Abstract
The mouse gene Recql is a member of the RecQ subfamily of DEx-H-containing DNA helicases. Five members of this family have been identified in both humans and mice, and mutations in three of these, BLM, WRN, and RECQL4, are associated with human diseases and a cellular phenotype that includes genomic instability. To date, no human disease has been associated with mutations in RECQL and no cellular phenotype has been associated with its deficiency. To gain insight into the physiological function of RECQL, we disrupted Recql in mice. RECQL-deficient mice did not exhibit any apparent phenotypic differences compared to wild-type mice. Cytogenetic analyses of embryonic fibroblasts from the RECQL-deficient mice revealed aneuploidy, spontaneous chromosomal breakage, and frequent translocation events. In addition, the RECQL-deficient cells were hypersensitive to ionizing radiation, exhibited an increased load of DNA damage, and displayed elevated spontaneous sister chromatid exchanges. These results provide evidence that RECQL has a unique cellular role in the DNA repair processes required for genomic integrity. Genetic background, functional redundancy, and perhaps other factors may protect the unstressed mouse from the types of abnormalities that might be expected from the severe chromosomal aberrations detected at the cellular level.
DNA helicases are ubiquitous enzymes that unwind DNA in an ATP-dependent and directionally specific manner. The unwinding of double-stranded DNA is essential for the processes of DNA repair, recombination, transcription, and DNA replication (1, 19, 30, 35, 45). One family of helicases is the RecQ subfamily of DEx-H-containing DNA helicases, of which Escherichia coli RecQ is the prototype member. All members of this family contain an approximately 450-amino-acid domain that contains seven helicase motifs of the DExH box superfamily (see Fig. S1 in the supplemental material). E. coli RecQ is a 3′→5′ helicase involved in the homologous recombination (HR) and double-strand break (DSB) repair mediated by the RecF pathway and suppression of illegitimate recombination (51). Other RecQ-related proteins include SGS1 in Saccharomyces cerevisiae, which plays a role in recombination, chromosome partitioning, and genome stability (32, 53), and RQH1 in Schizosaccharomyces pombe, which is required to prevent recombination and suppression of inappropriate recombination (47).
Unlike E. coli, S. cerevisiae, and S. pombe, which only have one RecQ helicase, certain bacteria (e.g., Bacillus subtilis [12] and Porphyromonas gingivalis [34]) and higher eukaryotes express multiple RecQ-related helicases (45). Five such helicases have been described in humans. The first described was RECQL or RECQL1 (39, 40, 43), followed by BLM (11, 44), WRN (59), RECQL4 (20, 21), and RECQL5 (21). Three of these have been shown to be implicated in distinct heritable diseases. BLM is mutated in Bloom syndrome, which is characterized by growth retardation, sunlight sensitivity, immunodeficiency, genomic instability, and a high incidence of cancer (11). WRN is mutated in Werner syndrome, which is characterized by premature aging, genomic instability, and a high incidence of cancer (59). RECQL4 is mutated in Rothmund-Thomson syndrome, which is characterized by skin rash, small stature, skeletal dysplasias, chromosomal instability, premature aging, and a high incidence of cancer (22). All three of these syndromes involve genome instability and a predisposition to cancer. Although these three human members of the RecQ helicase family share the conserved helicase domain, they are involved in clinically distinct syndromes, suggesting that the human RecQ homologues can participate in some nonoverlapping physiological functions.
All of the RecQ helicases contain a helicase domain that is closely related to that of the E. coli RecQ helicase, suggesting that they may share common biochemical functions. However, very little is known about the prototype mammalian enzyme, RECQL, in the physiology of the intact animal and no human syndrome has been associated with its deficiency. To begin to address the question of its physiological function, we disrupted Recql in mice. Mice deficient in RECQL appeared to be normal and fertile, with normal telomere lengths. However, we found that RECQL had a profound role in the maintenance of chromosomal stability in primary fibroblasts derived from RECQL-deficient embryos, suggesting that RECQL has a unique function distinct from that of the other RecQ helicases. Absence of RECQL led to the accumulation of DNA strand breaks and persistent Rad51 foci. Moreover, RECQL deficiency resulted in cellular sensitivity to ionizing radiation (IR). These findings are consistent with a role of RECQL in DNA repair, to process inappropriate recombination intermediates that might arise during replication and maintain genomic stability.
MATERIALS AND METHODS
Construction of targeting vector and generation of chimeric mice.
A 129SV mouse genomic library in lambda FIXII (Stratagene, La Jolla, CA) was screened by standard techniques (29) with 5′ and 3′ human RECQL cDNA probes (39, 40). A 4.6-kb NsiI/NotI fragment of the original 18-kb Recql clone was found to contain several exons and was used to make a homologous targeting vector. A 2-kb 3′ EcoRI/HindIII fragment from the 4.6-kb fragment was subcloned into the BamHI site of pJN52 (kindly provided by B. Koller, University of North Carolina) after the addition of BamHI linkers. A 1.5-kb 5′ StuI/Asp718 fragment of the 4.6-kb fragment was subcloned into the NotI site of pJN52. The resulting targeting vector, pJN52/RECQL, had PGKNeo replacing a 474-bp portion of the gene that included most of the exon encoding all of helicase domain IV and part of helicase domain V. This deletion in the mouse gene was at the approximate midpoint of the protein-coding region, based on sequence homology with the human RECQL cDNA.
The targeting vector was linearized with HindIII and electroporated into R1 embryonic stem (ES) cells. Genomic DNA from ES cells was analyzed by PCR to detect HR. Primers for the 5′ end of Recql (F1; 5′-GCCAGGAATGTTTCAGTCAAGAGG-3′) and Neo (R4; 5′-GTTGTGCCCAGTCATAGCCGAATAG-3′) were used to detect targeted ES cells (2.7-kb PCR product). PCR assay conditions included a 25-μl total reaction volume containing 50 ng genomic DNA, 16 mM ammonium sulfate, 67 mM Tris-HCl (pH 8.8), 0.01% (vol/vol) Tween 20, 5 mM β-mercaptoethanol, 1.5 mM MgCl2, 1 mg/ml bovine serum albumin, 0.2 mM deoxynucleoside triphosphates, 10 mM F1, 10 mM R4, 10% (vol/vol) dimethyl sulfoxide, and 1 U GeneChoice Taq DNA polymerase (PGC Scientifics Corp., Gaithersburg, MD). PCR cycling conditions were 93°C for 2 min; 30 cycles of 93°C for 30 s, 55°C for 30 s, and 65°C for 3 min; and a final incubation at 65°C for 10 min. Two targeted ES cell lines were injected into C57BL/6 blastocysts to generate chimeric mice. Chimeric mice were crossed with C57BL/6 females, and offspring heterozygous (+/−) for Recql were interbred to generate homozygous (−/−) knockout mice (B6;129-Recqltm1Pjb). Genomic DNA was isolated from mouse tail pieces and initially analyzed by Southern blot hybridization. Genomic DNA was digested with EcoRI, fractionated on a 0.7% agarose gel, transferred to NytranPlus (Schleicher and Schuell, Keene, NH), and hybridized with an 877-bp EcoRI Recql probe (see Fig. 2). Routine genotyping of offspring was done by PCR with Recql primers F2 (5′-GGAGAAGGAGGGTAAAGTTGGGAG-3′) and R1 (5′-GGAGTTATACATCTCCAAGCCCTG-3′) to detect the endogenous gene (0.4-kb PCR product), primers F2 and R4 to detect the Neo-interrupted gene (1.09 kb), and primers F2 and R2 to detect both the endogenous (0.8-kb) and Neo-interrupted (2.4-kb) genes. All mouse experiments were conducted according to the U.S. Public Health Service policy on the humane care and use of laboratory animals. All animal procedures used in this study were approved by the National Institute of Environmental Health Sciences Institutional Animal Care and Use Committee.
