Abstract
The mouse ephA8 gene is expressed in a rostral-to-caudal gradient in the developing superior colliculus, and these EphA gradients may contribute to the proper development of the retinocollicular projection. Thus, it is of considerable interest to elucidate how the ephA8 gene expression is controlled by upstream regulators during the development of the mesencephalon. In this study, we employed in vivo expression analysis in transgenic mouse embryos to dissect the cis-acting DNA regulatory region, leading to the identification of a CGGTCA sequence critical for the ephA8 enhancer activity. Using this element as the target in a yeast one-hybrid system, we identified a Meis homeobox transcription factor. Significantly, DNA binding sites for Pbx, another TALE homeobox transcription factor, were also identified in the ephA8 enhancer region. Meis2 and Pbx1/2 are specifically expressed in the entire region of the dorsal mesencephalon, where specific colocalization of EphA8 and Meis is restricted to a subset of cells. Meis2 and Pbx2 synergistically bind the ephA8 regulatory sequence in vitro, and this interaction is critical for the transcriptional activation of a reporter construct bearing the ephA8 regulatory region in the presence of histone deacetylase inhibitor. More importantly, when expressed in the embryonic midbrain, the dominant-negative form of Meis down-regulates endogenous ephA8. Interestingly, we found that both Meis2 and Pbx2 are constitutively bound in the ephA8 regulatory region in the dorsal mesencephalon. These studies strongly suggest that Meis and Pbx homeobox transcription factors tightly associate with the ephA8 regulatory sequence and require an additional unidentified regulator to ensure the specific activation of ephA8.
Eph family receptors and their ligands are dynamically expressed during embryonic development and play important roles in many developmental processes, including axonal guidance, cell migration, organization of cell boundaries, synaptogenesis, and formation of the cardiovasculature system (6, 11, 15, 39, 44, 58, 61). A common feature of Eph and ephrin expression in the nervous system is reciprocal graded expression of the receptor and ligand, which presents an efficient way to demarcate locations, as well as specific subsets, of cells (5, 10, 13, 41, 44). In addition, temporal changes in expression levels may also be important in controlling aspects of cell movement processes, such as guidance of growth cones and the migration of neural-crest cells. In this respect, an important and largely unresolved question is the mechanism by which the correct expression domains of these guidance molecules are arranged at the right time and place during embryogenesis.
Eph receptor and ephrin genes have been implicated as the downstream target genes of many different transcription factors, including homeodomain proteins in various developing tissues. In the hindbrain, Krox-20 and Hox transcription factors are direct transcriptional activators of ephA4 and ephA2, respectively (4, 54). More evidence has been obtained for the genetic regulation of Eph receptors and ephrins in retinal development. CBF1, a chick winged-helix transcription factor, plays a role in establishing the nasal and temporal specificity in the retina by which its specific expression in the nasal retina represses ephA3 while inducing ephrin-A2 and ephrin-A5 by upregulating either of two homeobox-containing genes, SOHo1 or GH6 (46, 53). In addition, recent studies have implicated the zinc finger transcription factor Zic2 as a possible upstream regulator of ephB1 in establishing an ipsilateral pathfinding program unique to the ventral temporal crescent retina (14, 59). In contrast to Zic2, the Lim homeodomain transcription factor encoded by Isl2 represses the expression of Zic2 and ephB1, marking only contralaterally projecting retinal-ganglion cells (35). It is also well known that topographic motor projections in the limb are established by Lim homeodomain protein control of Eph receptors and ephrin-A ligands. In the lateral motor column (LMC) of the developing spinal cord, Lim homeodomain proteins are implicated in the regulation of ephA4 expression (20). For example, Isl1 expression in the medial LMC represses ephA4 expression, whereas Lim1 expression in the lateral LMC upregulates EphA4. Additionally, Lmx1b represses ephrin-A gene expression in the dorsal limb mesenchyme, the target of EphA4-containing motor axons derived from the lateral LMC. Recently, it was shown that ephA7 is a direct downstream target of Hoxd13 and Hoxa13 in developing limbs (43). These studies indicate that cell-specific transcription factors are closely linked with Eph receptors and ephrins to control axonal guidance, neural patterning, and other developmental processes.
The superior colliculus (SC) is the most prominent midbrain target of retinal ganglion cell axons and has been the predominant model system for studying the development of topographic maps and the gradients of guidance molecules. Eph receptors and ephrins have been shown to be major players in the development of the retinocollicular topographic map (2, 5, 8, 10, 41). For example, Eph and ephrins are expressed in countergradients in the SC, with high expression of Ephs in the anterior SC and ephrins in the posterior SC (5, 13, 24, 41). In the retinocollicular projection, axons with a high EphA receptor concentration (e.g., from the temporal retina) project to areas with a low ephrin-A concentration (e.g., the anterior SC), while axons with a high ephrin-A concentration (e.g., from the nasal retina) project to areas having low EphA concentrations (e.g., the posterior SC). It has been very well documented that these axonal connections are achieved mainly by the repulsive interactions between EphA receptors and ephrin-A ligands (10, 13, 58). Importantly, the dynamically changing expression patterns of these Eph and ephrin genes may provide additional mechanisms to control retinocollicular mapping in the superior colliculus. Many studies have indicated that En, Pax2, and Pax5 are critically involved in the regionalization of the superior colliculus through the formation of a positive feedback loop for their expression, suggesting that these nuclear factors may regulate eph and ephrin gene expression (28, 29, 33, 34, 47). Indeed, it has been demonstrated that ectopic expression of En induces ephrin-A2 and ephrin-A5 expression (34, 51). Although it is conceivable that En and Pax5 are regulators for establishing the ephrin-A gradient, little is known about the upstream regulation of the EphA receptor genes in the anterior superior colliculus.
Our previous studies demonstrated that a 1-kb ephA8 enhancer region is critical for directing reporter expression to the anterior region of the superior colliculus (24). In this study, we used a transgenic approach to identify the cis-acting DNA regulatory element within this 1-kb enhancer region and found that a CGGTCA-containing sequence is critical for the ephA8 enhancer activity. This sequence was used as a yeast one-hybrid bait to screen a human fetal-brain cDNA library, leading to identification of a Meis homeobox transcription factor as a possible DNA binding protein. Three genes, which encode homeoproteins of the three-amino acid loop extension (TALE) class, constitute the mammalian Meis family. Meis proteins have a conserved N-terminal domain through which they interact with members of a second TALE family, the Pbx class (3, 7, 22, 32, 55, 56). Meis-Pbx dimerization is required for nuclear localization of both of the proteins, and this Meis-Pbx interaction is also considered to be critical for penetrating repressive chromatin to mark specific genes for activation (1, 40, 45). The sum of our transgenic and biochemical studies presented in this study highlights the critical role of the Meis-Pbx interaction in the regulation of ephA8 gene expression during the development of the superior colliculus. In addition, our results support a role for a Meis-Pbx complex in marking specific genes for activation in establishing a specific developmental potential.
MATERIALS AND METHODS
Construction of transgenic and expression vectors.
A restriction map of the 10-kb genomic DNA upstream of the 5′ end of the ephA8 coding region was previously described (19, 24). For construct 1 (Cn1), a 1-kb XbaI/NheI fragment isolated from an ephA8 genomic clone was directly subcloned into the βgnLacZpA vector containing a human β-globin minimal promoter (a gift from J. E. Johnson) (27). For Cn2, a 180-bp sequence was amplified with primers 5′-CCGTCTAGATGTGTCGAGCAGGCACAG-3′ and 5′-CCGACTAGTGGAAGGGGCCCGGGGGCT-3′, and the PCR products were digested with XbaI/SpeI and self-ligated to generate four tandem repeat products, which were digested with XbaI/SpeI and subcloned into the pSDKLacZpA vector containing a 3.5-kb ephA8 promoter (19, 24). For Cn3, the same insert as in Cn2 was subcloned into the βgnLacZpA vector. For Cn4 to -8, the XbaI/NheI fragment in Cn1 was subcloned into the pBluescript vector (Stratagene) and subjected to mutagenesis using overlapping PCR methods. The mutated sequences in each primer sequence indicated below are represented by boldface letters. To construct Cn4, a 630-bp fragment was amplified with primers 5′-AATTAACCCTCACTAAAGGG-3′ (T3 universal primer) and 5′-GCAGCCGGACTTACTGTAGCTCCAGCCATTA-3′, and a 433-bp fragment was amplified with primers 5′-GCTACAGTAAGTCCGGCTGTCAACCGGGAT-3′ and 5′-GTAATACGACTCACTATAGGGC-3′ (T7 universal primer). The two partially complementary PCR fragments thus generated were annealed and used as the templates in another PCR with the 5′ T3 and 3′ T7 universal primers. The resulting 1,239-bp product was digested with XbaI/NheI and subcloned into the corresponding region of Cn1. The same procedures were used to generate Cn5 to -8, except that different primers were used: for Cn5, a 668-bp fragment was amplified with the T3 universal primer and 5′-TGAAATCTATACGAACAGCCGTGACCGCTGTA-3′, and a 409-bp fragment was amplified with 5′-CGGCTGTTCGTATAGATTTCATGGGTTCGTTT-3′ and the T7 universal primer; for Cn6, a 704-bp fragment was amplified with the T3 primer and 5′-GCTAGGTACGCGGGAGAACCCATCAAATCCCG-3′, and a 385-bp fragment was amplified with 5′-GTTCTCCCGCGTACCTAGCACCGTCCTTCTCT-3′ and the T7 universal primer; for Cn7, a 741-bp fragment was amplified with the T3 universal primer and 5′-GGCTGGGCATGATGAAGTCACACGAGAAGGACGGTGCTAAAGAT-3′, and a 375-bp fragment was amplified with 5′-ATCTTTAGCACCGTCCTTCTCGTGTGACTTCATCATGCCCAGCC-3′ and the T7 universal primer; for Cn8, a 757-bp fragment was amplified with the T3 universal primer and 5′-AGGGAAGGGGCCCGAAAACTGGGCATGATGAAGT-3′, and a 349-bp fragment was amplified with 5′-ACTTCATCATGCCCAGTTTTCGGGCCCCTTCCCC-3′ and the T7 universal primer. For M1 to M6, the 10-kb insert in the wild-type (WT) construct was directly subjected to mutagenesis using overlapping PCR methods. The mutated nucleotide in each primer sequence indicated below is represented by a boldface letter. To construct M1, a 1,620-bp fragment was amplified with the T7 universal primer and 5′-ACAGCCGTGACCACTGTAGCTCCAGCCATTA-3′, and a 422-bp fragment was amplified with primers 5′-GCTACAGTGGTCACGGCTGTCAACCGGGAT-3′ and 5′-ACCCTGCTAGCAACGCTGGCTCTTC-3′. The two partially complementary PCR fragments thus generated were annealed and used as the template in another PCR with the T7 universal primer and 5′-ACCCTGCTAGCAACGCTGGCTCTTC-3′. The resulting 2,095-bp product was digested with SfiI/NheI and subcloned into the corresponding region of the WT construct containing a 10-kb ephA8 genomic DNA. The same procedures were used to generate M2 to -6, except that different primers were utilized: for M2, a 1,620-bp fragment was amplified with the T7 universal primer and 5′-ACAGCCGTGACTGCTGTAGCTCCAGCCATTA-3′, and a 422-bp fragment was amplified with primers 5′-ACCCTGCTAGCAACGCTGGCTCTTC-3′ and 5′-GCTACAGCAGTCACGGCTGTCAACCGGGAT-3′; for M3, a 1,620-bp fragment was amplified with the T7 universal primer and 5′-ACAGCCGTGATCGCTGTAGCTCCAGCCATTA-3′, and a 422-bp fragment was amplified with primers 5′-GCTACAGCGATCACGGCTGTCAACCGGGAT-3′ and 5′-ACCCTGCTAGCAACGCTGGCTCTTC-3′; for M4, a 1,620-bp fragment was amplified with the T7 universal primer and 5′-ACAGCCGTGCCCGCTGTAGCTCCAGCCATTA-3′, and a 422-bp fragment was amplified with primers 5′-GCTACAGCGGGCACGGCTGTCAACCGGGAT-3′ and 5′-ACCCTGCTAGCAACGCTGGCTCTTC-3′; for M5, a 1,620-bp fragment was amplified with the T7 universal primer and 5′-ACAGCCGTAACCGCTGTAGCTCCAGCCATTA-3′, and a 422-bp fragment was amplified with primers 5′-GCTACAGCGGTTACGGCTGTCAACCGGGAT-3′ and 5′-ACCCTGCTAGCAACGCTGGCTCTTC-3′; for M6, a 1,620-bp fragment was amplified with the T7 universal primer and 5′-ACAGCCGGGACCGCTGTAGCTCCAGCCATTA-3′, and a 422-bp fragment was amplified with primers 5′-GCTACAGCGGTCCCGGCTGTCAACCGGGAT-3′ and 5′-ACCCTGCTAGCAACGCTGGCTCTTC-3′. The regions amplified by PCR were intensively sequenced to exclude the possibility of errors introduced by the polymerase. Expression vectors for human Meis2b (hMEIS2b), -2d, and -2e have been described elsewhere (60), and mouse Pbx1 (accession no. BC002244) and Pbx2 (accession no. BC010287) were obtained from the 21C Frontier Gene Bank in Korea.
