts generally use CARE programs to extract data after first identifying a focused cohort of patients using Fast Retrieval. This allows CARE to run on a small subset of patients and requires less CPU than otherwise would be required to open and close two million patient records looking for relevant observations. Such a query can take hours to days to run, depending on its size. CARE allows the analyst to precisely define and calculate parameters such as hypertension (diagnosis of hypertension with a prescription for an antihypertensive or two consecutive systolic blood pressures ≥140 mm Hg or diastolic blood pressures ≥90 mm Hg), body mass index, and estimated creatinine clearance (based on age, sex, weight, and serum creatinine). CARE can find all values within a date range or relative time period (30 days after the last prescription for a drug) or it can restrict its extraction to the last, first, any, all, minimum, or maximum of values. Derived values may be computed such as the mean, sum, counts, percent change over time, the slope of a longitudinal set of values, or the area under the curve of those values. Finally, an enormous advantage of CARE is that a longitudinal vector of values (all body weights or blood pressure measurements within a data range) may be extracted with a single command.
▶ shows an example of a simple CARE query. The purpose of this query is to identify older adult patients (≥65 years of age) who have been prescribed a specific drug (Drug A). After the program begins, the patient's current age is calculated, and the program continues if age criteria are met and the patient has used Drug A at any time. If these criteria are not met, the program stops processing, and the next patient's record is opened. The dates of first and last drug use are then determined, and any emergency department diagnoses or prescriptions for any Drug B within that date range are defined.
Table 1.
A Sample CARE Query
| CARE Program Statement | Interpretation of the Statement |
|---|---|
| Begin block whole | Open a programming block. |
| Define “age” as ex: (today-“birth”)/365.25 | Compute patient's current age. |
| If “age” is ge 65 then save “one” as “oldpt” {integer} | Define a variable “oldpt” that is one if the patient is 65 years of age or older the day the program runs. |
| If any “oldpt” exists then continue else exit whole | If a patient is at least 65 years of age, then continue collecting the data listed below; otherwise, close the programming block and move on to the next patient's record. |
| If any “Drug A” [>0] exists then continue else exit whole | If a patient has been prescribed Drug A in a dose greater than zero, then continue collecting the data listed below; otherwise, close the programming block and move on to the next patient's record. |
| Define “drug_fst” as first “Drug A” [>0] exists | Find the date of first use of Drug A. |
| Define “drug_lst” as last “Drug A” [>0] exists | Find the date of last use of Drug A. |
| If any “er dx” [on_after “fst_date” & before “lst_date”] exists then save “one” {integer} as “evr_er” else save “zero” {integer} as “evr_er” | If the patient has received a diagnosis from the emergency department on the date of the prescription for Drug A but before the date of the last prescription, then set to one the variable “evr_er”; otherwise, set the variable to zero. |
| If any “Drug B” [after “fst_date” & >0] exists then save “one” as “Drug B” {integer} else save “zero” as “Drug B” {integer} | If the patient has received a prescription for any Drug B after the date of the first prescription for Drug A, then set to one the variable “Drug B”; otherwise, set the variable to zero. |
| End block whole | Close the programming block. |
Some investigators choose not to learn how to use Fast Retrieval or CARE or may not have access privileges. Even when they do have such access, investigators often ask for assistance from one of Regenstrief Institute's data analysts who are most familiar with the nuances of all of the retrieval programs as well as the data fields contained within the RMRS. The steps used for requesting data are delineated below:
Determine whether data are needed for a research or quality improvement project.
Complete a Data Distribution Form for research or quality improvement (see Appendix, available as an online data supplement at <www.jamia.org>).
- Within a week of the receipt of the request, a data analyst contacts the investigator by telephone or e-mail to discuss the proposed project. The data analyst verifies the:
- a. research question or issue being addressed
- b. inclusion and exclusion criteria
- c. variables or parameters of interest (dependent and independent variables, confounders, and effect modifiers)
Before proceeding with extraction of data, the data analyst labels a manila folder to contain all documents pertaining to the project. This folder must contain a copy of the signed IRB approval letter containing the IRB study number, a copy of the Summary Safeguard Statement or other documents used by the investigator to describe the data needs of the project, and the data distribution form completed in #2 above.
