Abstract
In this study α-lactalbumin was converted from the regular, native state to a folding variant with altered biological function. The folding variant was shown to induce apoptosis in tumor cells and immature cells, but healthy cells were resistant to this effect. Conversion to HAMLET (human α-lactalbumin made lethal to tumor cells) required partial unfolding of the protein and a specific fatty acid, C18:1, as a necessary cofactor. Conversion was achieved with α-lactalbumin derived from human milk whey and with recombinant protein expressed in Escherichia coli. We thus have identified the folding change and the fatty acid as two key elements that define HAMLET, the apoptosis-inducing functional state of α-lactalbumin. Although the environment in the mammary gland favors the native conformation of α-lactalbumin that serves as a specifier in the lactose synthase complex, the conditions under which HAMLET was formed resemble those in the stomach of the nursing child. Low pH is known to release Ca2+ from the high-affinity Ca2+-binding site and to activate lipases that hydrolyze free fatty acids from milk triglycerides. We propose that this single amino acid polypeptide chain may perform vastly different biological functions depending on its folding state and the in vivo environment. It may be speculated that molecules like HAMLET can aid in lowering the incidence of cancer in breast-fed children by purging of tumor cells from the gut of the neonate.
Proteins adopt a series of conformations during their synthesis and transport through human cells. According to the classical view, this process is determined by the amino acid sequence, and thermodynamics force the protein to adopt the conformation with the lowest free energy. During the last decade this classical view has been challenged, because proteins have been shown to adopt several stable conformations, depending on which kinetic pathway is followed (1). The high-energy barriers between the different conformations can be overcome with the help of chaperones (2, 3), but in many systems, the factors that cause or relieve such kinetic barriers remain to be specified.
Stable conformational variants of a protein may expose distinct functional regions and thus differ in biological activity. The most striking example is the prion protein that can change from the normal α-helix-rich to a β-sheet-rich, disease-causing isoform (4, 5). In a previous study we proposed α-lactalbumin as a second example of a protein that can acquire different functions depending on its folding state (6). An α-lactalbumin complex from acid-precipitated human milk casein was shown to induce apoptosis in tumor cells and immature cells, but not in mature, differentiated cells (7). Surprisingly, native α-lactalbumin (Fig. 1A) isolated from its traditional source, human milk whey, did not induce apoptosis. The difference in activity between the two forms of the protein was not caused by a change in secondary structure, but the active complex was found to have undergone a change in tertiary structure, with a conformational switch toward a molten globule-like state (6).
Figure 1.
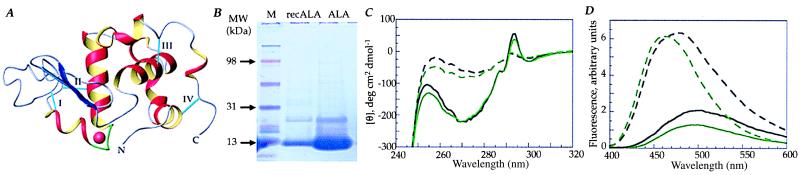
α-Lactalbumin has been studied extensively as a model of protein-folding intermediates (12–15). On lowering the pH the acidic side chains are protonated and the protein adopts the A state or molten globule state (16–18). A similar partially unfolded state, the apo state, is formed at neutral pH if the Ca2+ ion is removed (19, 20). Although these conformations essentially have retained secondary structure, they have fluctuating tertiary structure (18), exposed hydrophobic surfaces, and tryptophan residues accessible to solvent. (A) Three-dimensional structure of native human α-lactalbumin. α-Lactalbumin (14 kDa) is shown with four α-helices (red and yellow, residues 1–34 and 86–123) and an antiparallel β-sheet (blue, residues 38–82). The high-affinity Ca2+-binding site (green) is coordinated by the side chain carboxylates of Asp-82, Asp-87, and Asp-88, the carbonyl oxygens of Lys-79 and Asp-84, and two water molecules. Four disulfide bonds (cyan) are indicated with roman numerals: I, 61–77; II, 73–91; III, 28–111; and IV, 6–120. Crystal structure coordinates are from Acharya et al. (11), and the structure was created with molmol 2.6.1 (21). (B) SDS/PAGE of whey-derived and recombinant α-lactalbumin; SDS/PAGE on 4–20% polyacrylamide precast gels in a Bio-Rad Mini Protean II cell. Lanes: 1, molecular mass standard (Multimark Multicolored Standard; NOVEX, San Diego); 2, recombinant α-lactalbumin (14 and 30 kDa); 3, whey-derived α-lactalbumin (14 and 30 kDa). (C) Near-UV CD spectra of α-lactalbumin. Native whey-derived (solid, black line) or recombinant (solid, green line) α-lactalbumin showed the characteristic 270-nm tryptophan maximum and the 294-nm tyrosine minimum. The EDTA-treated whey-derived (dashed, black line) and recombinant (dashed, green line) apo α-lactalbumin controls showed the characteristic loss of signal in the tyrosine and tryptophan region. (D) ANS fluorescence spectra of α-lactalbumin. Whey-derived (solid, black line) or recombinant (solid, green line) α-lactalbumin in the native state did not bind ANS, but after EDTA treatment (whey: dashed, black line; recombinant: dashed, green line) ANS binding increased and the intensity maximum shifted to 480 nm.