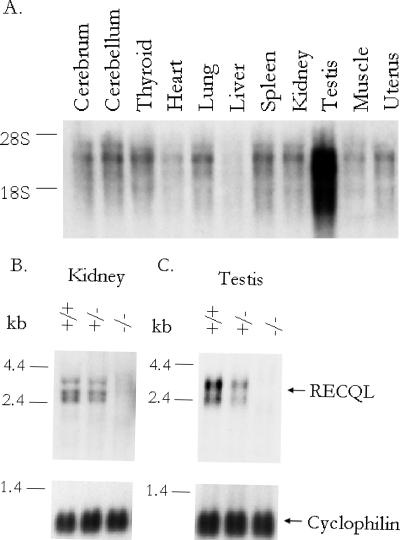
FIG. 2.
Tissue distribution of RECQL mRNA. (A) Total cellular RNA (15 μg) isolated from various mouse tissues was fractionated on a 1.2% formaldehyde-agarose gel, transferred to NytranPlus, and hybridized with a mouse RECQL cDNA probe. The 28S and 18S markers show the positions of the major rRNA species on the stained gel. (B and C) Expression of RECQL mRNA in the kidneys (B) and testes (C) of Recql+/+, Recql+/−, and Recql−/− mice. Total RNA (15 μg) was isolated from the indicated adult mouse tissues and fractionated on a 1.2% formaldehyde-agarose gel. The blots were hybridized either with the mouse RECQL cDNA probe or with cyclophilin. The locations of RNA molecular size markers are indicated to the left of each blot.
RNA isolation and Northern analysis.
Total cellular RNA was isolated from various tissues from mixed genetic background mice (50% C57BL/6 and 50% 129) by a modification of the single-step guanidinium thiocyanate procedure (7, 48), fractionated (15 μg) on a 1.2% formaldehyde-agarose gel, transferred to NytranPlus, and hybridized with random-primed, α-32P-labeled probes as previously described (49). Northern blots were hybridized with a 602-bp mouse RECQL cDNA or with a mouse cyclophilin cDNA (3) to monitor gel loading. The 602-bp mouse RECQL cDNA probe was generated by PCR with an NIH 3T3 mouse cDNA library in lambda ZAPII (Stratagene, La Jolla, CA) and Recql primers F3 (5′-GATGGAATTCGCTCTTGGCATCTTGAAGCGC-3′) and R3 (5′-CTAGTCTAGACTGCCCCACGTTCTCCATGAC-3′). Some Northern blots were also hybridized with cDNA probes for mouse WRN, BLM, RECQL4, and RECQL5. Mouse BLM and RECQL4 probes were generated by reverse transcription-PCR. Reverse transcription was carried out with 1 μg of mouse testis total RNA, an anchored oligo(dT) primer (T18VN), and Superscript II RNase H− reverse transcriptase (Invitrogen Life Technologies, Carlsbad, CA). PCR was performed with primers based on the sequence of mouse BLM (accession number Z98263; forward primer 5′-ATGAAGCTTGTATCTTCTCACTACTTTG-3′ and reverse primer 5′-GATCGGTACCCAAGACTAACAGCTTTCTATTCAC-3′) and human RECQL4 (accession number AB006532; forward primer 5′-GATGAAGCTTGTGCTTGTGGAGTTCAGTGAG-3′ and reverse primer 5′-GATCGGTACCGTATTTTCTCCAGAAGCGTCGGTCCTG-3′). Mouse WRN (1.4-kb SalI/StuI fragment of accession number BE569718) and RECQL5 (1.3-kb NotI/EcoRI fragment of accession number AW824665) probes were isolated from expressed sequence tags obtained from the IMAGE consortium. All PCR and expressed sequence tag clones were confirmed by sequencing with the ABI Prism dRhodamine Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA).
Isolation and analysis of MEFs.
Mouse embryo fibroblasts (MEFs) were isolated from fetal mice (75% C57BL/6 and 25% 129) at day 15.5 of gestation (E15.5), where E0.5 was the date of detection of the vaginal plug. MEFs were prepared from individual fetuses as previously described (50), and genotypes were determined from DNA isolated from the tails of the fetuses. To determine proliferation rates for the fibroblasts, 5 × 103 cells were plated in triplicate into 96-well plates in Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% (vol/vol) fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. On the days indicated, the cell proliferation reagent WST-1 (Roche Applied Science, Indianapolis, IN) was added to the wells and incubated for 2 h at 37°C and the amount of formazan dye generated by metabolically active cells was measured with a scanning multiwell spectrophotometer (Bio-Rad, Hercules, CA) at a wavelength of 450 nm (reference wavelength, 630 nm). Each time point represents the average of triplicate wells from two independent cultures of primary cells used at passage 4, and the error bars represent the standard error of the mean.
Histopathology and clinical pathology.
For a histopathological analysis of RECQL-deficient mice, 4-month-old littermates (one male and one female wild type [+/+] and one male and one female knockout [−/−]) of mixed (50% C57BL/6 and 50% 129) genetic background were euthanized. Gross examination of all organs was conducted at autopsy, and the carcasses and internal organs were fixed in 10% neutral buffered formalin and used for paraffin embedding, sectioning, and staining with hematoxylin and eosin. For hematological analysis, 9-week-old littermates (one male and two female +/+; one male and two female −/−) of mixed genetic background (75% C57BL/6 and 25% 129) were euthanized by CO2 inhalation, and blood was collected by cardiac puncture and transferred to EDTA-containing collection tubes. Blood samples were used for complete blood and white blood cell differential counts.
Telomere length analysis.
Splenocytes and thymocytes were isolated from Recql+/+, Recql+/−, and Recql−/− mice that had been backcrossed five times into 129SvEv (3% C57BL/6 and 97% 129) mice. Telomere lengths were measured by fluorescence in situ hybridization (FISH) and flow cytometry as previously described (2). Two 3-month-old animals of each genotype were analyzed.
DNA damage survival assays.
Passage 2 primary MEFs were seeded in quadruplicate at a density of 500 cells/well in 96-well plates. Cells were grown in DMEM (GIBCO-BRL) supplemented with 10% FBS (HyClone) and 1× penicillin-streptomycin (GIBCO-BRL). Cells were treated with increasing doses of IR with Gammacell 40 (Nordion International, Inc.), a 137Cs source emitting at a fixed dose rate of 0.82 Gy/min. Following treatment, cells were allowed to grow at 37°C for 5 days in 5% CO2. Plates were frozen at −80°C. Total DNA was quantified with CyQuant (Molecular Probes, Eugene, OR) and compared with untreated controls as an indication of cell growth as previously described (5, 16). Quantification of DNA with CyQuant was performed with a Fluorstar plate reader (B&L Systems) according to the manufacturer's instructions.
Rad51 and γH2AX focus formation in MEFs.
Primary wild-type and Recql−/− MEFs were grown to subconfluence on glass coverslips in 15-mm dishes in DMEM containing 10% FBS. For the detection of Rad51 foci, cells that had been either left untreated or treated with IR (12 Gy) were fixed 6 h after treatment with 4% paraformaldehyde, permeabilized with a 0.3% Triton X-100 solution, blocked with 10% normal goat serum in 1× phosphate-buffered saline (PBS), and incubated with anti-HsRad51 polyclonal antibody (1:1,200; Calbiochem) overnight at 4°C. Cells were washed with five changes of 1× PBS containing 0.1% Triton X-100 and incubated with a fluorescein isothiocyanate-labeled goat anti-rabbit immunoglobulin G secondary antibody (1:300; Vector Laboratories) for 1 h at room temperature. Cells were washed four times for 5 min in 1× PBS containing 0.1% Triton X-100, and coverslips were mounted with 4′,6′-diamidino-2-phenylindole (DAPI)-containing Vectashield mounting medium (Vector Laboratories) and analyzed by fluorescence microscopy. All images were captured on a Zeiss microscope with a black-and-white charge-coupled device (CCD) camera. The results shown are the means of two independent experiments. Cells were scored positive if they contained greater than five Rad51 foci per nucleus (untreated, 152 Recql+/+ cells and 212 Recql−/− cells; treated with 12 Gy, 137 Recql+/+ cells and 156 Recql−/− cells).