When the data analyst has a good grasp on the data needs of a project, data extraction begins.
Data Extraction
As a courtesy, the data analyst will contact the investigator intermittently to provide information on how the data extraction is proceeding and to discuss the format of the output data. Some investigators desire more input and control throughout this phase of the study, whereas others are satisfied to simply receive the requested data when they are available. Regardless, the data analyst provides information about each variable so the investigator is familiar with how data were captured and stored so they can better understand these variables. Frequently, data analysts must check with clinicians and programmers to understand a particular stored field or variable. Data analyst access to a study investigator or project coordinator greatly facilitates both communication and the overall data retrieval process.
The amount of time required for the data extraction and formatting varies widely but predominantly depends on the type and volume of data being requested and whether similar data have been extracted previously. When data analysts are not familiar with the data that have been extracted, careful verification must be conducted on the data. This increases the turnaround time on work but is a critical step. We have found that, ultimately, data validation reduces the amount of time involved in the analysis of data for a given project.
Data verification is done at two general levels. The first level is a simple verification of extracted data to determine whether the data pass face validity: Are the extracted data appropriately text or numeric? Are text data complete and not truncated? Are numeric data of the right length and type (real numbers vs. integers)? The second level of verification involves basic descriptive reporting such as cross-tabulation directly on an AlphaServer or after transferring the data to SAS, SPSS, or S-Plus. Means, standard deviations, percentiles, normality profiles, and ascertainment of outliers are determined. Failure to verify data at the first two levels results in a need to determine how data have been stored (by visiting with a programmer) or editing and rerunning the program to re-extract the data. More complicated data validation routines sometimes are necessary and generally are conducted by study biostatisticians.
Analysis
Data analysts may conduct simple descriptive analyses for investigators. However, most often, analytic data sets are transferred to biostatisticians who are well versed in sophisticated statistical procedures needed to analyze large data sets (such as multiple variable analysis, derivation of propensity scores, repeated measures, or time-series analysis). To illustrate this process, we reviewed our studies of the renal effects of nonsteroidal anti-inflammatory drugs (NSAIDs). We were particularly interested in learning the incidence of renal impairment in patients prescribed ibuprofen and risk factors for its development. To study this issue we used simple queries to first determine that there were sufficient numbers of patients who (1) had received prescriptions for ibuprofen and acetaminophen and (2) had serum creatinine and blood urea nitrogen testing. These initial queries allowed us to form active NSAID (ibuprofen) and control (acetaminophen) cohorts to conduct a retrospective cohort study.11 Using logistic regression, we found that 18% of 1,908 patients prescribed ibuprofen had renal impairment and that compared with the control group, users of ibuprofen in whom renal impairment had developed had a greater likelihood of being elderly (≥65 years of age) and having coronary artery disease. We also used longitudinal linear modeling to ascertain the effects of ibuprofen over time using 17,839 serum creatinine measurements on 1,482 patients.12 These confirmatory studies led to prospective acute interventional studies conducted in our general clinical research center to determine the differences in renal functional changes in older adult subjects compared with young subjects, many of whom were mostly identified using the RMRS.13,14 More recently, we used propensity score methods to control for important background characteristics to study the renal effects of ibuprofen, naproxen, tolmetin, piroxicam, and sulindac in cohorts of patients prescribed these drugs.15 Thus, this exemplifies our ability to access, query, extract, and analyze observations within target cohorts using data contained within the RMRS to provide relevant answers to real-world issues.