If folding variants of α-lactalbumin differ in biologic activity, it should be possible to convert native α-lactalbumin to the active, apoptosis-inducing form by changing its conformation. In the present study, we prove that α-lactalbumin, indeed, can be converted from the native conformation to the apoptosis-inducing form and define the requirements for this structural and functional change. HAMLET (human α-lactalbumin made lethal to tumor cells) is shown to consist of partially unfolded α-lactalbumin that has integrated a cofactor, which stabilizes the conformation. The cofactor has been identified as a specific fatty acid.
Materials and Methods
Purification of Human α-Lactalbumin.
Native α-lactalbumin was purified from human milk whey by ammonium sulfate precipitation followed by phenyl-Sepharose chromatography (8) and size-exclusion chromatography on a Sephadex G-50 column. The purity of the protein was controlled by SDS/PAGE and agarose gel electrophoresis and by spectroscopic techniques.
Recombinant α-lactalbumin was purified from Escherichia coli (BL21 DE3 pLysS), carrying the vector pALA with the entire human α-lactalbumin gene inserted between the NdeI (site 100) and EcoRI (site 499) sites of the pAED4 vector (pALA was a kind gift from P. S. Kim, Howard Hughes Medical Institute, Cambridge, MA), after induction with isopropyl β-d-thiogalactoside (1 mM). Inclusion bodies were isolated from 1 liter of culture medium, dissolved in 40 ml of buffer (8 M urea/10 mM Tris⋅HCl/10 mM reduced glutathione, pH 8.0), and applied to a DEAE cellulose column. The protein was eluted with 10 mM Tris/7 M urea/1 M NaCl/1 mM CaCl2, pH 8.0. The protein was reduced with 10 mM reduced glutathione added dropwise (2 ml/h) to 500 ml of folding buffer (10 mM Tris⋅HCl/1 mM CaCl2/100 mM KCl/10 mM reduced glutathione/1 mM oxidized glutathione/20% glycerol, pH 8.0, at room temperature) (9). When folding was complete, 10 mM EDTA was added, the folding suspension was applied to a phenyl-Sepharose column, and α-lactalbumin was eluted with 1 mM CaCl2 as described (8). The native fold was confirmed by 8-anilinonaphthalene-1-sulfonic acid (ANS) fluorescence and near-UV CD spectroscopy, with the characteristic 270-nm tyrosine minimum and the 294-nm tryptophan maximum (Fig. 1C).
Apo α-lactalbumin was generated from 25 mg of native α-lactalbumin dissolved at 1.8 mM in Tris (10 mM Tris⋅HCl, pH 8.5) by adding 3.5 mM EDTA to remove bound Ca2+. The conformational change was confirmed by near-UV CD and ANS spectroscopy. The near-UV CD spectrum showed the characteristic loss of signal in the tyrosine and tryptophan regions, and the apo α-lactalbumin-bound ANS, as shown by increased intensity and an intensity maximum, shifted to 480 nm (Fig. 1 C and D).
Spectroscopic Analyses.
CD spectra were obtained by using a JASCO J-720 spectropolarimeter with a JASCO PTC-343 Peltier-type thermostated cell holder. Quartz cuvettes were used with 1-cm path length, and spectra were recorded between 320 and 240 nm. The wavelength step was 1 nm, the response time was 4 s, and the scan rate was 10 nm per minute. Six scans were recorded and averaged for each spectrum. Baseline spectra were recorded with pure buffer in each cuvette and subtracted from the protein spectra.
ANS fluorescence emission spectra were recorded at 25°C on a Perkin–Elmer LS-50B spectrometer by using a quartz cuvette with 1-cm excitation path length, between 400 and 600 nm (step, 1 nm), with excitation at 385 nm. Both the excitation and emission bandpass were set to 5 nm.
Stock solutions were prepared by dissolving lyophilized samples in 10 mM potassium phosphate buffer at pH 7.5. The concentrations were determined by amino acid analysis after acid hydrolysis, and spectra were recorded on aliquots diluted in 10 mM potassium phosphate buffer at pH 7.5.
1H NMR Spectra.
1H NMR spectra were recorded by using an Omega 500 spectrometer at 500 MHz in D2O with 0.15 M NaCl at 37°C for 1–2 mM solutions of protein. Lyophilized HAMLET or native or apo α-lactalbumin (15 mg) was dissolved in 500 μl of D2O, and the pH was set to 7.0 by using NaOD. Oleic acid (4 mg) was dissolved in 75 μl of ethanol-d6, and 10 μl was added to 500 μl of D2O. Apo α-lactalbumin for 1H NMR was generated by dissolving α-lactalbumin in doubly distilled water containing 10-fold molar excess of EGTA at pH 8.0. The sample was applied to a G-25 gel-filtration column after an aliquot of saturated NaCl (calcium-depleted) and eluted by doubly distilled water. The sample was passed through the saturated NaCl to reduce binding of EGTA to the protein, and EGTA-free protein was eluted in the water.