Generation of transgenic embryos and analysis of transgene expression.
Inbred C57BL/6 mice were used to produce transgenic embryos and mouse lines as described (24). Transgenic embryos and mice were identified by PCR analysis of DNA extracted from yolk sacs or tail biopsy specimens, using specific primers: for Cn1 and Cn4 to -8, a 427-bp sequence was amplified with primers 5′-ATTACCAGTTGGTCTGGTGTC-3′ and 5′-AGTTCTCTGAGTTTGTCTGAAATCG-3′; for Cn2, a 650-bp sequence was amplified with primers 5′-ACCGTATGAACAGAGAATCCTTGGC-3′ and 5′-TTGTTTATTGCAGCTTATAATGG-3′; for Cn3, a 654-bp sequence was amplified with primers 5′-CGAGGCTAGAAGCAAATGTAAG-3′ and 5′-TTGTTTATTGCAGCTTATAATGG-3′; for M1 to M6, a 569-bp sequence was amplified with primers 5′-ATTACCAGTTGGTCTGGTGTC-3′ and 5′-ACGGATTCCAAAAGCCTCCAA-3′; and for Meis-En, a 575-bp sequence was amplified with primers 5′-TTGTAATGGACGGTCAGCAG-3′ and 5′-ATCATCCACATCCACATCAAT-3′. The nestin promoter used for expressing Meis1-En was described previously (31). Transgene expression patterns were analyzed primarily at 11.5 days postcoitus (dpc) in generation 0 (G0) embryos or in transgenic lines at postnatal (P0). The day on which a vaginal plug was observed was designated day 0.5 of gestation.
Yeast one-hybrid screening.
To prepare the reporter strain bearing the target sequences, the oligomers containing two repeats of 30 bp (see Fig. 4A), were annealed and subsequently inserted into the EcoRI-XbaI site of vector pHISi-1 and into the EcoRI-XhoI site of the pLacZi reporter vector (Clontech) upstream of the reporters HIS3 and LacZ, respectively. The resulting pHISi-1 and pLacZi reporter plasmids were sequentially integrated into the genome of the YM4271 yeast strain at the HIS3 and URA3 loci, respectively. The resulting strain was transformed with a human fetal-brain cDNA library (Clontech) and plated on the selective medium (minus His/minus Leu/minus Ura) containing 10 mM 3-amino-triazole. The plates were incubated at 30°C for 4 to 6 days. A total of 3.0 × 106 transformants were screened; approximately 600 His+ clones were assayed for blue coloring by colony lift 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining, and 100 LacZ+ clones were selected for plasmid recovery and sequencing.
FIG. 4.
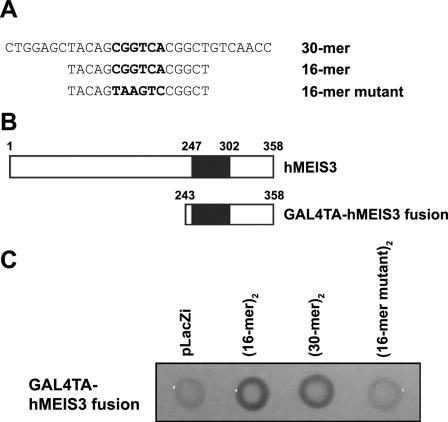
Cloning of Meis3 through its interaction with the CGGTCA motif. (A) The top-strand sequences of the double-stranded oligonucleotides used as baits for the yeast one-hybrid assay are listed, with the putative Meis binding sites in boldface. The mutant oligonucleotide is also shown at the bottom, with nucleotide changes in boldface. (B) Schematic diagrams of a full-length human Meis3 (top) and the interacting clone of Meis3 (bottom) identified in the yeast one-hybrid screen are shown. The homeodomain is represented by a black box in each diagram. (C) LEU-marked GAL4-human Meis3 plasmid was tested to activate lacZ transcription in yeast containing a reporter plasmid with two copies of each oligonucleotide shown in panel B. Yeast transformed with the control vector, pLacZi, served as a negative control for this yeast one-hybrid system.
EMSA.
Proteins for DNA binding assays were produced using an SP6 TNT-coupled in vitro transcription-translation kit (Promega). Reactions containing [35S]methionine were performed in parallel to verify translational efficiency. For binding reactions, 32P-labeled DNA (50,000 cpm/binding reaction) was incubated with 2 μl of each translation product in a 20-μl total volume with 75 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 10 mM Tris-HCl (pH 7.5), 6% glycerol, 2 μg of bovine serum albumin, and 16 ng of poly(dI-dC) plus 0.1 μg of salmon sperm DNA. Each oligomer was 32P end labeled with Klenow enzyme and added to the DNA binding mixture. The DNA binding reaction mixtures were incubated at room temperature for 30 min. The reaction mixtures were then separated on a 6% polyacrylamide gel in 0.25× Tris-borate-EDTA buffer. In some experiments, antiserum to the appropriate epitope was incubated with the reaction mixture for an additional 30 min at room temperature. The sequences of the double-stranded oligomers used for the electrophoretic mobility shift assay (EMSA) are indicated in the figures (see Fig. 6C and 8A).
FIG. 6.
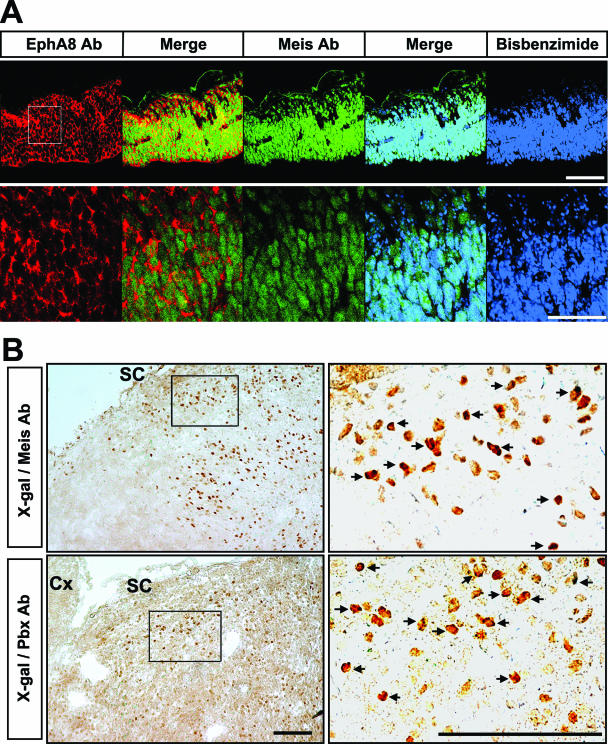
Colocalization of Meis, Pbx, and EphA8 proteins in the anterior region of the mesencephalon. (A, top) Immunohistochemical analyses of EphA8 (red) and Meis (green) proteins in the dorsal mesencephalic tissues at E11.5. Scale bar, 100 μm. (Bottom) Enlarged views of each image shown in panel A (the area of the enlargement is boxed in the left-hand upper image). Scale bar, 50 μm. (B) X-Gal staining of sagittal sections of the superior colliculus of P0 ephA8+/lacZ mouse, followed by incubation with Meis (top row) or Pbx (bottom row) antibody. The right-hand images are enlarged views of each box shown in the left-hand images. The arrows mark a subset of cells coexpressing either Meis or Pbx with the EphA8 receptor. SC, superior colliculus; Cx, cortex. Scale bar, 10 μm.
FIG. 8.
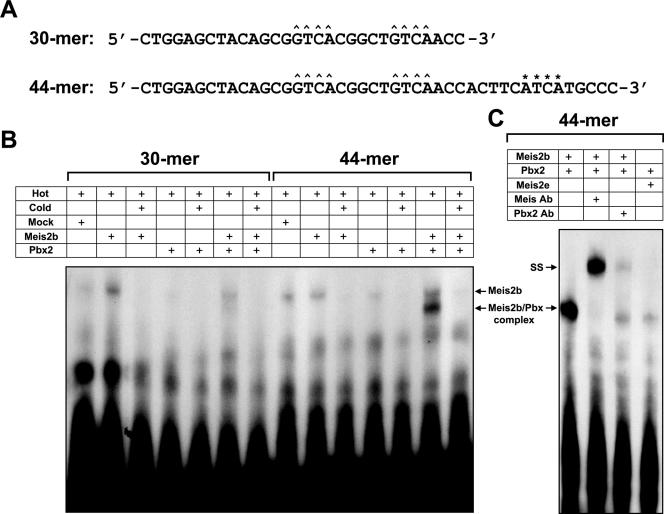
Meis2b protein binds the ephA8 regulatory sequence cooperatively as heterodimers with Pbx protein. (A) Nucleotide sequences of the ephA8 regulatory sites used in this study. Core recognition elements for Meis and Pbx are indicated by ^ and *, respectively, above their sequences. (B) Full-length Meis2b binds weakly to 30-bp and 44-bp probes (lanes 2 and 9 from left). Pbx2 protein does not bind the 30-bp probe (lane 4 from left) but does bind very weakly to the 44-bp probe (lane 11 from left). Meis2b and Pbx2 together form a strong cooperative complex with a 44-bp probe (lane 13 from left), but not the 30-bp probe (lane 6 from left). Mock lysate was used to normalize the levels of lysate in the reactions where one of the translated proteins was missing. (C) The binding specificities of proteins in the multimeric DNA complexes were tested using specific antibodies or Meis2e protein as indicated above each lane (+). SS, antibody-protein complexes resulting from supershift analyses.
Reverse transcription (RT)-PCR analysis.