Immunostaining to detect γH2AX foci was performed with a phosphospecific anti-H2AX mouse monoclonal antibody. Cells that had been either left untreated or treated with IR (5 Gy) were fixed 30 min after treatment by incubation for 10 min in 3% formaldehyde and permeabilized with 0.1% Triton X-100. After blocking with 10% normal goat serum in 1× PBS, cells were incubated with mouse monoclonal anti-γ H2AX antibody (1:1,000; Upstate) in blocking solution overnight at 4°C. Cells were rinsed with three changes of 1× PBS containing 0.1% Triton X-100 and incubated for 1 h at room temperature with a rhodamine-conjugated goat anti-mouse immunoglobulin G secondary antibody (1:500; Vector Laboratories). Following four washes for 5 min in 1× PBS containing 0.1% Triton X-100, coverslips were mounted with Vectashield mounting medium with DAPI (Vector Laboratories) and images were captured as described above. The results shown are the means of two independent experiments. Cells were scored positive if they contained greater than five γH2AX foci per nucleus (untreated, 186 Recql+/+ cells and 232 Recql−/− cells; treated with 5 Gy, 187 Recql+/+ cells and 152 Recql−/− cells).
Cytogenetic analysis.
For chromosome analysis, primary wild-type and RECQL-deficient MEFs of passage 3 were used. Cells were treated with Colcemid (0.5 μg/ml Karyomax; Invitrogen) for 2 h and harvested by trypsinization. Metaphase spreads were prepared by swelling trypsinized cells in a hypotonic (0.57%) KCl solution for 12 min at 37°C. Cells after hypotonic treatment were fixed in 3:1 methanol-acetic acid, and the cells were washed twice in fresh fixative. An aliquot of fixed cells was dropped onto clean glass sides and air dried. For the detection of chromosomal anomalies such as chromosome number and aberrations, metaphase spreads were stained with 2% Giemsa (Sigma) and mounted with Vectashield containing DAPI. Images of randomly chosen metaphase spreads were captured with a Zeiss epifluorescence microscope equipped with a CCD camera.
For multicolor FISH (M-FISH) analyses, chromosome spreads prepared as described above were aged for a week at 4°C. M-FISH and posthybridization washings were performed according to the manufacturer's (MetaSystems, Belmont, MA) specifications. A cocktail of DNA probes specific for all the mouse chromosomes (24X Cyte; Metasystems) was used for M-FISH. After pepsin (0.005% in 0.01 N HCl) treatment for 2 min at 37°C and postfixation of the slides for 10 min in formaldehyde (3%), the slides were heat denatured together with the DNA probes (12 μl/slide) at 75°C for 5 min and incubated for 2 to 4 days at 37°C in a moist chamber. The slides were briefly washed in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) at 75°C to remove the nonspecific binding of the probe. The chromosomes were counterstained with DAPI. Images were captured with the Zeiss Axioplan 2 imaging fluorescence microscope and analyzed with ISIS imaging software (Metasystems). At least 40 metaphase spreads were analyzed for each cell line to characterize their chromosome constitution.
SCE assays.
For sister chromatid exchange (SCE) analysis, primary wild-type and Recql−/− MEFs (passage 3) in the exponential growth phase were grown for 48 h in the presence of 10 μM bromodeoxyuridine (BrdU; Sigma). Metaphase chromosomes were prepared as described before. Slides were aged for 3 days and stained with 5 μg/ml Hoechst 33258 (Sigma) for 10 min. After rinsing in water, slides were mounted and placed under a 120-W Plant Lite (General Electric Inc.) at a distance of 10 cm for 2 h. Slides were stained in 2% Giemsa (Sigma) for 20 min, washed, and examined under a Zeiss microscope with a black-and-white CCD camera. Digitally captured images of 20 differentially stained metaphase chromosome spreads per genotype were scored in a blinded fashion.
RESULTS
Generation and analysis of RECQL-deficient mice.
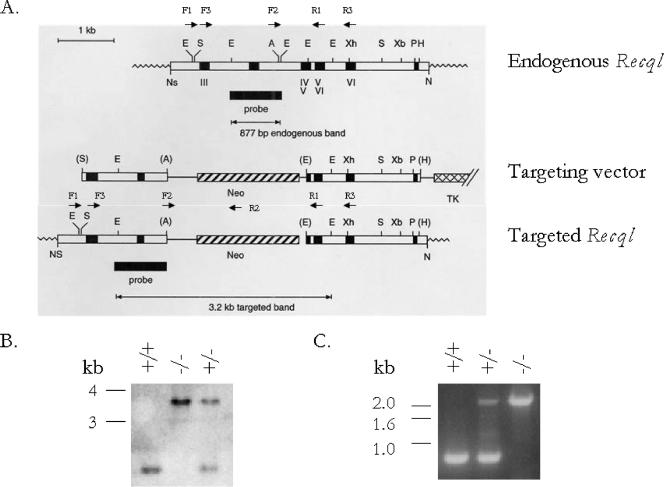
The targeting vector for HR was designed to replace a 474-bp portion of the gene that includes most of the exon encoding all of helicase domain IV and part of helicase domain V with a PGKNeo cassette (Fig. 1A). ES cells targeted with this vector (Fig. 1A) were used to make chimeric mice. Chimeric mice were bred to C57BL/6 mice, and heterozygous offspring were intercrossed. Matings of heterozygous mice of mixed genetic background (C57BL/6 and 129Sv) resulted in offspring with a Mendelian distribution of +/+, +/−, and −/− genotypes. Typical Southern blot and PCR genotyping results are shown in Fig. 1B and C, respectively. Wild-type and RECQL-deficient mice were indistinguishable in appearance and observable behavior. In addition, both male and female RECQL-deficient mice displayed normal fertility.
FIG. 1.
Targeted disruption of Recql. (A) Genomic structures of the endogenous Recql allele, the targeting vector, and the disrupted allele generated by HR. Exons are indicated by black boxes, and introns are indicated by white boxes. The helicase domains are indicated by Roman numerals. The position of the probe used for Southern blot analysis is shown. This probe recognizes an 877-bp EcoRI fragment in the wild-type allele and a 3.2-kb EcoRI fragment in the homologously recombined allele. PCR primers F1 and R2 were used to identify homologously recombined ES cells (2.7 kb). PCR primers F2, R1, R2, and R3 were used to genotype offspring. Abbreviations for restriction sites: A, Asp718; E, EcoRI; H, HindIII; P, PstI; S, StuI; Xb, XbaI; Xh, XhoI. (B) Southern blot analysis of EcoRI-digested genomic DNA isolated from Recql+/+ (wild type), Recql+/− (heterozygous), and Recql−/− (knockout) mice. (C) PCR analysis of genomic DNAs isolated from Recql+/+ (0.8 kb), Recql+/− (1.09 kb), and Recql−/− (2.4 kb) mice.