Anion-Exchange Chromatography.
α-Lactalbumin in the native or apo state was subjected to ion-exchange chromatography as described (6). Briefly, the column (14 cm × 1.6 cm) was packed with DEAE-Trisacryl M (BioSepra, Villeneuve-la-Garonne, France) attached to a Bio-Logic chromatography system (Bio-Rad) and eluted with a NaCl gradient (buffer A: 10 mM Tris⋅HCl, pH 8.5; buffer B: buffer A containing 1 M NaCl). The eluted protein fractions were desalted by dialysis (Spectra/Por; Spectrum Medical Industries, Laguna Hills, CA; membrane cut-off, 3.5 kDa) against distilled water, with at least four changes of water, and lyophilized.
Casein Conditioning of the Ion-Exchange Matrix.
Casein was isolated from human milk as described (6). The casein (50 mg) was dissolved in 10 ml of 10 mM Tris⋅HCl (pH 8.5), applied to the column, and run as described. This procedure yielded apoptosis-inducing material (6), and the matrix thereafter was denoted as casein-conditioned. α-Lactalbumin in the native or in the apo state was applied to the casein-conditioned matrix (25 mg dissolved in 10 ml of 10 mM Tris⋅HCl, pH 8.5, with or without EDTA) and eluted as described above.
Identification of the Cofactor on the Casein-Conditioned Column Matrix.
To elute protein cofactors, the casein-conditioned matrix was sequentially washed with 10 mM EDTA, 4 M urea, and 20% ethanol. Eluted protein fractions were analyzed by SDS/PAGE. This procedure yielded residual α-lactalbumin, but no other proteins were detected.
Lipids were eluted from the ion-exchange matrix using organic solvents (10). Casein-conditioned matrix (2 ml) was dissolved in chloroform/methanol/water (1:2:0.8 vol/vol/vol) and incubated at 37°C for 1.5 h. Chloroform/water (1:1 vol/vol) was added, the solution was mixed thoroughly, the two phases were separated overnight, and the organic phase was collected and dried under nitrogen. Lipids were dissolved in chloroform (3 mg/30 μl) and 10 μl was applied in triplicates to silica gel glass plates (1 mg on each lane) and developed by using petroleum ether/dietyl ether/acetic acid/methanol (80:20:1:2 vol/vol/vol/vol). The lipids were visualized by iodine vapor, and defined lipid species were scraped off the plates and eluted from the silica gel with chloroform/methanol (2:1, 1:1, 1:2 vol/vol). The fatty acids were analyzed further on a Varian gas chromatograph (model 3500) equipped with a split/splitless injector and a flame ionization detector. The fatty acids were separated on a fused silica capillary column (0.25-mm inner diameter) (Chrompack, Stockholm). The column temperature was programmed from 140°C to 240°C at 8°C per min. Chromatograms were evaluated by using a Varian integrator model 4290.
Fatty Acid Conditioning of the Ion-Exchange Matrix.
Ten milligrams of palmitic acid (16:0), steric acid (18:0), myristic acid (14:0), or oleic acid (18:1) was dissolved in 500 μl of 99.5% ethanol by sonication (3 min using a Branson 2200 bath sonicator; Branson). After addition of 10 ml of 10 mM Tris⋅HCl, pH 8.5, the different lipid solutions were applied to four separate, newly packed DEAE-Trisacryl M matrices. Native or apo α-lactalbumin isolated from milk or E. coli was applied to each matrix and eluted with NaCl, as described above.
Bioassays of Apoptosis.
The L1210 (ATCC, CCL 219), A549 (ATCC, CLL 185), A-498 (ATCC, HTB 44) and Jurkat (European Cell Culture Collection, no. 88042803) cell lines were cultured as described (7). The cells were harvested by centrifugation (200 × g for 10 min), resuspended in cell culture medium, and seeded into 24-well plates (Falcon/Becton Dickinson) at a density of 2 × 106 per well. The different molecular forms of α-lactalbumin, lipid extracts, or individual fatty acids were dissolved in cell culture medium, without FCS, and added to the cells (final volume, 1 ml per well). Plates were incubated at 37°C in 5% CO2 atmosphere with addition of 100 μl of FCS to each well after 30 min. Cell culture medium served as a control. Cell viability and DNA fragmentation were determined after 6 h of incubation, as described (7).
Subcellular Localization Studies.
L1210 cells (2 × 106 cells/ml, 490 μl) were incubated at 25°C with 12 μl of the biotinylated protein preparations, and surface-bound protein was visualized after counterstaining with FITC-conjugated streptavidin. Intracellular protein was detected after fixation in 4% paraformaldehyde and permeabilization with 0.1% saponin to allow entry of FITC-conjugated streptavidin. Cells were analyzed in Bio-Rad 1024 laser-scanning confocal equipment attached to a Nikon Eclipse E800 microscope.
Results
Apo α-Lactalbumin Can Be Converted to a Folding Variant with Apoptosis-Inducing Activity.