Poly(A)+ RNA was isolated from 10.5-dpc embryonic midbrain using a Poly(A) Purist mRNA purification kit (Ambion) and then used to synthesize the first-strand cDNA using oligo(dT) primers and Omniscript reverse transcriptase (QIAGEN). The resulting cDNA was then used in PCR to amplify Meis2 isoforms. According to our PCR strategy, at least four different splicing variants of Meis2 were amplified with primers 5′-CCAGAAGAAAAGAGGCATATTCCCCAAAGT-3′ (located within exon 8 of Meis2) and 5′-CTTATACTATTGGGCATGAATGT-3′ (located within exon 13 of Meis2); a 708-bp DNA fragment for Meis2a, a 687-bp DNA fragment for Meis2b, a 613-bp DNA fragment for Meis2c, and a 592-bp DNA fragment for Meis2d. For Meis-En cDNA, a 313-bp product was amplified with primers 5′-TGACCATCAAGGAGGAGGAAAG-3′ and 5′-GTGGCAGTCTGGGAACGACTC-3′. For ephA8 cDNA, a 492-bp product was amplified with primers 5′-ATCAAGGCTCTCAAAGCTGGCTACACAGA-3′ and 5′-CCACATGACCACACCAAAGCTCCACACATCAC-3′. For β-actin cDNA, a 397-bp product was amplified with primers 5′-CCAGATCATGTTTGAGACCT-3′ and 5′-GTTGCCAATAGTGATGACCT-3′. PCR was performed using HotStar Taq DNA polymerase (QIAGEN) with 28 cycles of amplification using a 1-min denaturation step at 94°C, a 0.5-min annealing step at 58°C, and a 1.5-min extension step at 72°C. The PCR products were separated in a 1.5% agarose gel, and the two major PCR products were subcloned for DNA-sequencing analysis. More than 20 different clones of each PCR product were analyzed by sequencing. Real-time PCR analysis was performed using a Platinum SYBR green qPCR SuperMix-UDG (Invitrogen) and the Rotor-Gene (Corbett Research; RG-3000A).
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed according to the protocol from Upstate Biotechnology. Briefly, embryos were dissected in 1× phosphate-buffered saline (PBS) at 10.5 dpc, and the brain tissues were subdivided into the diencephalon, anterior mesencephalon, and posterior mesencephalon (see Fig. 6D). Each tissue was cross-linked in 1% formaldehyde for 30 min at 37°C. Approximately 10 mg of each tissue was resuspended in 0.2 ml sodium dodecyl sulfate (SDS) lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris-HCl, pH 8.1) and sonicated to reduce the chromatin DNA length to a range from 500 bp to 1.5 kbp. The lysates were diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, and 167 mM NaCl) and precleared with salmon sperm DNA-protein A-agarose beads (Upstate Biotechnology) at 4°C for 1 h. Following incubation with 1 μg of anti-Meis, anti-Pbx2, or anti-Pbx antibodies overnight, immune complexes were immobilized with salmon sperm DNA-protein A-agarose beads for 1 h at 4°C. The DNA-protein-antibody-protein A complex was extensively washed with washing buffer I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl), washing buffer II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 500 mM NaCl), washing buffer III (1% Nonidet P-40 (NP-40), 0.25 M LiCl, 1% deoxycholic acid, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1), and Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). After elution with 1% SDS and 0.1 M NaHCO3, cross-links were reversed by incubation at 65°C for 4 h in the presence of 0.2 M NaCl, followed by treatment with 10 μg/ml proteinase K for 1 h at 45°C. The released DNA was phenol-chloroform purified, and the ephA8 enhancer, ephA8 basal promoter, and gapdh promoter sequences were detected by PCR and agarose gel visualization. The ChIP primers for the gapdh promoter were 5′-AACGACCCCTTCATTGAC-3′ and 5′-TCCACGACATACTCAGCAC-3′. The ephA8 enhancer primers were 5′-GATCAAACATGGCCTGGAGT-3′ (forward) and 5′-GGCATGATGAAGTCGTCAGA-3′ (reverse). The ephA8 basal promoter primers were 5′-GGCTCACTACATTTTTGAGG-3′ (forward) and 5′-GACTACCAGTGAGAATGGGA-3′ (reverse).
In situ hybridization and immunohistochemistry.
Whole-mount mRNA in situ hybridization was performed essentially as described previously (57). Single-stranded RNA probes labeled with digoxigenin-UTP were synthesized from linearized template DNA as directed by the manufacturer (Boehringer Manheim Biochemicals). Fragments matching nucleotides (nt) 1420 to 2639 of the mouse Meis1 cDNA sequence (GenBank accession no. BC023689), nt 1424 to 2900 of the mouse Meis2 cDNA sequence (GenBank accession no. BC017375), nt 1032 to 1721 of the mouse Meis3 cDNA sequence (GenBank accession no. BC003762), and nt 619 to 2132 of the mouse Pbx2 cDNA sequence (GenBank accession no. BC010287) were used as templates for riboprobe synthesis.
For immunohistochemistry, embryonic day 10.5 (E10.5) embryos were collected and immediately fixed in 4% paraformaldehyde overnight at 4°C, rinsed extensively in PBS, immersed in 30% sucrose in PBS overnight at 4°C, embedded in cryoembedding medium (OCT), sectioned (12 μm) on a cryostat, and collected on gelatin-coated slides. The slides were rinsed in PBS, treated with 100 mM glycine in PBS for 30 min, and washed in PBT (0.1% Tween 20 in PBS) for 5 min. For permeabilization, sections were treated with TBST (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.2% Triton X-100) for 10 min and washed in PBT for 5 min. A second permeabilization step was performed using PBS-dimethyl sulfoxide (1:1) for 8 min, followed by washing in PBT. The sections were blocked in blocking buffer I (10% goat serum in TBST) for 1 h at room temperature, incubated sequentially with the primary antibody overnight at 4°C, and washed three times in PBT for 5 min and then in blocking buffer II (10% horse serum in TBST) for 30 min. Subsequently, the slides were incubated with a suitable fluorophore secondary antibody for 1 h at room temperature and washed in PBT for 5 min. Raw images were acquired by confocal microscopy (Olympus; FV300) and processed for presentation with Adobe Photoshop. X-Gal staining of the P0 lacZ knock-in brain, followed by Meis or Pbx antibody staining, was carried out as described previously (36).
Antibodies.
Monoclonal anti-Meis1/2/3 antibody was purchased from Upstate Biotechnology. Polyclonal anti-Pbx2 and anti-Pbx1/2/3 antibodies were purchased from Santa Cruz Biotechnology. Polyclonal histone 3 (H3) antibody was purchased from Abcam. Polyclonal anti-EphA8 antibody specific for the EphA8 juxtamembrane was described previously (12). Goat anti-rabbit immunoglobulin G (IgG) conjugated with Alexa-594 and goat anti-mouse IgG conjugated with fluorescein isothiocyanate were purchased from Molecular Probes and Chemicon, respectively. Bis-benzimide (Hoechst 33258) was acquired from Molecular Probes.
Luciferase assay.
HEK293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with l-glutamine, 10% fetal bovine serum, and penicillin/streptomycin. A total of 0.9 μg of DNA containing 0.3 μg of luciferase reporter construct and 0.6 μg of expression constructs was transfected by Lipofectamine (Invitrogen). A cotransfected Renilla luciferase reporter (10 ng per transfection) was used to normalize transfection efficiency. The control luciferase reporter construct used in this assay contained a human β-globin minimal promoter inserted into the promoterless pGL3 vector (Promega). A luciferase reporter driven by the 1-kb XbaI/NheI ephA8 regulatory region was generated by subcloning the insert in Cn1 into the control luciferase reporter. Luciferase activities were measured using a Dual Luciferase Assay Kit (Promega), after which the cells were cultured in either the presence or absence of 2 μM trichostatin A (TSA) (Upstate Biotechnology) for 12 h.
RESULTS
The embryonic ephA8 midbrain regulatory sequences are conserved between mouse and human.
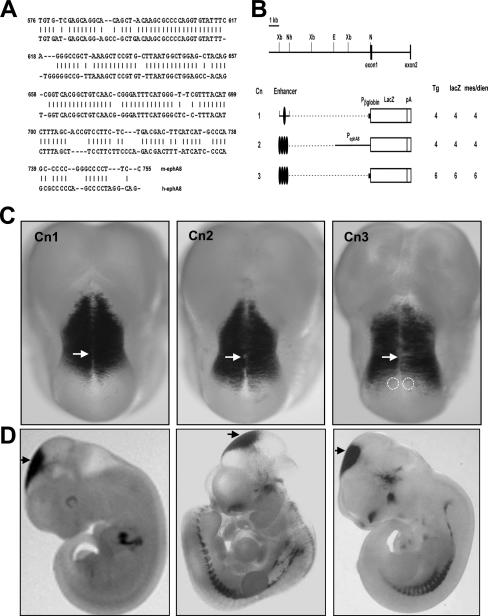
Our previous studies demonstrated that the ephA8 gene expression pattern in the developing midbrain is replicated by a lacZ reporter gene containing a 10-kb fragment of the ephA8 genomic-DNA sequence. In addition, we demonstrated that a 1-kb XbaI/NheI DNA fragment upstream of exon 1 (Fig. 1B, top) contains essential regulatory information for initiating and maintaining ephA8 gene expression in the developing midbrain (24). Human and mouse ephA8 genomic DNAs were compared to determine whether this 1-kb regulatory sequence is evolutionarily conserved (21). This analysis revealed a 180-bp region in the sequence in which the human ephA8 DNA fragment had 82% nucleotide identity to the corresponding mouse ephA8 DNA fragment (Fig. 1A). We tested whether this 180-bp DNA fragment had enhancer activity in vivo by cloning it into the lacZ reporter construct with either the 3.5-kb ephA8 promoter or the human β-globin basal promoter and then analyzing G0 transgenic embryos for lacZ expression at 11.5 dpc (Fig. 1B). Four tandem copies of the 180-bp mouse DNA fragment gave a reproducible but broader expression pattern in the anterior region of the mesencephalon (Cn2) (Fig. 1C and D, middle images), whereas one copy of the 180-bp mouse DNA fragment was insufficient to direct lacZ expression in the mesencephalon (data not shown). Interestingly, both the 1-kb XbaI/NheI fragment (Cn1) and the 180-bp fragment (Cn2) consistently gave a high level of lacZ expression in the diencephalon, suggesting that other regions of the 10-kb ephA8 genomic DNA, not present in the 1-kb XbaI/NheI fragment, contain elements that repress expression outside the anterior region of the midbrain. In contrast, expression of Cn3 in transgenic embryos at this stage became restricted to a narrow region at the anterior region of the mesencephalon, which was not posteriorly expanded compared with those of Cn1 and Cn2 (Fig. 1C, right image). This result suggests that a 1-kb XbaI/NheI DNA fragment in Cn1 or a 3.5-kb ephA8 promoter in Cn2 may contain an additional regulatory element critical for initiating and maintaining an anterior-to-posterior gradient expression of the ephA8 gene at the superior colliculus. Taken together, these results strongly suggest that the most critical ephA8 regulatory sequences required for embryonic ephA8 midbrain expression in the mouse are located in the 180-bp region of the 1-kb XbaI/NheI DNA fragment.
FIG. 1.