There were no differences seen in complete blood counts and white cell differential counts between the wild-type and knockout mice (data not shown). RECQL-deficient mice appeared to be normal at the time of autopsy. Routine staining of paraffin-embedded sections of all organs and tissues did not reveal any abnormalities at the level of hematoxylin-and-eosin staining and light microscopy (data not shown). A small group of mice was allowed to age and was monitored for tumor development. Animals that developed any visible or palpable masses or other evidence of disease were euthanized, and affected tissues were fixed in 10% neutral buffered formalin. Otherwise, groups of mice were euthanized between 15 and 35 months of age and histological analysis was performed. Similar percentages of mice from all three genotype groups developed tumors during this time. The types of tumors identified were also similar among the three groups, with lymphoma being the most common (data not shown).
Analysis of RECQL expression.
RECQL mRNA was readily detected in all mouse tissues examined. Three species of mRNA were seen in most tissues, with approximate sizes of 3.3, 2.8, and 2.6 kb (Fig. 2A). Levels of expression were highest in the testis and lowest in the liver when samples were normalized for total RNA concentration. To verify that Recql had been disrupted in the −/− mice, mRNA levels were determined in testis and kidney RNAs from +/+ and −/− mice. The three species of mRNA seen in the kidney tissue and the two species seen in the testis tissue (2.8 and 2.4 kb) were not detected in total cellular RNA from the −/− tissues and were expressed at decreased levels in tissues from the +/− animals (Fig. 2B and C). A longer exposure of the testis Northern blot revealed the appearance of a larger, 5.6-kb mRNA species in both the +/+ and the +/− mice and a smaller, 1.2-kb species present in the +/− and −/− samples but not in the +/+ sample (not shown). The larger mRNA species may represent incompletely spliced mRNA, and the smaller mRNA species most likely represents the truncated RECQL mRNA generated by inserting Neo into the gene at approximately the middle of the protein-coding region.
Analysis of other mouse RecQ family member mRNAs.
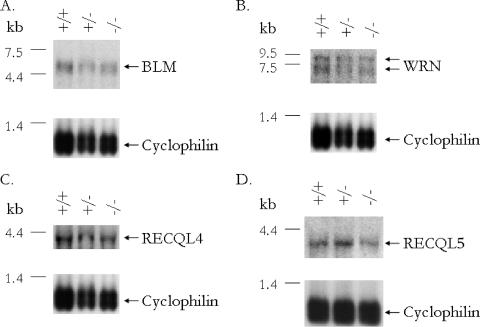
Northern blot analysis was performed to determine whether or not there were compensatory changes in the levels of mRNA for the other identified RecQ family members, since there may be functional redundancy among the family members. There were no apparent changes in the expression levels of BLM (Fig. 3A), WRN (Fig. 3B), RECQL4 (Fig. 3C), or RECQL5 (Fig. 3D) mRNA.
FIG. 3.
Expression of RecQ helicases in Recql+/+, Recql+/−, and Recql−/− mice. Total RNA (15 μg) was isolated from adult mouse testis tissue. Northern blots were hybridized with probes for mouse BLM (A), WRN (B), RECQL4 (C), RECQL5 (D), or cyclophilin. The locations of RNA molecular size markers are indicated to the left of each blot.
Growth characteristics of MEFs derived from RECQL knockout mice.
Fibroblasts isolated from Wrn knockout embryos have been shown to lose proliferative capacity (24). We established MEFs from four +/+ and four −/− E15.5 embryos (two +/+ and two −/− embryos from two different litters) and examined their growth rates after various times in culture and at different passage numbers. For the growth rates of passage 4 cells from one litter (from two +/+ and two −/− embryos), see Fig. S2 in the supplemental material. There were no observed differences in growth rates between the +/+ and −/− MEFs at this passage (see Fig. S2 in the supplemental material) or at passage 5 or 6. There were also no differences in the growth rates of the cells isolated from the second litter (data not shown).
Measurement of telomere lengths in Recql knockout mice.
Both WRN and BLM have been shown to be involved in telomere metabolism (6, 8, 10, 36, 58). The gradual shortening of telomeres can lead to premature senescence, as is seen in Werner syndrome fibroblasts. In addition, defects in telomere function may result in genetic instability that can promote malignant transformation. We examined whether or not RECQL was involved in the maintenance of telomere length by FISH and flow cytometry of Recql+/+, Recql+/−, and Recql−/− splenocytes and thymocytes. There were no differences in telomere length among the three genotypes (Table 1).
TABLE 1.
Telomere length in Recql+/+, Recql+/−, and Recql−/− splenocytes and thymocytes
| Genotype | Telomere length (kb)a
|
|
|---|---|---|
| Splenocytes | Thymocytes | |
| Recql+/+ | 38.7 ± 0.55 | 41.2 ± 0.25 |
| Recql+/− | 37.3 ± 0.66 | 38.9 ± 3.06 |
| Recql−/− | 39.7 ± 0.90 | 42.2 ± 0.10 |
The values shown represent the mean ± the standard error of the mean from splenocytes and thymocytes derived from two mice of each genotype.
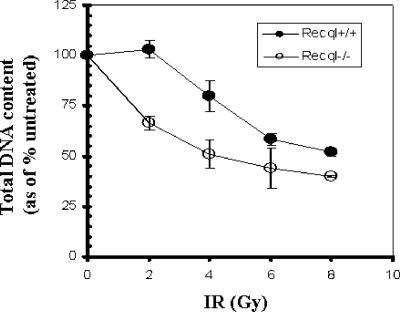
Cells deficient in RECQL show increased IR sensitivity.
Primary MEFs (passage 2 or 3) from Recql−/− knockout or wild-type mice were treated with increasing dose of IR and assayed for cell growth. Recql−/− MEFs displayed a greater sensitivity to IR than did wild-type MEFs (Fig. 4).
FIG. 4.
Cellular deficiency of RECQL leads to increased IR sensitivity. Primary MEFs from Recql−/− and wild-type mice were plated in quadruplicate and treated with increasing doses of IR. Total DNA content was measured as an indication of cell growth as described in Materials and Methods.
Elevated numbers of Rad51 and γH2AX foci in RECQ1-deficient cells.
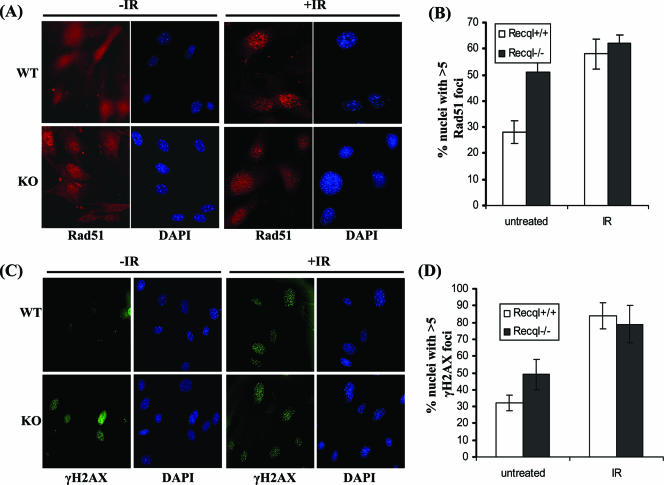
Single-strand breaks or DSBs can occur spontaneously from cellular processes or can be induced by exposure to DNA-damaging agents such as IR. A prominent pathway for the correction of DSBs is HR repair (54). To investigate if RECQL might have a role in this pathway, we examined the ability of Rad51, a major protein involved in the strand invasion step of HR repair, to form foci in primary mouse fibroblasts from wild-type and Recql−/− littermates. A diffuse nuclear staining of Rad51 was observed in the majority of unirradiated wild-type cells, consistent with the observations of others (16, 46); however, untreated Recql−/− MEFs exhibited an increased number of spontaneous Rad51 foci (Fig. 5A and B). Fifty-one percent of the Recql−/− MEFs contained greater than five Rad51 foci per nucleus, compared to only 29% for the wild-type MEFs. In contrast to the differences in untreated cells, Rad51 foci were induced to a comparable level in MEFs of Recql−/− and wild-type littermates (Fig. 5A and B). Normal induction of Rad51 foci in Recql−/− MEFs suggests that RECQL is not required for the recruitment of Rad51 to sites of IR-induced DNA damage. A significantly higher number of spontaneous Rad51 foci in Recql−/− cells may represent accumulation of unresolved recombination intermediates or a greater load of endogenous DNA damage in the absence of RECQL.