Whey-derived or recombinant α-lactalbumin was used as starting material in experiments aiming to convert the inactive, native protein to the active form (Fig. 1B). As a first step, the native proteins were partially unfolded by EDTA treatment, and the conformational change to the apo-state was confirmed by UV CD and ANS fluorescence spectroscopy (Fig. 1 C and D).
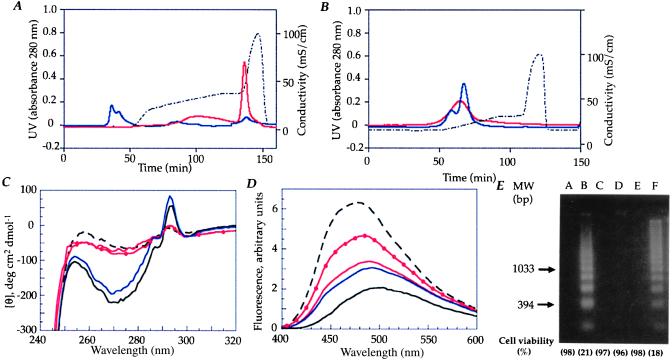
Apo α-lactalbumin then was subjected to ion-exchange chromatography on a column previously exposed to human milk casein. Eluted material was tested for its ability to kill tumor cells and to induce DNA fragmentation and was examined by spectroscopic techniques. Apo α-lactalbumin bound strongly to the ion-exchange matrix and eluted only after 1 M NaCl (Fig. 2A). Eluted protein remained in a partially unfolded conformation as shown by loss of signal in the near-UV CD spectrum and increased binding of ANS (Fig. 2 C and D), and it induced apoptosis with DNA fragmentation in tumor cells (Fig. 2E). α-Lactalbumin in the native conformation was used as a negative control. It could not be activated by these procedures (Fig. 2).
Figure 2.
Conversion of α-lactalbumin to the apoptosis-inducing form requires partial unfolding of the protein and the presence of a cofactor. (A) Apo α-lactalbumin was subjected to ion-exchange chromatography on a column previously exposed to human milk casein. More than 95% of the protein was retained on the column and eluted with 1 M NaCl (solid, red line). Native α-lactalbumin was not retained (solid, blue line). The NaCl gradient is shown by the dotted line. (B) Apo α-lactalbumin (solid, red line) and native α-lactalbumin (solid, blue line) eluted in the void volume when subjected to ion-exchange chromatography on a clean column. (C) CD spectra of the apo α-lactalbumin eluate (solid, red line) from the casein-conditioned matrix did not differ from the apo α-lactalbumin control (dashed, black line). The active fraction from casein was used as a control (solid, red line with solid circle). Native α-lactalbumin had similar spectra before (solid, black line) and after (solid, blue line) elution from the column. (D) ANS spectra of the apo α-lactalbumin eluate (solid, red line) from the casein-conditioned matrix did not differ from the apo α-lactalbumin control (dashed, black line). The native control did not bind ANS before (solid, black line) or after elution from the casein-conditioned matrix (solid, blue line). The active fraction from casein was used as a control (solid, red line with solid circle). (E) Loss of viability and DNA fragmentation of L1210 cells. Lanes: A, cell culture medium; B, the active fraction from human milk casein (0.2 mg/ml); C, native α-lactalbumin (1.0 mg/ml); D, void peak vol from the clean matrix (1.0 mg/ml); E, native α-lactalbumin eluate from the casein-conditioned matrix (1.0 mg/ml); and F, apo α-lactalbumin eluate from the casein-conditioned matrix (0.2 mg/ml). The proteins in lanes B and F were active.
Does Conversion Require a Cofactor?
The conversion of apo α-lactalbumin to the apoptosis-inducing form involved a cofactor from casein. This was first suspected when conversion could not be achieved on a clean column matrix (Fig. 2B). Furthermore, the eluted protein differed from the previously described partially denatured form of α-lactalbumin as Ca2+-depleted or acid-denatured protein revert to the native state when solvent conditions are brought back to normal, but the converted protein was preserved in a partially unfolded state even at neutral pH and in the presence of Ca2+. We also observed that the conversion efficiency of the casein-conditioned column gradually decreased with repeated runs. Thus, a cofactor from casein appeared to be required to maintain the protein in its altered state with apoptosis-inducing activity.
Identification of the Cofactor.
The cofactor was identified by chemical extraction of the casein-conditioned matrix under conditions suitable for proteins or lipids. Solvents known to elute proteins (1 M NaCl, 10 mM EDTA, 4 M urea, or 20% ethanol) released only residual α-lactalbumin; no other proteins were detected. Organic solvents (chloroform, methanol), on the other hand, released lipids from the column matrix. Individual lipid species were identified by GC/MS as C18:1, C16:0, and C14:0 fatty acids (data not shown).