Identification of mouse genomic-DNA fragments that direct ephA8-like brain expression. (A) Sequence conservation of the mouse and human ephA8 180-bp enhancer fragments. The bp 576 to 755 region of the murine ephA8 genomic DNA corresponding to the 1-kb XbaI/NheI fragment (see Cn1 in panel B) is shown, together with the evolutionarily conserved region of human ephA8 genomic DNA (the first nucleotide in the XbaI site is shown as +1). (B) (Top) Schematic diagram depicting the restriction map of the genomic region of the ephA8 locus. EphA8 exons are shown as dark boxes. Restriction sites: E, EcoRI; N, NotI; Nh, NheI; X, XbaI. (Bottom) Microinjected genomic-DNA fragments are indicated next to a table depicting the total number of G0 E11.5 transgenic embryos analyzed (Tg), the number of transgenics expressing lacZ (lacZ), and the number of transgenics expressing lacZ in the anterior mesencephalon (mes) or diencephalon (dien). The long ovals represent the 180-bp mouse ephA8 enhancer DNA conserved with human ephA8 DNA. Pβglobin indicates the human β-globin promoter; PephA8 indicates the mouse ephA8 promoter. The bracket indicates the 1-kb XbaI/NheI fragment. (C) Brain expression patterns of lacZ reporter genes directed by mouse ephA8 enhancer DNA fragments. Dorsal views of E11.5 transgenic embryos stained for β-galactosidase activity show lacZ expression in the anterior region of the mesencephalon or diencephalon. Anterior is at the top. The arrows indicate the diencephalon-mesencephalon junction region. The dotted circles mark the specific region lacking lacZ expression in the anterior region of the mesencephalon. (D) Lateral views of the embryos shown in panel C. Variable ectopic lacZ expression in the spinal cord or other regions was observed in some transgenic embryos without consistent patterns. The arrows indicate the diencephalon-mesencephalon junction region.
The CGGTCA-containing sequence is essential for the 1-kb ephA8 enhancer function.
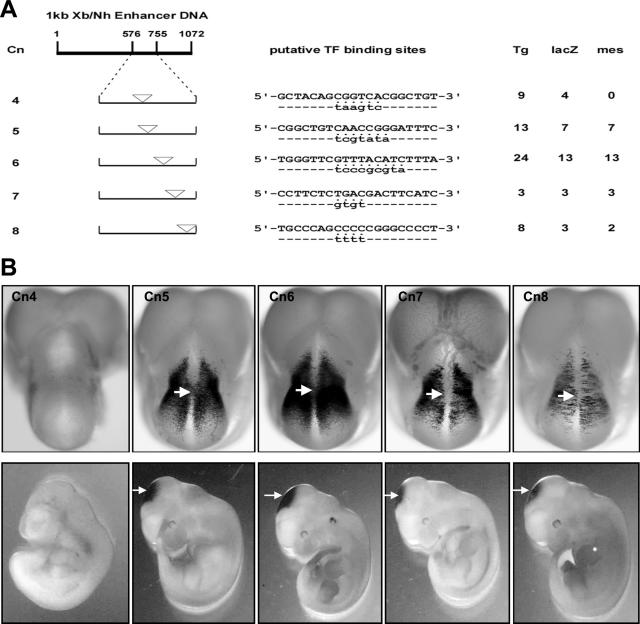
Sequence analysis using the Transcription Element Search System (TESS) program (http://www.cbil.upenn.edu/tess) program revealed that the 180-bp sequence displays at least five potential binding motifs for a variety of transcriptional modulators (Fig. 2A). The in vivo role of each putative transcription factor binding site was tested by mutagenesis of these sites within Cn1 (the 1-kb XbaI-NheI construct with the human β-globin basal promoter) at 11.5 dpc in transgenic embryos. Strikingly, a mutation replacing CGGTCA with TAAGTC from the 1-kb region led to a complete loss of lacZ expression in both the diencephalon and anterior region of the mesencephalon at 11.5 dpc in each lacZ-expressing G0 transgenic embryo analyzed (Fig. 2B, left image). In contrast, mutations in other putative transcription factor binding sites consistently resulted in variable mesencephalon expression patterns (Fig. 2, Cn5 to Cn8). These results strongly suggest that the CGGTCA-containing sequence is required for establishing and maintaining the ephA8-like transgene expression in the mouse midbrain.
FIG. 2.
Identification of a site essential for 180-bp enhancer activity. (A) The diagram at the top left shows a 1-kb ephA8 enhancer DNA fragment. The first nucleotide in the XbaI site is designated +1. Below are the positions of five different putative transcription factor binding sequences, indicated by inverted triangles. These sites were identified by the TESS program. Mutations introduced in each putative transcription factor binding sequence are shown below the wild-type sequences. The putative transcription factors are E2TF or Pax5 for Cn4, c-myb for Cn5, VBPF or GATA3 for Cn6, CREB for Cn7, and AP2 for Cn8. Tg, number of G0 E11.5 transgenic embryos analyzed; lacZ, number of transgenics expressing lacZ; mes, number of transgenics expressing lacZ in the anterior mesencephalon. (B, top row) Dorsal views of transgenic embryos stained for β-galactosidase activity at 11.5 dpc. Transgenic embryos carrying the reporter construct Cn4, which contains mutations in the CGGTCA sequence, show no ephA8-like lacZ expression in the anterior mesencephalon. Anterior is at the top. The arrows indicate the diencephalon-mesencephalon junction region. (Bottom row) Lateral views of the embryos shown in the top row.
The CGGTCA-containing sequence is also critical for the 10-kb ephA8 enhancer function.
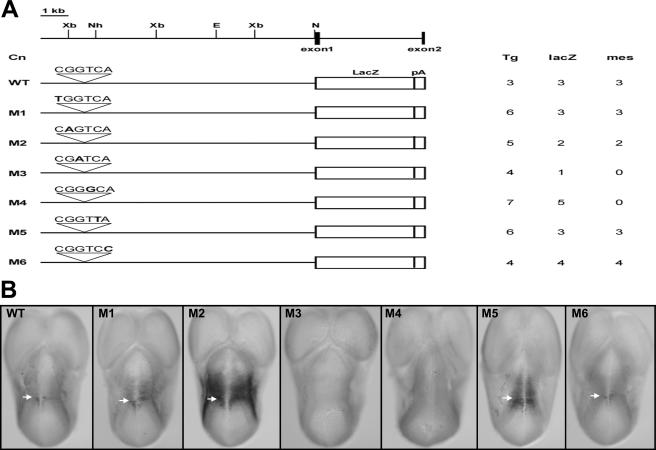
To assess the in vivo significance of the CGGTCA-containing sequence, we next introduced point mutations into each nucleotide of the CGGTCA sequence located in the 10-kb ephA8 genomic-DNA fragment. Figure 3A summarizes the generation and expression analyses of the different transgenic embryos at 11.5 dpc. Mutations at the third (M3) or fourth (M4) nucleotide resulted in complete abrogation of lacZ expression in the anterior region of the mesencephalon (Fig. 3B, fourth and fifth images from left). Mutations at either the second (M2) or fifth (M5) nucleotide also affected lacZ expression, since ectopic expression in the diencephalon was also observed (Fig. 3B, third and sixth images from left). In contrast, embryos carrying mutations at the first nucleotide (M1) or sixth nucleotide (M6) displayed lacZ staining in the anterior region of the midbrain that was indistinguishable from that of the wild-type 10-kb transgene (Fig. 3B, first, second, and seventh images from left). These results, therefore, confirmed that the CGGTCA motif is the critical regulatory element in the 10-kb ephA8 genomic DNA.
FIG. 3.
Functional analyses of a site essential for 180-bp enhancer activity in the context of the 10-kb ephA8 locus. (A) The top diagram depicts the restriction map of the genomic region of the ephA8 locus. EphA8 exons are shown as dark boxes. Restriction sites: E, EcoRI; N, NotI; X, XbaI. Below, microinjected genomic-DNA fragments are indicated as WT and M1 to M6. The sequences above the WT and M1 to M6 represent the wild-type and mutated sequences for a site essential for the 180-bp ephA8 enhancer activity, respectively. The introduced point mutation in each sequence is shown in boldface. Tg, number of G0 E11.5 transgenic embryos analyzed; lacZ, number of transgenics expressing lacZ; mes, number of transgenics expressing lacZ in the anterior mesencephalon. (B) Dorsal views of transgenic embryos (E11.5) stained for β-galactosidase activity. Anterior is at the top. The arrows indicate the diencephalon-mesencephalon junction region. Note that transgenic embryos carrying M3 and M4 show no lacZ expression in the anterior mesencephalon.
Fetal-brain cDNA library screening identifies Meis3 as a CGGTCA motif-binding protein.
A yeast one-hybrid screen was used to identify DNA binding proteins that specifically interact with the CGGTCA motif. We generated a recipient yeast strain stably transformed with two reporter constructs, each consisting of two copies of the CGGTCA-containing 30-mer sequences adjacent to a low-activity promoter directing either HIS3 or LacZ gene expression (Fig. 4A). A human fetal-brain cDNA library was then transformed into this recipient yeast strain, and 4 million transformants were plated on media lacking histidine. HIS+ colonies were subjected to X-Gal staining analysis, prey plasmids were rescued from approximately 100 HIS+ and LacZ+ clones, and the inserts were sequenced. From this analysis, only one clone containing a partial sequence of a DNA binding protein, human Meis3, was identified (Fig. 4B). The insert was in frame with the GAL4 transactivation domain (GAL4TA), enabling this clone to direct expression of a fusion protein between GAL4TA and amino acids 243 to 358 of human Meis3 containing a consensus homeodomain. To further confirm that the GAL4TA-hMEIS3 fusion protein interacts with the CGGTCA motif, yeast strains containing a similar reporter plasmid with two copies of a CGGTCA-containing 16-mer or its mutant were generated (Fig. 4B). As shown in Fig. 4C, expression of the GAL4TA-hMEIS3 fusion protein was able to induce LacZ expression in yeast containing either 16-mer or 30-mer regulatory sequences. In contrast, the same protein failed to induce LacZ expression in yeast containing the control reporter plasmid or the mutant 16-mer sequence containing TAAGTC instead of CGGTCA. These results strongly suggest that the Meis homeoprotein is a candidate DNA binding protein to directly interact with the CGGTCA motif.
Meis2 and Pbx1/2 are expressed in the dorsal region of the mesencephalon.