Protecting Privacy and Confidentiality
As a corollary, the research support section does not try to duplicate the expertise or efforts of our IRB. Instead, it keeps the IRB abreast of all aspects of study progress. Many studies of de-identified data are exempt from full IRB review. Under current federal regulations, studies are exempt from full review if personal identifiers such as names, addresses, social security numbers, and hospital numbers are not recorded. When the investigator needs such personal identifiers, the study undergoes expedited or full review. Our IRB requires filing of a simple checklist and brief description when the study is exempt but cautions investigators that “the ability to establish the veracity of research findings may be seriously jeopardized with the use of data that cannot be linked to subjects.”
We require copies of all materials pertaining to needed data that have been submitted to the IRB by the investigator. Materials may include the signed IRB approval letter, a description of data needs for the study, selection criteria, or electronic files such as documents or spreadsheets. Throughout the process of working with the study investigator, data analysts refer back to these initial documents and other materials to assure that the data to be retrieved agree with what was described to the IRB; deviations and disagreements between these documents may require a study amendment to the IRB. All study materials become part of a data analyst's records that are retained securely by Regenstrief Institute and are destroyed seven years after the study has closed. Recently, data analysts have begun maintaining a secure file containing the patients who have contributed their clinical data to research. This file will permit us to provide to patients a list of studies that have used their health care data. Before data are distributed to investigators or biostatisticians who are affiliated with the study, data analysts complete the Data Distribution Sign-off Sheet (see Appendix at <www.jamia.org>) to verify conformance with IRB regulations.
Recent deliberations over Health Insurance Portability and Accountability Act (HIPAA) requirements have cast a specter of concern over any such uses of secondary data from electronic medical record systems.16,17 Although a comprehensive description of HIPAA regulations relating to secondary data is beyond the scope of this report, it is important to note that all of the data analysts have had HIPAA awareness training, have special credentialing provided by the IRB, and have signed data confidentiality agreements with the Regenstrief Institute. Access to highly confidential data such as human immunodeficiency virus (HIV) test results are only available to two senior data analysts who have special access privileges. Moreover, desktop computers and special programs containing patient data (such as Inquiry and Fast Retrieval) have unique user identifiers, passwords, and time-out settings when programs have not been used within specified times, e.g., 5 minutes.
Throughout much of the debate between patient data privacy advocates and medical researchers about these regulations, little has been mentioned about the types of issues being addressed using electronic medical data. We felt that it would be instructive to describe the general types of issues being addressed using data from the RMRS. As shown below, we tracked and categorized the types of research projects conducted at the Regenstrief Institute for three and a half years using data from the RMRS.
Status Report
Activities
Beginning October 1998, data analysts began tracking the amount of time spent on all of their work. The original rationale for keeping track of time emanated from the need to provide better estimates of time spent on various funded grants and contracts. Along with the amount of time spent, data analysts kept track of work activities, the rationale for work, and the personnel requesting that work, within weekly reports that were subsequently stored in an Access Database. Time was tracked to the nearest 15 minutes. For the purposes of this report, we compiled the time and work-related descriptions from January 1, 1999, through July 31, 2002. We extracted the amount of time spent by analysts on research retrieval activities and categorized the request type. We also determined the requester's profession and primary academic department. We tried to classify work into mutually exclusive categories based on the primary reason for the request. However, there often were secondary aims of interest that could have had some overlap among categories. We classified work as clinical epidemiology when the primary aim was focused on determining the frequencies of multiple diseases, predicting disease or events, preventing disease or habits such as smoking, ascertaining prognosis, or assessing risk. Otherwise, work was classified under a specific disease category.
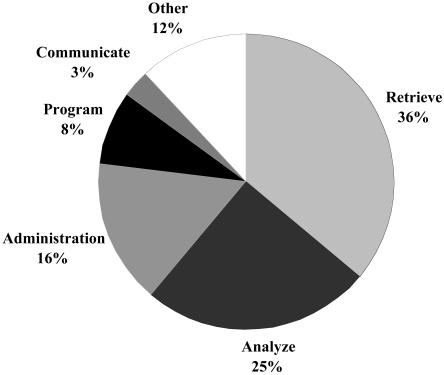
During the three-and-a-half-year study period, 47,559 hours of work were categorized. Training exercises and classes aimed at improving knowledge and skills involved 9% of data analysts' time and were included within the 47,559 hours. The proportions of time spent by data analysts on various activities appear in ▶. As expected, the greatest proportions of time were spent retrieving (36%) and analyzing (25%) data from the RMRS. After the extraction and preliminary analysis of data, data analysts reported (verbally and in writing) their summary data to key investigators working on the studies and performed other administrative and collaborative activities. Approximately three fourths of a data analyst's effort was spent working alone to extract, manage, analyze, and help interpret study data. However, data analysts also worked in coordination with others and frequently communicated with one another on data retrieval, analysis, and programming issues.