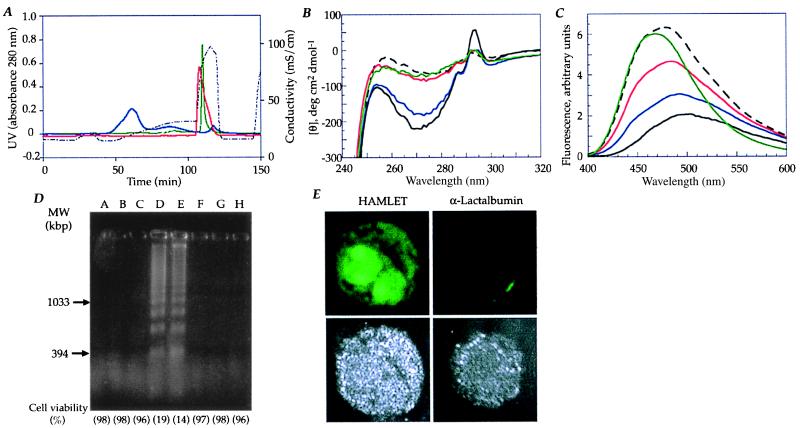
New column matrices then were conditioned with each of the fatty acids or with C18:0 and exposed to whey-derived or recombinant α-lactalbumin in the native or apo states. Fractions eluting after 1 M NaCl were tested for apoptosis-inducing activity. α-Lactalbumin from human milk whey and recombinant protein was shown to convert to the active complex only on the C18:1 fatty acid-preconditioned column and only when applied in the apo form. The proteins were retained on the C18:1-conditioned column, and each eluted as a sharp peak after 1 M NaCl, with a yield of about 90% (Fig. 3A). By near-UV CD and ANS spectroscopy (Fig. 3 B and C), the whey-derived and recombinant activated complexes strongly resembled the apo α-lactalbumin control, and both complexes induced apoptosis in L1210 cells (Fig. 3D). The C18:1-converted apo α-lactalbumin from human milk whey was named HAMLET, and the converted recombinant apo α-lactalbumin was named recombinant HAMLET.
Figure 3.
C18:1 is the fatty acid needed to convert apo α-lactalbumin to the apoptosis-inducing form. (A) Whey-derived or recombinant α-lactalbumin was subjected to ion-exchange chromatography by using a matrix preconditioned with C18:1 fatty acid. Whey-derived α-lactalbumin was added to the column in its native (solid, blue line) or apo state (solid, red line). The apo α-lactalbumin bound to the C18:1-conditioned matrix and eluted as a sharp peak after 1 M NaCl. Native α-lactalbumin bound poorly to the matrix, with >50% in the void. Recombinant apo α-lactalbumin bound to the C18:1-conditioned matrix (solid, green line) and eluted after 1 M NaCl. (B) Near-UV CD spectra of proteins eluting from the C18:1-conditioned matrix. The spectrum of HAMLET (solid, red line) and recombinant HAMLET (solid, green line) strongly resembled the apo α-lactalbumin control (dashed, black line). Native α-lactalbumin before (solid, black line) and after (solid, blue line) passage over the column had native properties. (C) ANS fluorescence spectra of material eluted from the C18:1-conditioned column. HAMLET (solid, red line) and recombinant HAMLET (solid, green line) resembled the apo α-lactalbumin control (dashed, black line). The native α-lactalbumin eluate off the C18:1-conditioned column (solid, blue line) and the native α-lactalbumin control (solid, black line) showed low ANS binding. (D) DNA fragmentation and loss of cell viability in L1210 cells. Lanes: A, cell culture medium; B, whey-derived native α-lactalbumin (1.0 mg/ml); C, recombinant, native α-lactalbumin (1.0 mg/ml); D, HAMLET (0.2 mg/ml); E, recombinant HAMLET (0.2 mg/ml); F, native α-lactalbumin eluate off a C18:1-conditioned column (5 mg/ml); G, lipids extracted from casein-conditioned matrix (0.05 mg/ml); and H, 18:1 fatty acid (0.025 mg/ml). Material in lanes D and E induced apoptosis. (E) Subcellular distribution of HAMLET in L1210 cells. Cell surface binding of HAMLET and recombinant HAMLET was detected after 30 min, followed by translocation into the cytoplasm and accumulation in the cell nuclei (Upper). Native α-lactalbumin bound weakly to the cell surface and did not enter the cells (Upper). The cellular outline is shown in blue reflection mode (Lower).
Conditioning of the ion-exchange matrix with the other fatty acids (C18:0, C16:0, and C14:0) did not convert apo α-lactalbumin to HAMLET, and native α-lactalbumin could not be activated on the C18:1-conditioned column. Most of the material eluted in the void volume with a small peak after 1 M NaCl that showed native three-dimensional structure and lacked apoptosis-inducing activity (Fig. 3D). These results identified the C18:1 fatty acid as the responsible cofactor required to convert α-lactalbumin to HAMLET.
There was no direct cellular effect of the C18:1 fatty acid or of lipid extracts from the casein-conditioned matrix at concentrations found in HAMLET (Fig. 3D). Furthermore, simple mixing experiments with C18:1 fatty acid and apo α-lactalbumin showed that the mixtures had lower activity than HAMLET at similar protein and lipid concentrations.
1H NMR Spectroscopy.