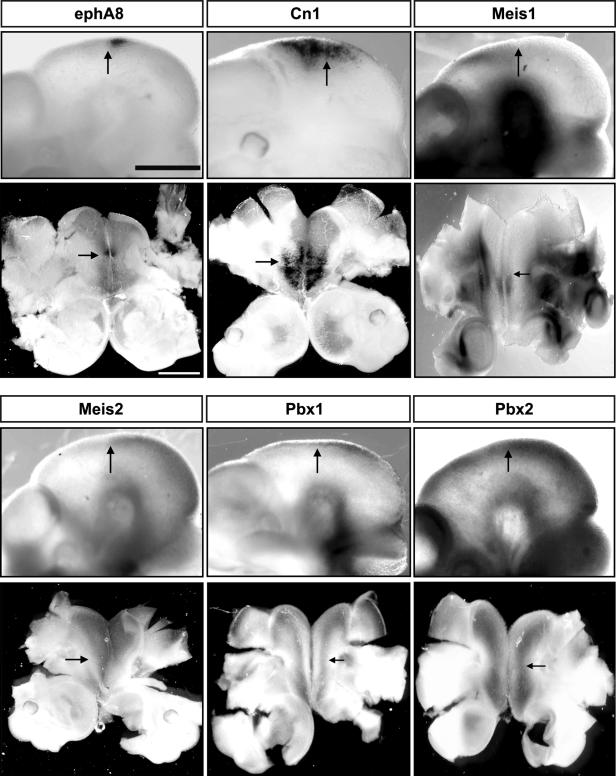
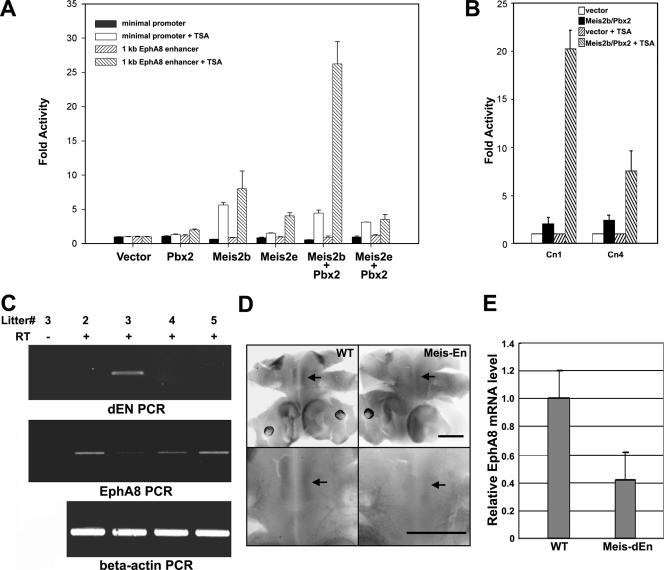
The Meis homeoproteins belong to the TALE superclass of homeobox proteins and are comprised of three different members, Meis1, Meis2, and Meis3. Pbx proteins, another group of the TALE superclass of homeobox proteins, are important cofactors for transcriptional regulation mediated by Hox or Meis proteins (32, 52, 55, 60). To ascertain whether Meis1, Meis2, Meis3, Pbx1, and Pbx2 are expressed at the correct time and place to be endogenous regulators of ephA8 during the development of the mesencephalon, we performed whole-mount RNA in situ hybridization in E10.5 mouse embryos. At E10.5, ephA8 transcripts were specifically restricted to the anterior region of the developing midbrain, as determined by X-Gal histochemistry in embryos heterozygous for the ephA8-lacZ allele previously reported (Fig. 5, top left images) (24, 36). A Cn1 transgenic embryo is also shown to compare its lacZ expression with those of Meis and Pbx (Fig. 5, top middle images). We did not observe specific expression of Meis1 in the mesencephalon, despite fairly specific expression elsewhere, including the diencephalon (Fig. 5, top right images) (16). Consistent with previous reports, Meis2 is specifically expressed in the developing midbrain, and its expression partially overlaps with ephA8 gene expression in the anterior region of the mesencephalon (Fig. 5, bottom left images) (24, 30). However, no evidence of Meis3 expression was detected in the brain at this age (data not shown). Pbx1 and Pbx2 transcripts were found in both the diencephalon and the mesencephalon, where endogenous Meis proteins were expressed (Fig. 5, bottom middle and right images). In addition, Meis1, Meis2, and Pbx2 expressions were persistently maintained in ephA8−/− embryos null for EphA8 function (data not shown).
FIG. 5.
Expression patterns of Meis and Pbx genes in the diencephalon/mesencephalon and relationships with the ephA8 expression domain. Each embryo (E10.5) subjected to whole-mount mRNA in situ hybridization or X-Gal staining is shown in both lateral (top) and dorsal (bottom) views. The arrows indicate the diencephalon-mesencephalon junction region. For the dorsal view, each embryonic brain was gently flattened onto the slide prior to taking a photo. For the lateral views, anterior is at the left. For side views, anterior is at the bottom. Scale bar, 50 μm.
Next, we examined the localization of the Meis and EphA8 proteins in the anterior region of the mesencephalon. Cryosections of E10.5 mouse embryo were colabeled with an EphA8 antibody and a pan-Meis antibody. The data shown in Fig. 6A clearly indicate that Meis proteins are present as nuclear factors in all of the cells expressing EphA8 receptor in the anterior region of the midbrain. However, these staining patterns were not observed in the diencephalon and myencephalon (see Fig. S1C posted at http://sookmyung.ac.kr/∼scpark/Paper/MCB/MCB.zip). To more convincingly demonstrate that the EphA8 receptor is present in cells expressing Meis and Pbx, sagittal sections from a P0 ephA8+/lacZ brain were subjected to X-Gal staining, followed by Meis or Pbx antibody staining. As shown in Fig. 6B, most LacZ-expressing superior colliculus cells reacted with Meis or Pbx antibody. Taken together, these results strongly suggest that Meis2 and Pbx1/2 are likely to be an upstream regulator of ephA8 gene expression during the development of the mesencephalon.
Meis2 protein binds to the CGGTCA-containing sequence in vitro.
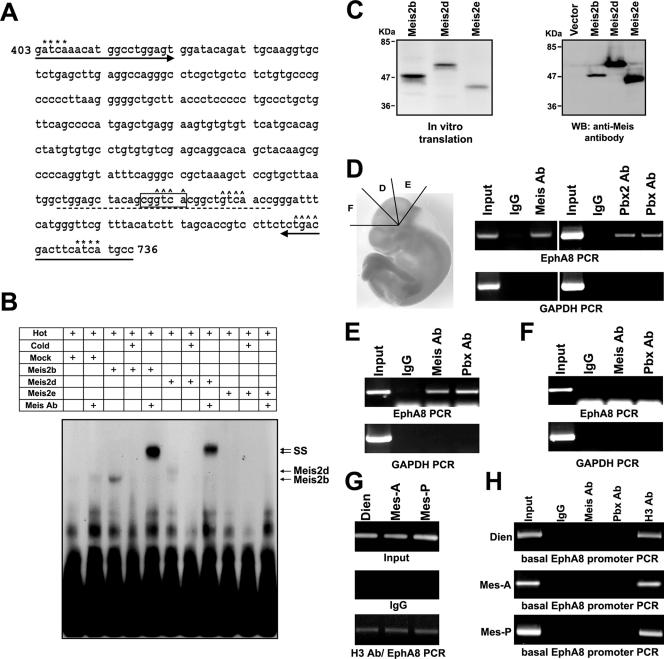
Unlike the majority of Hox proteins, which bind a TAAT core motif, the consensus DNA binding site for Meis homeoproteins contains a 5′ TGAC core motif. The 30-bp bait DNA sequence used for the yeast one-hybrid assay contained two Meis binding core motifs (Fig. 7A). To investigate whether Meis2 could indeed bind to the CGGTCA-containing 30-mer double-stranded DNA, gel mobility shift assays were carried out using in vitro-translated Meis2 isoforms. The abilities of three Meis2 cDNA isoforms, Meis2b, Meis2d, and Meis2e, to encode proteins was verified by SDS-polyacrylamide gel electrophoresis analysis of in vitro translation products (Fig. 7C, left) or Western blot analysis of whole-cell lysates from transiently transfected HEK293 cells using pan-Meis antibody (Fig. 7C, right). Meis2e protein has a truncated homeodomain due to alternative splicing, which places a termination codon within the homeodomain and results in both the deletion of exons 9 to 13 and a frameshift (60). Oligonucleotides containing a CGGTCA motif are specifically shifted by Meis2b and Meis2d, although the shifted complex is very weak (Fig. 7B, lanes 3 and 6). Cold-competitor oligonucleotides eliminated the binding of both Meis2b and Meis2d proteins (lanes 4 and 7). In contrast, labeled oligonucleotides mixed with Meis2e containing a truncated homeodomain failed to produce the shifted product (lane 9). When Meis antibody was mixed with preformed Meis-DNA complexes, a supershifted band of slower mobility, representing the ternary complex of the Meis protein-labeled DNA probe-antibody, was observed only with Meis2b or Meis2d, but not with Meis2e (Fig. 7B, lanes 5, 8, and 11). The use of preimmune serum did not affect the mobility of the Meis complex, and oligonucleotides containing mutations in two Meis core motifs eliminated the ability of the Meis2b protein to bind DNA (data not shown).
FIG. 7.
Meis protein directly binds to the ephA8 regulatory sequence. (A) The nt 403 to 736 region of the mouse ephA8 XbaI/NheI fragment depicted in Fig. 2A (the XbaI site is +1). The critical ephA8 regulatory element is boxed, and the 30-bp DNA for EMSA is indicated by a dashed underline. Putative Meis and Pbx binding sites are indicated by ^ and *, respectively. The arrows underline the sequences of primers used in the ChIP assays shown in panels D to G. (B) Electrophoretic mobility shift assays demonstrating binding of Meis2 isoforms with the 30-bp element containing a CGGTCA motif. Meis2b and Meis2d bind weakly (lanes 3 and 6 from left), while Meis2e does not bind (lane 9 from left). Cold competitor (100×) totally inhibits Meis binding to the labeled probe (lanes 4 and 7 from left). The addition of Meis antibody to the Meis2b and Meis2d protein supershifts (ss) the Meis-DNA complex, which is much stronger when bound by Meis antibody (Ab) (lanes 5 and 8). +, addition of the indicated reagent. (C) In vitro-translated Meis2 proteins were analyzed by SDS-polyacrylamide gel electrophoresis (left). Meis antibody specificity was tested in a Western blot (WB) analysis of total cell lysates from HEK293 cells transiently transfected with the indicated Meis expression constructs (right). (D to F) The ChIP assay was performed to examine the association of Meis and Pbx proteins with the ephA8 regulatory region in different brain regions. The left image in panel D is a schematic representation showing how the brain tissue was divided for ChIP assays. D, anterior region of dorsal mesencephalon; E, posterior region of dorsal mesencephalon; F, dorsal diencephalon. (G) The ChIP assay was performed to examine the association of H3 protein with the ephA8 regulatory region in different brain regions. Dien, diencephalon; Mes-A and -P, anterior and posterior regions, respectively, of the mesencephalon. (H) The ChIP assay was performed to examine the association of Meis, Pbx, and H3 proteins with the ephA8 basal promoter in different brain regions.
Interestingly, during the sequence analysis of the ephA8 regulatory region, we identified putative Pbx binding sites that could be important for the transcriptional regulation of ephA8 gene expression (Fig. 7A). To further determine whether Meis or Pbx homeoproteins are indeed bound to the ephA8 regulatory sequences shown in Fig. 7A, the 334-bp region including the Meis and Pbx binding sites was examined by ChIP assays. Tissue derived from E10.5 brain was cross-linked, and chromatin was immunoprecipitated with specific antibodies, followed by PCR analysis of the ephA8 regulatory sequences (Fig. 7D to F). For tissue derived from the anterior mesencephalon, a positive PCR signal was detected in the samples immunoprecipitated with Meis, Pbx, or Pbx2 antibodies (Fig. 7D, top, lanes 3, 6, and 7), while in control IgG samples, only background signals were observed (lanes 2 and 5). Interestingly, ChIP assays using brain tissue from the posterior region of the mesencephalon also gave positive PCR signals (Fig. 7E). However, a positive ChIP result was not observed in the immunoprecipitated sample from E10.5 mouse diencephalon (Fig. 7F). PCR of the glyceraldehyde-3-phosphate dehydrogenase promoter or the ephA8 basal promoter served as a control for the specificity of Meis or Pbx binding in the ChIP analysis (Fig. 7D to F, bottom, and H, respectively). In addition, PCR of the ephA8 regulatory sequences in the immunoprecipitated samples using H3 antibody gave all positive results for three different dissected tissues (Fig. 7G). These ChIP results strongly indicate that the Meis and Pbx homeoproteins do indeed bind to this ephA8 regulatory sequence in the embryonic midbrain.
Meis2b is the predominant isoform in the mesencephalon and cooperates with Pbx2 to bind the ephA8 regulatory sequence.
The Meis2 gene is comprised of 13 exons, and at least five different alternative splicing variants were identified in both mouse and human (see Fig. S2 posted at http://sookmyung.ac.kr/∼scpark/Paper/MCB/MCB.zip). To investigate which isoforms of Meis2 homeoproteins are predominantly expressed in the developing midbrain, RT-PCR analysis was carried out using mRNAs extracted from the E10.5 embryonic midbrain. Two major PCR products were reproducibly obtained (see Fig. S2D, lane 2, posted at http://sookmyung.ac.kr/∼scpark/Paper/MCB/MCB.zip). According to our PCR strategy, the larger PCR product represented either Meis2a or Meis2b, while the smaller one was either Meis2c or Meis2d (see Fig. S2B and C). To determine which isoforms were present, each PCR product was subcloned and subjected to DNA-sequencing analysis, revealing that the PCR products corresponded to Meis2b and Meis2d (see Fig. S2D, lanes 3 and 4). Since the level of the Meis2b PCR product was threefold higher than that of the Meis2d product (see Fig. S2E), it is likely that Meis2b is the isoform predominantly expressed in the developing midbrain.