Figure 2.
Activities of data analysts on projects requested by various investigators from January 1, 1999, to July 31, 2002.
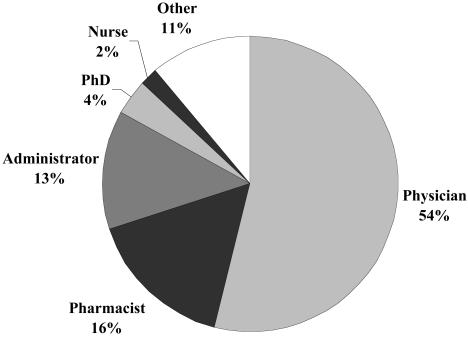
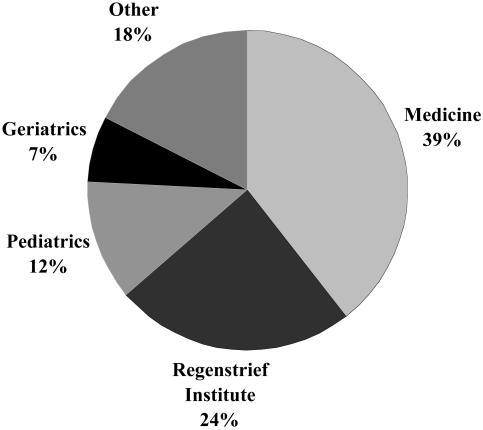
Investigators had varied backgrounds (▶). Physicians requested the majority of data from the RMRS. Administrators and pharmacists were closely tied for second followed by smaller proportions of retrievals requested by PhD researchers and nurses. As shown in ▶, most of the investigators were from the Department of Medicine at the Indiana University School of Medicine (39%) or Regenstrief Institute (24%). Faculty members from Riley Children's Hospital have been the fastest growing group of requesters, largely owing to a recently established Pediatric Health Services Research Department. Although geriatricians are academically affiliated with the Department of General Internal Medicine and Geriatrics, we chose to distinguish studies of older adults (as was done for pediatricians).
Figure 3.
Professions of investigators for whom data analysts extracted data from the Regenstrief Medical Records System from January 1, 1999, to July 31, 2002.
Figure 4.
Academic affiliations of investigators requesting data from the Regenstrief Medical Records System (January 1, 1999, to July 31, 2002).
▶ shows the classification of data analysts' work. Work related to specific diseases took 35.4% of total time. The largest proportion of work was conducted on guideline implementation (13.1% of total time), followed by work on drug-related studies at (12.2% of total time). Analyses within the realm of clinical epidemiology required 5.7% effort, whereas work involving clinical trials required 3.7%. It should be noted that continuing education/training (alone and as a group) was a critical activity involving 8.5% of total data analyst time.
Table 2.