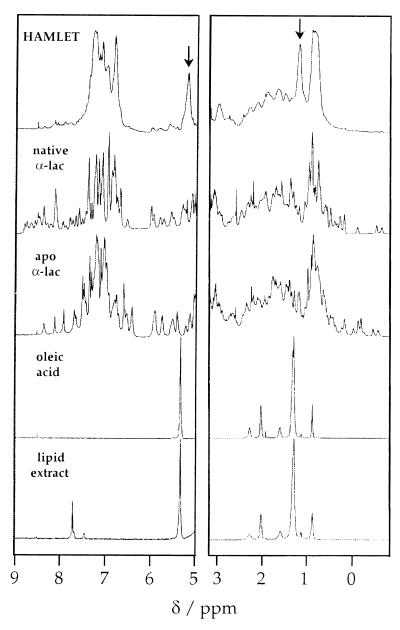
1H NMR spectra of the native α-lactalbumins were characteristic of folded and well-ordered proteins with narrow lines and significant shift dispersion, a large number of sharp signals in the aromatic region (around 7 ppm), and several out-shifted methyl signals (between 0.7 and −0.6 ppm) (Fig. 4). The spectra of the apoproteins displayed narrow lines and significant shift dispersion, with significant variations relative to the native state in the chemical shifts of a large number of resonances (Fig. 4).
Figure 4.
1H NMR spectra of HAMLET, native α-lactalbumin, apo α-lactalbumin, oleic acid, and a lipid extract from HAMLET. The aromatic and methylated regions are shown Left and Right, respectively. Virtually identical spectra were obtained for whey-derived and recombinant α-lactalbumin. The broad lines and lack of out-shifted methyl signals suggest that HAMLET is in a partially unfolded state that is significantly different from the native form of the protein, which displays narrow lines and a large shift dispersion. Furthermore, signals arising from oleic acid are much broader in the HAMLET spectrum (arrows) than in the oleic acid spectrum.
There were marked differences between HAMLET and native α-lactalbumin. The spectrum of HAMLET showed broader lines and little shift dispersion. The lines in the aromatic region were more clustered, with no outshifted methyl signals below 0.7 ppm (Fig. 4). The spectrum of HAMLET did not change between low and physiological salt concentrations. Despite the similarities seen by optical spectroscopy, there were, by NMR, distinct spectral differences between HAMLET and the apo form of α-lactalbumin, suggesting additional conformational changes because of the interaction with the lipid cofactor. Although C18:1 fatty acid was detected in the spectrum of HAMLET (as indicated by arrows), the signals were much broader compared with the signals of free oleic acid, suggesting that the fatty acid has become an integral part of HAMLET.
HAMLET Induces Apoptosis in Tumor Cell Lines and Targets Cell Nuclei.
When isolated from human milk casein the α-lactalbumin folding variant has broad activity against transformed cells of both human and animal origin (7). HAMLET was shown to induce apoptosis in several human and murine tumor cell lines, including the Jurkat and L1210 leukemia cell lines, the A549 lung carcinoma line, and the A-498 kidney carcinoma line. Native α-lactalbumin had no effects on these cells.
The apoptosis-inducing activity of the converted fractions was compared by using the L1210 mouse leukemia cell line. The loss of cell viability and induction of DNA fragmentation were used as end points (Figs. 2E and 3D). The L1210 cells died rapidly when exposed to whey-derived or recombinant HAMLET (200 μg/ml), and DNA fragmentation was induced. The L1210 cells survived exposure to native α-lactalbumin both before and after the protein had been eluted from the ion-exchange column.
By confocal microscopy we observed striking differences in subcellular localization and nuclear uptake depending on the folding state of α-lactalbumin. Whey-derived and recombinant HAMLET bound to the cell surface, passed through the cytoplasm to the nucleus, and accumulated in the cell nucleus (Fig. 3E). Native α-lactalbumin bound weakly to the cell surface but was not seen to translocate into the cytoplasm or to reach the cell nuclei (Fig. 3E).
Discussion
The present study demonstrated that α-lactalbumin can alter its biological function depending on the conformational state. We also showed that a specific fatty acid, C18:1, was a necessary cofactor. We thus have identified the folding change and the fatty acid as the two key elements that define HAMLET, the apoptosis-inducing functional state of α-lactalbumin.
The conversion to HAMLET was achieved first by changing the conformation of α-lactalbumin from the native to a partially unfolded state. EDTA treatment was chosen because it releases calcium and opens up the three-dimensional structure of the protein, and the conformational change was confirmed by spectroscopic techniques. By near-UV CD spectroscopy native α-lactalbumin had a minimum at 270 nm arising from tyrosine residues and a maximum at 294 nm arising from tryptophan residues. The EDTA-treated protein showed the characteristic loss of signal, indicating partial unfolding with less restrained tyrosines and tryptophans. Removal of Ca2+ made the protein more hydrophobic as probed in ANS fluorescence spectroscopy, and HAMLET had increased ANS fluorescence intensity and a shift to a shorter wavelength compared with the native protein.