FIG. 5.
Recql−/− MEFs display elevated numbers of spontaneous Rad51 and γH2AX foci. (A) Immunofluorescent detection of spontaneous and irradiation (12 Gy)-induced Rad51 focus (red) formation in wild-type (top) and Recql−/− (bottom) primary MEFs. DAPI staining of the same nuclei is shown in blue. (B) Quantitation of the percentage of cells showing five or more distinct Rad51 foci. (C) Immunofluorescent detection of γH2AX foci (green). Wild-type (top) and Recql−/− (bottom) MEFs were stained for γH2AX foci following either mock treatment (0 Gy) or irradiation (5 Gy). DAPI staining of the same nuclei is shown in blue. (D) The percentage of cells exhibiting five or more γH2AX foci is shown for each of the indicated genotypes. All counts represent averages, and standard deviations are indicated by error bars. KO, knockout; WT, wild type.
We next assessed the load of unrepaired, spontaneously occurring DSBs in Recql−/− versus wild-type MEFs. The histone H2A variant H2AX, which becomes rapidly phosphorylated following the induction of DSBs, can serve as a marker for the presence of DSBs (41, 42). The majority of unirradiated wild-type MEFs showed little detectable γH2AX staining (Fig. 5C and D). In contrast, ∼50% of unirradiated Recql−/− MEFs were positive for γH2AX foci, indicating a markedly elevated load of unrepaired, spontaneously occurring DSBs in RECQL-deficient cells (Fig. 5C and D). Following treatment with IR (5 Gy), γH2AX foci were similarly induced in wild-type and Recql null cells at 30 min postirradiation (Fig. 5C and D).
Elevated SCEs in Recql−/− MEFs.
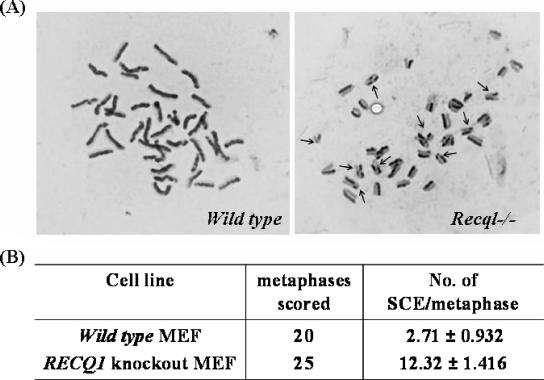
The elevated levels of spontaneous Rad51 and γH2AX foci in Recql−/− cells may be a consequence of aberrant recombination at sites of DSBs. A phenotype associated with defective HR is elevated SCE (15). Metaphase chromosome preparations from BrdU-labeled primary wild-type or Recql−/− fibroblasts were differentially stained for detection of SCEs (Fig. 6A). Recql−/− MEFs displayed a 4.5-fold greater frequency of spontaneous SCE compared to the wild-type MEFs (Fig. 6B). These results suggest that RECQL is involved in the resolution of HR intermediates and its absence leads to accumulation of DNA damage.
FIG. 6.
Elevated SCEs in Recql−/− MEFs. (A) Spontaneous SCEs were assayed in BrdU-labeled, Giemsa-stained chromosome spreads from primary wild-type and Recql−/− MEFs. A representative spread is shown for wild-type and Recql−/− MEFs, and the SCEs are marked with arrows. (B) Quantitative representation of the number of SCEs per metaphase.
Chromosomal instability in Recql−/− cells.
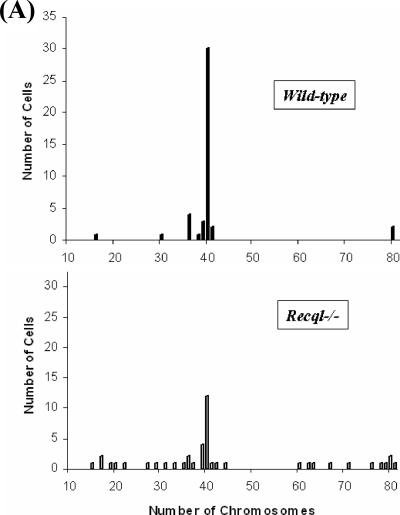
Chromosomal instability is often reflected by either an increase (hyperdiploid) or a decrease (hypodiploid) in the number of diploid chromosomes. To determine the role of RECQL in chromosome stability, chromosome numbers were determined in wild-type and Recql−/− MEFs. For this purpose, metaphase chromosomes were prepared and stained with Giemsa. The number of aneuploid cells was much higher in RECQL-deficient MEFs, with 73% of the Recql−/− metaphases showing deviation from a normal diploid chromosome number of 40 while only 32% of the metaphases from wild-type RECQL cells showed aneuploidy (Fig. 7A). The average number of chromosomes per diploid cell was found to be 40.6 for wild-type cells and 44 for Recql null cells, demonstrating the importance of RECQL in the segregation of chromosomes during mitosis.
FIG. 7.
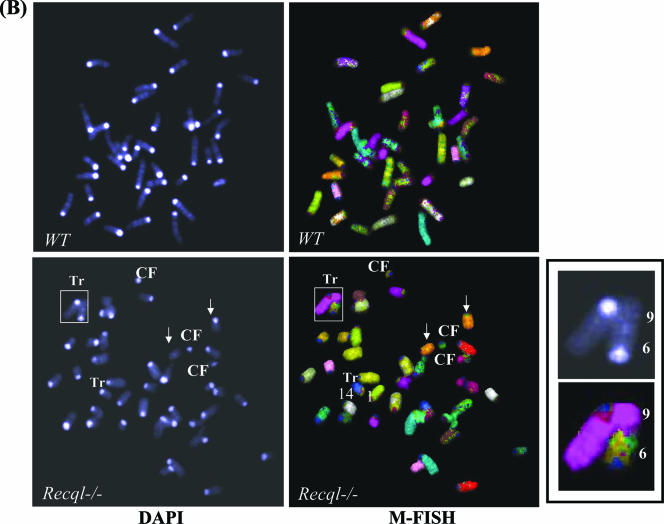
Chromosomal instability in primary Recql−/− MEFs. (A) Aneuploidy in Recql−/− MEFs. Giemsa-stained chromosomes were counted from the metaphase spreads (n = 44) of wild-type and Recql−/− MEFs. Chromosomal number distribution in primary wild-type and Recql−/− MEFs is shown. (B) M-FISH analyses of chromosomal aberrations in metaphase spreads from primary wild-type (WT) and Recql−/− MEFs. Representative spreads from wild-type and Recql−/− MEFs are shown. A translocation between chromosomes 9 and 6 from the metaphase spread of Recql−/− MEFs is further enlarged in the inset. Arrows indicate two copies of chromosome 8, one without a centromere (centromere loss). CF, centromeric fragment, Tr, translocation.