Classification of Data Analyst Work during 3.5 Years
| Category | Time (Hours) | Percent of Total Time | Amount of Time per Job (Hours) Median (Interquartile Range) |
|---|---|---|---|
| Administration | |||
| Meeting | 656 | 1.4 | 12 (2, 45) |
| Reporting | 853 | 1.8 | 4 (1, 4) |
| % subtotal | 3.2 | ||
| Clinical epidemiology | |||
| Prediction | 1,450 | 3.0 | 23 (3, 36) |
| Prevention | 482 | 1.0 | 24 (16, 38) |
| Prognosis | 196 | 0.4 | 11 (10, 23) |
| Risk | 628 | 133 | 16 (5, 79) |
| % subtotal | 5.7 | ||
| Clinical trials | |||
| Preliminary data | 583 | 1.2 | 6 (2, 13) |
| Subject monitoring | 1,194 | 2.5 | 23 (8, 90) |
| % subtotal | 3.7 | ||
| Disease | |||
| Asthma | 4,097 | 8.6 | 16 (4, 64) |
| Bone | 232 | 0.5 | 4 (3, 17) |
| Cancer | 1,188 | 2.5 | 8 (4, 35) |
| Diabetes | 2,026 | 4.3 | 15 (7, 58) |
| Gastrointestinal/hepatic | 406 | 0.8 | 41 (11, 46) |
| General | 1,220 | 2.6 | 5 (13, 48) |
| Heart | |||
| Coronary artery disease | 144 | 0.3 | 14 (6, 35) |
| Chronic heart failure | 3,642 | 7.7 | 8 (3, 37) |
| Hypertension | 530 | 1.1 | 9 (3, 41) |
| HIV | 510 | 1.1 | 5 (2, 10) |
| Kidney | 312 | 0.7 | 20 (10, 28) |
| Neurologic | 196 | 0.4 | 7 (3, 19) |
| Obstetric/gynecologic | 518 | 1.1 | 11 (5, 127) |
| Sexually transmitted disease | 1,586 | 3.3 | 169 (76, 248) |
| Skin | 49 | 0.1 | 24 (21, 28) |
| Thyroid | 94 | 0.2 | 18 (3, 23) |
| Tuberculosis | 51 | 0.1 | 51 (—, —) |
| % subtotal | 35.4 | ||
| Drug-related | |||
| Adverse drug event | 4,662 | 9.8 | 45 (12, 97) |
| General | 919 | 1.9 | 8 (12, 22) |
| Therapeutic drug monitoring | 217 | 0.5 | 7 (6, 204) |
| % subtotal | 12.2 | ||
| Economic | 257 | 0.5 | 25 (11, 52) |
| Geriatrics | 1,418 | 3.0 | 16 (4, 68) |
| Guideline implementation | 6,225 | 13.1 | 17 (5, 64) |
| International programs | 652 | 1.4 | 122 (73, 196) |
| Continuing education and training | |||
| Alone | 1,118 | 2.4 | 8 (2, 19) |
| Group | 2,888 | 6.1 | 29 (10, 112) |
| % subtotal | 8.5 | ||
| Pediatrics | 97 | 0.2 | 6 (1, 16) |
| Radiology | 73 | 0.2 | 21 (8, 21) |
| Surgery | 271 | 0.6 | 19 (10, 110) |
| Programming | |||
| Microsoft Access | 422 | 0.9 | 30 (2, 72) |
| VMS | 201 | 0.4 | 5 (3, 6) |
| % subtotal | 1.3 | ||
| Other | 5,297 | 11 | 71 (7, 144) |
| Total | 47,559 | 100 |
Outcomes
We were able to estimate benefits of the section using funded grant applications and the number of research papers that included data analyst support. From July 1, 2000, to June 30, 2002, the research support section received $600,000 direct funding from research grants and contracts for requested work. However, because many other factors play a role in grant and contract award decisions, determining the number of grant awards that are directly attributable to this research support is not realistically possible. However, it is clear that many data would not be readily available without the assistance of these data analysts.
During December 2002, we surveyed Regenstrief Institute investigators to determine the number of grants written requesting such support, the number of papers written using data analyst support, and the qualitative value investigators put on the services provided by Regenstrief Institute's data analysts. The investigators were told that their responses would be anonymous and used only in aggregate in this report. Of 18 investigators who had used data from the RMRS as provided by a data analyst, 15 investigators (83%) responded to the survey. We excluded from analysis the results of one new investigator who had not yet conducted an independent study. The 14 investigators worked with data analysts a mean (± SD) of 10.0 ± 7.2 years (range, 1 to 23 years) during which time investigators wrote 117 grant applications (mean, 8.4 ± 9.0; range, 0 to 33 grants) and 139 peer-reviewed papers (mean, 9.9 ± 9.3; range, 0 to 33 papers) that specifically included data extracted by a data analyst. Overall, one of every three research papers written by these investigators included data extracted from the RMRS by a Regenstrief Institute data analyst.