The second requirement for the stable conversion of native α-lactalbumin was the presence of the C18:1 fatty acid on the ion-exchange matrix. In the absence of this cofactor, the apo form of the protein was unstable and reverted to the native, inactive form at neutral pH and in the presence of Ca2+. The cofactor initially was identified by GC/MS in a lipid extract from the spent ion-exchange matrix. Several fatty acids present in casein eluted as part of the active complex, but only the unsaturated form of the C:18 fatty acid was able to precondition the column matrix for conversion of apo α-lactalbumin to HAMLET. Other structurally related fatty acids like the C18:0, the C16:0, and the C14:0 fatty acids were inactive, suggesting that only the unsaturated C18:1 could act as the necessary cofactor. It may be speculated that the increased hydrophobicity of the apo α-lactalbumin-folding variant enhances binding of C18:1 to exposed hydrophobic regions of the molecule, but the molecular details of these interactions remain to be defined.
α-Lactalbumin is the most abundant protein in human milk (22). The nursing child ingests around 2 g of α-lactalbumin, which travels from the mammary gland through the gastrointestinal tract of the baby. The environmental conditions in the mammary gland favor the native state of the protein with the tightly bound Ca2+ ion, and α-lactalbumin functions as a specifier protein in lactose synthesis (23). Through lactose, the water content of milk is controlled. This is crucial for successful breast-feeding, and it has been shown that α-lactalbumin knockout mice produce extremely viscous milk and fail to nurse their offspring (24).
Quite different environmental conditions meet α-lactalbumin in the stomach of the breast-fed child. The low pH is known to favor the release of Ca2+ (16–18), and casein is precipitated. The acid lipase hydrolyses triglycerides, and fatty acids are released (25, 26). By chance, we happened to purify the apoptosis-inducing form of α-lactalbumin from human milk after precipitation of casein at low pH, thus mimicking the conditions in the stomach of the breast-fed child (7, 27). When these conditions were reproduced in vitro, α-lactalbumin was shown to change to the partially unfolded state and to form HAMLET in complex with the obligatory C18:1 fatty acid.
The purification of proteins from biological fluids has a long history of uncertainty because of the potential for contamination. We used two approaches to exclude that the activity of our complex was due to some contaminating component other than the predominant protein. First, we used α-lactalbumin from human milk whey rather than from casein, showed that it was inactive, and then activated it by partial unfolding and exposure to oleic acid. Second, we expressed the native protein in E. coli and showed that the product was inactive in the native state and that it could be converted equally to the active form through the same procedures. Thus, we feel confident that contamination with other biologically active milk ingredients does not explain our results.
There are obvious parallels between α-lactalbumin and the prion system. Both proteins have multiple activities depending on their folding state, and both require a cofactor for the functional transition. The prion protein first changes to a molten globule-like state and then proceeds to a nonreversible β-sheet-rich form (4, 5), which is the disease-causing isoform. Like the prion protein, α-lactalbumin changes its fold to a molten globule-like state but stays in this intermediate form rather than altering its secondary structure to a β-sheet-rich state. The prion protein has been postulated to require a second protein called “protein X” for its functional switch, but despite many attempts, this cofactor has not been identified (28, 29). In the α-lactalbumin system, we demonstrate that a specific fatty acid can promote switching and/or stabilize the active complex. In the α-lactalbumin system the cofactor was not a protein but a C18:1 fatty acid.
Breast-feeding has been proposed to protect both mother and child against cancer. The overall incidence of childhood cancer is reduced in breast-fed children, with an especially strong epidemiological association for lymphomas (crude odds ratio, 8.9) (30). These differences imply that human milk furnishes the baby with molecules that have a beneficial effect on tissue development and homeostasis, but the mechanisms have not been investigated. It may be speculated that molecules such as HAMLET can have a protective function and that HAMLET is one of several naturally occurring surveillance molecules that purge unwanted cells from the local tissues, thereby driving the intestinal mucosa toward maturity. By inducing apoptosis, HAMLET may reduce the pool of potentially malignant cells that could serve as nuclei for future tumor development and explain the reduced frequency of cancer in breast-fed individuals. Further studies are required to understand the in vivo relevance of these findings.
Taken together, these observations suggest that proteins may respond to different environments by changing their fold and that this process allows a single polypeptide chain to exert vastly different and beneficial biologic functions in different tissue compartments.
Acknowledgments
We thank Hanna Nilsson and Jonas Fast for help with purification of recombinant α-lactalbumin and Lennart Larsson for the GC/MS analysis of the lipid extracts. This work was supported by The Swedish Cancer Society (Grant 3807-B97-01XAB to C.S.), The American Cancer Society (Grant RPG 97-157-01 to C.S.), the Swedish Medical Research Council (Grant K97-03X-11552-02BK to S.L.), and the Swedish Natural Science Research Council (Grant K-AA/KU 10178-300 to S.L.).