In order to determine whether or not structural chromosomal aberrations accumulate in the absence of RECQL, M-FISH was used to screen for genome-wide chromosomal alterations. This technique has been widely used to characterize the chromosomal instability in different mouse knockout models. M-FISH analysis revealed a low but detectable level of spontaneous chromosomal instability in wild-type cells, whereas Recql−/− MEFs exhibited a significantly much higher frequency of chromosomal aberrations (Fig. 7B and Table 2). Notably, Recql−/− MEFs displayed a substantial increase in the incidence of fragmented chromosomes relative to wild-type MEFs and also incurred events in which sister chromatids were visibly broken or fragmented (Fig. 7B and Table 2). In contrast, no chromatid breaks were detected in the metaphase spreads of wild-type MEFs. In addition to chromosomal fragmentation, Recql−/− cells had a significantly elevated incidence of spontaneous random chromosomal translocations. Other complex abnormalities observed in the chromosome spreads of the Recql−/− cells included a ring chromosome that may have resulted from a breakage-and-fusion event (Fig. 7B and Table 2). These results demonstrate a significantly elevated frequency of spontaneous chromosome aberrations in the Recql−/− primary MEFs compared to wild-type cells.
TABLE 2.
Increased genomic instability of RECQL-deficient primary MEFs versus wild-type MEFs
| Instability | Wild type at 40 metaphases | Recql−/− at 64 metaphases |
|---|---|---|
| No. of fragmented chromosomes | 5 | 19 |
| No. of chromatid breaks | 0 | 3 |
| No. of chromosome breaks | 1 | 2 |
| No. of translocations | 0 | 8 |
| No. of fusions | 0 | 3 |
| No. of multiradial chromosomes | 0 | 4 |
| Total no. of anomalies (no. per metaphase) | 6 (0.15) | 42 (0.65) |
| % of metaphases with abnormalities | 5 | 61 |
DISCUSSION
The genomic instability prevalent in human RecQ helicase disorders is characterized at the cellular level by gross chromosomal rearrangements. Studies of prokaryotes and eukaryotes indicate the importance of RecQ helicases to function in the DNA damage response by maintaining legitimate HR during replication of damaged DNA. In this study, we have investigated the importance of RECQL, one of the less-well-characterized RecQ helicases, for the preservation of genomic stability in mammalian cells. Embryonic fibroblasts from Recql knockout mice display chromosomal structural aberrations and aneuploidy, demonstrating for the first time that RECQL plays an important role in genome stability maintenance.
RECQL deficiency resulted in an increased sensitivity of the MEFs to IR which primarily induces DSBs. The elevated SCE observed in RECQL-deficient mouse cells (this study) and human cells (our unpublished results and reference 25) may be due to unsuccessful attempts to “repair” damaged replication forks by HR at DSBs. The persistence of abundant spontaneously forming Rad51 foci and γH2AX, an early marker of DSB formation, in RECQL-deficient cells adds further support to a proposed role for RECQL in HR repair at sites of chromosomal DNA damage. Constitutively high numbers of Rad51 foci have also been detected in Werner syndrome and Bloom syndrome cells (38, 56). The presence of Rad51 foci corresponding to nucleoprotein filaments that are necessary for early strand invasion during HR suggests incomplete resolution of recombination intermediates as a consequence of the absence of RECQL.
The gross chromosomal rearrangements, translocations, and aneuploidy in cells of Recql−/− but not wild-type littermates indicates that the helicase is necessary for the proper maintenance of genomic stability. In spite of this, RECQL is not required for cell viability or proliferation and Recql knockout mice are viable, fertile, and apparently not tumor prone under normal colony conditions. However, it should be emphasized that small numbers of mice were evaluated in each group and no genetic or environmental carcinogenic stimuli were used in these initial studies.
The genetic background and/or tissue specificity of RECQL function may protect the mouse from severe abnormalities that might be expected from the magnitude of chromosomal aberrations detected at the cellular level. It will be critically important to evaluate RECQL deficiency in other genetic backgrounds, including mutations in genes encoding other RecQ helicases or DNA replication and repair proteins. In addition, challenging Recql knockout mice with DNA-damaging agents may expose phenotypes that will be helpful to elucidate a disease or a predisposition to cancer associated with RECQL deficiency.
One potential reason for the apparent lack of a phenotype in RECQL-deficient mice is functional redundancy, in which one or more of the other RecQ DNA helicases is able to compensate for the absence of RECQL. Expression levels of the known family members, WRN, BLM, RECQL4, and RECQL5, were not altered in Recql knockout mice, suggesting that if one or more of these helicases compensates for the absence of RECQL, the normal level of expression is sufficient for this compensation. Recent studies carried out with the chicken B-lymphocyte DT40 cell line support the notion that there may be redundancy of function among the various RecQ helicases (52).
The mixed genetic background of the mice may also have contributed to the apparent lack of a phenotype, illustrated by the finding that a WRN helicase domain knockout mouse induced accelerated tumorigenesis only in a p53 null background (23). Finally, it remains possible that RECQL function is not critical under normal physiological conditions in the mouse but could be important in the response to external stressors such as environmental factors or genotoxins. The nearly 85% sequence homology of the mouse and human RECQL proteins suggests the two proteins are likely to have conserved functions; however, intrinsic differences between mice and humans can contribute to differences in tumor susceptibility. For example, WRN-deficient mice lack a disease phenotype (24, 28); however, late-generation telomerase- and WRN-deficient mice (mTerc−/− Wrn−/−) displayed clinical symptoms of premature aging and the types of tumors typically observed in Werner syndrome patients (4, 10). It is plausible that the manifestation of disease phenotypes of RECQL deficiency in mice is related to telomere maintenance or some other event critical for genomic stability.
Having demonstrated that RECQL is important for chromosomal stability, we now face the challenge of understanding the potentially unique molecular functions of RECQL helicase to prevent illegitimate recombination during replicational stress. It is highly likely that RECQL cooperates with other nuclear DNA metabolic factors in this capacity. Strong candidates for protein interactors with RECQL that serve to suppress crossover of sister chromatids are recombination proteins that are involved in DNA replication or mismatch repair factors already known to regulate genetic recombination. In the absence of a functional RECQL helicase, DNA replication forks may not be maintained properly, giving rise to elevated levels of SCE (33).
In addition to recombination proteins, type IA topoisomerases have been implicated genetically and biochemically in collaboration with RecQ helicases to preserve chromosomal integrity. BLM and Sgs1 helicases are proposed to act with type IA topoisomerases to regulate recombination levels in vivo by suppressing the formation of crossover products that accumulate from resolution of Holliday junction (HJ) intermediates (17, 57). A model for BLM to suppress crossovers was proposed on the basis of in vitro studies that demonstrated the unique ability of BLM with Topo IIIα to catalyze double HJ dissolution (57). Although RECQL was reported to be associated with Topo IIIα in human cells (18), the purified recombinant protein failed to substitute for BLM in the in vitro double-HJ dissolution reaction (55). It is conceivable that in the cellular setting, RECQL acts upon a key DNA intermediate of HR with additional protein factors. The posttranslational state or protein interactions of RECQL with mismatch repair factors (e.g., MSH2/6, EXO-1) (9) may be important for the role of RECQL in processing structures to prevent illegitimate recombination. The constitutively high number of Rad51 nuclear foci in RECQL-deficient cells further supports a role for RECQL in the disruption of inappropriately paired DNA recombination intermediates.
Although a genetic disorder has not been linked to a mutation in Recql, recent analyses of Recql single-nucleotide polymorphisms (SNPs) have identified an association of RECQL with a reduced survival of pancreatic cancer patients (26, 27). Only one RECQL A159C SNP allele is required to significantly decrease overall pancreatic cancer survival, suggesting a predictive and prognostic role for RECQL SNPs. RECQL SNPs displayed significant genetic interaction with SNPs in the HR repair genes ATM, RAD54L, XRCC2, and XRCC3. A role for RECQL in HR is further suggested by the observation that SNPs in RECQL result in a very poor response to the anticancer drug gemcitabine-induced radiosensitization that selectively requires functional HR (26, 27). Our results suggest that the chromosomal instability arising from RECQL deficiency may contribute to a predisposition to cancer. Microsatellite instability in the polyguanine repeat (G)9 in the Recql gene is frequently observed in mismatch repair-deficient human nonpolyposis colorectal cancer (37).