Previous studies have shown that Meis proteins are major in vivo DNA binding partners for Pbx proteins (3, 18, 22, 25, 45, 50). As a prelude to examining the synergistic binding of Meis2b with Pbx2 on the ephA8 regulatory DNA sequence, Meis2b-Pbx2 heterodimers were assessed by EMSAs for their ability to bind 30-mer oligonucleotides containing only Meis binding sites (Fig. 8A). In vitro-produced Meis2b or Meis2b-Pbx2 complexes bind weakly to this DNA (Fig. 8B, lanes 2 and 6), whereas Pbx2 alone does not bind (lane 4). Since a consensus site for Pbx was mapped to a position 55 bp downstream of the CGGTCA sequence, we designed a 44-bp synthetic oligonucleotide containing two Meis and one Pbx core motifs (Fig. 8A; see Fig. S3A posted at http://sookmyung.ac.kr/∼scpark/Paper/MCB/MCB.zip). Incubation of in vitro-translated Meis2b and Pbx2 proteins gave rise to a faster-migrating product, presumably the Meis2b-Pbx2 complex bound to DNA (Fig. 8B, lane 13), which is eliminated by cold-competitor oligonucleotides (lane 14). In addition, the labeled oligonucleotides failed to produce the faster-migrating product when mixed with a combination of Pbx2 and Meis2e (Fig. 8C, right lane). When Meis antibody was mixed with preformed Meis/Pbx/DNA complexes, a supershifted band of slower mobility was observed (Fig. 8C, second lane from left). The supershift of Meis-Pbx-DNA complexes was much weaker when incubated with Pbx2 antibody (Fig. 8C, third lane from left), perhaps due to its inhibitory effect on complex formation. Taken together, these results strongly support a model in which Meis2b and Pbx2 proteins cooperatively bind the ephA8 regulatory sequences.
The Meis2b-Pbx2 complex mediates transcriptional activation through the ephA8 regulatory sequence in the presence of an HDAC inhibitor.
To assess whether the Meis2b-Pbx2 complex directly induces transcriptional activation through interaction with the ephA8 regulatory sequence, we examined the activity of a luciferase reporter driven by the 1-kb XbaI/NheI ephA8 regulatory region in transiently transfected HEK293 cells. This 1-kb XbaI/NheI DNA fragment (Fig. 1B) contains several Meis- and Pbx-binding core motifs and was previously shown to be essential for initiating and maintaining ephA8 gene expression in the developing midbrain. As shown in Fig. 9A, the 1-kb ephA8 regulatory sequence is barely responsive to transfection with either Meis2b or Pbx2, suggesting that Meis2b/Pbx-dependent transcriptional activation requires certain cofactors and that these important factors are not present in HEK293 cells (Fig. 9A, bars 7, 11, and 19 from left). Previous studies have demonstrated that Meis-Pbx complexes are recruited to a target site and induce transcriptional activation in response to the histone deacetylase (HDAC) inhibitor TSA (17, 42). Consistent with these studies, we found that cotransfection with expression vectors for Meis2b and Pbx2 synergistically increased luciferase activity by at least 27-fold in TSA-treated cells (Fig. 9A, lane 20 from left). Meis2b alone also increased reporter activity by about sevenfold (lane 12 from left), whereas either Meis2e or Pbx2 did not significantly activate the reporter assay (lanes 8, 16, and 24 from left). Taken together, these results suggest that the Meis2b-Pbx2 complex is able to activate the ephA8 regulatory sequence through interaction with other specific cofactors mediating histone acetylation. To further test whether the mutant 1-kb ephA8 DNA depicted in Fig. 2 showed any defect for reporter activity in response to cotransfection of Meis2/Pbx2, the Cn4 construct was modified to replace lacZ with luciferase coding sequence. As shown in Fig. 9B, this modified Cn4 construct revealed approximately threefold-reduced reporter activity compared to the wild-type construct (Cn1) in cells cotransfected with Meis2 and Pbx2. This result was only observed in TSA-treated cells, partially supporting a functional significance of Meis and Pbx in the regulation of ephA8 expression. However, this result is rather different from in vivo reporter data for the Cn4 transgenic embryo, possibly suggesting that there are quite different levels of transcriptional complexity in the transfected HEK293 cells and the developing midbrain.
FIG. 9.
(A) Meis2b-Pbx2 complexes contribute to TSA-induced transcriptional activation through the ephA8 regulatory DNA. HEK293 cells were transiently transfected with reporter and expression constructs as indicated. Luciferase activities were measured after the cells were cultured in the presence of dimethyl sulfoxide or TSA (2 μM; 24 h). The values are expressed as activity over transfection of the respective reporter plasmids alone. All of the transfections were repeated at least three times, and the bars and error bars represent the means plus standard deviations (SD). (B) The Cn1 or Cn4 construct depicted in Fig. 2A was modified so that its lacZ gene was replaced with luciferase coding sequence. Transfections and luciferase activity measurements were performed as described in the legend to Fig. 8. All of the transfections were repeated at least three times; the bars and error bars represent the means plus SD. (C) EphA8 expression is down-regulated in the transgenic embryos expressing Meis1-En under the nestin promoter. The dorsal midbrain tissues were isolated from E12.5 littermates after the Meis1-En construct was injected to generate the transgenic embryo. RT-PCR analysis was utilized to examine the expression of Drosophila En (dEN) (top) or endogenous ephA8 (middle). The β-actin RT-PCR is also shown as a control (bottom). (D) Each embryo (E10.5) subjected to the whole-mount mRNA in situ hybridization with ephA8 probe is shown as a dorsal view, as described in the legend to Fig. 5 (top row). Enlarged views of each embryo are shown in the bottom row. Signal of ephA8 mRNA is marked by arrows. Anterior is at the bottom. Scale bar, 100 μm. (E) RNA quantitation by real-time RT-PCR shows that ephA8 expression is down-regulated by approximately twofold in Meis1-En transgenic embryos (n = 5) relative to wild-type embryos. The expression level of ephA8 is shown relative to β-actin RNA expression.
EphA8 gene expression is perturbed in transgenic embryos expressing Meis1-En under the nestin promoter.
To investigate the functional significance of Meis2/Pbx binding to the ephA8 regulatory sequence, we took advantage of an in vivo approach using a full-length Meis1-Engrailed repressor construct (Meis1-En). It was previously shown that Meis1-En specifically represses Pax6 expression in vivo, suggesting its dominant-negative role for Meis function in vivo (62). In order to test whether Meis1-En perturbs Meis2-ephA8 genetic interaction, we generated five different transgenic embryos expressing Meis1-En under the nestin promoter (26, 31). As shown in Fig. 9C and D, RT-PCR or whole-mount in situ analyses of the transgenic embryos expressing Meis1-En revealed that the ephA8 expression level in the mesencephalon was significantly reduced. The expression levels of ephA8 normalized to that of β-actin for the transgenic embryo (n = 5) are estimated to be approximately twofold less than those of wild-type embryos (n = 5) (Fig. 9E). Taken together, these in vivo experiments strongly suggest that ephA8 expression is indeed perturbed in the embryos deficient for Meis function and that Meis2/Pbx does functionally regulate ephA8 expression in vivo.
DISCUSSION
The present study provides several lines of evidence demonstrating that the homeobox transcription factors Meis2 and Pbx are possible direct upstream regulators of ephA8 gene expression in the developing midbrain. These are (i) a Meis binding site is required for ephA8 enhancer activity in transgenic embryos; (ii) Meis2 and Pbx proteins are coexpressed, along with the EphA8 receptor, in the anterior region of the dorsal mesencephalon; (iii) Meis2 and Pbx2 bind the ephA8 regulatory sequence in vitro and in vivo; (iv) Meis2-Pbx2 complexes enhance the transcriptional activation of a reporter construct bearing the ephA8 regulatory region in a certain cellular context where HDACs are inhibited; and (v) EphA8 gene expression is significantly down-regulated in transgenic embryos expressing Meis1-En under the nestin promoter.
Although Meis3 was isolated from a human fetal-brain cDNA library, subsequent experiments have indicated that Meis2 transcripts are more abundant in the developing midbrain than either Meis1 or Meis3. However, the ability of Meis1 and Meis3 to bind the 30-bp ephA8 regulatory element was almost equivalent to that of Meis2 when tested using EMSA (data not shown). On the other hand, both the 1-kb XbaI/NheI fragment and the 180-bp fragment of the ephA8 regulatory region consistently gave high levels of lacZ expression in the diencephalon in E11.5 transgenic embryos. This result may be consistent with the possibility that in the diencephalon, Meis1 and Pbx synergistically bind the ephA8 regulatory sequence to exert transcriptional activation of the lacZ reporter gene. In contrast, embryos carrying the wild-type 10-kb ephA8 transgene showed specific lacZ staining only in the anterior region of the midbrain. These results strongly suggest that other regions of the 10-kb ephA8 genomic DNA, not present in the 1-kb XbaI/NheI fragment, contain elements that repress expression outside of the midbrain. Consistent with these data, our ChIP assay demonstrated that neither Meis nor Pbx is recruited to the ephA8 regulatory region in the diencephalon. Although the negative regulatory element responsible for this repression remains to be identified, it seems plausible that this region is critical for inducing changes in the chromatin organization of the ephA8 regulatory sequence and its transcriptional silencing during the development of the diencephalon.
Our RT-PCR analysis suggests that Meis2b is the predominantly expressed isoform of Meis2 in the developing midbrain. In contrast to the exclusive expression of Meis2 in the mesencephalon, Meis1 does not appear to be expressed in the mesencephalon, based on our in situ hybridization and RT-PCR analyses. This finding is also consistent with the published data, in which lacZ expression from the Meis1 promoter was not detected in the mesencephalons of heterozygous or homozygous mutant embryos (16). Other members of the TALE family of proteins may be involved in the regulation of ephA8 gene expression during the development of the mesencephalon. Although Pbx2 was investigated as a cofactor for Meis2b in this study, it is possible that Pbx1 is involved in the regulation of ephA8 gene expression, since it is highly expressed in the mesencephalon. Likewise, Pbx3 may be a candidate to act as an upstream regulator, since it has previously been shown to be expressed in the mesencephalon at similar stages (7). Prep1, a member of another TALE subclass, is also expressed in the diencephalon and mesencephalon (see Fig. S1B posted at http://sookmyung.ac.kr/∼scpark/Paper/MCB/MCB.zip) (9). However, Prep1 is not able to effectively bind an ephA8 point mutant as well as wild-type DNA in EMSA analysis, although it effectively binds the probe used as a positive control (see Fig. S3B and C). In addition, we have not been able to demonstrate the in vivo binding of Prep1 to the ephA8 regulatory sequence using a ChIP assay (data not shown). These results suggest that Prep1 is not an authentic ephA8 regulatory-sequence binding factor. Taken together, these expression analyses and biochemical studies led us to conclude that Meis2 and Pbx1/2 are the most likely candidates to act as an upstream regulator for the regulation of ephA8 gene expression.