We asked investigators to give their qualitative impression of the value of the data analysts' work using a five-point scale (extremely valuable to my research, moderately valuable, helpful but not critical, not helpful at all, or harmful to my research). Of 13 investigators who felt that they had worked a sufficient amount of time with data analysts to provide their assessment, all rated the data analysts' services as extremely valuable to their research. These results suggest that the support provided by the data analysts has a high overall value to researchers in many of their grants and research publications.
Conclusion
The computerization of medicine has provided scientists with rich sources of electronic data for research and quality improvement.18 Most sectors of the health care system now perform many of their primary work functions using computer applications, which, in turn, generate data for capture and storage in large repositories of clinical data. When secondary data from disparate sources (such as clinics, laboratories, radiology, and pharmacy) for the same patients can be merged using either unique identifiers or valid and reliable algorithms, the research questions that can be addressed are innumerable.19,20,21,22,23 Especially numerous are research applications of electronic medical record systems.24 The breadth of this research is apparent in the recent review of three decades of informatics research that was supported by the Agency for Healthcare Research and Quality by Fitzmaurice et al.25 Investigators from Regenstrief Institute have been front and center with much of this research. The research support section described in this report has contributed to many grant applications and peer-reviewed research publications. Further, this research support is perceived as extremely valuable by Regenstrief Institute investigators. We hope that others will benefit by our approach to providing this important service. This approach has evolved over the last 30 years molded by the changing needs of our investigators and our attempts to provide them with timely and accurate data while protecting patients' privacy.
Supplementary Material
The authors are grateful to the patients they serve and who contribute health care data to the Regenstrief Medical Records System for their projects that are aimed at improving their care. The authors thank the IUPUI Institutional Review Board for their timely and guiding reviews and the administrators and staff at Regenstrief Institute who support their efforts. The authors thank Beverly Clark for her kind assistance in preparing the manuscript.
Footnotes
In compliance with JAMIA's policy on HIPAA compliance, patient data have been modified to protect patient privacy and the confidentiality of data.
References
- 1.McDonald CJ, Overhage JM, Tierney WM, et al. The Regenstrief Medical Record System: a quarter century experience. Int J Med Inform. 1999;54:225–53. [DOI] [PubMed] [Google Scholar]
- 2.McDonald CJ, Tierney WM. Computer-stored medical records: their future role in medical practice. JAMA. 1982;259:3433–40. [PubMed] [Google Scholar]
- 3.McDonald C, Tierney W. The Medical Gopher—a microcomputer system to help find, organize and decide about patient data. West J Med. 1986;145:823–9. [PMC free article] [PubMed] [Google Scholar]
- 4.McDonald C, Wheeler LA, Glazener T, Blevins L. A data base approach to laboratory computerization. Am J Clin Pathol. 1985;83:707–15. [DOI] [PubMed] [Google Scholar]
- 5.McDonald CJ, Overhage JM, Dexter P, et al. Canopy computing. Using the Web in clinical practice. JAMA. 1998;280:1325–9. [DOI] [PubMed] [Google Scholar]
- 6.Tierney WM, McDonald CJ. Practice databases and their uses in clinical research. Stat Med. 1991;10:541–57. [DOI] [PubMed] [Google Scholar]