Loss of heterozygosity of 12p12, the chromosomal location of the Recq1 gene, is a frequent event in a wide range of hematological malignancies and solid tumors, suggesting the presence of a tumor suppressor locus. Allelic losses on chromosome 12p12-13 are associated with childhood acute lymphoblastic leukemia and several solid neoplasms (31). Chromosome 12p12 deletion has been reported in a rare chronic myeloid leukemia-like syndrome case in a Li-Fraumeni syndrome family (14). RECQL is highly expressed in the lungs (21), and deletions at chromosome 12p12 have been reported in bronchial epithelia of patients with primary non-small-cell lung cancer (13), suggesting that RECQL sequence alterations may influence the risk of lung cancer.
RECQL has not been directly linked to a human disease or cancer; however, the present study demonstrates that RECQL deficiency leads to spontaneous chromosomal instability and aneuploidy, both characteristics of cancer cells, suggesting a role for RECQL in a predisposition to cancer. Results obtained with RECQL-deficient mice provide a foundation from which to investigate the interactions of RECQL with other genetic factors or tumor suppressors that exist in vivo to preserve chromosomal integrity and prevent carcinogenesis.
Supplementary Material
Acknowledgments
We are very grateful to Julie Horton and Manas Ray for reviewing the manuscript, to Robert Maronpot for analysis of histological sections, and to Irma Vulto for help in the telomere length analysis.
This research was supported by the Intramural Research Program of the NIH (NIEHS and NIA).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 11 December 2006.
REFERENCES
- 1.Bachrati, C. Z., and I. D. Hickson. 2003. RecQ helicases: suppressors of tumorigenesis and premature ageing. Biochem. J. 374:577-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baerlocher, G. M., J. Mak, T. Tien, and P. M. Lansdorp. 2002. Telomere length measurement by fluorescence in situ hybridization and flow cytometry: tips and pitfalls. Cytometry 47:89-99. [DOI] [PubMed] [Google Scholar]
- 3.Carballo, E., W. S. Lai, and P. J. Blackshear. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001-1005. [DOI] [PubMed] [Google Scholar]
- 4.Chang, S., A. S. Multani, N. G. Cabrera, M. L. Naylor, P. Laud, D. Lombard, S. Pathak, L. Guarente, and R. A. DePinho. 2004. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat. Genet. 36:877-882. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, N. C., H. J. van de Vrugt, M. A. van der Valk, A. B. Oostra, P. Krimpenfort, Y. de Vries, H. Joenje, A. Berns, and F. Arwert. 2000. Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum. Mol. Genet. 9:1805-1811. [DOI] [PubMed] [Google Scholar]
- 6.Choi, D., P. S. Whittier, J. Oshima, and W. D. Funk. 2001. Telomerase expression prevents replicative senescence but does not fully reset mRNA expression patterns in Werner syndrome cell strains. FASEB J. 15:1014-1020. [DOI] [PubMed] [Google Scholar]
- 7.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 8.Davis, T., S. K. Singhrao, F. S. Wyllie, M. F. Haughton, P. J. Smith, M. Wiltshire, D. Wynford-Thomas, C. J. Jones, R. G. Faragher, and D. Kipling. 2003. Telomere-based proliferative lifespan barriers in Werner-syndrome fibroblasts involve both p53-dependent and p53-independent mechanisms. J. Cell Sci. 116:1349-1357. [DOI] [PubMed] [Google Scholar]
- 9.Doherty, K. M., S. Sharma, L. Uzdilla, T. M. Wilson, S. Cui, A. Vindigni, and R. M. Brosh, Jr. 2005. RECQ1 helicase interacts with human mismatch repair factors that regulate genetic recombination J. Biol. Chem. 280:28085-28094. [DOI] [PubMed] [Google Scholar]
- 10.Du, X., J. Shen, N. Kugan, E. E. Furth, D. B. Lombard, C. Cheung, S. Pak, G. Luo, R. J. Pignolo, R. A. DePinho, L. Guarente, and F. B. Johnson. 2004. Telomere shortening exposes functions for the mouse Werner and Bloom syndrome genes. Mol. Cell. Biol. 24:8437-8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellis, N. A., J. Groden, T. Z. Ye, J. Straughen, D. J. Lennon, S. Ciocci, M. Proytcheva, and J. German. 1995. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83:655-666. [DOI] [PubMed] [Google Scholar]
- 12.Fernández, S., A. Sorokin, and J. C. Alonso. 1998. Genetic recombination in Bacillus subtilis 168: effects of recU and recS mutations on DNA repair and homologous recombination. J. Bacteriol. 180:3405-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grepmeier, U., W. Dietmaier, J. Merk, P. J. Wild, E. C. Obermann, M. Pfeifer, F. Hofstaedter, A. Hartmann, and M. Woenckhaus. 2005. Deletions at chromosome 2q and 12p are early and frequent molecular alterations in bronchial epithelium and NSCLC of long-term smokers. Int. J. Oncol. 27:481-488. [PubMed] [Google Scholar]
- 14.Guran, S., C. Beyan, O. Nevruz, C. Yakicier, and Y. Tunca. 2005. A chronic myeloid leukemia-like syndrome case with del (12) (p12) in a Li-Fraumeni syndrome family. Clin. Lab. Haematol. 27:135-138. [DOI] [PubMed] [Google Scholar]
- 15.Hickson, I. D. 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3:169-178. [DOI] [PubMed] [Google Scholar]
- 16.Houghtaling, S., C. Timmers, M. Noll, M. J. Finegold, S. N. Jones, M. S. Meyn, and M. Grompe. 2003. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 17:2021-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, F. B., D. B. Lombard, N. F. Neff, M. A. Mastrangelo, W. Dewolf, N. A. Ellis, R. A. Marciniak, Y. Yin, R. Jaenisch, and L. Guarente. 2000. Association of the Bloom syndrome protein with topoisomerase IIIα in somatic and meiotic cells. Cancer Res. 60:1162-1167. [PubMed] [Google Scholar]
- 19.Khakhar, R. R., J. A. Cobb, L. Bjergbaek, I. D. Hickson, and S. M. Gasser. 2003. RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol. 13:493-501. [DOI] [PubMed] [Google Scholar]
- 20.Kitao, S., N. M. Lindor, M. Shiratori, Y. Furuichi, and A. Shimamoto. 1999. Rothmund-Thomson syndrome responsible gene, RECQL4: genomic structure and products. Genomics 61:268-276. [DOI] [PubMed] [Google Scholar]
- 21.Kitao, S., I. Ohsugi, K. Ichikawa, M. Goto, Y. Furuichi, and A. Shimamoto. 1998. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics 54:443-452. [DOI] [PubMed] [Google Scholar]
- 22.Kitao, S., A. Shimamoto, M. Goto, R. W. Miller, W. A. Smithson, N. M. Lindor, and Y. Furuichi. 1999. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 22:82-84. [DOI] [PubMed] [Google Scholar]
- 23.Lebel, M., R. D. Cardiff, and P. Leder. 2001. Tumorigenic effect of nonfunctional p53 or p21 in mice mutant in the Werner syndrome helicase. Cancer Res. 61:1816-1819. [PubMed] [Google Scholar]