Consistent with previous reports, it appeared that binding of Meis2b alone to the ephA8 regulatory element is very weak (3, 18, 45, 48, 55). In contrast, incubation with an antibody specific for the N terminus of the Meis protein produced stable supershifted complexes, which were much more enhanced than Meis-DNA complexes. Previous studies have shown that Meis and Pbx interact through their N termini for their synergistic binding to target DNA sequences (48, 49). Since the Meis antibody used for EMSA recognizes the N terminus of Meis2b, it is conceivable that the Meis antibody mimics Pbx, leading to stable supershifted complexes. Interestingly, we have consistently observed that Meis2b and Pbx2 together form faster-migrating complexes on a 44-bp ephA8 regulatory sequence and that DNA binding is absolutely required for the formation of this complex. The faster-migrating structure is likely to represent a heteromeric DNA complex in which both Meis2b and Pbx2 proteins are bound on the same double-stranded oligonucleotide. It appears that a DNA-bound Meis-Pbx heterocomplex migrates faster in EMSA, as previously shown. However, we were not able to observe the higher-order complexes reported by others. Together, these findings are perfectly consistent with previous reports that Meis proteins are highly preferred endogenous DNA binding partners for Pbx proteins (3, 18, 25, 45, 48). An important question is whether the in vitro binding of the Meis-Pbx complex is consistent with the in vivo reporter activity. As shown in Fig. 3, mutations at the third (M3) or fourth (M4) nucleotide in the CGGTCA sequence caused complete abrogation of in vivo reporter activity in the anterior region of the mesencephalon. Consistent with the in vivo effects of M2 and M4 point mutations, the Meis2b-Pbx2 complex did not effectively bind M4 DNA, although it bound M2 (see Fig. S3C posted at http://sookmyung.ac.kr/∼scpark/Paper/MCB/MCB.zip). These results strongly suggest that our in vivo reporter data correlate well with in vitro binding data, although we have not tested all of the point mutants depicted in Fig. 3. The transcriptional activity of the Cn4 construct compared to the Cn1 construct in the transfected HEK293 cells was attenuated but not completely eliminated, and these data are partially consistent with the essential roles of the CGGTCA sequence for ephA8 gene regulation. However, this result is not correlated with the in vivo evidence that the single Meis binding site in the CGGTCA sequence is essential for in vivo reporter activity. This paradox may result from the possibility that the activity of the ephA8 regulatory sequence could be dependent upon the nature of the proteins bound at the different sites, the natures of the coactivators or corepressors recruited, and the specific chromatin structure.
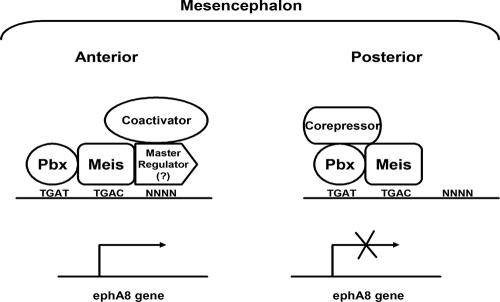
The ChIP assays of mesencephalic cell extracts described here clearly indicate that Meis and Pbx are recruited to the ephA8 regulatory sequence throughout the dorsal mesencephalon. These results suggest that the Meis and Pbx proteins serve a broader and more general role as transcription cofactors during the development of the mesencephalon. Interestingly, it has been demonstrated that Meis and Pbx may penetrate repressive chromatin to mark specific genes for activation (1). For example, Meis and Pbx act as cofactors to Hox proteins and MyoD, suggesting that they may act as pioneer factors, penetrating repressive chromatin and marking specific genes for activation by master regulators, such as Hox proteins and MyoD (1, 17, 45, 48, 49, 55, 56). Might Meis and Pbx play similar roles in the ephA8 regulatory sequence? This may be possible, although we do not yet have direct and conclusive evidence on the issue. This is complicated by the fact that Meis and Pbx are expressed in the posterior region of the dorsal mesencephalon, where the ephA8 gene is silent. It is well known that Pbx proteins bind several corepressors or HDACs (17, 42), and consistent with these reports, we found that coexpression of Meis2 and Pbx2 in HEK293 cells appears inert in their ability to induce transcriptional activation through the ephA8 regulatory element. By contrast, the HDAC inhibitor TSA induced reporter gene expression through the ephA8 regulatory sequence. Additionally, because they are known to associate with Meis or Pbx and are expressed in the posterior region of the mesencephalon, En proteins may be responsible for silencing ephA8 gene expression in the posterior region of the mesencephalon (23, 37, 38). More importantly, a dominant-negative mutant form of Meis, Meis1-En, consistently down-regulated the endogenous ephA8 gene expression. Thus, it seems plausible that Meis-Pbx-En heterotrimeric complexes mediate negative regulation of ephA8 gene expression, although this hypothesis remains to be further tested. Also, what are the master regulators critical for the transcriptional activation of the ephA8 gene in the anterior region of the mesencephalon, in addition to the Meis/Pbx “mark”? Interestingly, the Meis2-VP16-Pbx2 complex did not mediate transcriptional activation in transfected cells (see Fig. S4A posted at http://sookmyung.ac.kr/∼scpark/Paper/MCB/MCB.zip), although it bound to the 44-mer DNA and produced a supershifted band of slower mobility when mixed with Meis antibody (see Fig. S4B). These results suggest that a sequence-specific cofactor may interact with the Meis-Pbx complex to mediate histone acetylation at the ephA8 locus and that this factor is not present in 293 cells. This is partially consistent with our Cn3 transgenic embryo, showing that its lacZ expression is not posteriorly expanded in the anterior region of the mesencephalon compared with those of Cn1 and Cn2, possibly due to a lack of additional regulatory sequences. This sequence-specific transcription factor might bind both Pbx/Meis and an adjacent specific element. Although this master regulator remains to be identified, it may recruit coactivators to relieve any repressor activity associated with the Meis/Pbx mark in the anterior region of the mesencephalon. Taken together, our current model, based on the MyoD and Hox transcription factor paradigm described above, is that Meis and Pbx tightly associate with the ephA8 regulatory sequence and require an additional, as yet unidentified, master regulator to ensure the specific activation of the ephA8 gene (Fig. 10). The identification of this master regulator and its specific binding sequence should illuminate the essential roles of Meis and Pbx for the regulation of the ephA8 gene, as well as vertebrate mesencephalic development.
FIG. 10.
A model for transcriptional regulation of the ephA8 gene by Meis-Pbx complexes and a putative master regulator. Meis-Pbx complexes penetrate and specifically bind the ephA8 regulatory sequences in the dorsal mesencephalon. However, the ephA8 promoter remains inactive in the posterior region, perhaps as a result of corepressors or HDACs recruited by Meis-Pbx. By contrast, in the anterior region, an unidentified master transcription factor binds a specific sequence around Meis and Pbx binding sites and recruits coactivators to activate ephA8 transcription.
Acknowledgments
We are indebted to Todd Kroll and Todd McLaughlin for their valuable discussions, to Young Yang for Meis2 isoform constructs, and to Richard Maas for the Meis1-En construct.
This research was supported by a grant (M103KV010008 04K2201 00840) from the Brain Research Center of the 21st Century Frontier Research Program funded by the Ministry of Science and Technology (MOST), a grant (R11-2005-017-01002-0) from the SRC program funded by MOST/KOSEF (Research Center for Women's Diseases), and the Korea Research Foundation Grant funded by the Korean Government (MOEHRD, Basic Research Promotion Fund) (KRF-2005-015-C00373). Sungbo Shim was supported by a Korea Research Foundation Grant funded by the Korean Government (MOEHRD) (KRF-2005-037-C00039). Jieun Kim and Yujin Kim were supported by the Brain Korea 21 project in 2006.
Footnotes
Published ahead of print on 18 December 2006.
REFERENCES
- 1.Berkes, C., D. A. Bergstrom, B. H. Penn, K. J. Seaver, P. S. Knoepfler, and S. J. Tapscott. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14:465-477. [DOI] [PubMed] [Google Scholar]
- 2.Brown, A., P. A. Yates, P. Burrola, D. Ortuno, A. Vaidya, T. M. Jessell, S. L. Pfaff, D. D. M. O'Leary, and G. Lemke. 2000. Topographic mapping from the retina to the midbrain is controlled by relative but not absolute levels of EphA receptor signaling. Cell 102:77-88. [DOI] [PubMed] [Google Scholar]
- 3.Chang, C. P., Y. Jacobs, T. Nakamura, N. Jenkins, N. G. Copeland, and M. L. Cleary. 1997. Meis proteins are major in vivo DNA binding partners for wt but not chimeric Pbx proteins. Mol. Cell. Biol. 17:5679-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, J., and H. E. Ruley. 1998. An enhancer element in the EphA2(Eck) gene sufficient for rhombomere-specific expression is activated by HOXA1 and HOXB1 homeobox proteins. J. Biol. Chem. 273:24670-24675. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, H., M. Nakamoto, A. D. Bergemann, and J. G. Flanagan. 1995. Complementary gradients in expression and binding of ELF-1 and Mek4 in development of the topographic retinotectal projection map. Cell 82:371-381. [DOI] [PubMed] [Google Scholar]
- 6.Cooke, J. E., H. A. Kemp, and C. B. Moens. 2005. EphA4 is required for cell adhesion and rhombomere-boundary formation in the zebrafish. Curr. Biol. 15:536-542. [DOI] [PubMed] [Google Scholar]
- 7.Di Giacomo, G., M. Koss, T. D. Capellini, A. Brendolan, H. Popperl, and L. Selleri. 2006. Spatio-temporal expression of Pbx3 during mouse organogenesis. Gene Expr. Patterns 6:747-757. [DOI] [PubMed] [Google Scholar]
- 8.Dufour, A., J. Egea, K. Kullander, R. Klein, and P. Vanderhaeghen. 2006. Genetic analysis of EphA-dependent signaling mechanisms controlling topographic mapping in vivo. Development 133:4415-4420. [DOI] [PubMed] [Google Scholar]
- 9.Ferretti, E., F. Cambronero, S. Tumpel, E. Longobardi, L. M. Wiedemann, F. Blasi, and R. Krumlauf. 2005. Hoxb1 enhancer and control of rhombomere 4 expression: complex interplay between PREP1-PBX1-HOXB1 binding sites. Mol. Cell. Biol. 25:8541-8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flanagan, J. G. 2006. Neural map specification by gradients. Curr. Opin. Neurobiol. 16:59-66. [DOI] [PubMed] [Google Scholar]
- 11.Foo, S. S., C. J. Turner, S. Adams, A. Compagni, D. Aubyn, N. Kogata, P. Lindblom, M. Shani, D. Zicha, and R. H. Adams. 2006. Ephrin-B2 controls cell motility and adhesion during blood-vessel-wall assembly. Cell 124:161-173. [DOI] [PubMed] [Google Scholar]
- 12.Gu, C., S. Shim, J. Shin, J. Kim, J. Park, K. Han, and S. Park. 2005. The EphA8 receptor induces sustained MAP kinase activation to promote neurite outgrowth in neuronal cells. Oncogene 24:4243-4256. [DOI] [PubMed] [Google Scholar]
- 13.Hansen, M. J., G. E. Dallal, and J. G. Flanagan. 2004. Retinal axon response to ephrin-As shows a graded, concentration-dependent transition from growth promotion to inhibition. Neuron 42:717-730. [DOI] [PubMed] [Google Scholar]