Abbreviations
- HAMLET
human α-lactalbumin made lethal to tumor cells
- ANS
8-anilinonaphthalene-1-sulfonic acid
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Booth D R, Sunde M, Bellotti V, Robinson C V, Hutchinson W L, Fraser P E, Hawkins P N, Dobson C M, Radford S E, Blake C C, et al. Nature (London) 1997;385:787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- 2.Herrmann J M, Malkus P, Schekman R. Trends Cell Biol. 1999;9:5–7. doi: 10.1016/s0962-8924(98)01414-7. [DOI] [PubMed] [Google Scholar]
- 3.Martin J, Hartl F U. Curr Opin Struct Biol. 1997;7:41–52. doi: 10.1016/s0959-440x(97)80006-1. [DOI] [PubMed] [Google Scholar]
- 4.Safar J, Roller P, Gajdusek D, Gibbs C J. J Biol Chem. 1993;268:20276–20284. [PubMed] [Google Scholar]
- 5.Safar J, Roller P, Gajdusek D, Gibbs C J. Biochemistry. 1994;33:8375–8383. doi: 10.1021/bi00193a027. [DOI] [PubMed] [Google Scholar]
- 6.Svensson M, Sabharwal H, Hakansson A, Mossberg A K, Lipniunas P, Leffler H, Svanborg C, Linse S. J Biol Chem. 1999;274:6388–6396. doi: 10.1074/jbc.274.10.6388. [DOI] [PubMed] [Google Scholar]
- 7.Håkansson A, Zhivotovsky B, Orrenius S, Sabharwal H, Svanborg C. Proc Natl Acad Sci USA. 1995;92:8064–8068. doi: 10.1073/pnas.92.17.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindahl L, Vogel H J. Anal Biochem. 1984;140:394–402. doi: 10.1016/0003-2697(84)90184-2. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhuri T K, Horii K, Yoda T, Arai M, Nagata S, Terada T P, Uchiyama H, Ikura T, Tsumoto K, Kataoka H, et al. J Mol Biol. 1999;285:1179–1194. doi: 10.1006/jmbi.1998.2362. [DOI] [PubMed] [Google Scholar]
- 10.Bligh E G, Dyer W J. Can J Biochem Physiol. 1959;37:911–916. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 11.Acharya K R, Ren J S, Stuart D I, Phillips D C, Fenna R E. J Mol Biol. 1991;221:571–581. doi: 10.1016/0022-2836(91)80073-4. [DOI] [PubMed] [Google Scholar]
- 12.Kuwajima K. Proteins. 1989;6:87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- 13.Dobson C M. CIBA Found Symp. 1991;161:167–181. doi: 10.1002/9780470514146.ch11. [DOI] [PubMed] [Google Scholar]
- 14.Wu L C, Kim P S. J Mol Biol. 1998;280:175–182. doi: 10.1006/jmbi.1998.1825. [DOI] [PubMed] [Google Scholar]
- 15.Kataoka M, Kuwajima K, Tokunaga F, Goto Y. Protein Sci. 1997;6:422–430. doi: 10.1002/pro.5560060219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu L C, Schulman B A, Peng Z Y, Kim P S. Biochemistry. 1996;35:859–863. doi: 10.1021/bi951408p. [DOI] [PubMed] [Google Scholar]
- 17.Schulman B A, Redfield C, Peng Z Y, Dobson C M, Kim P S. J Mol Biol. 1995;253:651–657. doi: 10.1006/jmbi.1995.0579. [DOI] [PubMed] [Google Scholar]
- 18.Dolgikh D A, Gilmanshin R I, Brazhnikov E V, Bychkova V E, Semisotnov G V, Venyaminov S, Ptitsyn O B. FEBS Lett. 1981;136:311–315. doi: 10.1016/0014-5793(81)80642-4. [DOI] [PubMed] [Google Scholar]
- 19.Pfeil W. Biochim Biophys Acta. 1987;911:114–116. doi: 10.1016/0167-4838(87)90277-9. [DOI] [PubMed] [Google Scholar]
- 20.Kuwajima K. FASEB J. 1996;10:102–109. doi: 10.1096/fasebj.10.1.8566530. [DOI] [PubMed] [Google Scholar]
- 21.Koradi R, Billeter M, Wuthrich K. J Mol Graph. 1996;14:29–32. doi: 10.1016/0263-7855(96)00009-4. , 51–55. [DOI] [PubMed] [Google Scholar]
- 22.Heine W E, Klein P D, Reeds P J. J Nutr. 1991;121:277–283. doi: 10.1093/jn/121.3.277. [DOI] [PubMed] [Google Scholar]
- 23.Brodbeck U, Ebner K E. J Biol Chem. 1966;241:762–764. [PubMed] [Google Scholar]
- 24.Stinnakre M G, Vilotte J L, Soulier S, Mercier J C. Proc Natl Acad Sci USA. 1994;91:6544–6548. doi: 10.1073/pnas.91.14.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarles J, Moreau H, Verger R. Acta Paediatr Scandinavica. 1992;81:511–513. doi: 10.1111/j.1651-2227.1992.tb12284.x. [DOI] [PubMed] [Google Scholar]
- 26.Bernback S, Blackberg L, Hernell O. J Clin Invest. 1990;85:1221–1226. doi: 10.1172/JCI114556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maffei H V, Nobrega F J. Gut. 1975;16:719–726. doi: 10.1136/gut.16.9.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaneko K, Zulianello L, Scott M, Cooper C M, Wallace A C, James T L, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Telling G C, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen F E, DeArmond S J, Prusiner S B. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 30.Davis M K, Savitz D A, Graubard B I. Lancet. 1988;2:365–368. doi: 10.1016/s0140-6736(88)92835-8. [DOI] [PubMed] [Google Scholar]