- 24.Lebel, M., and P. Leder. 1998. A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc. Natl. Acad. Sci. USA 95:13097-13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LeRoy, G., R. Carroll, S. Kyin, M. Seki, and M. D. Cole. 2005. Identification of RecQL1 as a Holliday junction processing enzyme in human cell lines. Nucleic Acids Res. 33:6251-6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, D., M. Frazier, D. B. Evans, K. R. Hess, C. H. Crane, L. Jiao, and J. L. Abbruzzese. 2006. Single nucleotide polymorphisms of RecQ1, RAD54L and ATM genes are associated with reduced survival of pancreatic cancer. J. Clin. Oncol. 24:1720-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, D., H. Liu, L. Jiao, D. Z. Chang, G. Beinart, R. A. Wolff, D. B. Evans, M. M. Hassan, and J. L. Abbruzzese. 2006. Significant effect of homologous recombination DNA repair gene polymorphisms on pancreatic cancer survival. Cancer Res. 66:3323-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lombard, D. B., C. Beard, B. Johnson, R. A. Marciniak, J. Dausman, R. Bronson, J. E. Buhlmann, R. Lipman, R. Curry, A. Sharpe, R. Jaenisch, and L. Guarente. 2000. Mutations in the WRN gene in mice accelerate mortality in a p53-null background. Mol. Cell. Biol. 20:3286-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Mohaghegh, P., and I. D. Hickson. 2001. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum. Mol. Genet. 10:741-746. [DOI] [PubMed] [Google Scholar]
- 31.Montpetit, A., J. Larose, G. Boily, S. Langlois, N. Trudel, and D. Sinnett. 2004. Mutational and expression analysis of the chromosome 12p candidate tumor suppressor genes in pre-B acute lymphoblastic leukemia. Leukemia 18:1499-1504. [DOI] [PubMed] [Google Scholar]
- 32.Myung, K., A. Datta, C. Chen, and R. D. Kolodner. 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27:113-116. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama, H. 2002. RecQ family helicases: roles as tumor suppressor proteins. Oncogene 21:9008-9021. [DOI] [PubMed] [Google Scholar]
- 34.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Opresko, P. L., W. H. Cheng, and V. A. Bohr. 2004. At the junction of RecQ helicase biochemistry and human disease. J. Biol. Chem. 279:18099-18102. [DOI] [PubMed] [Google Scholar]
- 36.Ouellette, M. M., L. D. McDaniel, W. E. Wright, J. W. Shay, and R. A. Schultz. 2000. The establishment of telomerase-immortalized cell lines representing human chromosome instability syndromes. Hum. Mol. Genet. 9:403-411. [DOI] [PubMed] [Google Scholar]
- 37.Park, J., D. Betel, R. Gryfe, K. Michalickova, N. Di Nicola, S. Gallinger, C. W. Hogue, and M. Redston. 2002. Mutation profiling of mismatch repair-deficient colorectal cancers using an in silico genome scan to identify coding microsatellites. Cancer Res. 62:1284-1288. [PubMed] [Google Scholar]
- 38.Pichierri, P., A. Franchitto, P. Mosesso, and F. Palitti. 2001. Werner's syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol. Biol. Cell 12:2412-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puranam, K. L., and P. J. Blackshear. 1994. Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J. Biol. Chem. 269:29838-29845. [PubMed] [Google Scholar]
- 40.Puranam, K. L., E. Kennington, S. N. Sait, T. B. Shows, J. M. Rochelle, M. F. Seldin, and P. J. Blackshear. 1995. Chromosomal localization of the gene encoding the human DNA helicase RECQL and its mouse homologue. Genomics 26:595-598. [DOI] [PubMed] [Google Scholar]
- 41.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 43.Seki, M., H. Miyazawa, S. Tada, J. Yanagisawa, T. Yamaoka, S. Hoshino, K. Ozawa, T. Eki, M. Nogami, and K. Okumura. 1994. Molecular cloning of cDNA encoding human DNA helicase Q1 which has homology to Escherichia coli Rec Q helicase and localization of the gene at chromosome 12p12. Nucleic Acids Res. 22:4566-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seki, T., W. S. Wang, N. Okumura, M. Seki, T. Katada, and T. Enomoto. 1998. cDNA cloning of mouse BLM gene, the homologue to human Bloom's syndrome gene, which is highly expressed in the testis at the mRNA level. Biochim. Biophys. Acta 1398:377-381. [DOI] [PubMed] [Google Scholar]
- 45.Sharma, S., K. M. Doherty, and R. M. Brosh, Jr. 2006. Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem. J. 398:319-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smiraldo, P. G., A. M. Gruver, J. C. Osborn, and D. L. Pittman. 2005. Extensive chromosomal instability in Rad51d-deficient mouse cells. Cancer Res. 65:2089-2096. [DOI] [PubMed] [Google Scholar]
- 47.Stewart, E., C. R. Chapman, F. Al-Khodairy, A. M. Carr, and T. Enoch. 1997. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16:2682-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stumpo, D. J., C. B. Bock, J. S. Tuttle, and P. J. Blackshear. 1995. MARCKS deficiency in mice leads to abnormal brain development and perinatal death. Proc. Natl. Acad. Sci. USA 92:944-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stumpo, D. J., J. M. Graff, K. A. Albert, P. Greengard, and P. J. Blackshear. 1989. Molecular cloning, characterization, and expression of a cDNA encoding the “80- to 87-kDa” myristoylated alanine-rich C kinase substrate: a major cellular substrate for protein kinase C. Proc. Natl. Acad. Sci. USA 86:4012-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor, G. A., E. Carballo, D. M. Lee, W. S. Lai, M. J. Thompson, D. D. Patel, D. I. Schenkman, G. S. Gilkeson, H. E. Broxmeyer, B. F. Haynes, and P. J. Blackshear. 1996. A pathogenetic role for TNFα in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4:445-454. [DOI] [PubMed] [Google Scholar]
- 51.Umezu, K., K. Nakayama, and H. Nakayama. 1990. Escherichia coli RecQ protein is a DNA helicase. Proc. Natl. Acad. Sci. USA 87:5363-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, W., M. Seki, Y. Narita, T. Nakagawa, A. Yoshimura, M. Otsuki, Y. Kawabe, S. Tada, H. Yagi, Y. Ishii, and T. Enomoto. 2003. Functional relation among RecQ family helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol. Cell. Biol. 23:3527-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watt, P. M., I. D. Hickson, R. H. Borts, and E. J. Louis. 1996. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144:935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.West, S. C. 2003. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4:435-445. [DOI] [PubMed] [Google Scholar]
- 55.Wu, L., K. L. Chan, C. Ralf, D. A. Bernstein, P. L. Garcia, V. A. Bohr, A. Vindigni, P. Janscak, J. L. Keck, and I. D. Hickson. 2005. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 24:2679-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, L., S. L. Davies, N. C. Levitt, and I. D. Hickson. 2001. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem. 276:19375-19381. [DOI] [PubMed] [Google Scholar]
- 57.Wu, L., and I. D. Hickson. 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426:870-874. [DOI] [PubMed] [Google Scholar]
- 58.Wyllie, F. S., C. J. Jones, J. W. Skinner, M. F. Haughton, C. Wallis, D. Wynford-Thomas, R. G. Faragher, and D. Kipling. 2000. Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nat. Genet. 24:16-17. [DOI] [PubMed] [Google Scholar]
- 59.Yu, C. E., J. Oshima, Y. H. Fu, E. M. Wijsman, F. Hisama, R. Alisch, S. Matthews, J. Nakura, T. Miki, S. Ouais, G. M. Martin, J. Mulligan, and G. D. Schellenberg. 1996. Positional cloning of the Werner's syndrome gene. Science 272:258-262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.