- 14.Herrera, E., L. Brown, J. Aruga, R. A. Rachel, G. Dolen, K. Mikoshiba, S. Brown, and C. A. Mason. 2003. Zic2 patterns binocular vision by specifying the uncrossed retinal projection. Cell 114:545-557. [DOI] [PubMed] [Google Scholar]
- 15.Hirashima, M., and T. Suda. 2006. Differentiation of arterial and venous endothelial cells and vascular morphogenesis. Endothelium 13:137-145. [DOI] [PubMed] [Google Scholar]
- 16.Hisa, T., S. E. Spence, R. A. Rachel, M. Fujita, T. Nakamura, J. M. Ward, D. E. Devor-Henneman, Y. Saiki, H. Kutsuna, L. Tessarollo, N. A. Jenkins, and N. G. Copeland. 2004. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 23:450-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, H., M. Rastegar, C. Bodner, S. L. Goh, I. Rambaldi, and M. Featherstone. 2005. MEIS C termini harbor transcriptional activation domains that respond to cell signaling. J. Biol. Chem. 280:10119-10127. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs, Y., C. A. Schnabel, and M. L. Cleary. 1999. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol. Cell. Biol. 19:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong, J., S. Choi, C. Gu, H. Lee, and S. Park. 2000. Genomic structure and promoter analysis of the mouse EphA8 receptor tyrosine kinase gene. DNA Cell Biol. 19:291-300. [DOI] [PubMed] [Google Scholar]
- 20.Kania, A., and T. M. Jessell. 2003. Topographic motor projections in the limb imposed by LIM homeodomain protein regulation of ephrin-A:EphA interactions. Neuron 38:581-596. [DOI] [PubMed] [Google Scholar]
- 21.Kimura, C., N. Takeda, M. Suzuki, M. Oshimura, S. Aizawa, and I. Matsuo. 1997. cis-acting elements conserved between mouse and pufferfish Otx2 genes govern the expression in mesencephalic neural crest cells. Development 124:3929-3941. [DOI] [PubMed] [Google Scholar]
- 22.Knoepfler, P. S., K. R. Calvo, H. Chen, S. E. Antonarakis, and M. P. Kamps. 1997. Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc. Natl. Acad. Sci. USA 94:14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi, M., M. Fujioka, E. N. Tolkunova, D. Deka, M. Abu-Shaar, R. S. Mann, and J. B. Jaynes. 2003. Engrailed cooperates with extradenticle and homothorax to repress target genes in Drosophila. Development 130:741-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koo, J., S. Shim, C. Gu, O. Yoo, and S. Park. 2003. Identification of an enhancer region in the mouse ephA8 locus directing expression to the anterior region of the dorsal mesencephalon. Dev. Dyn. 226:596-603. [DOI] [PubMed] [Google Scholar]
- 25.Kurant, E., C.-Y. Pai, R. Sharf, N. Halachmi, Y. H. Sun, and A. Salzberg. 1998. Dorsothonal/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning the embryonic PNS. Development 125:1037-1048. [DOI] [PubMed] [Google Scholar]
- 26.Lothian, C., N. Prakash, U. Lendahl, and G. M. Wahlström. 1999. Identification of both general and region-specific embryonic CNS enhancer elements in the nestin promoter. Exp. Cell Res. 248:509-519. [DOI] [PubMed] [Google Scholar]
- 27.Lumpkin, E. A., T. Collisson, P. Parab, A. Omer-Abdalla, H. Haeberle, P. Chen, A. Doetzlhofer, P. White, A. Groves, N. Segil, and J. E. Johnson. 2003. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr. Patterns 3:389-395. [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Barbera, J. P., M. Signore, P. P. Boyl, E. Puelles, D. Acampora, R. Gogoi, F. Schubert, A. Lumsden, and A. Simeone. 2001. Regionalisation of anterior neuroectoderm and its competence in responding to forebrain and midbrain inducing activities depend on mutual antagonism between OTX2 and GBX2. Development 128:4789-4800. [DOI] [PubMed] [Google Scholar]
- 29.Matsunaga, E., I. Araki, and H. Nakamura. 2000. Pax6 defines the di-mesencephalic boundary by repressing En1 and Pax2. Development 127:2357-2365. [DOI] [PubMed] [Google Scholar]
- 30.Mercader, N., E. M. Tanaka, and M. Torres. 2005. Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development 132:4131-4142. [DOI] [PubMed] [Google Scholar]
- 31.Mignone, J. L., V. Kukekov, A. S. Chiang, D. Steindler, and G. Enikolopov. 2004. Neural stem and progenitor cells in nestin-GFP transgenic mice. J. Comp. Neurol. 469:311-324. [DOI] [PubMed] [Google Scholar]
- 32.Moskow, J. J., F. Bullrich, K. Huebner, I. O. Daar, and A. M. Buchberg. 1995. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol. Cell. Biol. 15:5434-5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura, H. 2001. Regionalization of the optic tectum: combinations of gene expression that define the tectum. Trends Neurosci. 24:32-39. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura, H., and S. Sugiyama. 2004. Polarity and laminar formation of the optic tectum in relation to retinal projection. J. Neurobiol. 59:48-56. [DOI] [PubMed] [Google Scholar]
- 35.Pak, W., R. Hindges, Y. S. Lim, S. L. Pfaff, and D. D. O'Leary. 2004. Magnitude of binocular vision controlled by islet-2 repression of a genetic program that specifies laterality of retinal axon pathfinding. Cell 119:567-578. [DOI] [PubMed] [Google Scholar]
- 36.Park, S., J. Frisen, and M. Barbacid. 1997. Aberrant axonal projections in mice lacking EphA8 (Eek) tyrosine protein kinase receptors. EMBO J. 16:3106-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peltenburg, L. T. C., and C. Murre. 1996. Engrailed and Hox homeodomain proteins contain a related Pbx interaction motif that recognizes a common structure present in Pbx. EMBO J. 15:3385-3393. [PMC free article] [PubMed] [Google Scholar]
- 38.Peltenburg, L. T. C., and C. Murre. 1997. Specific residues in the Pbx homeodomain differentially modulate the DNA-binding activity of Hox and Engrailed proteins. Development 124:1089-1098. [DOI] [PubMed] [Google Scholar]
- 39.Poliakov, A., M. Cotrina, and D. G. Wilkinson. 2004. Diverse roles of Eph receptors and ephrins in the regulation of cell migration and tissue assembly. Dev. Cell. 7:465-480. [DOI] [PubMed] [Google Scholar]
- 40.Qin, P., J. M. Haberbusch, Z. Zhang, K. J. Soprano, and D. R. Soprano. 2004. Pre-B cell leukemia transcription factor (PBX) proteins are important mediators for retinoic acid-dependent endodermal and neuronal differentiation of mouse embryonal carcinoma P19 cells. J. Biol. Chem. 279:16263-16271. [DOI] [PubMed] [Google Scholar]
- 41.Rashid, T., A. L. Upton, A. Blentic, T. Ciossek, B. Knoll, I. D. Thompson, and U. Drescher. 2005. Opposing gradients of ephrin-As and EphA7 in the superior colliculus are essential for topographic mapping in the mammalian visual system. Neuron 47:57-69. [DOI] [PubMed] [Google Scholar]
- 42.Saleh, M., I. Rambaldi, X. J. Yang, and M. S. Featherstone. 2000. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 20:8623-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salsi, V., and V. Zappavigna. 2006. Hoxd13 and Hoxa13 directly control the expression of the EphA7 ephrin tyrosine kinase receptor in developing limbs. J. Biol. Chem. 281:1992-1999. [DOI] [PubMed] [Google Scholar]
- 44.Santiago, A., and C. A. Erickson. 2002. Ephrin-B ligands play a dual role in the control of neural crest cell migration. Development 129:3621-3632. [DOI] [PubMed] [Google Scholar]
- 45.Schnabel, C. A., Y. Jacobs, and M. L. Cleary. 2000. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene 19:608-616. [DOI] [PubMed] [Google Scholar]
- 46.Schulte, D., and C. L. Cepko. 2000. Two homeobox genes define the domain of EphA3 expression in the developing chick retina. Development 127:5033-5045. [DOI] [PubMed] [Google Scholar]
- 47.Shamim, H., R. Mahmood, C. Logan, P. Doherty, A. Lumsden, and I. Mason. 1999. Sequential roles for Fgf4, En1 and Fgf8 in specification and regionalisation of the midbrain. Development 126:945-959. [DOI] [PubMed] [Google Scholar]
- 48.Shanmugam, K., N. C. Green, I. Rambaldi, H. U. Saragovi, and M. S. Featherstone. 1999. PBX and MEIS as Non-DNA-binding partners in trimeric complexes with HOX proteins. Mol. Cell. Biol. 19:7577-7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen, W. F., J. C. Montgomery, S. Rozenfeld, J. J. Moskow, H. J. Lawrence, A. M. Buchberg, and C. Largman. 1997. AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol. Cell. Biol. 17:6448-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen, W. F., S. Rozenfeld, A. Kwong, L. G. Kömüves, H. J. Lawrence, and C. Largman. 1999. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol. Cell. Biol. 19:3051-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shigetani, Y., J. I. Funahashi, and H., Nakamura. 1997. En-2 regulates the expression of the ligands for Eph type tyrosine kinases in chick embryonic tectum. Neurosci. Res. 27:211-217. [DOI] [PubMed] [Google Scholar]
- 52.Swift, G. H., Y. Liu, S. D. Rose, L. J. Bischof, S. Steelman, A. M. Buchberg, C. V. Wright, and R. J. MacDonald. 1998. An endocrine-exocrine switch in the activity of the pancreatic homeodomain protein PDX1 through formation of a trimeric complex with PBX1b and MRG1 (MEIS2). Mol. Cell. Biol. 9:5109-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi, H., T. Shintani, H. Sakuta, and M. Noda. 2003. CBF1 controls the retinotectal topographical map along the anteroposterior axis through multiple mechanisms. Development 130:5203-5215. [DOI] [PubMed] [Google Scholar]
- 54.Theil, T., M. Frain, P. Gilardi-Hebenstreit, A. Flenniken, P. Charnay, and D. G. Wilkinson. 1998. Segmental expression of the EphA4(Sek-1) receptor tyrosine kinase in the hindbrain is under direct transcriptional control of Krox-20. Development 125:443-452. [DOI] [PubMed] [Google Scholar]
- 55.Vlachakis, N., S. K. Choe, and C. G. Sagerstrom. 2001. Meis3 synergizes with Pbx4 and Hoxb1 in promoting hindbrain fates in the zebrafish. Development 128:1299-1312. [DOI] [PubMed] [Google Scholar]
- 56.Waskiewicz, A. J., H. A. Rikhof, R. E. Hernandez, and C. B. Moens. 2001. Zebrafish Meis functions to stabilize Pbx proteins and regulate hindbrain patterning. Development 128:4139-4151. [DOI] [PubMed] [Google Scholar]
- 57.Wilkinson, D. G. 1998. In situ hybridization: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 58.Wilkinson, D. G. 2001. Multiple roles of Eph receptors and ephrins in neural development. Nat. Rev. Neurosci. 2:155-164. [DOI] [PubMed] [Google Scholar]
- 59.Williams, S. E., F. Mann, L. Erskine, T. Sakurai, S. Wei, D. J. Rossi, N. W. Gale, C. E. Holt, C. A. Mason, and M. Henkemeyer. 2003. Ephrin-B2 and EphB1 mediate retinal axon divergence at the optic chiasm. Neuron 39:919-935. [DOI] [PubMed] [Google Scholar]
- 60.Yang, Y., C. K. Hwang, U. M. D'Souza, S. Lee, E. Junn, and M. M. Mouradian. 2000. Three-amino acid extension loop homeodomain proteins Meis2 and TGIF differentially regulate transcription. J. Biol. Chem. 275:20734-20741. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, J., and S. E. Hughes. 2006. Role of the ephrin and Eph receptor tyrosine kinase families in angiogenesis and development of the cardiovascular system. J. Pathol. 208:453-461. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, X., A. Friedman, S. Heaney, P. Purcell, and R. L. Maas. 2002. Meis1 homeoprotein directly regulates Pax 6 during vertebrate lens morphogenesis. Genes Dev. 16:2097-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]