- 7.McDonald CJ, Tierney WM. Research uses of computer-stored practice records in general medicine. J Gen Intern Med. 1986;1(suppl 4):S19–24. [PubMed] [Google Scholar]
- 8.Weinberger M, Murray MD, Marrero DG, et al. Effectiveness of pharmacist care for patients with reactive airways disease: a randomized controlled trial. JAMA. 2002;288:1594–602. [DOI] [PubMed] [Google Scholar]
- 9.Weinberger M, Murray MD, Marrero DG, et al. Pharmaceutical care program for patients with reactive airways disease. Am J Health Syst Pharm. 2001;58:791–6. [DOI] [PubMed] [Google Scholar]
- 10.McDonald CJ, Blevins L, Glazener T, Haas J, Lemmon L, Meeks-Johnson J. Data base management, feedback control, and the Regenstrief medical record. J Med Syst. 1983;7:111–25. [DOI] [PubMed] [Google Scholar]
- 11.Murray MD, Brater DC, Tierney WM, Hui SL, McDonald CJ. Ibuprofen-associated renal impairment in a large general internal medicine practice. Am J Med Sci. 1990;299:222–9. [DOI] [PubMed] [Google Scholar]
- 12.Murray MD, Brater DC. Renal toxicity of the nonsteroidal anti-inflammatory drugs. Annu Rev Pharmacol Toxicol. 1993;33:435–65. [DOI] [PubMed] [Google Scholar]
- 13.Murray MD, Black PK, Kuzmik DD, et al. Acute and chronic effects of nonsteroidal antiinflammatory drugs on glomerular filtration rate in elderly patients. Am J Med Sci. 1995;310:188–97. [DOI] [PubMed] [Google Scholar]
- 14.Murray MD, Lazaridis EN, Brizendine E, Haag K, Becker P, Brater DC. The effect of nonsteroidal antiinflammatory drugs on electrolyte homeostasis and blood pressure in young and elderly persons with and without renal insufficiency. Am J Med Sci. 1997;314:80–8. [DOI] [PubMed] [Google Scholar]
- 15.Perkins SM, Tu W, Underhill MG, Zhou XH, Murray MD. The use of propensity scores in pharmacoepidemiologic research. Pharmacoepidemiol Drug Safety. 2000;9:93–101. [DOI] [PubMed] [Google Scholar]
- 16.Turkington RC. Medical record confidentiality law, scientific research, and data collection in the information age. J Law Med Ethics. 1997;25(2–3):113–29. [DOI] [PubMed] [Google Scholar]
- 17.Kulynych J, Korn D. The effect of the new federal medical-privacy rule on research. N Engl J Med. 2002;346:201–4. [DOI] [PubMed] [Google Scholar]
- 18.Collen MF. Clinical research databases—a historical review. J Med Syst. 1990;14:323–44. [DOI] [PubMed] [Google Scholar]
- 19.Paty D, Studney D, Redekop K, Lublin F. MS COSTAR: a computerized patient record adapted for clinical research purposes. Ann Neurol. 1994;36(Suppl 5):S134–S135 [DOI] [PubMed] [Google Scholar]
- 20.Carpenter PC. The electronic medical record: perspective from Mayo Clinic. Int J Biomed Comput. 1994;34(1–4):159–71. [DOI] [PubMed] [Google Scholar]
- 21.Tamblyn R, Lavoie G, Petrella L, Monette J. The use of prescription claims databases in pharmacoepidemiological research: the accuracy and comprehensiveness of the prescription claims database in Quebec. J Clin Epidemiol. 1995;48:999–1009. [DOI] [PubMed] [Google Scholar]
- 22.Evans JM, MacDonald TM. Record-linkage for pharmacovigilance in Scotland. Br J Clin Pharmacol. 1999;47(1):105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Safran C, Chute CG. Exploration and exploitation of clinical databases. Int J Biomed Comput. 1995;39(1):151–6. [DOI] [PubMed] [Google Scholar]
- 24.van der Lei J. Closing the loop between clinical practice, research, and education: the potential of electronic patient records. Methods of Inf Med. 2002;41(1):51–4. [PubMed] [Google Scholar]
- 25.Fitzmaurice J, Adams K, Eisenberg JM. Three decades of research on computer applications in health care: medical informatics support at the Agency for Healthcare Research and Quality (AHRQ). J Am Med Inform Assoc. 2002;9:144–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.